Abstract
The drug pentamidine inhibits calcium-dependent complex formation with p53 (CaS100B•p53) in malignant melanoma (MM), and restores p53 tumor suppressor activity in vivo. However, off-target effects associated with this drug were problematic in MM patients. Structure-activity relationship (SAR) studies were therefore completed here with 23 pentamidine analogues, and X-ray structures of CaS100B•inhibitor complexes revealed that the C-terminus of S100B adopts two different conformations, with location of Phe-87 and Phe-88 being the distinguishing feature and termed the “FF-Gate”. For symmetric pentamidine analogues (CaS100B•5a, CaS100B•6b) a channel between Sites 1 and 2 on S100B was occluded by residue Phe-88, but for an asymmetric pentamidine analogue (CaS100B•17), this same channel was open. The CaS100B•17 structure illustrates, for the first time, a pentamidine analog capable of binding the “open” form of the “FF-gate” and provides a means to block all three “hot spots” on CaS100B, which will impact next generation CaS100B•p53 inhibitor design.
INTRODUCTION
Long-time survival (> 3 years) remains extremely poor for most malignant melanoma (MM) patients (>70% mortality), and side effects from treatments are often severe1-3. Prognosis is even more problematic after the onset of metastasis and/or when drug-resistant mechanisms arise4, 5. While advances in immune checkpoint inhibitors can have remarkable outcomes for some MM patients, these therapies have limited or small effects in others (50%), so trials with immune checkpoint inhibitors are often done in drug combination trials including with chemotherapy regimens, radiation, and/or with other targeted approaches 6, 7. One alternative target in MM is a calcium-binding protein, S100B, which is an important biomarker for this and other cancers8. S100B is elevated in >90% of MM patients and its protein level correlates directly with poor survival (<1 yr.) and relapse, and it is especially predictive as a biomarker when used in combination with other diagnostic indicators9-11. For example, MM vaccines are only effective in a small number of patients (5-10%) and they nearly all have low levels of S100B12, 13. While this correlation is not fully understood, mechanistic studies show that lowering S100B levels via siRNA or via small molecule inhibitors restore p53 levels and its tumor suppressor activities including UV-activated apoptosis pathways found in normal melanocytes14-18. Studies suggest that S100B blocks p53 oligomerization, promotes p53 phosphorylation in its C-terminus, and contributes to p53 degradation in concert with the E3 ubiquitin ligase, hdm28. Elevated S100B and overexpression of the receptor for advanced glycation end products (RAGE) confers a metastatic phenotype19, so learning how S100B affects tumor progression and metastasis are ongoing including the development of novel S100B inhibitors (termed SBiXs) to ablate S100B-dependent effects in cancer.
One of the first compounds found to inhibit S100B was the FDA-approved drug pentamidine16. In these studies, pentamidine, 4b (SBi1) binds two pockets in the p53 binding site on Ca2+-S100B (termed Sites 2 & 3, Fig. 1), and it was repurposed for use in a phase II clinical trial for treatment of relapsed and/or refractory melanoma in patients with detectable levels of S100B protein (www.clinicaltrials.gov Identifier: NCT00729807). While p53 levels were often restored in these patients after treatment, the toxicity profile for pentamidine was not ideal, likely in part since 4b binds to several cellular targets, including DNA20-22. For these reasons, structure-activity relationship (SAR) studies are ongoing to improve the affinity and selectivity of pentamidine analogues for binding Ca2+-S100B. Thus far, modifying the chemical scaffold of pentamidine first yielded heptamidine, 4a (SBi4211), which occupies the same two persistent binding sites as pentamidine (i.e. Sites 2 & 3), only it does so with a single molecule bound to the protein versus two pentamidines/monomer 23. However, improvements to this drug scaffold are still needed to increase its affinity and a provide a meaningful therapeutic effect 23. Therefore, in the present study, structure-activity relationships (SARs) of novel pentamidine/heptamidine derivatives were explored.
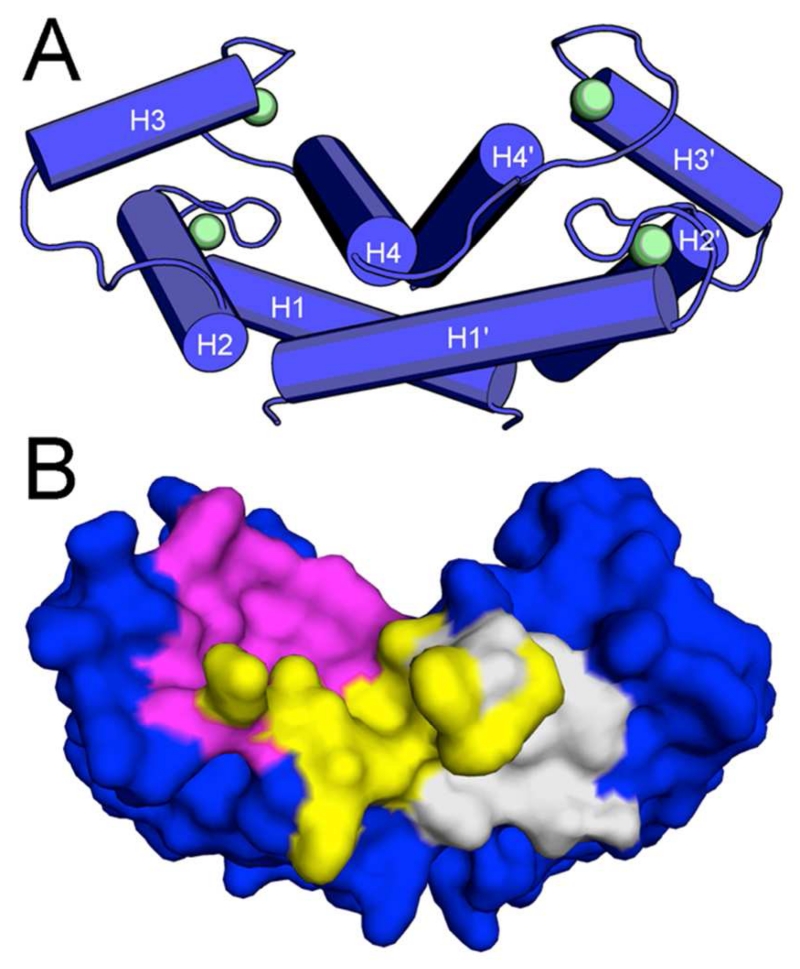
Figure 1. Overall Structure of S100B.
(A) Homodimer of S100B (blue tubes) with Ca2+ bound (green spheres). (B) Surface rendering of S100B with hot spots 1 (magenta), 2 (yellow), and 3 (gray) highlighted.
The SBiX compounds synthesized here bind Sites 2 and 3, as found for pentamidine and heptamidine; however, few such analogues interact with Site 1 (Fig. 1), or showed potential for efficient extension into Site 1. This is due to conformational exchange in this region involving Phe87 and Phe88, which serve as a gate to access a narrow channel linking Sites 1 & 2 (termed the “FF-gate”). In one case, structural results with compound 17 (SBi29), indicate that the Site 2/3 interactions in the S100B•17 complex are made in a unique way, as compared to the other pentamidine analogues. Importantly, the FF-gate is “open” in the S100B•17 complex, such that it can accommodate modifications that would possibly extend into Site 1 from Site 2 efficiently while retaining an amidine moiety like those in the pentamidine diamidine scaffold. Importantly, the gained understanding of the FF-gate now permits a more rational design of novel S100B inhibitor(s) that can simultaneously occupy Sites 1 & 2 and perhaps all three target-binding pockets in the CaS100B•p53 complex (i.e. Sites 1, 2, and 3). Although chemical scaffolds occupying Site 1 have been successfully identified24, 25, combining moieties or tethering with pentamidine derivatives towards Sites 2 and 3 have not yet been achieved. Therefore, the results described here represent a significant advance towards developing such SBiX molecules that achieves a threshold required for a useful therapeutic window.
LIGAND DESIGN STRATEGY
The development of small molecule inhibitors that block the CaS100B•p53 interaction requires inhibition of large protein-protein interfaces (PPIs) that were once thought to be difficult, if not impossible, to block. It is now recognized that the interfaces of most PPIs have smaller amino acid domain(s), termed “hot spots” or “persistent sites” that contribute more energetically to the PPIs than the surrounding protein surface 26. Successful strategies to target small molecule PPI inhibitors were developed and, as a result, several PPIs have been successfully inhibited, including those involving bcl-6, bcl-2, p53, ERK, tubulin, and several SH2 domains, to name but a few27, 28. For PPI’s involving S100B, two inhibitors, pentamidine (4b) and heptamidine (4a), were discovered to block the CaS100B•p53 complex and occupy persistent binding Sites 2 and 3 simultaneously within Ca2+-S100B23, 29 (Fig. 1). Efforts were made here to find the most efficient means to extend the heptamidine scaffold towards Site 1 and create compounds capable of binding all three persistent hot spots in S100B simultaneously (i.e. Sites 1, 2, and 3).
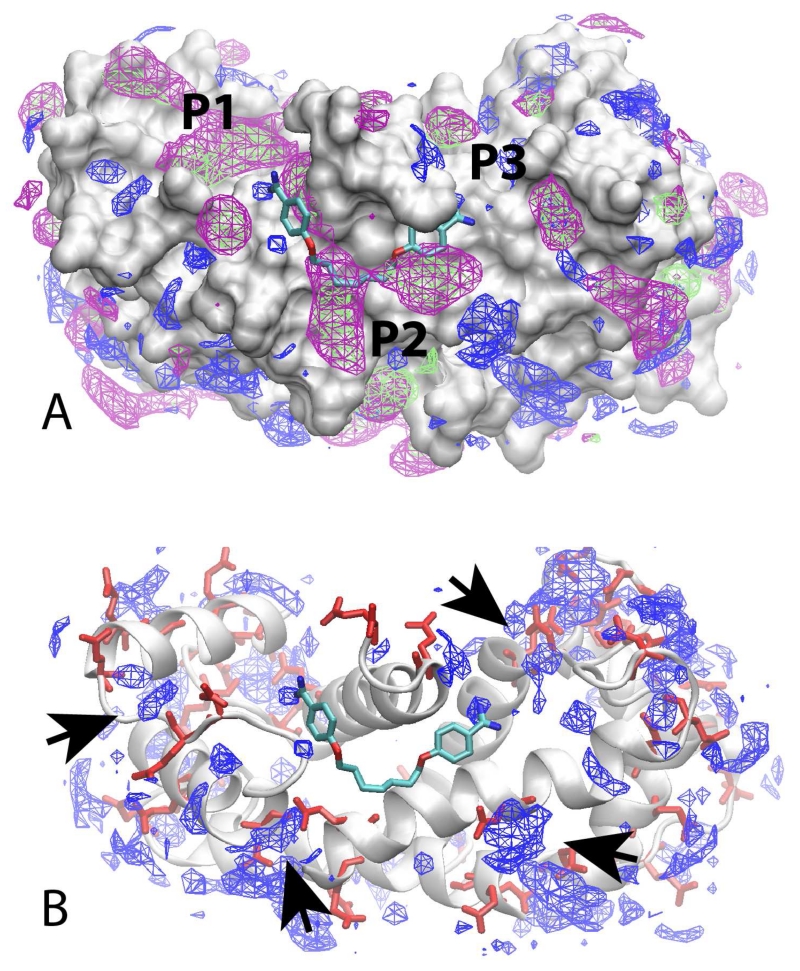
To facilitate this design strategy, the site identification by ligand competitive saturation (SILCS) approach was applied to map the functional group requirements of the three-targeted sites. From the SILCS calculations, 3D FragMaps are obtained that indicate the different functional group requirements of the protein, information that can be used to identify moieties of a compound that are contributing to binding as well as to be used to indicate how modifications of a compound can be used to improve affinity. During the design stage, what is termed a “Tier 1” SILCS approach was used. This approach includes aromatic maps based on benzene, aliphatic maps based on propane, and hydrogen bond donors and acceptors maps based on water hydrogen atoms and oxygen atoms, respectively. Shown in Figure 2 are the SILCS FragMaps overlaid on the crystal structure of the CaS100B•SBi132 complex (PDB 3GK1) along with the crystallographic orientation of heptamidine (PDB 4FQO). The approximate locations of the three hot spots are labeled. The occupancy of Sites 2 and 3 by heptamidine is evident, with aromatic and aliphatic FragMaps in the vicinity of the aromatic rings and aliphatic linker of heptamidine indicating the contribution of those moieties to ligand binding. The aliphatic and aromatic FragMaps cover a relatively wide range indicating that variations in the linker length between the aromatic rings could be tolerated. Notably, only small hydrogen bond donor FragMaps are directly adjacent to either amidine moiety, indicating that the positive charges may not be essential for binding. In addition, in regions beyond the amidine moieties in Sites 1 and 3 are hydrogen bond donor FragMaps that are adjacent to acidic residues (Fig. 2B), suggesting that extending those moieties with additional positively charged groups with linkers of varying lengths may lead to improved binding. Therefore, functionalities were added to the amidine moieties of both heptamidine (4a) and pentamidine (4b)23 in an attempt to improve affinity. In addition, the extended aromatic/aliphatic FragMaps around the linker region in the P2 site, supported by other CaS100B•SBiX X-ray structures23, 29, 30, indicated that increased diversity of the linker could facilitate binding.
