Abstract
The widespread and improper use of pyrethroid insecticides, such as deltamethrin, has resulted in the evolution of resistance in many mosquito species, including Culex pipiens pallens. With the development of high-throughput sequencing, it is possible to massively screen pyrethroid resistance-associated gene. In this study, we used Illumina-Solexa transcriptome sequencing to identify genes that are expressed differently in deltamethrin-susceptible and -resistant strains of Culex pipiens pallens as a critical knowledge base for further studies. A total of 4,961,197,620 base pairs and 55,124,418 reads were sequenced, mapped to the Culex quinquefasciatus genome and assembled into 17,679 known genes. We recorded 1,826 significantly differentially expressed genes (DEGs). Among them, 1,078 genes were up-regulated and 748 genes were down-regulated in the deltamethrin-resistant strain compared to -susceptible strain. These DEGs contained cytochrome P450s, cuticle proteins, UDP-glucuronosyltransferases, lipases, serine proteases, heat shock proteins, esterases and others. Among the 1,826 DEGs, we found that the transcriptional levels of CYP6AA9 in the laboratory populations was elevated as the levels of deltamethrin resistance increased. Moreover, the expression levels of the CYP6AA9 were significantly higher in the resistant strains than the susceptible strains in three different field populations. We further confirmed the association between the CYP6AA9 gene and deltamethrin resistance in mosquitoes by RNA interfering (RNAi). Altogether, we explored massive potential pyrethroid resistance-associated genes and demonstrated that CYP6AA9 participated in the pyrethroid resistance in mosquitoes.
Keywords: Mosquitoes, Pyrethroids, RNA-seq, CYP6AA9, RNAi
Introduction
Mosquitoes spread numerous human infectious diseases, including malaria, West Nile fever, yellow fever, dengue fever, encephalitis and filariasis (Reiner et al. 2013). Chemical control, via the use of insecticides, is the most important part of the global strategy for managing the mosquito-borne diseases (Liu 2015), as it is rapid, simple and economical. Since the dichlorodiphenyltrichloroethane (DDT) was introduced in 1939, four classes of chemical insecticides have been developed: DDT and other organochlorines, organophosphorus, carbamates and pyrethroids. Among them, pyrethroids are most commonly used for the indoor residual spraying (IRS) and impregnation of bednets (ITNs) due to its high efficacy and low mammalian toxicity (Edi et al. 2014). However, their excessive and improper application has resulted in the evolution of pyrethroid resistance (Rivero et al. 2010), which has become a major obstacle for the mosquito-borne diseases management (Hemingway et al. 2002).
Three mechanisms responsible for pyrethroid resistance have previously been acknowledged. Target resistance occurs via mutations in the target sites of the insect nervous system, such as voltage-gated sodium channel (VGSC) (Dong et al. 2014). Metabolic resistance occurs via alterations in either the protein levels or the activity of detoxification enzyme that resist insecticides. For example, cytochrome P450s (CYP or P450) (Edi et al. 2014), esterases (EST) (Santo-Orihuela et al. 2013) and glutathione S-transferases (GST) (Koou et al. 2014) have all been considered to be involved in metabolic resistance. Moreover, pyrethroid insecticides cause the physiological changes of mosquitoes, such as the epidermal thickening, which subsequently reduces the absorption or penetration of insecticides (Wood et al. 2010). Other pyrethroid resistance-related genes have also been identified, such as UDP-glucuronosyltransferases (Bozzolan et al. 2014) and serine proteases (Reid et al. 2012). However, the pyrethroid resistance results from polygenic inheritance (Ffrench-Constant et al. 2004). None of the currently known genes can entirely explain the molecular mechanisms of pyrethroid resistance. Hence, identifying novel genes associated with pyrethroid resistance and elucidating the manner in which they regulate this process is critical for effective control of mosquitoes.
Previous studies screening pyrethroid resistance-related genes used suppression subtractive hybridization (SSH) combined with cDNA microarray (Liu et al. 2007; Wu et al. 2004). Subsequently, gene cloning and sequencing were used to obtain the whole sequences, a process that was time-consuming and laborious. Fortunately, new high-throughput sequencing technologies, such as those based on the transcriptome, avoid the cloning process and are quick and efficient (Ozsolak and Milos 2011). The inexpensive production of large volumes of sequence data is the primary advantage over conventional methods (Metzker 2010). The transcriptome is the set of all transcripts within a cell or tissue that can reflect the mRNA expression levels of different genes in a sample (Wang et al. 2009). RNA-seq, developed in 2008, achieves single-base-level resolution and a much higher dynamic range to quantify gene expression levels and is currently the most widely used transcriptome analysis technology (Martin and Wang 2011).
Several researchers have used transcriptome sequencing to study pyrethroid resistance in mosquitoes, including Anopheles gambiae (Bonizzoni et al. 2012), Anopheles sinensis (Zhu et al. 2014), Aedes aegypti (David et al. 2014), and Culex quinquefasciatus (Reid et al. 2012). However, the mosquitoes from different genetic background evolve different mechanisms of resistance to pyrethroids (Bonizzoni et al. 2012; Zhong 2011). In northern China, Cx. pipiens pallens is the dominant mosquito species. The pyrethroid resistance is widely reported (Wang et al. 2012). However, there is very limited genomic information of Cx. pipiens pallens. The incompleteness of the database has hindered the development of resistance management and mosquito-borne disease control.
In this study, we used Illumina-Solexa sequencing to identify genes that were expressed differently in deltamethrin-susceptible and -resistant strains as a critical theoretical basis for further molecular studies investigating mechanisms of pyrethroid resistance. In addition, we detected the relative expression levels of several likely genes by quantitative real-time PCR (qRT-PCR), and further verified the functionality of a key candidate gene by RNAi.
Materials and methods
Mosquito strains
The laboratory deltamethrin-susceptible strain (Lab-DS strain) of Cx. pipiens pallens was collected from Tangkou (N 35.12; E 116.50) town of Shandong Province in 2009 and reared in our laboratory, without exposure to any insecticide. The deltamethrin-resistant strain (Lab-DR, Lab-DR1 and Lab-DR2 strain) was selected from early fourth-instar larvae of the DS strain exposed to deltamethrin for > 30 generations. Before selection, the fifty percent larval lethal concentration (LC50) to deltamethrin was determined by larval bioassay (Chen et al. 2010), and used as the screening concentration. The LC50 of Lab-DS, Lab-DR, Lab-DR1 and Lab-DR2 strains were 0.03, 0.85, 3.7 and 7.0 mg/L, respectively. All laboratory populations were maintained at 28°C, 70 – 80% humidity and a 16:8h light:dark photoperiod. The three field populations of Cx. pipiens pallens were collected from Shanghe (N 37.31; E 117.16), Pingyin (N 36.30; E 116.42) and Dongping (N 36.09; E 116.21) towns of Shandong Province in 2011. To distinguish resistant from susceptible strains, non-blood-fed adult females were exposed to 0.05% deltamethrin-impregnated drug membranes by the standard WHO susceptibility tube bioassay (WHO 2013). The mosquitoes that knocked down after one-hour exposure were classified as deltamethrin susceptible, and those survived after the 24-hour recovery period were classified as deltamethrin resistant (Bonizzoni et al. 2012; Zhu et al. 2014). The knocked-down mosquitoes were immediately preserved in Eppendorf tubes after one-hour exposure to avoid the RNA degradation in the following 24-hour recovery period.
