Abstract
Elevated expression of NF-E2-related factor 2 (Nrf2), a nuclear transcription factor, is a frequent genetic abnormality seen in this malignancy and is an important contributor to chemoresistance in cancer therapy. In the present study, we investigated if Nrf2 was associated with drug resistance in tamoxifen-resistant MCF-7 (MCF-7/TAM) cells, and whether EGCG, major flavonoid isolated from green tea, could reverse drug resistance in MCF-7/TAM cells. Our results showed that the endogenous expression of Nrf2 as well as its target proteins heme oxygenase-1, NADP (H):quinone oxidoreductase in MCF-7/TAM cells was higher than that in MCF-7 cells. Epicatechin gallate (EGCG) significantly sensitizes MCF-7/TAM cells to tamoxifen and dramatically reduced Nrf2 expression at both the messenger RNA and protein, leading to a reduction of Nrf2-downstream genes. In addition, using siRNA technique, we found that the intracellular Nrf2 protein level was significantly decreased in MCF-7/TAM cells and tamoxifen resistance was partially reversed by Nrf2 siRNA. Combination of siRNA-directed gene silencing with EGCG downregulated the Nrf2-dependent response and partly reversed tamoxifen resistance in MCF-7/TAM cells in a synergic manner. These results suggested that combining the chemotherapeutic effect of EGCG with siRNA-mediated Nrf2 knock-down results in the feasibility of using Nrf2 inhibitors to increase efficacy of chemotherapeutic drugs.
Keywords: Apoptosis, Epigallocatechin-3-gallate, MCF-7 breast cancer, Nrf2 siRNA, Tamoxifen
Introduction
Breast cancer is the most common invasive malignancy among women in the USA and the leading cause of death in women worldwide [1]. The treatment of breast cancers resistant to current standard therapies poses significant clinical challenges. Cancers intrinsically possess or develop mechanisms to evade the death-inducing effects of cytotoxic agents, as well as targeted therapies [2]. The ability to reduce breast tumor growth through the administration of anti-estrogen agents has played a key role in the endocrine therapy of breast cancer. A non-steroidal antiestrogen, tamoxifen (TAM), is the most widely used in estrogen receptor-positive breast cancer patients [3]. Although most patients are initially responsive, resistance to TAM is a critical problem for anti-estrogen therapy [4]. Therefore, it is urgently needed to develop new adjuvants that enhance the efficacy of TAM-based chemotherapy and circumvent chemoresistance.
Erythroid transcription factor NF-E2 (Nrf2) plays a critical role against oxidative and electrophilic stress [5]. Under normal conditions, majority of Nrf2 molecules are found to be in the cytoplasm. When the stable environment inside the cell is destroyed, Nrf2 translocates into the nucleus [6, 7], forms a heterodimer with small Maf protein [8], which then binds to Antioxidant Response Element (ARE), and enhances transcription of its target genes including NAD(P)H: quinone oxidoreductase (NQO1) and antioxidant proteins, such as hemeoxygenase-1 (HO-1) [9]. Many investigations have shown that the Nrf2-Keap1 pathway protects cells from many diseases including neurodegenerative disease and cancers [10, 11]. Recently, several studies indicated that activation of Nrf2 is associated with tumorigenesis and resistance of chemotherapeutics in lung cancer, colon cancer, prostate cancer, and type II endometrial cancer [12–15]. Elevated expression of Nrf2 and its downstream genes contributed to MCF-7-derived tamoxifen resistance cell line, and using Nrf2 siRNA to block the expression of Nrf2 can reverse resistance of the cell lines to tamoxifen [16].
Flavonoids are a group of natural compounds with variable phenolic structures and are found in plants, showing strong anti-proliferative activity against many types of cancers and sensitized cancer cells to anticancer agents [17, 18]. Green tea is one of the most popular and widely consumed beverages. Many studies suggest a protective role of green tea consumption against development of various types of cancers [19]. Thus, tea drinking has been associated with a reduced risk of stomach [20], pancreatic and colorectal cancers [21], and also with decreased recurrence of stage I and II breast cancer [22].