Figure 2. SILCS FragMaps.
Crystal orientation of heptamidine (licorice, atom-type coloring) overlaid on the SILCS FragMaps calculated using the SBi132•S100B (PDB 3GK1) crystal structure. A) FragMaps are shown for aliphatic (green), aromatic (purple), and hydrogen bond donors (blue) with the protein show in surface representation. The approximate locations of the Site 1, 2 and 3 pockets are indicated. B) Protein in cartoon representation with acidic residues in red licorice along with the hydrogen bond donors (blue). Arrow heads indicate hydrogen bond acceptor FragMaps that are adjacent to acidic residue sidechains.
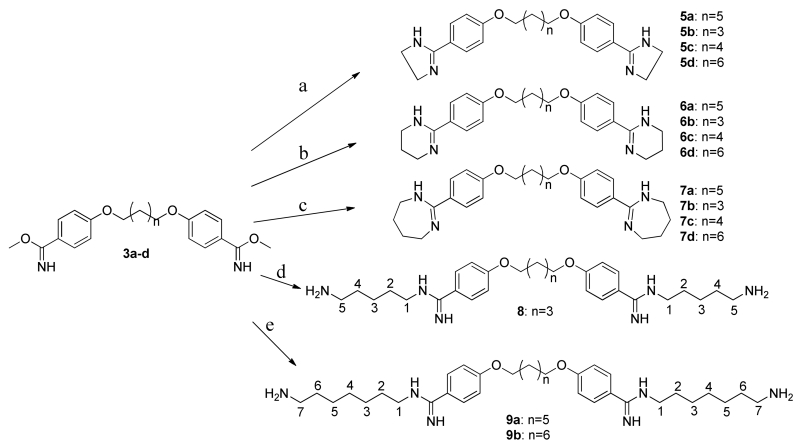
Based on these design criteria and synthetic considerations, a number of novel analogs were developed. Compounds of series 5, 6 and 7 (Scheme 2) include modification of the amidine moiety, yielding neutral species in aliphatic rings ranging in size from 5 to 7 non-hydrogen atoms that may have favorable interactions as evidenced by the aliphatic FragMaps in the vicinity of the amidine moieties and the adjacent aromatic rings. Alternatively, the amidine moieties were substituted with phenylamine and amide in 10 and 11. In addition, in the 5, 6 and 7 series the linker length between the aromatic rings was varied from n = 3, as in pentamidine, to n = 6, which is one methylene group longer than heptamidine. To take advantage of the donor FragMaps in the vicinity of acid groups (Fig. 2B) compounds 8 and 9 were designed. These include a primary amino group terminating aliphatic linkers of 5 and 7 methylene groups in 8 and 9, respectively, with different linker lengths between the aromatic rings, such that 8 represents a shorter compound as compared to the two 9 analogs. To address the presence of the aliphatic and aromatic FragMaps in the vicinity of the linker in heptamidine (Fig. 2A), compounds 16a and b were designed, in which a linker of the same length as that in heptamidine is present that contains a tertiary amine to which a benzyl or propyl moiety is attached. An additional compound, 17, was identified later in the project based on the developed SAR model, as described below.
Scheme 2. Preparation of cyclic and fatty amine substituted diamidine analogs.
Reagents and conditions: (a) 1,2-diaminoethane, DMF, rt; (b) 1,3-diaminopropane, DMF, rt; (c) 1,4-diaminobutane, DMF, rt; (d) 1,2-diaminopentane, DMF, rt; (e) 1,2-diaminoheptane, DMF, rt.
CHEMICAL SYNTHESIS
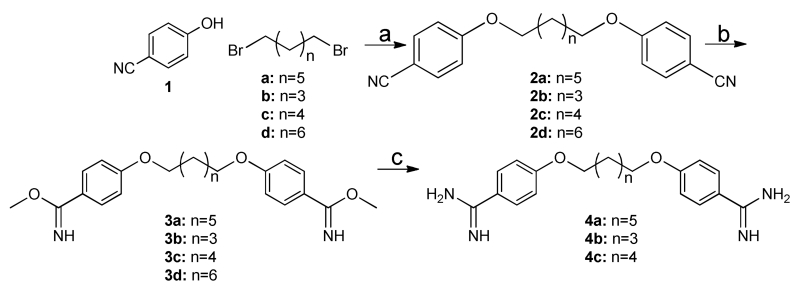
Symmetric molecules flanked by diamidine moieties were synthesized using short, convergent syntheses strategies as shown in schemes 1 to 4. For example, pentamidine derivatives 4a-c were synthesized by the Pinner reaction as described (Scheme 1) starting from 4-cyanophenol23.
Scheme 1. Preparation of diamidines 4a-c.
Reagents and conditions: (a) CsCO3, acetone, reflux, 14 hrs.; (b) methanol:dioxane (1:1), HCl gas; (c) ethanolic ammonia, ammonium chloride, 80 °C, 32 hrs. Cyclic amidines (5a-d, 6a-d, and 7a-d) were synthesized according to literature with some modification (Scheme 2). Specifically, treatment of dibenzimidate with diaminoethane, diaminopropane and diaminobutane at room temperature in DMF accomplished cyclic amidine in satisfactory yield31-33. Increasing the chain length of the diamine results in open chain products (8 and 9a, 9b), which were useful for testing aliphatic chain derivatives of varying lengths (Scheme 2).
Scheme 4. Preparation of analogs with central amine substitutions.
Reagents and conditions: (a) For 13a, Benzylamine, 13b, Propylamine, K2CO3, acetone; (b) 1, CsCO3, acetone; (c) methanol, HCl gas; (d) ethanolic ammonia, ammonium chloride. One additional commercial compound, 17, was included in the study. Commercially available compound 17 maintains many of the advantageous functionalities of the pentamidine scaffold but is a more rigid scaffold since it has fewer rotatable bonds.
For further synthesis of N-derived diamidine compound 10, compound 4a was coupled with phenyl isocyanate to furnish final diamidine compound in good yield. Compound 11 was obtained through the carbomylation of compound 4a with N-succinamidyl-N-methyl carbamate at 40 °C for 2 hours (Scheme 3)34.
Scheme 3. Preparation of N-acyl substituted diamidine analogs.
Reagents and conditions: (a) phenyl isocyanate, DBU, DMF, 40 °C; (b) N-succinamidyl-N-methyl carbamate, DBU, DMF, 40 °C.
Compounds 16a-b were synthesized to further diversify the structure-activity relationship series, N-ligated. For these compounds, 1,3-dibromopropane was reacted with benzylamine and propylamine to obtain compound 13a-b, which then reacted with 4-hydroxybenzonitrile to furnish the intermediate 14a-b. After that, the well-known Pinner reaction was used to prepare the target compounds 16a-b (Scheme 4)35, 36.
RESULTS & DISCUSSION
Characterization of cyclic diamidine analogs bound to Ca2+-S100B (CaS100B)
Twelve compounds prepared in Scheme 2 retained the linker scaffold as found for compounds in Scheme 1; however, the diamidine moiety for these twelve molecules was modified to form a five membered ring (compounds 5a-5d), a six membered ring (compounds 6a-6d), or a seven membered ring (compounds 7a-7d; Scheme 2). These compounds (Scheme 1) were designed to retain hydrogen bond donor capacity and to include aliphatic hydrocarbon regions on the compounds that could potentially provide hydrophobic interactions with residues Leu44, Ile80, and Ala83 of Ca2+-S100B at or nearby the lower portion of Site 1 as indicated by the aliphatic FragMaps adjacent to the amidine moiety (Fig. 2).
Using an established fluorescent polarization competition assay (FPCA)1, compounds 5a-5d, 6a-6d, and 7a-7d were tested to determine if they were able to compete with a probe, TAMRA-TRTK, which binds Site 1 in Ca2+-S100B (Table 1). Of the twelve compounds in this series of cyclic diamidine analogues, they all interacted with Ca2+-S100B as measured using NMR; however, none were able to compete for binding with the TAMRA-TRTK probe at low concentrations in the FPCA (i.e. < 1mM). In cellular assays (Table 2), EC50 values for these compounds were all in the micromolar range (EC50: 5a-d ≈ 20-60 μM; 6a-6b ≈ 200-600 μM; 6c-6d ≈ 25-45 μM; 7a, 7b, 7d ≈ 5-20 μM; 7c ≈ 250-300 μM; Table 2) but the EC50 values for the compounds in all four of these series were all higher than those observed for comparable studies with pentamidine (EC50: 4b=17±2 μM; Table 2) and none showed improvement over that observed for heptamidine, which also demonstrated an S100B specific affect (High S100BEC50: 4b=11±2 μM; Low S100BEC50: 4b=26±4 μM; Table 2). While the NMR spectra of Ca2+-S100B upon binding to 5a, 6a, 6c, and 6d showed similar chemical shift perturbations as pentamidine and heptamidine (Table 3), the binding of the other compounds in this series (5b, 5c, 5d, 6b, 7a-7d) caused significant broadening to the NMR spectra either due to an intermediate exchange binding regime and/or because of protein aggregation. The NMR and FPCA results provided indication that the cyclic diamidine moieties in 5a-5d, 6a-6d, or 7a-7d did not interact with Site 1 of Ca2+-S100B. Nonetheless, X-ray crystallography experiments were initiated and structure determinations were completed for Ca2+-S100B complexes with compounds 5a (SBi4225) and 6b (SBi4214) to further explore this possibility.
Table 1. Tabulation of Binding Assay Results a.
| I.D. | Internal I.D. SBiX |
HSQC b | FPCA IC50 (μM) | Calc. Kd (μM) c |
|---|---|---|---|---|
|
| ||||
| Mean ± S.D. | Mean ± S.D. | |||
| 5a | 4225 | + | 222.1 ± 213.5 | 202.6 ± 19.5 |
| 8 | 4236 | + | 51.1 ± 2.3 | 4.6 ± 0.2 |
| 9a | 4232 | A | 6.7 ± 0.1 | 0.5 ± 0.0 |
| 9b | 4230 | A | 18.3 ± 0.5 | 1.6 ± 0.0 |
| 11 | 4212 | + | 981.7 ± 31.1 | 89.5 ± 2.8 |
| 17 | 29 | + | 66.6 ± 2.0 | 6.0 ± 0.2 |
4a Kd: 1D NMR 23 = 6.9±0.9 μM. CellsEC50 = 26±4 μM; 4b Kd: ITC29 = 50±5μM; Fluorescence29 = 44±4 μM; 1D NMR 23 = 0.13±0.01 μM; CellsEC50 = 26±4 μM; 4c
A = signal lost likely due to formation of soluble aggregates; + = binding; − = no binding.
The Kd was calculated from the FPCA IC50 using the Nikolovska-Coleska equation.
Table 2. Tabulation of Cellular Assay Results.
| Prep. I.D. | Internal I.D. SBiX |
High S100B EC50 (μM) a |
Low S100B EC50 (μM) a |
|---|---|---|---|
|
| |||
| Mean ± S.D. | Mean ± S.D. | ||
| 4a | 4211 | 11.5 ± 2.4 | 25.6 ± 4.0 |
| 4b | 1 | 16.3 ± 1.7 | 16.7 ± 2.4 |
| 4c | 4210 | 245.2 ± 15.5 | 234.0 ± 7.9 |
| 5a | 4225 | 32.2 ± 1.5 | 21.2 ± 1.5 |
| 5b | 4224 | 56.2 ± 7.0 | 38.9 ± 6.8 |
| 5c | 4226 | 52.0 ± 4.5 | 34.5 ± 4.6 |
| 5d | 4221 | 21.8 ± 4.4 | 24.3 ± 5.6 |
| 6a | 4213 | 612.5 ± 232.9 | 288.9 ± 106.0 |
| 6b | 4214 | 203.1 ± 147.7 | 283.6 ± 221.1 |
| 6c | 4218 | 44.1 ± 35.0 | 42.7 ± 43.2 |
| 6d | 4217 | 24.8 ± 8.9 | 37.9 ± 32.9 |
| 7a | 4227 | 7.8 ± 1.6 | 9.2 ± 2.1 |
| 7b | 4223 | 20.0 ± 3.0 | 19.0 ± 3.7 |
| 7c | 4228 | 264.2 ± 107.3 | 301.2 ± 139.2 |
| 7d | 4222 | 5.4 ± 0.9 | 4.8 ± 0.9 |
| 8 | 4236 | 37.1 ± 3.6 | 33.3 ± 0.5 |
| 9a | 4232 | 24.3 ± 6.3 | 25.4 ± 6.8 |
| 9b | 4230 | 46.3 ± 5.6 | 45.8 ± 6.4 |
| 10 | 4235 | 0.4 ± 0.1 | 0.5 ± 0.1 |
| 11 | 4212 | 130.8 ± 18.1 | 146.1 ± 17.4 |
| 16a | 4239 | 51.8 ± 6.0 | 41.0 ± 7.0 |
| 16b | 4238 | 863.6 ± 103.2 | 500.1 ± 128.6 |
| 17 | 29 | <1.0 | <1.0 |
The SYBR Green cell proliferation assay was done using WM115 melanoma cells transfected with shRNAscrambled (i.e. high S100B) or shRNAS100B (low S100B).