RNA extraction and quality assessment
Total RNA was extracted from four developmental stages (fourth instar larvae, pupae, adult males and females) (Xie et al. 2012) of the Lab-DS and the Lab-DR strains by RNAiso plus (Takara, Japan). Recombinant DNase I (Takara, Japan) was used to remove potential genomic DNA. The RNA integrity was assessed by denaturing agarose gel electrophoresis, while the purity and concentration of samples were authenticated using a NanoDrop spectrophotometer (NanoDrop, USA). Five micrograms of RNA from each developmental stage of the Lab-DS and the Lab-DR strains were respectively mixed together. Then the RNA was immediately stored at − 80°C until it was shipped to BGI (Beijing Genomics Institute, China) for Illumina-Solexa sequencing. The quality of the two RNA mixtures was further confirmed by the Agilent 2100 Bioanalyzer (Agilent, USA) in BGI.
cDNA library construction and Illumina-Solexa sequencing
After the total RNA extraction and DNase I treatment, magnetic beads with Oligo-dT were used to isolate Poly (A) mRNA. The mRNA was fragmented into short fragments by the fragmentation buffer. First-strand cDNA was synthesized using fragmented mRNA templates and random hexamer primers. Subsequently, we synthesized second-strand cDNAs using DNA polymerase I, RNaseH, dNTPs, and buffer. We purified short fragments, resolved them with EB buffer for end reparation and single nucleotide A (adenine) addition. After that, the short fragments were connected with adapters. The suitable fragments were selected as templates for PCR amplification via agarose gel electrophoresis. We used 2100 Bioanaylzer (Agilent, USA) and StepOnePlus Real-Time PCR System (ABI, USA) to quantify and qualify the sample libraries. Finally, the two libraries could be carried out high-throughput transcriptome sequencing using Illumina-Solexa HiSeq™ 2000 platform.
Quality control (QC) and alignment
The quality control (QC) of raw reads included base composition analysis and base sequence quality analysis. In addition, the “dirty” raw reads, which contained the sequence of adapter, high content of unknown bases and low quality reads, needed to be removed before downstream bioinformatics analyses. The clean reads filtering from raw reads were aligned to the reference sequences by SOAP aligner/SOAP2 software (Li et al. 2008). We chose the Cx. quinquefasciatus sequences (https://www.vectorbase.org/organisms/culex-quinquefasciatus) as the reference genome (assembly version: CpipJ1) and the reference genes (geneset version: CpipJ1.3), since the genome of Cx. pipiens pallens is not available. We calculated the distribution of reads on reference genes to perform gene coverage statistics and to assess the sequencing randomness. The value of gene coverage is equal to the ratio of the base number in a gene covered by unique mapping reads to the total base number of coding region in that gene. We used the distribution of reads on the genes to evaluate the randomness. Since reference genes had different lengths, the read location on each gene was standardized to a relative position (which was calculated as the ratio between read location on the gene and the gene length), and then the number of reads in each relative position was counted.
Analysis of differentially expressed genes (DEGs)
The reads per kilobase per million mapped reads (RPKM) method was used for the gene expression level analysis (Mortazavi et al. 2008). The RPKM method is able to eliminate the influence of different gene length and sequencing discrepancy on the calculation of gene expression. Therefore, the calculated gene expression can be directly used for comparing the difference of gene expression among samples (Audic and Claverie 1997). The criteria of significant difference expression was |log2Ratio| ≥ 1 (Wang et al. 2010) and False Discovery Rate (FDR) ≤ 0.001 (Benjamini and Yekutieli 2001). If there was more than one transcript for a gene, the longest transcript was used to calculate its expression level and coverage (Zhang et al. 2013).
Gene ontology enrichment analysis and pathway enrichment analysis of DEGs
The GO enrichment analysis provides all GO terms that significantly enriched in a list of DEGs, comparing to a genome background, and filter the DEGs that correspond to specific biological functions. This method firstly mapped all DEGs to GO terms in the database (http://www.geneontology.org/), calculating gene numbers for every term, then used the hypergeometric test to find significantly enriched GO terms in the input list of DEGs, based on ‘GO::TermFinder’ (http://search.cpan.org/dist/GO-TermFinder/lib/GO/TermFinder.pm).
Kyoto Encyclopedia of Genes and Genomes (KEGG, the major public pathway-related database) (Kanehisa et al. 2008) is used to perform pathway enrichment analysis of DEGs. This analysis identified significantly enriched metabolic pathways or signal transduction pathways in DEGs comparing with the whole genome background.
Novel transcript units (NTUs) prediction
We assembled transcripts with reads using Cufflink (Roberts et al. 2011). According to the distribution of reads on reference genome, continuous and overlapping reads would form a transcriptional activity area (TAR). We linked different TARs to form an assembled transcript. We used a Support Vector Machine-based classifier, named Coding Potential Calculator (CPC, http://cpc.cbi.pku.edu.cn/), to assess the protein-coding potential of NTUs.
RNA-seq data validation
To validate the results of gene expression difference analysis, we selected 22 DEGs to quantify their relative expression levels in the Lab-DS and the Lab-DR strains by qRT-PCR using the same premixed RNA templates as were used in the RNA-seq analysis. The first-strand cDNA was synthesized from total RNA using PrimeScript™ RT Master Mix (Takara, Japan). Gene-specific qRT-PCR primers were designed according to Cx. pipiens pallens transcriptome sequences (Table S2) by Primer Premier 3.0 (Premier Biosoft International). The qRT-PCR reactions were performed in a 7300 FAST Real-Time PCR System (ABI, USA) using Power SYBR Green PCR Master Mix (ABI, USA). The PCR products were used for melting curve and agarose gel electrophoresis analysis to confirm their amplification specificity. We chose β-actin (Canales et al. 2009) and RsP7 (Leal et al. 2013) as the internal normalization. The relative expression levels of each gene were calculated using the 2−(ΔΔCt) method (Livak and Schmittgen 2001). Three technical and three biological replicates were performed.
On the other hand, to evaluate the accuracy of Illumina-Solexa sequencing, thirteen NTUs were randomly chosen for reverse transcription PCR (RT-PCR) validation using the same cDNA templates as qRT-PCR. Primers were designed according to Cx. pipiens pallens NTUs sequences (Table S2) by Primer Premier 5.0 software (Premier Biosoft International). The PCR products were sequenced in BGI. Three biological replicates were performed for RT-PCR amplification. All primers sequences for qRT-PCR and RT-PCR are presented in Table S5.
Full-length cloning and sequencing of CYP6AA9
The full-length cDNA of CYP6AA9 from Cx. pipiens pallens was amplified in three sections: the open reading frame (ORF) and the 5′ and 3′-cDNA ends (5′ and 3′-RACE). We prepared the templates for 5′-RACE and 3′-RACE with a SMART™ RACE cDNA Amplification Kit (Clontech, USA). The PCR reactions were carried out using Advantage 2 Polymerase Mix (Clontech, USA). We separated the PCR products by agarose gel electrophoresis, and then purified them with a MiniBEST Agarose Gel DNA Extraction Kit Ver.4.0 (Takara, Japan). The purified products were then inserted into the pMD19-T simple vector (Takara, Japan) to be sequenced in BGI. We finally assembled the sequences of three sections to generate the full-length cDNA. We used the standard protein/protein BLAST sequence comparison programs (http://beta.uniprot.org/?tab=blast) to search sequences with similarities to the translated sequences of CYP6AA9 in Cx. pipiens pallens in the SWISS-PROT databases. We aligned deduced amino acid sequences using the ClustalW2 computer program (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and constructed the phylogenetic tree using the neighbor-joining method of the MEGA5.1 program. All primers sequences for 5′ and 3′ RACE are presented in Table S5.