Green tea is rich in a variety of catechin polyphenols such as (−)-epicatechin, (−)-epicatechin-3-gallate, (−)-epigallocatechin, and (−)-epigallocatechin-3-gallate (EGCG) [23]. Among these components, EGCG is the most abundant polyphenol with potent antioxidant and chemopreventive activities. EGCG has been shown to be protective against experimentally induced lung, forestomach, colon, breast, prostate, and skin cancer [24–27].
Recently, it was shown that epicatechin gallate (EGCG) inhibited HO-1 expression through blocking Nrf2 in non-small cell lung cancer A549 cells [28]. Based on these data, we are interested to answer this question whether combining the chemotherapeutic effects EGCG with siRNA-mediated Nrf2 knock-down can sensitize breast cancer cells to tamoxifen or reverse drug resistance by antagonizing the Nrf2/ARE signaling pathway.
Materials and methods
Reagents
EGCG was obtained from Sigma–Aldrich (St. Louis, MO) and dissolved in DMSO (the DMSO concentration in all drug-treated cell culture media was 0.1 %) and were used in all experiments unless otherwise indicated. Tamoxifen and 3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide (MTT) were obtained from Sigma–Aldrich (St. Louis, MO). Primary antibodies for NQO1, HO-1, β-actin, α-tubulin, Nrf2 siRNA, and control siRNA were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). All other reagents used in this study were of analytical grade.
Cell culture
The human breast adenocarcinoma cells (MCF-7) were purchased from the National Cell Bank of Iran (NCBI), Pasteur Institute of Iran (Tehran, Iran), and maintained in DMEM medium supplemented with 10 % fetal bovine serum (FBS) and 100 U/ml penicillin and 100 μg/ml streptomycin. The cells were grown as monolayers at 37 °C in humidified atmosphere with 5 % CO2 and 95 % air. Tamoxifen-resistant MCF-7 cells (MCF-7/TAM) were established using the methodology reported elsewhere [29]. Briefly, MCF-7 cells were washed with PBS, and the culture medium was changed to phenol-red-free DMEM containing 10 % charcoal-stripped, steroid-depleted fetal bovine serum (Hyclone, Logan, UT) and tamoxifen (0.1 μM). The cells were continuously exposed to this treatment regimen for 2 weeks and the concentration of tamoxifen was gradually increased to 3 μM over a 9-month period. Initially, the cell growth rates were reduced. However, after exposure to the medium for 9 months, the rate of cell growth gradually increased, showing the establishment of a tamoxifen-resistant cell line [30].
Cell viability assay
Cell viability was determined using the MTT assay. Briefly, 2.5 × 104 cells in logarithmic phase were seeded in 96-well plates at 37 °C with 5 % CO2 for overnight incubation and treated with appropriate concentrations of test samples for the indicated times. The cells were then incubated with a serum-free medium containing MTT at a final concentration of 0.5 mg/ml for 4 h. The dark blue formazan crystals formed in intact cells were solubilized in dimethyl sulfoxide, and the absorbance was measured at 570 nm. Results were expressed as the percentages of reduced MTT, assuming the absorbance of control cells as 100 %.
Determination of drug resistance
To determine the drug resistance of MCF-7/TAM cells to tamoxifen, MCF-7 and MCF-7/TAM cells were plated into 96-well plates at approximately 8000 cells/well in 100 μl medium, then treated with various concentrations of tamoxifen for 24 h. Cell viability was assessed with MTT assay and cell survival ratio was calculated using Atreated/Acontrol × 100 %, where Atreated and Acontrol were the absorbance from treated and control cells after 24 h of incubation, respectively. The IC50 was taken as the concentration that caused 50 % inhibition of cell proliferation, and the degree of resistance was estimated by resistance index (RI), which was calculated by IC50 of MCF-7/TAM cells/IC50 of MCF-7 cells [16].