Table 3. Chemical Shift Perturbations for Residues in caS100B upon the Addition of Inhibitorsa.
| I.D. | Helix 1 | Loop 2 (hinge) | Helix 3 | Helix 4 | C-terminal loop |
|---|---|---|---|---|---|
| 4b 16 | A9 ILL F14 H15 | S41 L44 E45 E46 | V52 T59 | M74 V77 A83 | E89 H90 |
| 11 | K5 A9 | L40 K48 | A75 V77 S78 T81 T82 A83 C84 H85 | ||
| 6a | S1 | H42 E45 | S78 T81 T82 C84 H85 | E86 F88 E89 | |
| 6b | S1 | H42 | S78 T81 T82 C84 H85 | E86 F88 E89 | |
| 16b | S1 ILL F14 | H42 E45 | S78 T81 T82 | E89 H90 | |
| TRTK16 | L40 H42 L44 E45 I47 | V52 K55 V56 T59 | M79 V80 A83 C84 | F87 | |
| p5316 | L44 | V52 V56 | M79 V80 | F87 |
Mapping of the binding site on caS100B using chemical shift perturbations (Δ1H + Δ 15N) for residues upon addition of inhibitors.
Global Conformation
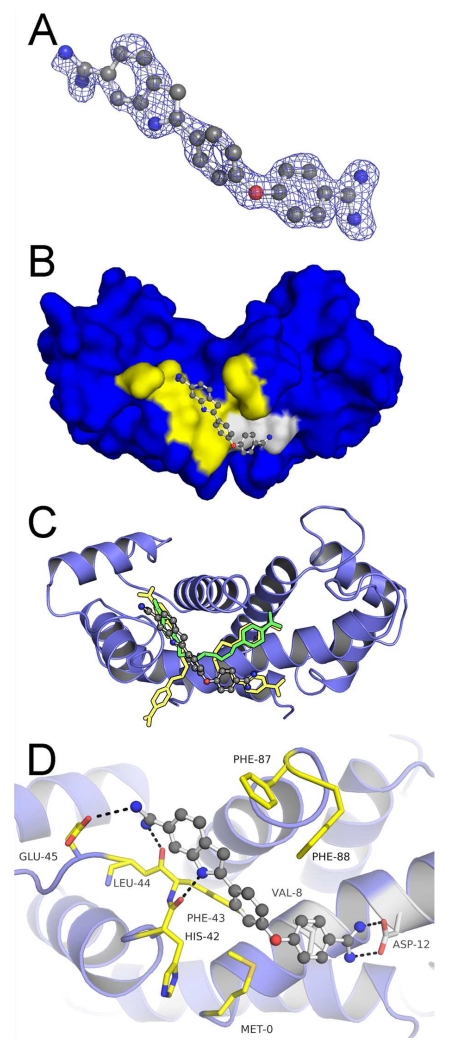
X-ray structures of Ca2+-S100B bound to either 5a or 6b consists of four helixes (helix 1, E2-G19; helix 2, K28-L40; helix 3, E49-D61; helix 4, D69-F86) and is assembled as an active symmetric homodimer (Fig. 1a), as found previously in other structures of S100B. Both the typical and non-canonical EF-hand calcium binding domains in each subunit were found to coordinate a Ca2+ ion in the two structures29, 30, 37, 38. A comparison with the structure of Ca2+-S100B in the absence of compound (PDB ID: 1MHO39) shows a strong conservation of the global fold (see Supporting Information Table S1) with the only significant deviation observed at C-terminal residues His85-Glu91 involving, most notably, residues Phe87 and Phe88 (Fig. 3d,4d). This segment of the polypeptide consistently conforms to accommodate the binding of the SBiX molecules, as similarly found for the binding of pentamidine and heptamidine.
Figure 3. Crystallographic Structure of the S100B•5a Complex (PDB ID: 5DKN).
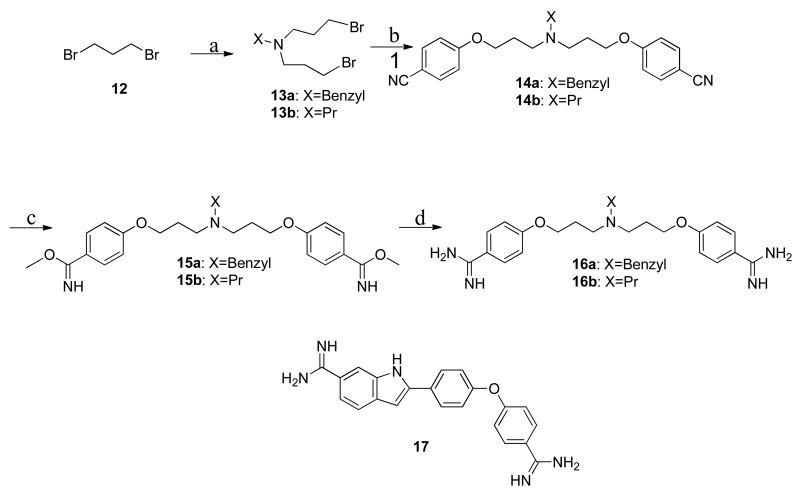
(A) |Fo|-|Fc| electron density omit map (blue mesh) of 5a (ball and stick) contoured at 2.5σ levels. (B) 5a within Sites 2 (yellow highlight) and 3 (gray highlight) of the S100B homodimer (surface in blue). (C) Modeled orientation of 5a overlaid on the previously elucidated compound-bound structures of pentamidine and heptamidine. (D) The specific interactions of 5a within dimeric S100B (blue ribbons) are rendered with residues (sticks) within 4Å of the 5a highlighted in yellow (Site 2) or gray (Site 3). 5a is situated within a hydrophobic pocket with atoms within hydrogen bond distance at the backbone carbonyl of His85 and the sidechain of Cys84.
Figure 4. Crystallographic Structure of the S100B•6b Complex (PDB ID: 5DKQ).
(A) |Fo|-|Fc| electron density omit map (blue mesh) of 6b (ball and stick) contoured at 2.5σ levels. (B) 6b within Sites 2 (yellow highlight) and 3 (gray highlight) of the S100B homodimer (surface in blue). (C) Modeled orientation of 6b overlaid on the previously elucidated compound-bound structures of pentamidine and heptamidine. (D) The specific interactions of 6b with dimeric S100B (blue ribbons) is rendered with residues (sticks) within 4Å of the 6b highlighted in yellow (Site 2) or gray (Site 3). 6b is situated within a hydrophobic pocket with atoms within hydrogen bond distance at the backbone carbonyl of His85 and the sidechain of Cys84.
CaS100B•5a
The X-ray structure of 5a bound, calcium loaded S100B (CaS100B•5a) was solved at 1.53 Å resolution (Fig. 3a). The asymmetric unit contained one S100B monomer with the second monomer forming the biologically active dimeric form of S100B contained within the crystal symmetry. Residues Met0 to His90 were modeled. The monomer in the CaS100B•5a structure bound two calcium ions and a 5a molecule. In the final refined model (Rcrys/Rfree of 0.195/0.216), all residues were in the favored (100%) region of the Ramachandran Plot with no outliers (see Table 4).
Table 4. Statistics of Reflection Data and Structure Refinements. a.
| Ligand | 5a | 6b | 17 |
| Space Group | P41212 | P41212 | P212121 |
| Unit Cell Dimension (Å) | 64.0, 64.0. 47.6 | 63.5, 63.5, 48.3 | 45.7, 47.4, 90.3 |
| Unit Cell Angles(°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution Range (Å) | 47.56 – 1.53 | 38.43 – 1.59 | 33.30 – 1.74 |
| Reflections Observed | 74,444 (10,810) | 97,324 (4,162) | 82,661 (3,111) |
| Unique Reflections | 15,494 (2,197) | 13,794 (663) | 20,971 (951) |
| Reflections Rfree Set | 1,546 | 1,376 | 1,994 |
| Completeness (%) | 99.8 (99.6) | 99.9 (99.3) | 98.7 (83.4) |
| Redundancy | 4.8 (4.9) | 7.1 (6.3) | 3.9 (3.3) |
| <I/σ> | 26.1 (3.8) | 23.3 (1.5) | 11.8 (1.2) |
| CC1/2 (%) | 99.9 (90.5) | 99.9 (51.7) | 99.7 (68.2) |
| Rsymb | 0.048 (0.347) | 0.046 (1.28) | 0.055 (0.693) |
| Rcrysb | 0.195 | 0.207 | 0.209 |
| Rfreeb | 0.216 | 0.230 | 0.247 |
| No. of Amino Acids | 91 | 91 | 179 |
| No. of Protein Atoms | 747 | 743 | 1,457 |
| No. of Hetero Atoms | 33 | 33 | 33 |
| No. of Waters | 89 | 108 | 172 |
| RMSDc Bond Lengths (Å) | 0.012 | 0.006 | 0.013 |
| Angles (°) | 0.937 | 0.971 | 1.217 |
| Mean B Factor | 18.59 | 31.69 | 30.20 |
| Protein Atoms (Å2) | 17.18 | 30.32 | 29.13 |
| Hetero Atoms (Å2) | 21.54 | 38.29 | 31.58 |
| Water Atoms (Å2) | 25.92 | 39.05 | 39.03 |
| Ramachandran Outliers (%) | 0.0 | 0.0 | 0.0 |
| Ramachandran Favored (%) | 100.0 | 100.0 | 100.0 |
The numbers in parentheses represent values from the highest resolution shell: caS100B•5a (1.61-1.53Å), CaS100B•6b (1.62-1.59Å), and CaS100B•17 (1.77-1.59Å).
Rsym = Σ h(Σ j|Ihj - <Ih>|/ Σ Ihj), where h=set of Miller indices, j=set of observations of reflection h, and <Ih>=the mean intensity. Rcrys= Σh||Fo,h| – |Fc,h||/ Σh|Fo,h|. Rfree was calculated using a percentage of the complete data set excluded from refinement.
RMSD values are deviation from ideal values
CaS100B•6b
The X-ray structure of 6b bound, calcium loaded S100B (CaS100B•6b) was solved at 1.59 Å resolution (Fig. 4a). The asymmetric unit contained one S100B monomer with the second monomer forming the biologically active dimeric form of S100B contained within the crystal symmetry. Residues Met0 to His90 were modeled. The monomer in the CaS100B•6b structure bound two calcium ions and a 6b molecule. In the final refined model (Rcrys/Rfree of 0.207/0.230), all residues were in the favored (100%) region of the Ramachandran Plot with no outliers (see Table 4).
Structural comparisons of CaS100B•5a, CaS100B•6b, CaS100B•Pentamidine, and CaS100B•Heptamidine
As both 6b and 5a are derivative compounds based on the previously published SBiX molecules pentamidine (4b) and heptamidine (4a), they share similar binding sites and interactions as their predecessors23, 29 (Fig. 3b,c and Fig. 4b,c). These derivatives bind within close proximity of several residues of S100B in a pocket formed by Helix 4, Loop2, and Helix1 ’ (Fig. 3d and Fig. 4d). The diazacyclohexene and the imidazoline groups of 6b and 5a, respectively, are able to participate in hydrogen bonds with the backbone carbonyl His85 (Helix 4) while participating in stacking interactions with Phe88 and Phe87 (Helix4). Such interactions were previously described in pentamidine and heptamidine structures. Similar to both pentamidine and heptamidine, 5a and 6b occupy both binding Sites 2 and 3. However, like heptamidine and unlike pentamidine, the derivatives occupy both sites with a single molecule. When compared to the preceding compounds, the larger substituent groups of 5a and 6b within Site 2 do not extend into Site 1, consistent with the FPCA results above. Rather, in both cases, the ring starts to protrude into solvent with few if any interactions with the protein. On the other hand, within Site 3 the larger ring displaces the benzyl moiety 0.9-1.2 Å from the center in both cases (i.e. for 5a and 6b) while the second benzyl moieties occupy the same hydrophobic site formed by the sidechains of Leu40, Phe43, and Phe87. However, neither derivative compound makes the hydrogen bond with the backbone carbonyl of Phe43 as observed with heptamidine.
Characterization of non-cyclic aliphatic amine substituted diamidine analogs
Three additional compounds (8, 9a, 9b) prepared in Scheme 2 retained the linker scaffold as in the Scheme 1 compounds; however, the diamidine moiety for these three molecules was modified to include long-chain aliphatic primary amine moieties. It was thought that these compounds would also retain a hydrogen bond donor as found in the original diamidines (in Scheme 1), but extend into Sites 1 and 3 to occupy regions indicated by donor SILCS FragMaps adjacent to acidic residues (Fig 2B). In addition, the linkers may provide hydrophobic interactions with residues of Ca2+-S100B at or nearby the lower portion of Site 1, also termed here as a channel connecting Sites 1 and Sites 2 (i.e. with residues Phe43, Leu44, Ile80, and Ala83) as indicated by the SILCS FragMaps (Fig. 2).
The in vitro fluorescence polarization competition assay (FPCA) was completed with these three compounds and they were all (8, 9a, 9b) found to compete with the Site 1 probe, so IC50’s were measured and the dissociation constants determined to be in the low micromolar range in all cases (<5 μM; see Table 1). However their EC50 values in cells were significantly higher than their KD values (EC50: 8 ≈ 35-40 μM; EC50: 9a-b ≈ 25-50 μM), so off-target affects are likely in each of these cases. Likewise, all three of these compounds interacted with Ca2+-S100B as measured using NMR and in the case of compound 8, it showed similar chemical shift perturbations as pentamidine and heptamidine, as well as numerous additional perturbations. The other two compounds (9a, 9b) caused significant broadening to the NMR spectra either due an intermediate exchange and/or because of protein aggregation. In these cases, the NMR and FPCA results provided indication that the long-chain primary amine moiety did indeed interact with Site 1 (Table 1). Nonetheless, X-ray crystallography experiments were initiated and structure determinations were attempted for Ca2+-S100B complexes with compounds 8, 9a, and 9b to further explore this possibility.