RNAi of CYP6AA9 and American CDC Bottle Bioassay
According to the full-length sequence of CYP6AA9, we designed and synthesized the siRNA432 (GenePharma, China). After one day post adult emergence, the non-blood-fed adult female mosquitoes from the Lab-DR2 strain were used for the microinjection experiments. According to standard methodology (Blandin et al. 2002), we microinjected 0.07 μL of the DEPC-water, negative control (NC) RNA or siRNA432 (5 μg/μL) into the thorax of mosquitoes. The sequences of siRNA432 and NC are presented in Table S5. Three days later, we verified the RNAi efficiency using qRT-PCR, and detected the relative mortality of adult female mosquitoes under deltamethrin exposure by American CDC Bottle Bioassay (Aizoun et al. 2013). The diagnostic dose of deltamethrin applied in our study was 6 mg per bottle (1 mL). The acetone control bottle was simultaneously employed to test the mosquitoes with siRNA432 microinjection. A total of 20–25 female mosquitoes after 3 days of microinjection were transferred into each bottle. The numbers of dead mosquitoes in bottles were recorded every 15 minutes, up to 2 hours. The mortality is equal to the ratio of the number of dead mosquitoes at each diagnostic time to the total number of mosquitoes before American CDC Bottle Bioassay in each bottle. Three biological replicates were performed for the American CDC Bottle Bioassay.
Database submission
All the reads generated in Illumina-Solexa sequencing were deposited in the NCBI Sequence Read Archive (SRA) database (accession number: PRJNA266574).
Results
RNA-seq and quality control (QC)
To obtain the overall transcriptional information of Cx. pipiens pallens, we sequenced the Lab-DS and the Lab-DR strains by high-throughput technology. Two premixed cDNA libraries were constructed separately and then deeply sequenced on an Illumina-Solexa HiSeq™ 2000 platform. We carried out the quality control (QC) of raw reads via base composition analysis and base sequence quality analysis. The A/T and G/C curves overlapped, indicating a uniform distribution of the base composition (Fig. S1). The low quality base (<20) ratio was low, again indicating that we achieved a relative high sequence quality (Fig. S2).
A total of 4,961,197,620 base pairs and 55,124,418 reads were generated from the Lab-DS and the Lab-DR strains (Table 1). For the Lab-DS strain, 62.63% (34,524,233) and 44.02% (24,264,446) reads were mapped to the reference genome and the reference genes of Cx. quinquefasciatus, respectively. The percentages of unique matched reads were 58.03% (31,987,234) and 41.96% (23,129,820). For the Lab-DR strain, 62.28% (34,332,448) and 44.69% (24,636,261) of the reads were mapped to the reference genome and the reference genes, respectively. The percentages of unique matched reads were 57.74% (31,827,840) and 42.57% (23,464,323).
Table 1.
Alignment statistics results of the Lab-DS strain and the Lab-DR strain
| Sample | Lab-DS | Lab-DR | ||
|---|---|---|---|---|
| Total Reads | 55124418 | 55124418 | ||
| Total Base Pairs | 4961197620 | 4961197620 | ||
| Map to Genome | Reads number | Percentage | Reads number | Percentage |
| Total mapped reads | 34524233 | 62.63% | 34332448 | 62.28% |
| perfect match | 14519833 | 26.34% | 14083456 | 25.55% |
| <= 5 bp mismatch | 20004400 | 36.29% | 20248992 | 36.73% |
| unique match | 31987234 | 58.03% | 31827840 | 57.74% |
| multi-position match | 2536996 | 4.60% | 2504607 | 4.54% |
| Total Unmapped Reads | 20600185 | 37.37% | 20791970 | 37.72% |
| Map to Gene | Reads number | Percentage | Reads number | Percentage |
| Total mapped reads | 24264446 | 44.02% | 24636261 | 44.69% |
| perfect match | 9944281 | 18.04% | 9877334 | 17.92% |
| <= 5 bp mismatch | 14320165 | 25.98% | 14758927 | 26.77% |
| unique match | 23129820 | 41.96% | 23464323 | 42.57% |
| multi-position match | 1134626 | 2.06% | 1171938 | 2.13% |
| Total Unmapped Reads | 30859972 | 55.98% | 30488157 | 55.31% |
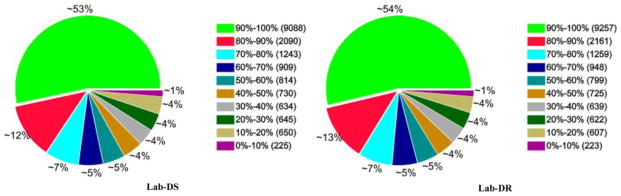
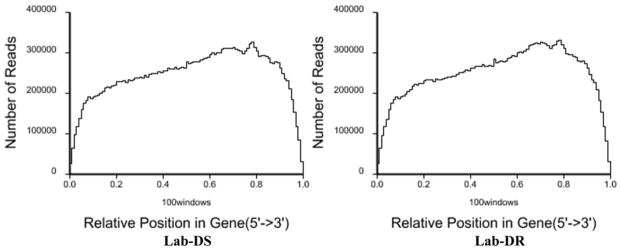
Gene coverage statistics for the Lab-DS and the Lab-DR strains showed that more than half the genes (53% and 54%, respectively) reached coverage of 90% to 100% (Fig. 1). A randomness assessment revealed the even distribution of reads in gene 5′ to the 3′ end of the Lab-DS and the Lab-DR strains (Fig. 2). The cDNA library construction by mRNA fragmentation provided more even coverage along the gene body (Wang et al. 2009).
Fig. 1. Gene coverage statistics for the Lab-DS and the Lab-DR strains.
Gene coverage is calculated as the percentage of a gene covered by reads. This value is equal to the ratio of the base number in a gene covered by unique mapping reads to the total base number of coding region in that gene. Different colors represent the proportions of genes with certain coverage in a pie chart. For the Lab-DS and the Lab-DR strains, more than half the gene (53% and 54%, respectively) reached coverage of 90% to 100%.
Fig. 2. Randomness assessment for the Lab-DS and the Lab-DR strains.
The distribution of reads on the genes was used to evaluate the randomness. Since reference genes had different lengths, the read location on each gene was standardized to a relative position, and then the number of reads in each relative position was counted. For the Lab-DS and the Lab-DR strains, the reads are evenly distributed in the 5′ to 3′ end of the reference genes.
Gene differential expression analysis and quantification via qRT-PCR
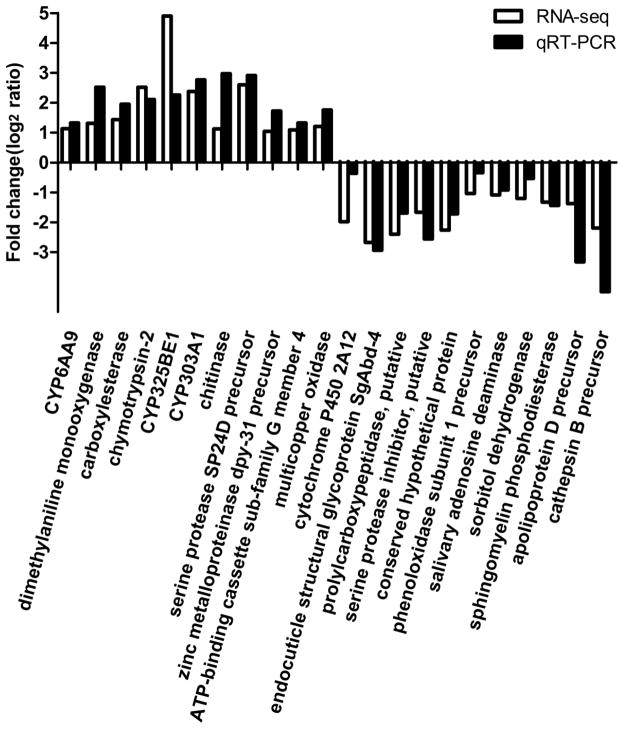
We next assembled all unique matched reads into 17,679 known genes, with Cx. quinquefasciatus database as the reference. Their length, coverage, RPKM and Gene Ontology (GO) classification are shown in Table S1. The assembled sequences of Cx. pipiens pallens transcriptome are presented in Table S2. The clean reads from the Lab-DS and the Lab-DR strains were mapped to Cx. pipiens pallens transcriptome for gene differential expression analysis. We detected a total of 1,826 significantly differentially expressed genes (DEGs). Among them, 1,078 genes were up-regulated and 748 genes were down-regulated in the Lab-DR strain (Table S3). These DEGs included P450s, cuticle proteins, UDP-glucuronosyltransferases, lipases, serine proteases, heat shock proteins, esterases, peptidases, ATP-binding cassette transporters and others. Subsequently, considering the RPKM and the fold change, we selected 22 DEGs to quantify their relative expression levels by qRT-PCR using the same premixed RNA templates as were used in the RNA-seq analysis. As shown in Fig. 3, the trend of the gene differential expression analysis by RNA-seq and qRT-PCR was consistent, again increasing the credibility of RNA-seq.