Nrf2 siRNA transfection
The transfection of siRNA was performed using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Briefly, a total of 20 × 104 cells were seeded into six-well plates and transfected the next day with a 100 nM final concentration of siRNA, using 5 μl Lipofectamine 2000. Cells were harvested 48 h after transfection for western blot analysis. To measure the effect of siRNA and EGCG treatment together, the cells were treated with EGCG for another 24 h before determining cell viability and apoptosis.
Quantitative real-time PCR analysis (qRT-PCR)
Total RNA was extracted from cultured cells using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol and quantified with Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA). First-strand cDNA synthesis and amplification were performed using reverse transcription reagents (SABiosciences) following the manufacturer’s instructions. The quantitative PCR reactions included 7.6 μl cDNA and 12.4 μl of SYBR Green Master Mix (SABiosciences) with a pair of primers. The reactions were monitored on a 7500 Real-Time PCR System with 7500 software, version 2.0.5 (Applied Biosystems, Foster City, CA). The primers were synthesized by Integrated DNA Technologies (Iowa, USA) and are listed in Table 1. The real-time PCR condition was as follows: one cycle of initial denaturation (95 °C for 10 min), 40 cycles of amplification (95 °C for 15 s and 60 °C for 60 s), and a cooling program (50 °C for 5 s). The data were expressed as relative mRNA levels and were normalized to GAPDH.
Table 1.
Real-time PCR primer sequences
| Genes | Primers |
|---|---|
| Nrf2 | Forward (ACACGGTCCACAGCTCATC) Reverse (TGTCAATCAAATCCATGTCCTG) |
| HO-1 | Forward (AACTTTCAGAAGGGCCAGGT) Reverse (CTGGGCTCTCCTTGTTGC) |
| NQO-1 | Forward (ATGTATGACAAAGGACCCTTCC) Reverse (TCCCTTGCAGAGAGTACATGG) |
| GAPDH | Forward (AGGGCTGCTTTTAACTCTGGT) Reverse (CCCCACTTGATTTGGAGGGA) |
Preparation of nuclear extracts
Cells in the dishes were washed with ice-cold PBS. The cells were then scraped, transferred to microtubes, and allowed to swell after adding 100 μl of a hypotonic buffer containing 10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.5 % Nonidet P-40, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonylfluoride. The lysates were incubated for 10 min on ice and centrifuged at 7200×g for 5 min at 4 °C. Pellets containing the crude nuclei were resuspended in 50 μl of an extraction buffer containing 20 mM HEPES (pH 7.9), 400 mM NaCl, 1 mM EDTA, 10 mM dithiothreitol, and 1 mM phenylmethylsulfonylfluoride and incubated for 30 min on ice. The samples were centrifuged at 15,800×g for 10 min to obtain supernatants containing the nuclear fractions. The nuclear fractions were stored at −80 °C until needed.
Lactate dehydrogenase (LDH) release assay
LDH activity was assayed spectrophotometrically following the decrease in absorbance of NADH at 340 nm by LDH assay kit (Pars azmoon, Tehran, Iran). The percentage of LDH released from the cells to the culture medium was calculated according to following formula: %LDH released = (LDH in culture medium/LDH in culture medium + LDH in cell lysates) × 100.
Oxidative stress parameters assay
Glutathione peroxidase (GSH-Px) activity was measured spectrophotometrically by a Randox laboratory kit (Randox, Lab. Ltd. Ireland) according to the method described by Pagila and Valentin [31]. SOD activity was measured based on inhibition of the formation of amino blue tetrazolium formazan in nicotineamide adenine dinucleotide, phenazine methosulfate, and nitroblue tetrazolium (NADH-PMS-NBT) system, according to the method of Kakkar et al. [32]. The mean MDA levels (nmol/mg protein) were measured by the double heating method of Draper and Hadley [33].