Co-crystals of 8, 9a, and 9b were obtained from conditions similar to that of 6b and 5a. Although an examination of electron density maps could confirm the presence of small-molecule ligands occupying the predicted binding sites, this sub-family of compounds maintained low occupancy despite various attempts at improvement. Amongst these compounds, the S100B•9a crystal diffraction data provided the best ligand electron density, and the atoms of benzamidine-like chemical groups could be accurately modeled. However, the acyl chains terminated with amino groups could not be tracked in the electron density with the same confidence. Therefore, in silico methods were used to predict the positions of atoms with weak and/or missing electron density (see Supporting Information Fig. S1). Both AutoDock and MC-SILCS sampling similarly place the linker alkyl chain. The location of one of the terminal alkyl chains predicted by AutoDock places the amino group such that it hydrogen bonds with Glu86 and His85. The location of the second amino group does not allow hydrogen bonding with the protein. The only favorable interactions would be with the hydrophobic environment provided by the sidechains of Leu44, Ala83, and Phe88. MC-SILCS, on the other hand, places the first amino group in the vicinity of Glu2 (of the other S100B chain), and the second group close to Glu46, forming hydrophilic interactions in both cases. These locations are associated with the positive donor SILCS FragMap adjacent to these residues leading to favorable placement of the basic group (see Supporting Information Fig. S1). The MC-SILCS docking also point to the diversity of conformations populated by the terminal groups. The additional hydrogen bonding predicted by AutoDock and/or MC-SILCS would explain the increased affinity of this sub-family for S100B as measured by FPCA. The diversity of orientations found by the two methods is also consistent with the alkyl tails not being resolved in the crystal structure. The small differences in affinity between the amino group containing compounds are likely due to the varying lengths of linkers and associated positions of the amino groups, which would likely impact the hydrogen bond network between the ligands and the protein. Importantly, the SILCS modeling nicely explains why these molecules compete with TRTK12 since an interaction at Glu46 would compete with the interactions between TRTK12 and S100B as seen in the co-crystal structure37.
Characterization of N-acyl substituted diamidine analogs
Two compounds (10, 11) were prepared as shown in Scheme 3 that retained the heptamidine linker scaffold; however, the diamidine moieties for these molecules were designed to retain two hydrogen bond donors as found in heptamidine, but also have some aliphatic character either via a phenyl ring (compound 10) or with a methyl group (compound 11). Both of these compounds had an affect the cellular assays with compound 10 having contributions from general toxicity effects (EC50: 10 < 1 μM; EC50: 11 ≈ 130-150 μM). The in vitro fluorescence polarization competition assay (FPCA) was completed with these compounds and neither was able to compete with TAMRA-TRTK indicating that they do not interact with Site 1 despite their ability to bind Ca2+-S100B as determined by NMR (see Supporting Information Fig. S2-5). 11 showed a significant number of chemical shift perturbations that mimicked those found for pentamidine and heptamidine (see Supporting Information Fig. S6). 10 did not perturb chemical shifts at the concentrations tested. Although, X-ray crystallography experiments were initiated, crystallization of Ca2+-S100B complexes with compounds 10 and 11 were not successful. While the atomic level detail of compounds 10 and 11 bound to Ca2+-S100B remains somewhat elusive, the NMR and FPCA data indicate that neither of these compounds extends into Site 1.
Characterization of diamidine analogs with a central substituted amine
Two compounds (16a, 16b) were prepared as shown in Scheme 4 that retained the symmetric diamidine moieties; however, the heptamidine linker was designed to include a substituted secondary amine. Such a substitution was designed to complement a hydrophobic pocket that is indicated in the SILCS FragMaps and observed in the heptamidine X-ray crystal structure 23. For these complexes, the in vitro fluorescence polarization competition assay (FPCA) was completed and as expected, neither was able to compete with TAMRA-TRTK. Unexpectedly, the compounds bound weakly and the chemical shift perturbations were unlike those observed for pentamidine and heptamidine suggesting an alternative-binding mode. The EC50 values for these compounds were measured in cellular assays and found to affect cell death quite differently with 16b having little if any effect on cellular functions (EC50: 16a ≈ 40-50 μM; EC50: 16b > 500 μM).
Characterization of an asymmetric diamidine molecule
In all of the structural studies completed thus far with symmetric diamidines, it was becoming clear that the position of Phe88 in the pocket between Sites 1 and 2 was a common feature. Such conformation of the protein target necessitated the synthesis of very large molecules to circumvent this occlusion and to compete with a Site 1 binding peptide (see Supporting Information Fig. S1). This requires a compound that eliminates the symmetric nature of the pentamidine/heptamidine scaffold such that only Site 3 interactions would occur. In addition, it was of interest to determine whether it was possible to have the channel between Sites 1 and 2 open while occupying Sites 2 and 3 with an inhibitor. Using chemical similarity searching with the program MOE (Chemical Computing Group) we searched an in house database of commercially available molecules for those that contain a single aromatic amidine moiety as present in heptamidine. One commercial molecule, compound 17, fit these criteria and was tested for its ability to bind to Ca2+-S100B. Remarkably, 17 was found to bind Ca2+-S100B via NMR, has a binding affinity in the low micromolar range (KD=6.0 ± 0.2 μM; Table 1), and had a low EC50 in cellular assays (EC50 < 1 μM; Table 2) likely due to contributions from general toxicity affects. Therefore, compound 17 is not itself an endpoint for a pharmaceutical, but it turned out to provide very important information for structure-based drug design. Specifically, 17 was found to inhibit TAMRA-TRTK binding into Site 1 in the FPCA, despite being designed to interact with Sites 2 and 3. Notably, this is the first small molecule with a pentamidine-like scaffold (M.W. < 500 Da) shown to block the Site 1 binding peptide TAMRA-TRTK. X-ray crystal determination of CaS100B•17 was thus undertaken to identify the binding site of 17.
CaS100B•17
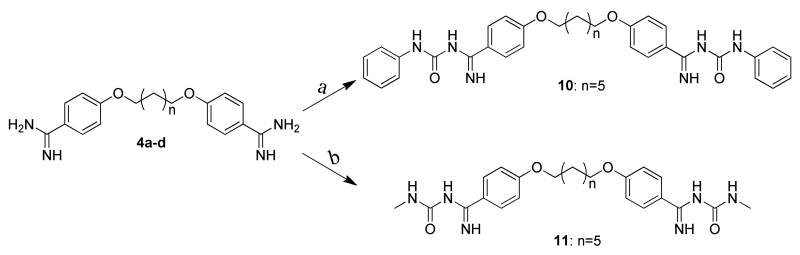
The X-ray structure of 17 bound, calcium loaded S100B (CaS100B•17) was solved at 1.74 Å resolution (Fig. 5a). The asymmetric unit contained two S100B monomers. Residues Met0 to Phe88 were modeled in monomer A while residues Met0 to Glu89 were modeled in monomer B. The monomers in the CaS100B•17 structure bound two calcium ions. Only one molecule 17 was modeled as the symmetrical binding cleft formed by the dimer remained vacant. In the final refined model (Rcrys/Rfree of 0.209/0.247), all residues were in the favored (100%) region of the Ramachandran Plot with no outliers (see Table 4). The occupation of the binding cleft by 17 in Sites 2 & 3 was discovered to indeed be the case, but it required an entirely different conformation of helix 4 versus that observed in the structures of the other pentamidine/heptamidine scaffold compounds. In fact, 17 binds a more native-like conformation (unliganded) of Ca2+-S100B while other pentamidine derivatives require a breaking of the last turn of helix 4 coinciding with a repositioning of Phe87 and Phe88 in stacking position with the cyclic moieties (Fig. 1c). Although the global positioning of 17 in Site 2 & 3 remains similar (Fig. 4b,c), there are other major differing interactions with the protein. The carbamimidoyl-phenoxyl group of 17, which would be expected to hydrogen bond with the backbone carbonyl of His85, as occurs with pentamidine, lies in the hydrophobic pocket formed by Val8, Phe88, and Met0, where it makes bidentate hydrogen bonds with the sidechain carbonyl of Asp12 (Fig. 5d). In addition, the indole-carboximidamide of 17 is in the hydrophobic pocket formed by Ala83, Phe43, and Phe87 and makes hydrogen bonds with not only the backbone carbonyl of Phe43 but also the backbone carbonyl His42 and the sidechain carbonyl of Glu45. Notably, the binding orientation of 17 is such that the FF-gate residue, Phe88, assumes an open conformation that allows access between Sites 1 and 2. Thus, when comparing all of the inhibitor complex structures, so far available for Ca2+-S100B, there are two major conformations observed for the position of the C-terminal residues. One is with Phe88 within the small channel connecting Sites 1 & 2 (i.e. the “closed” FF-gate) and the other is with this small channel open and Phe88 outside this narrow channel (i.e. the “open” FF-gate) and part of helix 4 (Fig. 5).
Figure 5. Crystallographic Structure of the S100B•17 Complex (PDB ID: 5DKNR).
(A) |Fo|-|Fc| electron density omit map (blue mesh) of 17 (ball and stick) contoured at 2.5σ levels. (B) 17 within Sites 2 (yellow highlight) and 3 (gray highlight) of the S100B homodimer (surface in blue). (C) Modeled orientation of 17 overlaid on the previously elucidated compound-bound structures of pentamidine and heptamidine. (D) The specific interactions of 17 with dimeric S100B (blue ribbons) is rendered with residues (sticks) within 4Å of the 17 highlighted in yellow (Site 2) or gray (Site 3). Compound 17 is situated within a hydrophobic pocket with atoms within hydrogen bond distance at the backbone carbonyls of His42 and Phe43, and the sidechains of Aps12 andGlu45.
CONCLUSIONS
Three binding sites or “hot spots” on Ca2+-bound S100B were recognized in structural studies with several different CaS100B•peptide and CaS100B•SBiX complexes38, 40, 41. Pentamidine, which binds to Sites 2 and 3, exhibits enough potency to be used in clinical trials for malignant melanoma. However, its low efficacy, poor specificity, and toxicity issues limit its therapeutic potential and needed to be addressed. The development of heptamidine was a reasonable improvement as indicated by increased efficacy in cellular assays, with the efficacy shown to be dependent on S100B levels (Table 2). In the evaluation of heptamidine, a more stable knockdown of S100B was developed and used, and selectivity in killing melanoma cells with high S100B over those with low S100B was clearly observed.
In this study, we attempted to extend the heptamidine scaffold from binding Sites 2 and 3 towards Site 1. The addition of aliphatic amine groups indeed showed significant competition with the Site 1 and can be used as a molecular probe of the S100B binding pocket. Also, the results here indicate the optimal length of linker region between the aromatic moieties and from the amidine to amine are seven methylene groups. From the available crystallographic structure of S100B in complex with heptamidine 16, the importance of hydrogen bonding made with the backbone carbonyl of Phe43 may be assumed. Although higher affinity binders were observed in vitro, it did not improve the efficacy in cellular assays when compared to many of the other diamidine derivatives (see Table 2); this result may be due to decreased cellular absorption or unfavorable solubility.