Fig. 3. Comparison of RNA-seq and qRT-PCR results.
RNA-seq and qRT-PCR based fold-changes in transcript levels. The x-axis shows the 22 selected genes, while the y-axis gives the degree of fold change observed for the Lab-DR vs the Lab-DS strain, presented as the log2ratio value. Three technical and three biological replicates were performed for qRT-PCR.
Gene ontology enrichment analysis and pathway enrichment analysis of DEGs
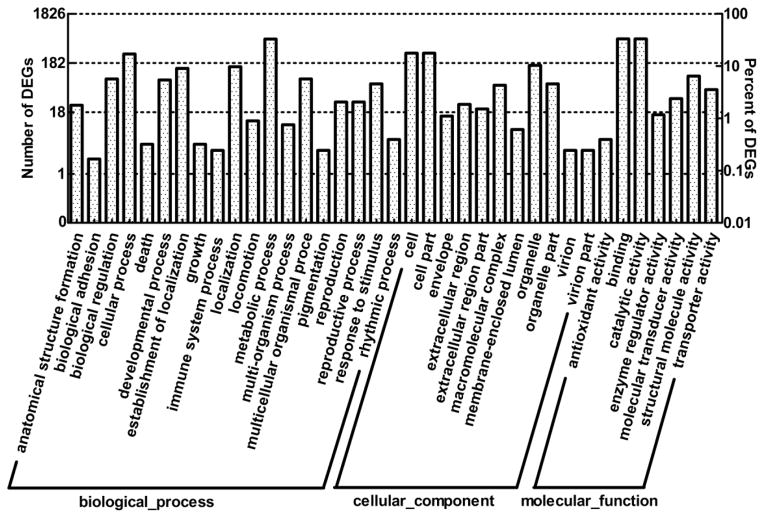
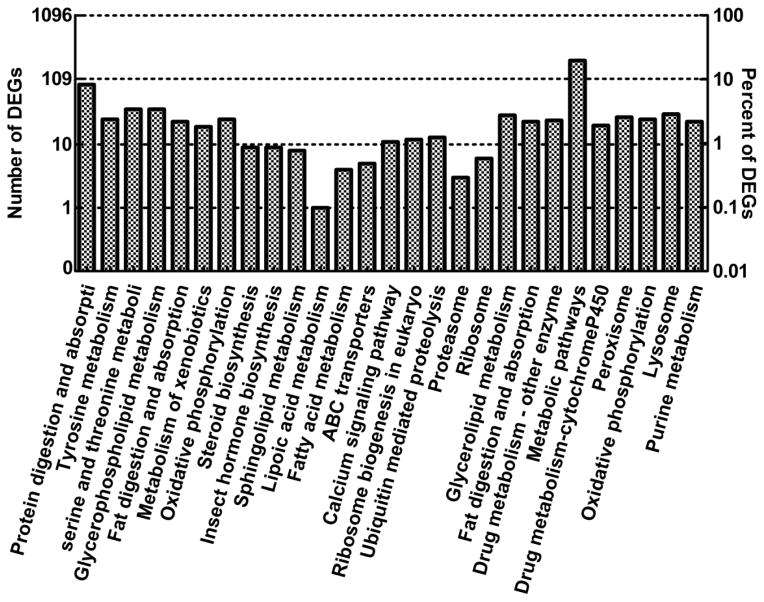
A total of 1,826 DEGs were classified into three main GO categories: biological processes, cellular components, and molecular functions (Fig. 4). For biological processes, the categories most represented were metabolic process (GO: 0008152, 558 out of 794 DEGs, 70.3%) and cellular process (GO: 0009987, 278 out of 794 DEGs, 35.0%). As for cellular components, genes involved in cell (GO: 0005623, 288 out of 412 DEGs, 69.9%) and cell part (GO: 0044464, 288 out of 412 DEGs, 69.9%) were the most. For molecular functions, binding (GO: 0005488, 563 out of 1041 DEGs, 54.1%) and catalytic activity (GO: 0003824, 564 out of 1041 DEGs, 54.2%) were the most highly represented categories. Pathway-based analysis allowed a greater understanding of their biological functions. A total of 1,096 DEGs were mapped into 232 KEGG pathways (Fig. 5). The largest category was metabolic pathways, containing 212 annotated DEGs (19.34%), followed by protein digestion and absorption, with 88 DEGs (8.03%).
Fig. 4. Results of the gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs).
A total of 1,826 DEGs were classified into three main categories: biological process, cellular component and molecular function. The left y-axis represents the number of DEGs in a GO term, and the right y-axis represents the corresponding percentage.
Fig. 5. Results of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes (DEGs).
A total of 1,096 DEGs were mapped into KEGG pathways. The left y-axis represents the number of DEGs in a KEGG pathway, and the right y-axis represents the corresponding percentage.
In general, the most represented GO and KEGG categories were “metabolic process” and “metabolic pathway”, which were widely acknowledged to involve in insecticide metabolism (Copley 2000; Sagri et al. 2014).
Novel transcript units (NTUs) prediction
A total of 4,957 and 5,048 novel transcript units (NTUs) were predicted in the Lab-DS and the Lab-DR libraries. In the Lab-DS library, 1,515 NTUs contained completed or partly CDS and 3,442 NTUs may be non-coding sequence. In the Lab-DR library, the numbers were 1,529 and 3,519, respectively (Table S4). All sequences are presented in Table S2. Next, we randomly selected 13 NTUs and designed respective primers for PCR amplification. We found that all primers could amplify the corresponding bands using the Cx. pipiens pallens cDNA templates (Fig. S3), which reflected the actual transcriptional expression rather than any technical noise from spurious sequences. The Sanger sequencing results of PCR products were exactly consistent with RNA-seq (data not shown), which provided further support that the RNA-seq was accurate, and that sequence assembly quality was excellent.
Screening candidate genes
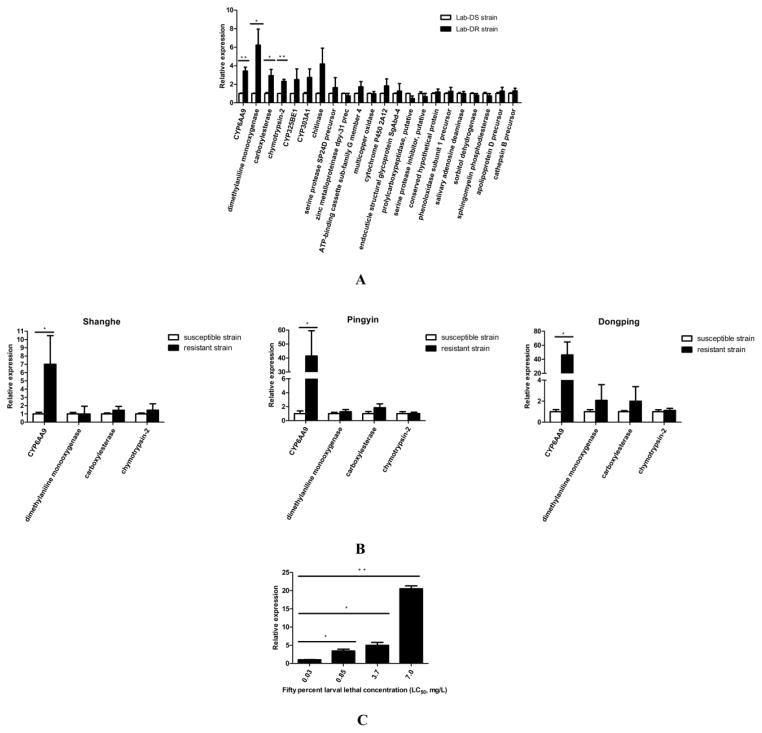
We used qRT-PCR to test the relative expression levels of the 22 DEGs (Fig. 3) between the Lab-DS and the Lab-DR strains in adult female mosquito stage. That we chose the adult female mosquito stage was because the mosquito-borne diseases are mainly transmitted by adult females (WHO 2013). As shown in Fig. 6A, four of the 22 genes (CYP6AA9, dimethylaniline monooxygenase, carboxylesterase, chymotrypsin-2) were significantly overexpressed in the Lab-DR strain compared to the Lab-DS strain (P < 0.05). The results of three field populations (Shanghe, Pingyin and Dongping) indicated that the expression levels of the CYP6AA9 were 7.0-, 41.4-, and 46.2-fold higher in the resistant strains than the susceptible strains (P < 0.05; Fig. 6B). Next, we quantified the relative expression levels of CYP6AA9 in adult female mosquitoes with different levels of deltamethrin resistance. The LC50 of Lab-DS, Lab-DR, Lab-DR1 and Lab-DR2 strains were 0.03, 0.85, 3.7 and 7.0 mg/L, respectively. As shown in Fig. 6C, the relative expression levels of CYP6AA9 were 3.4-, 5.0-, and 20.5-fold higher in the Lab-DR, Lab-DR1, and Lab-DR2 strains than the Lab-DS strain (P < 0.05; Fig. 6C). Thus, CYP6AA9 was selected as our candidate gene for the subsequent functional verification.
Fig. 6. Screening candidate genes via quantitative real-time PCR (qRT-PCR).
(A) The relative expression levels of 22 genes in the adult female mosquitoes of the Lab-DS and the Lab-DR strains. Four of the 22 genes (CYP6AA9, dimethylaniline monooxygenase, carboxylesterase, chymotrypsin-2) were significantly overexpressed in the Lab-DR strain compared to the Lab-DS strain.
(B) The relative expression levels of four genes in the adult female mosquitoes of the susceptible and resistant strains of three field populations (Shanghe, Pingyin and Dongping). The relative expression levels of the CYP6AA9 were 7.0-, 41.4- and 46.2-fold higher in the resistant strains than the susceptible strains.
(C) The relative expression levels of CYP6AA9 in the adult female mosquitoes with different levels of deltamethrin resistance. The LC50 of the Lab-DS, Lab-DR, Lab-DR1 and Lab-DR2 strains were 0.03, 0.85, 3.7 and 7.0 mg/L, respectively. The relative expression levels of CYP6AA9 were 3.4-, 5.0-, and 20.5-fold higher in the Lab-DR, Lab-DR1, and Lab-DR2 strains, respectively, than the Lab-DS strain.
Results are presented as mean ± standard deviation (SD) of three independent experiments (*P < 0.05, **P < 0.01).
Full-length cloning and sequencing of CYP6AA9
The full-length cDNA of CYP6AA9 contained 1871 base pairs (bp). The ORF region had 1548 bp and encoded a protein with 515 amino acids (GenBank accession number: KP162164). The start codon ATG was found at positions 184–186, while the same frame stop codon TAA was at positions 1729–1731 with a polyadenylation signal sequence “AATAAA” and a poly (A) tail present at the 3′-untranslated region. At the C-terminal, there was a conserved domain of heme binding site (FGDGPRNCIG), which was the characteristic sequence of P450 family members (Fig. 7A). The deduced amino acid of CYP6AA9 gene in Cx. pipiens pallens shared 99.22% identity with the ortholog in Cx. quinquefasciatus, 59.61% identity with Ae. aegypti, 55.43% identity with An. gambiae and 38.65% identity with Drosophila melanogaster (Fig. 7B). Phylogenetic relationships showed that CYP6AA9 of Cx. pipiens pallens had the highest homology with Cx. quinquefasciatus (Fig. 7C).
Fig. 7. Full-length cDNA of CYP6AA9 in Cx. pipiens pallens.
(A) The nucleotide and deduced amino acid sequences of CYP6AA9 from Cx. pipiens pallens. The initial codon “ATG” and the termination codon “TGA” are represented in bold letters and underlined. The conserved domain of heme binding site (FGDGPRNCIG) is boxed. The polyadenylation signal sequence “AATAAA” and a poly (A) at the 3′-untranslated region are underlined.
(B) Amino acid alignment of the CYP6AA9 gene in Cx. pipiens pallens and four other species. Asterisks and dots respectively indicate identical and similar amino acids.
(C) Phylogenetic relationships of the CYP6AA9 in Cx. pipiens pallens and four other species. Species name and GenBank Accession No.: Cx. pipiens pallens, KP162164; Cx. quinquefasciatus, XP_001847405.1; Ae. aegypti, XP_001662604.1; An. gambiae XP_312054.2; Drosophila melanogaster, NP_523628.1.
RNAi of CYP6AA9 and American CDC Bottle Bioassay
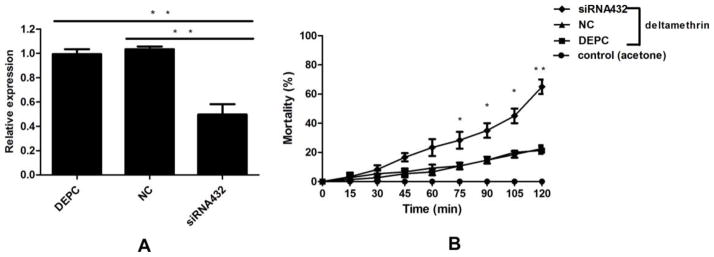
The siRNA432 was microinjected into the thorax of the adult female mosquitoes from the Lab-DR2 strain to knock down the expression of CYP6AA9. As shown in Fig. 8A, the gene expression level of CYP6AA9 could be reduced approximately 50% by siRNA432, compared with the DEPC-water and negative control (NC) RNA (P < 0.01). According to the results of the American CDC Bottle Bioassay, we found that the siRNA432 group had a significantly higher mortality rate than the control groups after 75 minutes of exposure time (P < 0.05; Fig. 8B). All mosquitoes in the acetone control bottles were alive. The knockdown of CYP6AA9 by RNAi increased the sensitivity of the adult female mosquitoes to deltamethrin.
Fig. 8. RNAi of CYP6AA9 and American CDC Bottle Bioassay.
(A) RNAi efficiency verification of CYP6AA9 by qRT-PCR.
The DEPC-water, negative control (NC) RNA and siRNA432 was respectively microinjected into the thorax of the adult female mosquitoes. The gene expression level of CYP6AA9 could be reduced approximately 50% by siRNA432, compared with the DEPC-water and negative control (NC) RNA.
(B) Mortality of microinjected mosquitoes after 120 minutes of exposure time in American CDC Bottles with deltamethrin (6 mg/ml).
The siRNA432 microinjected group had a significantly higher mortality rate than the DEPC and the negative control (NC) groups after 75 minutes of exposure time. All mosquitoes in the acetone control bottles were alive.
The results are presented as mean ± standard deviation (SD) of three independent experiments (*P < 0.05, **P < 0.01).