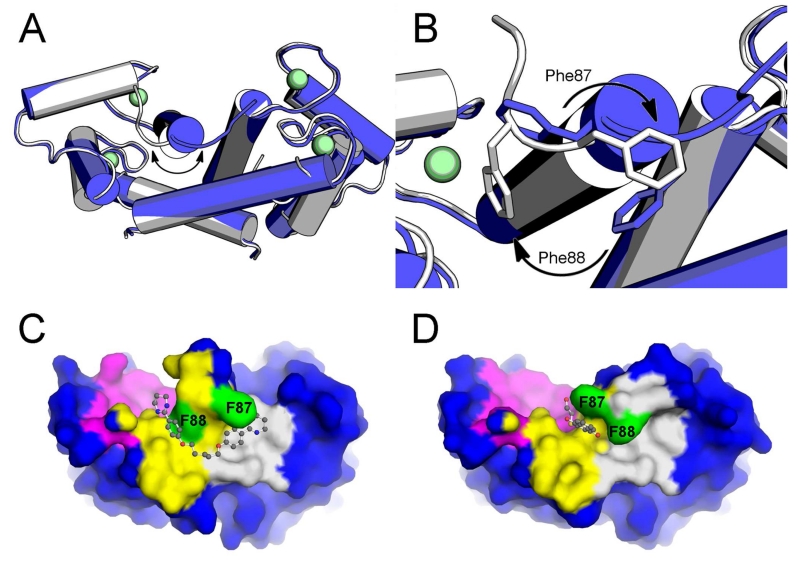
Further analysis of the crystal structures of S100B in complex with symmetric pentamidine analogs revealed a consistent reorganization of the C-terminal helix from the native state towards the closing of the “FF-gate”. This includes heptamidine even though such a reorganization of the C-terminal helix was not reported in the original coordinates deposited in the PDB. A reexamination of the deposited raw data for CaS100B-heptamidine (structure factors) and newly calculated maps, in light of these new structures, showed that the heptamidine-bound state of S100B does indeed exhibit the same reorganization of the C-terminal helix associated with a “closed” FF-gate, with Phe-88 blocking the cavity connecting Sites 1 & 2, as found here for structures of CaS100B•5a and CaS100B•6b. Thus, the protein conformational change necessary for the binding of symmetric pentamidine analogs leads to closing of the FF-gate, which pushes the inhibitors away from Site 1 and towards bulk solvent. As the occupation of Site 1 (the p53-peptide binding site) is predicted to be important for successful inhibition of the targeted PPI, the dependence on this conformation change for binding has proven to be problematic. The inability of pentamidine/heptamidine and many of the analogs to compete effectively with the peptide probe of Site 1 may, at least partially, be explained by the need for the FF-gate to be in the “open” conformation with a more native-like orientation of Phe-87 and Phe-88. Relaxation dispersion curves of S100B in the absence and presence of peptides or small molecule inhibitors indicate that the C-terminus of Ca2+-S100B exists in at least two slowly exchanging conformations in the millisecond time regime in solution, consistent with the FF-gating mechanism proposed here 42 (Fig. 6). Notably, by breaking the symmetry of heptamidine and its other analogs we identified a compound, 17, that binds to the “open” FF-gate while maintaining several desirable features of the pentamidine scaffold. Unfortunately, the killing efficacy of 17 far exceeds in binding affinity and shows no S100B-dependent killing in S100B-knockdown cell lines. Thus, compound 17 does not resolve the toxicity problems of pentamidine-like compounds, but its structure bound to CaS100B represents an important advance in S100B inhibitor design, perhaps as a linker, since it enables the design of inhibitors that can take advantage of all three hot spots in CaS100B for the first time (i.e. Sites 1, 2 and 3; Figure 1).
Figure 6. Small molecule inhibitors of Ca2+-S100B reveal two protein conformations.
(A) Overlaid crystallographic structures (tubes) of S100B in uninhibited (PDB ID: 1MHO, blue) and pentamidine (and analog, PDB ID: 5DKQ) inhibited conformations. A substantial conformational shift is observed in the C-Terminal Loop region. (B) A close-up rendering of the C-Terminal Loop region reveals that Phe87 and Phe88 (sticks) are the major contributors to the conformational shift. (C) Surface rendering showing that a bound pentamidine analog (ball and stick, PDB ID: 5DKQ) has difficulty accessing Site 1(magenta) from Site 2 (yellow) because of the obstruction caused by intruding phenylalanine sidechains (green). (D) Other non-pentamidine-like inhibitors (ball and stick, PDB ID: 4PE0) have ready access to Site 1 from Site 2 when the phenylalanine sidechains are in a native conformation30.
EXPERIMENTAL PROCEDURES
All reactions were performed under an atmosphere of nitrogen, and all solvents were removed on a rotary evaporator under reduced pressure. TLC was performed on plates coated with silica gel GHLF-0.25 mm plates (60 F254) (Analtech). Mass spectra were obtained on a ThermoFinnigan LCQ Classic. 1H and 13C NMR spectra were obtained using a 500 MHz Varian NMR Spectrometer. Melting points were determined in open capillary tubes using a Mel-Temp melting point apparatus; melting points are uncorrected. Combustion analysis was performed by Atlantic Microlab, Inc. (Norcross, GA). Synthesis follows that of Hisashi et al. (Scheme 1-4) Purity of all of the compounds synthesized and/or used in this study were determined to be >95% by HPLC analyses (see Supporting Information Fig. S7-29).
Heptamidine (4a), pentamidine (4b), hexamidine (4c)
Compounds 4a-c were synthesized by reported procedure23 as shown in scheme 1.
1,7-bis (4-(4, 5-dihydro-1H-imidazol-2-yl)phenoxy)heptane (5a)
3a (0.1 g, 0.25 mmol) and 1,2-diaminoethane (0.075 g, 1.25 mmol) were dissolved in DMF (2 mL) and stirred at room temperature for overnight. DMF was removed under vacuum (Scheme 2). The crude reaction mixture was purified on neutral alumina column (6:94 Methanol/Chloroform) to yield the pure product 5a as a white solid Yield 61%; Mp (250-252 °C). 1H NMR (400 MHz, DMSO-d6): δ 7.98 (d, J = 8.8 Hz, 4H), 7.11 (d, J = 8.8 Hz, 4H), 4.07 (t, J = 6.2 Hz, 4H), 3.85 (m, 8H), 1.72-1.74 (m, 4H), 1.41 (m, 6H); 13C NMR (100 MHz, DMSO-d6): δ 163.8, 162.6, 130.3, 116.0, 114.7, 67.9, 45.3, 28.3, 25.3; MS (ESI) m/z: 421 [M+H]
Compounds 5b-d were similarly synthesized as per the above procedure (Scheme 2).
1,5-bis(4-(4,5-dihydro-1H-imidazol-2-yl)phenoxy)pentane (5b)
White solid; Yield 61%; Mp (250-252 °C); 1H NMR (400 MHz, DMSO-d6): δ 8.01 (d, J = 8.8 Hz, 4H), 7.13 (d, J = 8.4 Hz, 4H), 4.11 (t, J = 6.4 Hz, 4H), 3.83 (m, 8H), 1.77-1.82 (m, 4H), 1.57-1.59 (m, 2H); 13C NMR (100 MHz, DMSO-d6): δ 164.3, 162.2, 130.9, 115.3, 68.4, 45.4, 28.5, 22.4; MS (ESI) m/z: 393 [M+H]
1,6-bis(4-(4,5-dihydro-1H-imidazol-2-yl)phenoxy)hexane (5c)
Off white solid, Yield 63%; Mp (221-223 °C); 1H NMR (400 MHz, DMSO-d6): δ 8.00 (d, J = 8.4 Hz, 4H), 7.12 (d, J = 8.4 Hz, 4H), 4.09 (t, J = 6.2 Hz, 4H), 3.85 (m, 8H), 1.76 (m, 4H), 1.48 (m, 4H); 13C NMR (100 MHz, DMSO-d6): δ 164.3, 163.3, 130.9, 115.8, 115.3, 68.4, 45.4, 28.8, 25.5; MS (ESI) m/z: 407 [M+H]
1,8-bis (4-(4, 5-dihydro-1H-imidazol-2-yl) phenoxy) octane (5d)
Off white solid, Yield 61%; Mp (198-200 °C); 1H NMR (400 MHz, DMSO-d6): δ 8.01 (d, J = 8.4 Hz, 4H), 7.14 (d, J = 8.4 Hz, 4H), 4.08 (t, J = 6.4 Hz, 4H), 3.91 (m, 8H), 1.71-1.75 (m, 4H), 1.35-1.42 (m, 8H); 13C NMR (100 MHz, DMSO-d6): δ 163.9, 163.1, 130.6, 114.9, 68.1, 44.6, 28.6, 28.3, 25.3; MS (ESI) m/z: 435 [M+H]
1,7-bis (4-(1,4,5,6-tetrahydropyrimidin-2-yl)phenoxy)heptane (6a)
3a (0.1 g, 0.25 mmol) and 1,3-diaminopropane (0.092 g, 1.25 mmol) were dissolved in DMF (2 mL) and stirred at room temperature for overnight. DMF was removed under vacuum (Scheme 2). The crude reaction mixture was purified on neutral alumina column (6:94 Methanol/Chloroform) to yield the pure product 6a as a white solid, Yield 65%; Mp (183-185 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.72 (d, J =8.4 Hz, 4H), 7.12 (d, J = 8.4 Hz, 2H), 4.06 (t, J = 6.4 Hz, 4H), 3.45-3.47 (m, 8H), 1.94 (m, 4H), 1.72-1.74 (m, 4H), 1.41 (m, 6H); 13C NMR (100 MHz, DMSO-d6): δ 162.8, 158.8, 129.8, 120.4, 115.1, 68.4, 28.8, 25.8, 18.3; MS (ESI) m/z: 449 [M+H]
Compounds 6b-d were similarly synthesized as per the above procedure (Scheme 2).
1,5-bis(4-(1,4,5,6-tetrahydropyrimidin-2-yl)phenoxy)pentane (6b)
White solid, Yield 62%; Mp (99-101 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.83 (d, J = 8.8 Hz, 4H), 7.23 (d, J = 8.4 Hz, 4H), 4.19 (t, J = 6.2 Hz, 4H), 3.55 (m, 8H), 1.92-2.05 (m, 4H), 1.87-1.92 (m, 4H), 1.67-1.9 (m, 2H); 13C NMR (100 MHz, DMSO-d6): δ 162.8, 158.8, 129.8, 120.5, 115.1, 68.3, 28.6, 22.5, 18.3; MS (ESI) m/z: 421 [M+H]
1,6-bis(4-(1,4,5,6-tetrahydropyrimidin-2-yl)phenoxy)hexane (6c)
3c (0.1 g, 0.26 mmol); and 1,3-diaminopropane (0.096 g, 1.3 mmol) were reacted using the same procedure as the one described for the synthesis of 6a, providing 6c as white solid, Yield 61%; Mp (274-276 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.72 (d, J = 8.4 Hz, 4H), 7.13 (d, J = 9.2 Hz, 4H), 4.08 (t, J = 6.2 Hz, 4H), 3.44-3.48 (m, 8H), 1.94-1.97 (m, 4H), 1.76 (m, 4H), 1.49 (m, 4H); 13C NMR (100 MHz, DMSO-d6): δ 162.8, 158.8, 129.8, 120.5, 115.1, 68.4, 28.8, 25.6, 18.3; MS (ESI) m/z: 435 [M+H]
1,8-bis(4-(1,4,5,6-tetrahydropyrimidin-2-yl)phenoxy)octane (6d)
White solid, Yield 64%; Mp (260-262 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.73 (d, J = 8.8 Hz, 4H), 7.13 (d, J = 8.8 Hz, 4H), 4.06 (t, J = 6.6 Hz, 4H), 3.34-3.47 (m, 8H), 1.94-1.96 (m, 4H), 1.72-1.75 (m, 4H), 1.36-1.42 (m, 8H); 13C NMR (100 MHz, DMSO-d6): δ 162.8, 158.8, 129.8, 115.1, 68.4, 29.1, 28.9, 25.8, 18.3; MS (ESI) m/z: 463 [M+H]
1,7-bis(4-(4,5,6,7-tetrahydro-1H-1,3-diazepin-2-yl)phenoxy)heptane (7a)
3a (0.1 g, 0.25 mmol) and 1,4-diaminobutane (0.11 g, 1.25 mmol) were dissolved in DMF (2 mL) and stirred at room temperature for overnight. DMF was removed under vacuum (Scheme 2). The crude reaction mixture was purified on neutral alumina column (6:94 Methanol/Chloroform) to yield the pure product 7a as a white solid, Yield 60%; Mp (169-171 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.71 (m, 4H), 7.1 (m, 4H), 4.06 (m, 4H), 3.62 (m, 8H), 1.92 (m, 8H), 1.73 (m, 4H), 1.42 (m, 6H); 13C NMR (100 MHz, DMSO-d6): δ 164.5, 163.3, 131.5, 121.6, 114.9, 68.4, 28.8, 25.9, 25.7; MS (ESI) m/z: 477 [M+H]
Compounds 7b-d were similarly synthesized as per the above procedure (Scheme 2).