Discussion
During the alignment process, we chose the Cx. quinquefasciatus sequences as the reference genome and the reference genes, since the genome of Cx. pipiens pallens is not available. Xu et al. (2013) similarly performed a comprehensive transcriptome analysis of Setaria viridis utilizing a reference-based assembly with Setaria italica genome as the reference (Xu et al. 2013). Biosystematic studies (including hybridization), morphological studies and gas chromatography analyses of cuticular hydrocarbons confirmed that Cx. pipiens pallens and Cx. quinquefasciatus as being subspecies of Culex pipiens (Cui et al. 2007). In our results, the percentages of alignment rate (Table 1) and gene coverage (Fig. 1) agreed with the RNA-seq results of other species (Fu et al. 2014; Peng et al. 2013; Wang et al. 2013; Zhang et al. 2013), indicating the feasibility of the alignment method in our study.
We detected 4,957 and 5,048 NTU in the Lab-DS and the Lab-DR libraries. Daines et al. (2011) identified 319 novel transcripts using broad RNA-seq samples in 10 developmental stages of Drosophila melanogaster (Daines et al. 2011). Xu et al. (2013) identified 32,304 known transcripts and 10,450 NTUs in Setaria viridis using Setaria italica genome as the reference (Xu et al. 2013). A great quantity of the NTUs identified in our RNA-seq analysis were likely attributed either to the use of multiple developmental stages in the premixed process (Roberts et al. 2011) or to actual differences between Cx. pipiens pallens and Cx. quinquefasciatus. We used the premixed samples of different developmental stages to increase the transcript coverage (Zhu et al. 2014), and to provide the most comprehensive Cx. pipiens pallens transcriptome database for subsequent analyses. Simultaneously, we found these NTUs sequences could be mapped to Cx. quinquefasciatus genome, but not to any known transcripts. These NTUs may represent potential novel isoforms or reads that fell within currently annotated introns (Bonizzoni et al. 2012), indicating that the transcribed portions of the genomes were larger and more complex than expected. However, complete annotation and functional verification of these NTUs is needed.
Our transcriptome sequencing identified 1,826 DEGs, with 1,078 genes up-regulated and 748 genes down-regulated. As mentioned before, researchers have identified massive pyrethroid resistance-related DEGs by the transcriptome sequencing of other mosquito species. By comparison, many of the DEGs belong to the same gene families, such as P450s, cuticle proteins, UDP-glucuronosyltransferases, lipases, serine proteases, heat shock proteins and others (Bonizzoni et al. 2012; David et al. 2014; Reid et al. 2012; Zhu et al. 2014).
In our results, 37 DEGs belonged to drug metabolism cytochrome P450 family, with 18 genes being up-regulated and 19 genes being down-regulated (Table S3). Up- or down-regulation of P450 genes may be responsible for the detoxification of insecticides and the homeostatic response of mosquitoes to changes in cellular environment (Yang and Liu 2011). It has been suggested that reduction in the expression of some metabolic detoxification genes may result from responses to various endogenous and exogenous compounds, or to pathophysiological signals (Marinotti et al. 2005). In this study, P450 genes whose expression levels changed significantly belonged primarily to the CYP4, CYP6 and CYP325 families. Many members of CYP4 and CYP6 families have previously been reported to participate in pyrethroid resistance (Hemingway et al. 2004; Zhu et al. 2000). Recently, the CYP325 family, which is similar to the CYP4 family in many insects, was also shown to be associated with pyrethroid resistance (David et al. 2005; Saavedra-Rodriguez et al. 2012). We found six CYP325 family genes in the DEGs, five of which were up-regulated in the Lab-DR strain. Their regulatory mechanism in pyrethroid resistance warrants further study.
The genes encoding cuticle protein were associated with physiological resistance, which is another main mechanism of pyrethroid resistance. A total of 90 of the 1,826 DEGs belonged to cuticle protein family (Table S3). Wood et al. (2010) reported that cuticle thickening was associated with pyrethroid resistance in Anopheles funestus (Wood et al. 2010). Bonizzoni et al. (2012) found that the dynamic balance of cuticle protein genes was also likely associated with pyrethroid resistance via the transcriptome sequencing of An. gambiae (Bonizzoni et al. 2012). We also found other genes related to cuticle protein, including chitinase, suggesting that association between cuticle proteins and pyrethroid resistance merits further research.
We detected five UDP-glucuronosyltransferases in the DEGs. Six UDP-glucuronosyltransferases were considered to be relevant to pyrethroid resistance by the transcriptome sequencing of Cx. quinquefasciatus (Reid et al. 2012). Members of this family had been shown in rats to catalyse the transfer of glucuronic acid to lipophilic molecules, suggesting a means by which they detoxify xenobiotics (including insecticides) (Okazaki and Katayama 2003).
Nineteen DEGs belonged to lipase family, with eighteen up-regulated genes. Trehalase and lipase are considered as the main enzymes responsible for the higher energy mobilization in the resistant maize weevil populations, which are required for maintaining active pyrethroid resistance mechanisms in the resistant insects (Araujo et al. 2008).
These DEGs also included serine proteases, heat shock proteins, esterases, peptidases, ATP-binding cassette transporters and others novel potential resistance-related genes. We provided a critical theoretical basis for further molecular studies investigating mechanisms of pyrethroid resistance, and presented massive candidate genes for field resistance detection.
Among 1,826 DEGs, we found that the transcriptional levels of CYP6AA9 in laboratory populations was elevated as the levels of deltamethrin resistance increased. Moreover, the expression levels of the CYP6AA9 were 7.0-, 41.4- and 46.2-fold higher in the resistant strains than the susceptible strains in three different field populations (Fig. 6). The orthologous genes of CYP6AA9 in Cx. quinquefasciatus (Yang and Liu 2011), CYP6AA5 in Ae. aegypti (Marcombe et al. 2009) and CYP6AA1 in An. gambiae (Kwiatkowska et al. 2013) were all transcriptionally overexpressed in the pyrethroid-resistant strains than the corresponding susceptible strains. Combining with our experimental results of RNAi, we predicted that CYP6AA9 played a role in regulating pyrethroid resistance in mosquitoes and was expected to become a potential marker to monitor and predict the pyrethroid resistance level of field mosquito populations.
In this study, we used Illumina-Solexa sequencing to identify 1,826 genes that were expressed differently in the Lab-DS and the Lab-DR strains as a critical theoretical basis for further molecular studies investigating mechanisms of pyrethroid resistance. In addition, we demonstrated that CYP6AA9 participated in the pyrethroid resistance in mosquitoes and was expected to become a target gene in field resistance detection.
Supplementary Material
On the x-axes, 1–90 bp represents the base position of read 1, while 91–180 bp represents the base position of read 2. The y-axes are the base composition percentage. The A/T and G/C curves overlapped, indicating a uniform distribution of the base composition.
On the x-axes, 1–90 bp represents the base position of read 1, while 91–180 bp represents the base position of read 2. The y-axes are the base sequence quality value. The low quality base (< 20) ratio was low, again indicating that we achieved a relative high sequence quality.
We assembled transcripts with reads using Cufflink. The NTUs were predicted using the Culex quinquefasciatus database as the reference. RT-PCR amplification on Culex pipiens pallens cDNA template was used to validate the NTUs.
M: DL2000 DNA Marker; 1–13: the thirteen RT-PCR amplified fragments of the predicted NTUs, with the lengths of 300, 261, 287, 566, 251, 233, 342, 231, 192, 415, 240, 102 and 381bp.