1,5-bis (4-(4,5,6,7-tetrahydro-1H-1,3-diazepin-2-yl)phenoxy)pentane (7b)
Off white solid, Yield 59%; Mp (153-155 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.74 (d, J = 6.00 Hz, 4H), 7.10 (d, J = 6.00 Hz, 4H), 4.10 (m, 4H), 3.63 (m, 8H), 1.92 (m, 8H), 1.80 (m, 4H), 1.57 (m, 2H); 13C NMR (100 MHz, DMSO-d6): δ 164.4, 163.2, 131.5, 121.6, 115.0, 68.4, 43.9, 28.5, 25.9, 22.4; MS (ESI) m/z: 449 [M+H]
1,6-bis(4-(4,5,6,7-tetrahydro-1H-1,3-diazepin-2-yl)phenoxy)hexane (7c)
White solid, Yield 57%; Mp (174-176 °C); 1H NMR (500 MHz, DMSO-d6): δ 7.57-7.71 (m, 4H), 7.09-7.21 (m, 4H), 4.06-4.21 (m, 4H), 3.56-3.69 (m, 4H), 3.12-3.22 (m, 4H), 1.81-2.01 (m, 4H), 1.62-1.79 (m, 6H), 1.51-1.59 (m, 2H), 1.40-1.51 (m, 4H). MS (ESI) m/z: 463 [M+H]
1,8-bis (4-(4,5,6,7-tetrahydro-1H-1,3-diazepin-2-yl)phenoxy)octane (7d)
3d (0.1 g, 0.24 mmol) and 1,4-diaminobutane (0.106 g, 1.21 mmol) were reacted using the same procedure as the one described for the synthesis of 7a, providing 7d as off white solid, Yield 62%; Mp (223-225 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.69 (d, J = 7.6 Hz, 4H), 7.11 (d, J = 8.8 Hz, 4H), 4.05-4.08 (m, 4H), 3.63 (m, 8H), 1.93 (m, 8H), 1.72-1.74 (m, 4H), 1.36-1.42 (m, 8H); 13C NMR (100 MHz, DMSO-d6): δ 164.5, 163.3, 131.5, 130.8, 121.6, 115.5, 114.9, 68.4, 43.9, 29.0, 28.8, 25.9, 25.7; MS (ESI) m/z: 491 [M+H]
4,4′-(pentane-1,5-diylbis(oxy))bis(N-(5-aminopentyl)benzimidamide) (8)
3b (0.1 g, 0.25 mmol) and 1,2-diaminopentane (0.128 g, 1.25 mmol) were dissolved in DMF (2 mL) and stirred at room temperature for overnight. DMF was removed under vacuum, the crude reaction mixture was washed with chloroform (5 mL each). Residue was extracted with water and chloroform, water layer was separated and evaporated to obtain solid residue (Scheme 2). It was dissolved in methanol (1 mL) and product was precipitated by the addition of di-ethyl ether (5 mL). Solvent was decanted and dried in vacuum to obtain pure product 8a as a white solid Yield 51%; Mp (109-111 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.80 (d, J = 7.2 Hz, 4H), 7.09 (d, J = 7.6 Hz, 4H), 4.08 (m, 4H), 3.41 (m, 4H), 2.73 (m, 4H), 1.40-1.79 (18); 13C NMR (100 MHz, DMSO-d6): δ 162.8, 162.2, 130.6, 120.9, 114.9, 68.3, 42.7, 28.6, 27.3, 27.2, 23.4, 22.4; MS (ESI) m/z: 511 [M+H]
4,4′-(heptane-1,7-diylbis(oxy))bis(N-(7-aminoheptyl)benzimidamide) (9a)
Procedure is similar as described for 8. White solid, Yield 47%; Mp (98-101 °C); (Scheme 2); 1H NMR (400 MHz, CD3OD): δ 7.89 (d, J = 8 Hz, 4H), 7.30 (d, J = 8.4 Hz, 4H), 4.28 (m, 4H), 3.3 (m, 4H), 2.99 (m, 2H), 1.64-2.03 (m, 32H); 13C NMR (100 MHz, DMSO-d6): δ 162.8, 162.1, 130.5, 121.1, 114.9, 79.6, 68.4, 42.8, 28.8, 28.0, 27.2, 25.8, 23.5; MS (ESI) m/z: 595 [M+H]
4,4′-(octane-1,8-diylbis (oxy))bis(N-(7-aminoheptyl)benzimidamide) (9b)
Solid, Yield 42%; Mp (111-113 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.79 (d, J = 8 Hz, 4H), 7.08 (d, J = 7.6 Hz, 4H), 4.04 (m, 4H), 2.68 (m, 4H), 1.53-1.70 (m, 12H), 1.25-1.39 (m, 24H); 13C NMR (100 MHz, DMSO-d6): δ 162.9, 162.1, 130.5, 120.8, 114.9, 68.4, 42.8, 40.3, 29.0, 28.8, 28.5, 28.4, 27.7, 26.4, 26.2, 26.1, 25.7; MS (ESI) m/z: 609 [M+H]
4,4′-(heptane-1,7-diylbis(oxy))bis(N-(phenylcarbamoyl)benzimidamide) (10)
4a (0.1 g, 0.27 mmol) and DBU (0.16 mL, 1.1 mmol) were dissolved in DMF (2 mL) and stirred at 40 °C. A solution of phenyl isocyanate (0.06mL, 0.54mmol) in DMF (0.5 mL) was added to the above reaction mixture and was stirred at 40 °C for 2 hours. DMF was removed under reduced pressure, chloroform (20 mL) was added to the residue and washed with water (10mL) (Scheme 3). The organic layer was separated, concentrated and purified on neutral alumina column (4:96 Methanol/Chloroform) to yield the pure product 11 as a white solid, yield 68%; Mp (136-138 °C); 1H NMR (400 MHz, DMSO-d6): δ 8.01 (d, J = 8.4 Hz, 4H), 7.65 (d, J = 8.0 Hz, 4H), 7.24 (t,J = 7.8 Hz, 4H) 7.03 (d, J = 9.6 Hz, 4H), 6.91-6.95 (m, 2H), 4.03-4.06 (m, 4H), 1.74 (m, 4H) 1.43 (m, 6H); 13C NMR (100 MHz, DMSO-d6): δ 163.9, 130.4, 129.1, 119.0, 114.7, 68.3, 28.9, 25.8; MS (ESI) m/z: 607 [M+H]
4,4′-(heptane-1, 7-diylbis (oxy))bis(N-(methylcarbamoyl)benzimidamide) (11)
4a (0.1 g, 0.27 mmol) and DBU (0.20 mL, 1.35 mmol) were dissolved in DMF (2 mL) and stirred at 40 °C. A solution of N-succinamidyl-N-methyl carbamate (0.093 g, 0.54 mmol) in DMF (0.5 mL) was added to the above reaction mixture and was stirred at 40 °C for 2 hours (Scheme 3). DMF was removed under reduced pressure, and chloroform (20 mL) was added to the residue and washed with water (10mL). The organic layer was separated, chloroform was evaporated and purified on neutral alumina column (5:95 Methanol/Chloroform) to yield the pure product 13a as a white solid, yield 61%; Mp (147-149 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.90 (d, J = 8.4 Hz, 4H), 6.97 (d, J = 8.4 Hz, 4H), 4.02 (t, J = 6.4 Hz, 4H), 2.60 (s, 6H), 1.71-1.74 (m, 4H), 1.41 (m, 6H); 13C NMR (100 MHz, DMSO-d6): δ 162.8, 161.5, 129.3, 127.5, 114.2, 68.0, 28.9, 26.6, 25.8; MS (ESI) m/z: 483 [M+H]
N-benzyl-3-bromo-N-(3-bromopropyl)propan-1-amine (13a)
Potassium carbonate (3.22 g/23.4 mmol/2.5 equivalents) and 1,3-dibromopropane (12) (4.32 g/21.5 mmol/2.3 equiv.) was added to benzylamine (1.0 mL/9.3 mmol/1 equivalent) in acetone (50mL). The reaction mixture was refluxed for 48 hours at 60 °C, and then cooled to room temperature. Acetone was removed under reduced pressure to give a solid residue. Water (75 mL) was added to the residue and extracted with dichloromethane (25 mL × 3). The organic layer was separated and dried with sodium sulfate, filtered, and evaporated to give pure solid 13a (2.15 g, 69%) (Scheme 4); 1H NMR (400 MHz, CDCl3): δ 7.14-7.54 (m, 5H), 3.29-3.61 (m, 6H), 2.21-2.46 (m, 4H), 1.92-2.14 (m, 4H); MS (ESI) m/z: 350 [M+H]
3-bromo-N-(3-bromopropyl)-N-propylpropan-1-amine (13b)
1H NMR (400 MHz, CDCl3): δ 3.53-3.41 (m, 4H), 2.59-3.46 (m, 4H), 2.21-2.29 (m, 2H), 1.91-1.99 (m, 4H), 1.40-1.51 (m, 2H), 0.91-0.99 (m, 3H); MS (ESI) m/z: 302 [M+H]
4,4′-(((benzylazanediyl)bis(propane-3,1-diyl))bis(oxy))dibenzonitrile (14a)
Cesium carbonate (10.9 g/33.6 mmoles/4 equivalents) and 15a (1.4 g/4.2 mmol/ 0.5 equiv.) was added to 1 (1 g/ 8.4 mmol/1 equiv.) in acetone (50mL). The reaction mixture was refluxed for 48 hours at 60 °C, and then cooled to room temperature. Acetone was removed under reduced pressure to give a solid residue (Scheme 4). Water (75 mL) was added to the residue and extracted with dichloromethane (25 mL × 3). The organic layer was separated and washed with 10% sodium hydroxide in water (10 mL × 3) and washed with water until neutral to pH paper. The organic layer was dried with sodium sulfate, filtered, and evaporated to give pure solid 14a; 1H NMR (400 MHz, CDCl3): δ 7.40-7.62 (m, 4H), 7.717-7.39 (M, 5H), 6.79-6.87 (m, 4H), 3.82-4.06 (m, 4H), 3.61 (s, 2H), 2.59-2.68 (m, 4H), 1.89-1.99 (m, 4H); MS (ESI) m/z: 425 [M+H].
4,4′-(((propylazanediyl)bis(propane-3,1-diyl))bis(oxy))dibenzonitrile (14b)
1H NMR (400 MHz, CDCl3): δ 7.53-7.61 (m, 4H), 6.80-6.92 (m, 4H), 4.09-4.19 (m, 4H), 2.42-2.53 (m, 4H), 1.82-1.99 (m, 4H), 1.39-1.51 (m, 2H), 0.92-0.98 (m, 3H); MS (ESI) m/z: 378 [M+H].
4,4′-(((benzylazanediyl)bis(propane-3,1-diyl))bis(oxy))dibenzimidamide (16a)
1.0 g of 14a (2.3 mmole) in anhydrous dioxane (25mL) and dry methanol (20mL) was saturated with hydrochloric acid gas at 0-5°C under nitrogen atmosphere. The reaction mixture was stirred at room temperature for 36 hours, after which the solvent was reduced by 50%. 30 mL of anhydrous ether was added and the resulting precipitate was filtered, washed with anhydrous ether (10 mL × 5) and dried under vacuum to yield 15a (0.83 g, 73% yield). Dry solid 15a (0.8 g, 1.6 mmol) was dissolved in dry ethanol (50mL) and 2 M ethanolic ammonia (50mL), to which ammonium chloride (0.025 g, 0.47 mmol) was added. This mixture was refluxed at 80°C for 32 hours under nitrogen atmosphere, after which it was cooled and concentrated (Scheme 4). The solid obtained was filtered and washed with cold ethanol (5 mL × 3) and ether (10 mL × 3) to give diamidine product 16a as brown solid, Yield 75%; Mp (183-185 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.82-7.84 (m, 4H), 7.18-7.25 (m, 5H), 7.01-7.3 (m, 4H), 4.04-4.05 (m, 4H), 3.56 (s, 2H), 2.55 (m, 4H), 1.88 (m, 4H); 13C NMR (100 MHz, DMSO-d6): δ 165.0, 163.4, 130.5, 129.7, 128.9, 128.4, 127.1, 119.5, 119.3, 115.0, 66.4, 58.4, 49.7, 26.5; MS (ESI) m/z: 460 [M+H].
Compounds 16b was similarly synthesized as per the above procedure (Scheme 4).
4,4′-(((propylazanediyl)bis(propane-3,1-diyl))bis(oxy))dibenzimidamide (16b)
Off white solid, Yield 67%; Mp (177-179 °C); 1H NMR (400 MHz, DMSO-d6): δ 7.88 (d, J = 8.4 Hz, 4H), 7.16 (d, J = 8.8 Hz, 4H), 4.21 (m, 4H), 3.25 (m, 4H), 3.07 (m, 2H), 2.24 (m, 4H), 1.73-1.75 (m, 2H), 0.92 (t, J = 6.4 Hz, 3H); 13C NMR (100 MHz, DMSO-d6): δ 165.0, 163.0, 130.6, 120.0, 115.2, 65.9, 23.2, 16.8, 11.3; MS (ESI) m/z: 412 [M+H].
2-(4-(4-carbamimidoylphenoxy)phenyl)-1H-indole-6-carboximidamide (17)
Purchased from Sigma-Aldrich (St. Louis, MO, Catalog No. A8675). A purity of >95% was confirmed HPLC and by HPLC-MS.
Cellular Assays
Cell lines and cell culture
WM115 (American Type Tissue Collection (ATCC)) malignant melanoma cells were cultured in Minimum Essential medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS). Cells were maintained in a 37°C incubator with 5% CO2. The WM115 have the activating BRAF V600E mutation and wild type NRAS19. As shown by Western blot analysis, the WM115 cells have elevated levels of endogenous S100B protein.
Growth inhibition assay
A collection of pentamidine analogs were tested for their ability to inhibit the growth of WM115 human melanoma cells expressing high levels of S100B (WM115shSCRAMBLED) and low levels of S100B (WM115shS100B)30, 43 using a modification of the high-throughput screening assay performed by Bachman et al 44 (Table 2). Using a Biomek FX Laboratory Automation Workstation (Beckman-Coulter) equipped with a 96-channel pipetting head, 20μl of MEM (Invitrogen) supplemented with 10% fetal bovine serum, were added to each well of a 384-well tissue culture plate (Corning) containing enough cells such that growing uninhibited they reach 80% confluence in ~5 days. The cells are propagated in the presence of 0.5 mg/ml puromycin to maintain the vector expressing S100B siRNA to knockdown (KD) the protein or the same vector containing a scrambled control siRNA. After 24 hours of growth at 37 °C in 5% CO2 humidity controlled incubator the cells were treated with 20 μl of compound in the same culture media, while in the control cultures, only an equivalent amount of DMSO added with the compound is added. Titrations of the compounds were performed in triplicate using the Biomek NXP Laboratory Automation Workstation (Beckman-Coulter) equipped with an 8-channel pipetting head before being added to the cell plates. After four additional days of incubation, the cells were lysed by the adding 20μl of lysis buffer consisting of 1.8% Igepal with 1:10,000 dilution of SYBR Green I (Invitrogen). The covered plates were incubated for another 24 hours at 37 °C in a 5% CO2 humidity controlled incubator. The fluorescence intensity was then read through the bottom of the plate using a POLARstar Optima fluorescent plate reader (BMG) with 485 nm excitation and 520 nm emission filters. The SYBR-green fluorescence is used to measure total DNA that in turn correlates with cell number as previously described45. The EC50 of the compounds were determined using serial dilutions and done in quadruplicate, as above, with no more than 1.0% DMSO or 1000 μM compound.