Details of 17,679 known genes along with the length, coverage, RPKM, and GO classification. (XLS)
Details of all assembled sequences of Cx. pipiens pallens transcriptome. The sequences of novel transcript units (NTUs) are underlined. (DOC)
List of all the significantly differentially expressed genes (DEGs) between the Lab-DR and the Lab-DS strains (FDR ≤ 0.001 AND |log2Ratio|≥1). (XLS)
Details of the predicted novel transcript units (NTUs) for the Lab-DS and the Lab-DR strains. Blocks represent exon number. Sizes are each exon size. Starts indicate starting positions of each exon in a chromosome. (XLS)
List of all primers sequences for RT-PCR, qRT-PCR, RACE and siRNA sequences for RNAi. (DOC)
Acknowledgments
This work was supported by the National Institutes of Health of the United states (NIH) (Grant No. 2R01AI075746), the National Natural Science Foundation of China (Grant Nos. 81171900, 81101279 and 81301458), the National S & T Major Program (Grant Nos. 2012ZX10004-219 and 2012ZX10004-220), the Specialized Research Fund for the Doctoral Program of Higher Education of China (Grant No. 20113234120007), and the Natural Science Foundation of Jiangsu Province (Grant No. 81101279).
Footnotes
Conflict of interest Authors declare no conflicts of interest.
Ethical approval All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
References
- Aizoun N, Aikpon R, Padonou GG, Oussou O, Oke-Agbo F, Gnanguenon V, Osse R, Akogbeto M. Mixed-function oxidases and esterases associated with permethrin, deltamethrin and bendiocarb resistance in Anopheles gambiae s.l. in the south-north transect Benin, West Africa. Parasit Vectors. 2013;6:223. doi: 10.1186/1756-3305-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo RA, Guedes RN, Oliveira MG, Ferreira GH. Enhanced activity of carbohydrate- and lipid-metabolizing enzymes in insecticide-resistant populations of the maize weevil, Sitophilus zeamais. Bull Entomol Res. 2008;98:417–424. doi: 10.1017/S0007485308005737. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO reports. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzoni M, Afrane Y, Dunn WA, Atieli FK, Zhou G, Zhong D, Li J, Githeko A, Yan G. Comparative transcriptome analyses of deltamethrin-resistant and -susceptible Anopheles gambiae mosquitoes from Kenya by RNA-Seq. PLoS One. 2012;7:e44607. doi: 10.1371/journal.pone.0044607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzolan F, Siaussat D, Maria A, Durand N, Pottier MA, Chertemps T, Maibeche-Coisne M. Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: diversity and putative function in odorant and xenobiotics clearance. Insect Mol Biol. 2014;23:539–549. doi: 10.1111/imb.12100. [DOI] [PubMed] [Google Scholar]
- Canales M, Naranjo V, Almazan C, Molina R, Tsuruta SA, Szabo MP, Manzano-Roman R, Perez de la Lastra JM, Kocan KM, Jimenez MI, Lucientes J, Villar M, de la Fuente J. Conservation and immunogenicity of the mosquito ortholog of the tick-protective antigen, subolesin. Parasitol Res. 2009;105:97–111. doi: 10.1007/s00436-009-1368-2. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhong D, Zhang D, Shi L, Zhou G, Gong M, Zhou H, Sun Y, Ma L, He J, Hong S, Zhou D, Xiong C, Chen C, Zou P, Zhu C, Yan G. Molecular ecology of pyrethroid knockdown resistance in Culex pipiens pallens mosquitoes. PLoS One. 2010;5:e11681. doi: 10.1371/journal.pone.0011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley SD. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem Sci. 2000;25:261–265. doi: 10.1016/s0968-0004(00)01562-0. [DOI] [PubMed] [Google Scholar]
- Cui F, Qiao CL, Shen BC, Marquine M, Weill M, Raymond M. Genetic differentiation of Culex pipiens (Diptera: Culicidae) in China. Bull Entomol Res. 2007;97:291–297. doi: 10.1017/S0007485307004968. [DOI] [PubMed] [Google Scholar]
- Daines B, Wang H, Wang L, Li Y, Han Y, Emmert D, Gelbart W, Wang X, Li W, Gibbs R, Chen R. The Drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 2011;21:315–324. doi: 10.1101/gr.107854.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JP, Faucon F, Chandor-Proust A, Poupardin R, Riaz MA, Bonin A, Navratil V, Reynaud S. Comparative analysis of response to selection with three insecticides in the dengue mosquito Aedes aegypti using mRNA sequencing. BMC Genomics. 2014;15:174. doi: 10.1186/1471-2164-15-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JP, Strode C, Vontas J, Nikou D, Vaughan A, Pignatelli PM, Louis C, Hemingway J, Ranson H. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc Natl Acad Sci U S A. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K, Du Y, Rinkevich F, Nomura Y, Xu P, Wang L, Silver K, Zhorov BS. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem Mol Biol. 2014;50:1–17. doi: 10.1016/j.ibmb.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edi CV, Djogbenou L, Jenkins AM, Regna K, Muskavitch MA, Poupardin R, Jones CM, Essandoh J, Ketoh GK, Paine MJ, Koudou BG, Donnelly MJ, Ranson H, Weetman D. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10:e1004236. doi: 10.1371/journal.pgen.1004236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant RH, Daborn PJ, Le Goff G. The genetics and genomics of insecticide resistance. Trends Genet. 2004;20:163–170. doi: 10.1016/j.tig.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Fu WQ, Zhao ZG, Ge XH, Ding L, Li ZY. Anatomy and transcript profiling of gynoecium development in female sterile Brassica napus mediated by one alien chromosome from Orychophragmus violaceus. BMC Genomics. 2014;15:61. doi: 10.1186/1471-2164-15-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemingway J, Field L, Vontas J. An overview of insecticide resistance. Science. 2002;298(5591):96–97. doi: 10.1126/science.1078052. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Hawkes NJ, McCarroll L, Ranson H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem Mol Biol. 2004;34(7):653–665. doi: 10.1016/j.ibmb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koou SY, Chong CS, Vythilingam I, Lee CY, Ng LC. Insecticide resistance and its underlying mechanisms in field populations of Aedes aegypti adults (Diptera: Culicidae) in Singapore. Parasit Vectors. 2014;7:471. doi: 10.1186/s13071-014-0471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska RM, Platt N, Poupardin R, Irving H, Dabire RK, Mitchell S, Jones CM, Diabate A, Ranson H, Wondji CS. Dissecting the mechanisms responsible for the multiple insecticide resistance phenotype in Anopheles gambiae s.s., M form, from Vallee du Kou, Burkina Faso. Gene. 2013;519:98–106. doi: 10.1016/j.gene.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS, Choo YM, Xu P, da Silva CS, Ueira-Vieira C. Differential expression of olfactory genes in the southern house mosquito and insights into unique odorant receptor gene isoforms. Proc Natl Acad Sci U S A. 2013;110:18704–18709. doi: 10.1073/pnas.1316059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Liu N. Insecticide resistance in mosquitoes: impact, mechanisms, and research directions. Annu Rev Entomol. 2015;60:537–559. doi: 10.1146/annurev-ento-010814-020828. [DOI] [PubMed] [Google Scholar]