Protein Purification
15N-labeled Bovine or Rat S100B was expressed and purified (> 99%) with methods similar to those previously46, 47. The concentrations of S100B stock solutions were determined using the Bio-Rad Protein Assay (Bio-Rad Inc., Hercules, CA) using bovine serum albumin as a standard. The S100B was concentrated to 10-15 mM using Amicon Ultra centrifugal filter units with a 10 kDa MWCO and stored at a concentration of ~ 10 mM in 0.25mM Tris, pH 7.2 with 0.25 mM DTT at −20 °C until use.
Protein NMR Spectrometry
The Ca2+-loaded S100B•SBiXs HSQC samples contained 0.1 mM S100B Rat subunit, 0.125 mM SBiX, 0.34 mM NaN3, 15 mM NaCl, 5% DMSO-d6, 10 mM CaCl2, 10% D2O, 0.2% TPEN and 10 mM HEPES, adjusted to pH 7.2 with HCl. Heteronuclear single-quantum coherence (HSQC) NMR data were collected at 37 °C with a Bruker Avance 800 US2 (800.27 MHz for protons) instrument equipped with pulsed-field gradients, four frequency channels, and a triple-resonance, z-axis gradient cryogenic probe. Data were processed with NMRPipe, and proton chemical shifts were reported with respect to the H2O or HDO signal taken as 4.608 ppm relative to external TSP (0.0 ppm). The 15N chemical shifts were indirectly referenced as previously described using the following ratio of the zero-point frequency: 0.10132905 for 15N to 1H. HSQC spectra were visually inspected for chemical shift perturbations (Table 1, Table 3, Fig. S2-6).
Fluorescence Polarization Competition Assay
The ability of the compounds to compete with a peptide probe was evaluated using an adaptation of Wilder et al, 2010 (Table 1). Briefly, changes is fluorescence polarization were measured upon competition with the TAMRA-labeled version of the peptide TRTK12, which is derived from the CapZ protein residues 265–276 (TRTKIDWNKILS), for Ca2+-loaded S100B. The S100B TAMRA-TRTK FPCA was performed in 384-well black polypropylene microplate with a final volume of 20 μL containing: 10 nM TAMRA-TRTK, 1 μM S100B Rat, 0.01% Triton X-100, 0.5% DMSO, 50 mM HEPEs, pH 7.2, 100 mM KCl, 15 mM NaCl, 10 mM CaCl2. Titrations of each compound were done beginning from the highest concentration possible. The fluorescence polarization was measured at room temperature in a BMG PHERAstar Plus multimode microplate equipped with dual detection PMTs with an excitation of 544 nm and emission of 590 nm.
X-ray Crystallography
Crystallization
Crystallization experiments were conducted using vapor diffusion methods. CaS100B•5a crystals were grown in hanging drops consisting of 3.0:3.0 μL protein solution (40mg/mL S100B Bovine; 10mM Cacodylate, pH 7.2; 7.5mM CaCl2; and 4mM 5a prepared in DMSO) and mother liquor (10% Peg 3,350; 0.1M Cacodylate, pH 6.4; 7.5mM CaCl2) with no additional volume of mother liquor left in the crystallization chamber. CaS100B•6b crystals were grown in sitting drops consisting of 0.75:0.75 μL protein solution (20mg/mL S100B Bovine; 10mM Cacodylate, pH 7.2; 7.5mM CaCl2; and 2mM 6b prepared in DMSO) and mother liquor (25% Peg 3,350; 0.1M Bis-Tris, pH 7.5; 7.5mM CaCl2; 5% glycerol). CaS100B•6b crystals were grown with mother liquor left in the crystallization chamber. CaS100B•17 crystals were grown in sitting drops consisting of 0.75:0.75μL protein solution (20mg/mL S100B bovine; 10mM Cacodylate, pH 7.2; 7.5mM CaCl2; and 2mM 10 prepared in DMSO) and mother liquor (40% 2-methyl-2,4-pentanediol; 0.1M Hepes, pH 7.0; and 7.5mM CaCl2). The crystals were grown over a period of one to fourteen days at a temperature of 295K. Crystals were not cryo-protected before being flash-cooled in liquid nitrogen.
Data Collection and Processing
Diffraction data for CaS100B•5a crystals were collected Northeastern Collaborative Access Team (NE-CAT) beamline at the Advanced Photon Source (Argonne National Laboratory). Data was recorded at 100K on an ADSC Q315 (315mm × 315mm) detector and processed by NE-CAT’s RAPD automated processing (https://rapd.nec.aps.anl.gov/rapd), which uses XDS 22 for integration and scaling. A 1.53Å dataset was collected at a wavelength of 0.97920Å while oscillating the crystal 1.0° each frame. The space group was determined to be P41212.
Diffraction data for CaS100B•6b and CaS100B•17 crystals were collected remotely48, 49 at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7-1. Data was recorded at 100K on an ADSC Q315 (315mm × 315mm) detector with collection strategies generated by BLU-ICE50. Datasets were processed and integrated by AUTOXDS51. A 1.59Å dataset was collected at a wavelength of 1.1271Å while oscillating the crystal 0.74° or 0.75°, respectively, each frame. The space groups were determined to be P41212 and P212121, respectively. Diffraction data statistics for both models are summarized in Table 4.
Structure Determination and Refinement
To determine the structure of S100B in complex with SBiXs, molecular replacement (MR) was performed. A search model from a previously elucidated S100B structure (PDB accession code 1MHO39) was generated by removing coordinates for ligands and water. Molecular replacement was carried out using within the AUTOMR52, 53 function of the PHENIX54 software suite. The models were finished by manual building within COOT55. The models was refined the PHENIX.REFINE56 function of the PHENIX54 software suite. Ligands and waters were incorporated into the models by referring to the |Fo|-|Fc| omit maps. The structure refinement statistics for both models are summarized in Table 4.
Coordinates and structure factors have been deposited in the Protein Data Bank with the accession numbers: CaS100B•5a (PDB ID:5DKN), CaS100B•6b (PDB ID: 5DKQ), CaS100B•17 (PDB ID: 5DKR).
In Silico Methods
Ligand design was performed using the Site Identification by Ligand Competitive Saturation (SILCS) approach57, 58. For ligand design the Tier 1 SILCS approach was applied in which the protein is immersed in an aqueous solution containing 1 M benzene and propane. SILCS simulations, initiated from the SBi132•S100B (PDB 3GK1) crystal structures, were performed as previously described yielding probability distributions for aromatic groups based on benzene carbons, aliphatic groups based on propane carbons, hydrogen bond donors based on water hydrogen waters and hydrogen bond acceptors based on water oxygens58. The probability distributions in the presence of the protein are normalized with respect to probability distributions in the absence of the protein, such the final distributions, termed FragMaps, take into account both functional group and protein desolvation as well and protein flexibility and protein-functional group interactions. FragMaps may be visualized as 3D contour maps that indicate the regions around the protein favorable for the different classes of functional groups (Fig. 2).
The prediction of the location of the missing amino-alkyl tails of 9a was accomplished by applying two independent methods. In both approaches the initiator Met0 residue was retained since this residue is present in the S100B co-crystal data with 9a. In the first method, molecular docking was performed using the AutoDock Vina program59. Standard parameters were applied to obtain 20 docked orientations. A consensus-binding mode was obtained based both on favorable AutoDock score and the overlap of the partially resolved part of the ligand with the crystal data. The second approach involved the more recent Tier2 SILCS approach57, 60. Tier2 SILCS simulations were performed on the protein conformation obtained from the crystal data with compound 9a following the removal of the partially resolved ligand. The 10 simulation trajectories spanned a cumulative sampling time of 400ns were used to calculate the protein functional group affinity FragMaps. In the Tier 2 approach a wider range of solute molecules are used in the aqueous solution including benzene, propane, methanol, formamide, acetaldehyde, imidazole, acetate and methylammonium at ~0.25 M. Notable is the inclusion of methylammonium allowing the calculation of positive FragMaps that are directly relevant for docking of the positively charged amines of 9a. Docking of 9a into the “field” of the FragMaps used the validated MC-SILCS approach that involves Monte Carlo (MC) simulation based sampling. MC moves included translational, rotational and dihedral sampling with Metropolis criteria determined using the Ligand Grid Free Energies obtained from the FragMaps60. Intramolecular energies of the ligand during MC sampling were calculated using the CHARMM General Force Field.61-63 The initial conformation of the ligand was seeded with the resolved atoms from the crystal data. The MC-SILCS docking revealed a consensus-binding mode, which differed in the location of the amino-alkyl tails from the consensus AutoDock conformation.
Supplementary Material
ACKNOWLEDGEMENTS
The Advanced Photon Source (APS) was used in this research, and it is supported, in part, by a grant from the National Institute of General Medical Sciences (P41 GM103403) in the National Institutes of Health and by a U.S. Department of Energy (DOE) contract (DE-AC02-06CH11357). In the DOE, the APS is part of the Office of Science Users Facility operated by the U.S. Office of Science at the Argonne National Laboratory. The U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences supports Stanford Synchrotron Radiation Light (SSRL) synchrotron source as part of the SLAC National Accelerator Laboratory (DE-AC02-76SF00515). Additional support is provided to the SSRL Structural Molecular Biology Program by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The NMR spectrometers used in these studies were purchased, in part, with funds from shared instrumentation grants from the National Institutes of Health [S10 RR10441, S10 RR15741, S10 RR16812, and S10 RR23447 (D.J.W.)] and from the National Science Foundation (DBI 1005795 to D.J.W.). The authors would also like to thank Jace W. Jones and the University of Maryland School of Pharmacy Mass Spectrometry Centre for his help with LCMS.
Funding Source: This work was supported by grants from the NIH (GM58888 and CA107331) to D.J.W. and (GM070855) to A.D.M. Jr. The training of M.C.C. is supported by a NIH T32 grant (CA154274).
Abbreviations Used
- MM
malignant melanoma
- SBiX
S100B inhibitor X=number
- CaS100B
Calcium-bound S100B
- PPI
Protein-Protein Interactions
- SILCS
Site Identification by Ligand Competitive Saturation
- FPCA
Fluorescence Polarization Competition Assay
Footnotes
Conflict of Interest Disclosure. A.D.M. Jr., is co-founder and CSO of Silcsbio LLC.
PDB ID Codes: CaS100B•5a (PDB ID:5DKN), CaS100B•6b (PDB ID: 5DKQ), CaS100B•17 (PDB ID: 5DKR).
Supporting Information. Additional details of crystal model RMSD, in silico data, 2D 1H-15N S100B HSQC NMR spectra, cellular assays, purities derived by HPLC, 1D 1H and 13C compound NMR spectra, and SMILES.