- Liu N, Li T, Reid WR, Yang T, Zhang L. Multiple Cytochrome P450 genes: their constitutive overexpression and permethrin induction in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS One. 2011;6:e23403. doi: 10.1371/journal.pone.0023403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Liu H, Zhu F, Zhang L. Differential expression of genes in pyrethroid resistant and susceptible mosquitoes, Culex quinquefasciatus (S.) Gene. 2007;394:61–68. doi: 10.1016/j.gene.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marcombe S, Poupardin R, Darriet F, Reynaud S, Bonnet J, Strode C, Brengues C, Yebakima A, Ranson H, Corbel V, David JP. Exploring the molecular basis of insecticide resistance in the dengue vector Aedes aegypti: a case study in Martinique Island (French West Indies) BMC Genomics. 2009;10:494. doi: 10.1186/1471-2164-10-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JM. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol Biol. 2005;14:365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Katayama T. Effects of dietary carbohydrate and myo-inositol on metabolic changes in rats fed 1,1,1-trichloro-2,2-bis (p-chlorophenyl) ethane (DDT) J Nutr Biochem. 2003;14:81–89. doi: 10.1016/s0955-2863(02)00279-6. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Zhang C, Zhang Y, Hu T, Mu S, Li X, Gao J. Transcriptome sequencing and analysis of the fast growing shoots of moso bamboo (Phyllostachys edulis) PLoS One. 2013;8:e78944. doi: 10.1371/journal.pone.0078944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid WR, Zhang L, Liu F, Liu N. The transcriptome profile of the mosquito Culex quinquefasciatus following permethrin selection. PLoS One. 2012;7:e47163. doi: 10.1371/journal.pone.0047163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner RC, Jr, Perkins TA, Barker CM, Niu T, Chaves LF, Ellis AM, George DB, Le Menach A, Pulliam JR, Bisanzio D, Buckee C, Chiyaka C, Cummings DA, Garcia AJ, Gatton ML, Gething PW, Hartley DM, Johnston G, Klein EY, Michael E, Lindsay SW, Lloyd AL, Pigott DM, Reisen WK, Ruktanonchai N, Singh BK, Tatem AJ, Kitron U, Hay SI, Scott TW, Smith DL. A systematic review of mathematical models of mosquito-borne pathogen transmission: 1970–2010. J R Soc Interface. 2013;10:20120921. doi: 10.1098/rsif.2012.0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero A, Vezilier J, Weill M, Read AF, Gandon S. Insecticide control of vector-borne diseases: when is insecticide resistance a problem? PLoS Pathog. 2010;6:e1001000. doi: 10.1371/journal.ppat.1001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Saavedra-Rodriguez K, et al. Transcription of detoxification genes after permethrin selection in the mosquito Aedes aegypti. Insect Mol Biol. 2012;21:61–77. doi: 10.1111/j.1365-2583.2011.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagri E, Reczko M, Gregoriou ME, Tsoumani KT, Zygouridis NE, Salpea KD, Zalom FG, Ragoussis J, Mathiopoulos KD. Olive fly transcriptomics analysis implicates energy metabolism genes in spinosad resistance. BMC Genomics. 2014;15:714. doi: 10.1186/1471-2164-15-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo-Orihuela PL, Carvajal G, Picollo MI, Vassena CV. Analysing deltamethrin susceptibility and pyrethroid esterase activity variations in sylvatic and domestic Triatoma infestans at the embryonic stage. Mem Inst Oswaldo Cruz. 2013;108:1031–1036. doi: 10.1590/0074-0276130184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tao X, Tang XM, Xiao L, Sun JL, Yan XF, Li D, Deng HY, Ma XR. Comparative transcriptome analysis of tomato (Solanum lycopersicum) in response to exogenous abscisic acid. BMC Genomics. 2013;14:841. doi: 10.1186/1471-2164-14-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZM, Li CX, Xing D, Yu YH, Liu N, Xue RD, Dong YD, Zhao TY. Detection and widespread distribution of sodium channel alleles characteristic of insecticide resistance in Culex pipiens complex mosquitoes in China. Med Vet Entomol. 2012;26:228–232. doi: 10.1111/j.1365-2915.2011.00985.x. [DOI] [PubMed] [Google Scholar]
- WHO. Test procedures for insecticide resistance monitoring in malaria vector mosquitoes. World Health Organization; Geneva, Switzerland: 2013. [Google Scholar]
- Wood O, Hanrahan S, Coetzee M, Koekemoer L, Brooke B. Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus. Parasit Vectors. 2010;3:67. doi: 10.1186/1756-3305-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HW, Tian HS, Wu GL, Langdon G, Kurtis J, Shen B, Ma L, Li XL, Gu Y, Hu XB. Culex pipiens pallens: identification of genes differentially expressed in deltamethrin-resistant and-susceptible strains. Pestic Biochem Physiol. 2004;79:75–83. [Google Scholar]
- Xie W, Meng QS, Wu QJ, Wang SL, Yang X, Yang NN, Li RM, Jiao XG, Pan HP, Liu BM, Su Q, Xu BY, Hu SN, Zhou XG, Zhang YJ. Pyrosequencing the Bemisia tabaci transcriptome reveals a highly diverse bacterial community and a robust system for insecticide resistance. PLoS One. 2012;7:e35181. doi: 10.1371/journal.pone.0035181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li Y, Ma X, Ding J, Wang K, Wang S, Tian Y, Zhang H, Zhu XG. Whole transcriptome analysis using next-generation sequencing of model species Setaria viridis to support C4 photosynthesis research. Plant Mol Biol. 2013;83:77–87. doi: 10.1007/s11103-013-0025-4. [DOI] [PubMed] [Google Scholar]
- Yang T, Liu N. Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS One. 2011;6:e29418. doi: 10.1371/journal.pone.0029418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang G, Wang J, Ji Z, Liu Z, Pi X, Chen C. Characterization and comparative analyses of muscle transcriptomes in Dorper and small-tailed Han sheep using RNA-Seq technique. PLoS One. 2013;8:e72686. doi: 10.1371/journal.pone.0072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D. Molecular and biochemical evidence for pyrethroid resistance mechanism in the malaria vector Anopehels sinesis mosquitoe. 60th ASTMH Meeting Poster LB-2263; 2011. [Accessed 2012 May 1]. May 1, [Google Scholar]
- Zhu CL, Li JM, Tian HS, Ma L, Li XL, Wu GL. Cloning and identification of cytochrome P450 resistance related genes in the mosquitoCulex pipiens pallens. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2000;18:263–268. [PubMed] [Google Scholar]
- Zhu G, Zhong D, Cao J, Zhou H, Li J, Liu Y, Bai L, Xu S, Wang MH, Zhou G, Chang X, Gao Q, Yan G. Transcriptome profiling of pyrethroid resistant and susceptible mosquitoes in the malaria vector, Anopheles sinensis. BMC Genomics. 2014;15:448. doi: 10.1186/1471-2164-15-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
On the x-axes, 1–90 bp represents the base position of read 1, while 91–180 bp represents the base position of read 2. The y-axes are the base composition percentage. The A/T and G/C curves overlapped, indicating a uniform distribution of the base composition.
On the x-axes, 1–90 bp represents the base position of read 1, while 91–180 bp represents the base position of read 2. The y-axes are the base sequence quality value. The low quality base (< 20) ratio was low, again indicating that we achieved a relative high sequence quality.
We assembled transcripts with reads using Cufflink. The NTUs were predicted using the Culex quinquefasciatus database as the reference. RT-PCR amplification on Culex pipiens pallens cDNA template was used to validate the NTUs.
M: DL2000 DNA Marker; 1–13: the thirteen RT-PCR amplified fragments of the predicted NTUs, with the lengths of 300, 261, 287, 566, 251, 233, 342, 231, 192, 415, 240, 102 and 381bp.
Details of 17,679 known genes along with the length, coverage, RPKM, and GO classification. (XLS)
Details of all assembled sequences of Cx. pipiens pallens transcriptome. The sequences of novel transcript units (NTUs) are underlined. (DOC)
List of all the significantly differentially expressed genes (DEGs) between the Lab-DR and the Lab-DS strains (FDR ≤ 0.001 AND |log2Ratio|≥1). (XLS)
Details of the predicted novel transcript units (NTUs) for the Lab-DS and the Lab-DR strains. Blocks represent exon number. Sizes are each exon size. Starts indicate starting positions of each exon in a chromosome. (XLS)
List of all primers sequences for RT-PCR, qRT-PCR, RACE and siRNA sequences for RNAi. (DOC)