References
- 1.Gogas H, Abali H, Ascierto PA, Demidov L, Pehamberger H, Robert C, Schachter J, Eggermont AM, Hauschild A, Espinosa E. Who Benefits Most From Adjuvant Interferon Treatment for Melanoma? Am. J. Ther. 2013 doi: 10.1097/MJT.0b013e31829e883d. [DOI] [PubMed] [Google Scholar]
- 2.Jang S, Atkins MB. Which drug, and when, for patients with BRAF-mutant melanoma? Lancet Oncol. 2013;14:e60–69. doi: 10.1016/S1470-2045(12)70539-9. [DOI] [PubMed] [Google Scholar]
- 3.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcus DM, Lowe M, Khan MK, Lawson DH, Crocker IR, Shelton JW, Melton A, Maynard N, Delman KA, Carlson GW, Rizzo M. Prognostic Factors for Overall Survival After Radiosurgery for Brain Metastases From Melanoma. Am. J. Clin. Oncol. 2013 doi: 10.1097/COC.0b013e318280d7be. [DOI] [PubMed] [Google Scholar]
- 5.Espinosa E, Grob JJ, Dummer R, Rutkowski P, Robert C, Gogas H, Kefford R, Eggermont AM, Martin Algarra S, Hauschild A, Schadendorf D. Treatment Algorithms in Stage IV Melanoma. Am. J. Ther. 2014 doi: 10.1097/MJT.0b013e31829e885c. [DOI] [PubMed] [Google Scholar]
- 6.Lee L, Gupta M, Sahasranaman S. Immune checkpoint inhibitors: An introduction to the next generation cancer immunotherapy. J. Clin. Pharmacol. 2015 doi: 10.1002/jcph.591. [DOI] [PubMed] [Google Scholar]
- 7.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discovery. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 8.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat. Rev. Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Hajj P, Journe F, Wiedig M, Laios I, Sales F, Galibert MD, Van Kempen LC, Spatz A, Badran B, Larsimont D, Awada A, Ghanem G. Tyrosinase-related protein 1 mRNA expression in lymph node metastases predicts overall survival in high-risk melanoma patients. Br. J. Cancer. 2013;108:1641–1647. doi: 10.1038/bjc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henry L, Fabre C, Guiraud I, Bastide S, Fabbro-Peray P, Martinez J, Lavabre-Bertrand T, Meunier L, Stoebner PE. Clinical use of p-proteasome in discriminating metastatic melanoma patients: comparative study with LDH, MIA and S100B protein. Int. J. Cancer. 2013;133:142–148. doi: 10.1002/ijc.27991. [DOI] [PubMed] [Google Scholar]
- 11.Wieder HA, Tekin G, Rosenbaum-Krumme S, Klode J, Altenbernd J, Bockisch A, Nagarajah J. 18 FDG-PET to assess recurrence and long term survival in patients with malignant melanoma. Nuklearmedizin. 2013;52:198–203. doi: 10.3413/Nukmed-0584-13-05. [DOI] [PubMed] [Google Scholar]
- 12.Schiltz PM, Dillman RO, Korse CM, Cubellis JM, Lee GJ, De Gast GC. Lack of elevation of serum S100B in patients with metastatic melanoma as a predictor of outcome after induction with an autologous vaccine of proliferating tumor cells and dendritic cells. Cancer Biother. Radiopharm. 2008;23:214–221. doi: 10.1089/cbr.2007.0432. [DOI] [PubMed] [Google Scholar]
- 13.O’Rourke MG, Johnson M, Lanagan C, See J, Yang J, Bell JR, Slater GJ, Kerr BM, Crowe B, Purdie DM, Elliott SL, Ellem KA, Schmidt CW. Durable complete clinical responses in a phase I/II trial using an autologous melanoma cell/dendritic cell vaccine. Cancer Immunol. Immunother. 2003;52:387–395. doi: 10.1007/s00262-003-0375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Yang Q, Wilder PT, Carrier F, Weber DJ. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. J. Biol. Chem. 2010;285:27487–27498. doi: 10.1074/jbc.M110.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin J, Yang Q, Yan Z, Markowitz J, Wilder PT, Carrier F, Weber DJ. Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. J. Biol. Chem. 2004;279:34071–34077. doi: 10.1074/jbc.M405419200. [DOI] [PubMed] [Google Scholar]
- 16.Markowitz J, Chen I, Gitti R, Baldisseri DM, Pan Y, Udan R, Carrier F, MacKerell AD, Jr., Weber DJ. Identification and characterization of small molecule inhibitors of the calcium-dependent S100B-p53 tumor suppressor interaction. J. Med. Chem. 2004;47:5085–5093. doi: 10.1021/jm0497038. [DOI] [PubMed] [Google Scholar]
- 17.Hartman KG, McKnight LE, Liriano MA, Weber DJ. The evolution of S100B inhibitors for the treatment of malignant melanoma. Future Med. Chem. 2013;5:97–109. doi: 10.4155/fmc.12.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartman KG, Wilder PT, Varney K, MacKerell AD, Jr., Coop A, Zimmer D, Lapidus RG, Weber DJ. Inhibiting S100B in Malignant Melanoma. In: Duc HT, editor. Melanoma - From Early Detection to Treatment. 2013. pp. 649–667. [Google Scholar]
- 19.Meghnani V, Vetter SW, Leclerc E. RAGE overexpression confers a metastatic phenotype to the WM115 human primary melanoma cell line. Biochim. Biophys. Acta. 2014;1842:1017–1027. doi: 10.1016/j.bbadis.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Briceland LL, Bailie GR. Pentamidine-associated nephrotoxicity and hyperkalemia in patients with AIDS. DICP, Ann. Pharmacother. 1991;25:1171–1174. doi: 10.1177/106002809102501102. [DOI] [PubMed] [Google Scholar]
- 21.Edwards KJ, Jenkins TC, Neidle S. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 1992;31:7104–7109. doi: 10.1021/bi00146a011. [DOI] [PubMed] [Google Scholar]
- 22.Pathak MK, Dhawan D, Lindner DJ, Borden EC, Farver C, Yi T. Pentamidine is an inhibitor of PRL phosphatases with anticancer activity. Mol. Cancer Ther. 2002;1:1255–1264. [PubMed] [Google Scholar]
- 23.McKnight LE, Raman EP, Bezawada P, Kudrimoti S, Wilder PT, Hartman KG, Godoy-Ruiz R, Toth EA, Coop A, Mackerell AD, Jr., Weber DJ. Structure-Based Discovery of a Novel Pentamidine-Related Inhibitor of the Calcium-Binding Protein S100B. ACS Med. Chem. Lett. 2012;3:975–979. doi: 10.1021/ml300166s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agamennone M, Cesari L, Lalli D, Turlizzi E, Del Conte R, Turano P, Mangani S, Padova A. Fragmenting the S100B-p53 Interaction: Combined Virtual/Biophysical Screening Approaches to Identify Ligands. ChemMedChem. 2010;5:428–435. doi: 10.1002/cmdc.200900393. [DOI] [PubMed] [Google Scholar]
- 25.Yoshimura C, Miyafusa T, Tsumoto K. Identification of small-molecule inhibitors of the human S100B-p53 interaction and evaluation of their activity in human melanoma cells. Bioorg. Med. Chem. 2013;21:1109–1115. doi: 10.1016/j.bmc.2012.12.042. [DOI] [PubMed] [Google Scholar]
- 26.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 27.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, Bunting KL, Polo JM, Fares C, Arrowsmith CH, Yang SN, Garcia M, Coop A, Mackerell AD, Jr., Prive GG, Melnick A. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samadani R, Zhang J, Brophy A, Oashi T, Priyakumar UD, Raman EP, St John FJ, Jung KY, Fletcher S, Pozharski E, MacKerell AD, Jr., Shapiro P. Small-molecule inhibitors of ERK-mediated immediate early gene expression and proliferation of melanoma cells expressing mutated BRaf. Biochem. J. 2015;467:425–438. doi: 10.1042/BJ20131571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charpentier TH, Wilder PT, Liriano MA, Varney KM, Pozharski E, MacKerell AD, Jr., Coop A, Toth EA, Weber DJ. Divalent metal ion complexes of S100B in the absence and presence of pentamidine. J. Mol. Biol. 2008;382:56–73. doi: 10.1016/j.jmb.2008.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavalier MC, Pierce AD, Wilder PT, Alasady MJ, Hartman KG, Neau DB, Foley TL, Jadhav A, Maloney DJ, Simeonov A, Toth EA, Weber DJ. Covalent small molecule inhibitors of Ca(2+)-bound S100B. Biochemistry. 2014;53:6628–6640. doi: 10.1021/bi5005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spychala J. 4-(Cyclic Amidino)phenols - Preparation and Use In A Diamidine Synthesis. Synth. Commun. 2000;30:1083–1094. [Google Scholar]
- 32.Pigini M, Brasili L, Cassinelli A, Giardina D, Gulini U, Quaglia W, Melchiorre C. Synthesis and Alpha-Blocking Activity of Some Analogs of Idazoxan. Eur. J. Med. Chem. 1987;22:273–276. [Google Scholar]
- 33.Manley PJ, Nanda KK, Trotter BW. In Google Patents. 2010. Imidazoisoindole neuropeptide s receptor antagonists. [Google Scholar]
- 34.Perner RJ, DiDomenico S, Koenig JR, Gomtsyan A, Bayburt EK, Schmidt RG, Drizin I, Zheng GZ, Turner SC, Jinkerson T, Brown BS, Keddy RG, Lukin K, McDonald HA, Honore P, Mikusa J, Marsh KC, Wetter JM, George KS, Jarvis MF, Faltynek CR, Lee CH. In vitro structure-activity relationship and in vivo characterization of 1-(Aryl)-3-(4-(amino)benzyl)urea transient receptor potential vanilloid 1 antagonists. J. Med. Chem. 2007;50:3651–3660. doi: 10.1021/jm070276i. [DOI] [PubMed] [Google Scholar]
- 35.Koyanagi T, Tsukuda S, 佃晋太朗, Nakamura Y, 中村行男, TSUNEMATSU K, 常松孝祜, Shimizu M. In Google Patents. 2013. Plant disease control agent containing arylamidine derivative or salt thereof. [Google Scholar]
- 36.de Jong MR, Engbersen JFJ, Huskens J, Reinhoudt DN. Cyclodextrin dimers as receptor molecules for steroid sensors. Chem. - Eur. J. 2000;6:4034–4040. doi: 10.1002/1521-3765(20001103)6:21<4034::aid-chem4034>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Charpentier TH, Thompson LE, Liriano MA, Varney KM, Wilder PT, Pozharski E, Toth EA, Weber DJ. The effects of CapZ peptide (TRTK-12) binding to S100B-Ca2+ as examined by NMR and X-ray crystallography. J. Mol. Biol. 2010;396:1227–1243. doi: 10.1016/j.jmb.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilder PT, Charpentier TH, Liriano MA, Gianni K, Varney KM, Pozharski E, Coop A, Toth EA, Mackerell AD, Weber DJ. In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction. Int. J. High Throughput Screen. 2010;2010:109–126. doi: 10.2147/IJHTS.S8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumura H, Shiba T, Inoue T, Harada S, Kai Y. A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain. Structure. 1998;6:233–241. doi: 10.1016/s0969-2126(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 40.Charpentier TH, Wilder PT, Liriano MA, Varney KM, Zhong S, Coop A, Pozharski E, MacKerell AD, Jr., Toth EA, Weber DJ. Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography. Biochemistry. 2009;48:6202–6212. doi: 10.1021/bi9005754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilder PT, Lin J, Bair CL, Charpentier TH, Yang D, Liriano M, Varney KM, Lee A, Oppenheim AB, Adhya S, Carrier F, Weber DJ. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim. Biophys. Acta. 2006;1763:1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 42.Liriano MA, Varney KM, Wright NT, Hoffman CL, Toth EA, Ishima R, Weber DJ. Target Binding to S100B Reduces Dynamic Properties and Increases Ca(2+)-Binding Affinity for Wild Type and EF-Hand Mutant Proteins. J. Mol. Biol. 2012;423:365–385. doi: 10.1016/j.jmb.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartman KG, Vitolo MI, Pierce AD, Fox JM, Shapiro P, Martin SS, Wilder PT, Weber DJ. Complex Formation between S100B Protein and the p90 Ribosomal S6 Kinase (RSK) in Malignant Melanoma Is Calcium-dependent and Inhibits Extracellular Signal-regulated Kinase (ERK)-mediated Phosphorylation of RSK. J. Biol. Chem. 2014;289:12886–12895. doi: 10.1074/jbc.M114.561613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachman KE, Sager J, Cheong I, Catto M, Bardelli A, Park BH, Vogelstein B, Carotti A, Kinzler KW, Lengauer C. Identification of compounds that inhibit growth of 2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine-resistant cancer cells. Mol. Cancer Ther. 2005;4:1026–1030. doi: 10.1158/1535-7163.MCT-05-0038. [DOI] [PubMed] [Google Scholar]
- 45.Myers MA. Direct measurement of cell numbers in microtitre plate cultures using the fluorescent dye SYBR green I. J. Immunol. Methods. 1998;212:99–103. doi: 10.1016/s0022-1759(98)00011-8. [DOI] [PubMed] [Google Scholar]
- 46.Drohat AC, Amburgey JC, Abildgaard F, Starich MR, Baldisseri D, Weber DJ. Solution structure of rat apo-S100B(beta beta) as determined by NMR spectroscopy. Biochemistry. 1996;35:11577–11588. doi: 10.1021/bi9612226. [DOI] [PubMed] [Google Scholar]
- 47.Amburgey JC, Abildgaard F, Starich MR, Shah S, Hilt DC, Weber DJ. 1 H, 13C and 15N NMR assignments and solution secondary structure of rat Apo-S100 beta. J. Biomol. NMR. 1995;6:171–179. doi: 10.1007/BF00211781. [DOI] [PubMed] [Google Scholar]
- 48.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact sample cassettes and a small-scale robot. J. Appl. Crystallogr. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soltis SM, Cohen AE, Deacon A, Eriksson T, Gonzalez A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, Miller M, Moorhead P, Phizackerley RP, Smith C, Song J, van dem Bedem H, Ellis P, Kuhn P, McPhillips T, Sauter N, Sharp K, Tsyba I, Wolf G. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008;64:1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez A, Tsai Y. AUTOXDS. Stanford University; 2010. http://smb.slac.stanford.edu/facilities/software/xds/#autoxds_script. [Google Scholar]
- 52.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afonine PV, Grosse-Kunstleve RW, Adams PD. The Phenix Refinement Framework. CCP4 Newsl. 2005;42 contribution 8. [Google Scholar]
- 57.Guvench O, MacKerell AD., Jr. Computational fragment-based binding site identification by ligand competitive saturation. PLoS Comput. Biol. 2009;5:e1000435. doi: 10.1371/journal.pcbi.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raman EP, Yu W, Guvench O, Mackerell AD. Reproducing crystal binding modes of ligand functional groups using Site-Identification by Ligand Competitive Saturation (SILCS) simulations. J. Chem. Inf. Model. 2011;51:877–896. doi: 10.1021/ci100462t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raman EP, Yu W, Lakkaraju SK, MacKerell AD., Jr. Inclusion of multiple fragment types in the site identification by ligand competitive saturation (SILCS) approach. J. Chem. Inf. Model. 2013;53:3384–3398. doi: 10.1021/ci4005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanommeslaeghe K, MacKerell AD., Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanommeslaeghe K, Raman EP, MacKerell AD., Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD., Jr. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.