Western blot assay
MCF-7 and MCF-7/TAM cells with different treatments were washed twice with PBS then collected for protein extract preparation. Briefly, the cells were lysed with RIPA buffer then nuclear and cytoplasmic extracts were separated using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific, Waltham, MA, USA). Equal amounts of lysate (based on the protein contents) were then separated using SDS–PAGE, blotted onto polyvinylidene di-fluoride membranes, reacted with specific primary antibodies, and then visualized with appropriate conjugated secondary antibodies. The immunoreactive bands were detected using the ECL method and visualized on Kodak Bio-MAX MR film. All blots were stripped and probed with polyclonal anti-β-actin antibody to ascertain equal loading of the proteins.
Cell colony formation assay
The inhibition of the colony formation of MCF-7/TAM cells following treatment with EGCG and Nrf2siRNA was measured by soft agar assay. Briefly, transfected cells (8 × 103 cells/ml) were treated with (100 μM) EGCG in 0.3 % Basal Medium Eagle (BME) agar containing 10 % FBS, 2 mM l-glutamine, and 25 μg/ml gentamicin. The cultures were maintained at 37 °C with 5 % CO2 atmosphere for 10 days. Cell colonies were scored using a conventional microscope.
Flow cytometry analysis
The extent of apoptosis was measured through annexin V-FITC/PI apoptosis detection kit (Invitrogen, CA, USA) as described by the manufacturer’s instruction. After treatment, the cells were collected, washed twice with PBS, gently re-suspended in annexin V binding buffer and incubated with annexin V-FITC/PI in the dark for 10 min, and analyzed by flow cytometry (Partec PAS, Germany).
Statistical analysis
All results shown represent means ± SD from triplicate experiments performed in a parallel manner unless otherwise indicated. Statistical analysis was performed using one-way ANOVA. See details of each statistical analysis used in figures and figure legends.
Results
Establishment of a tamoxifen-resistant breast cancer cell line
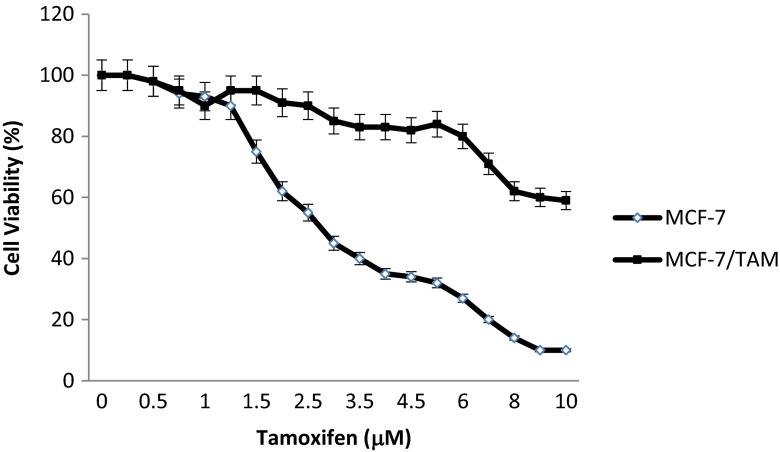
In this study, the acquisition of tamoxifen resistance in MCF-7/TAM cells was confirmed through MTT assay. The cell viability assay revealed that the percentage of surviving cells decreased significantly in a dose-dependent manner. Tamoxifen (10 μM) treatment in control MCF-7 cells significantly inhibited cell proliferation but not in MCF-7/TAM cells (more than 80 % of live cells) (Fig. 1a). The 50 % inhibition concentration (IC50) for tamoxifen in the MCF-7 and MCF-7/TAM cells was 3.0 ± 0.2 vs. 10.2 ± 0.9 μM, respectively (p < 0.05). To verify the drug resistance phenotype of MCF-7/TAM cells, the resistance index (RI) was calculated by IC50 of MCF-7/TAM cells/IC50 of MCF-7 cells. The RI was 3.4, indicating that MCF-7/TAM cell line is tamoxifen resistant. Our results manifested that MCF-7/TAM cells could be relatively resistant to tamoxifen treatment and suitable for our further studies.
Fig. 1.
The inhibitory effect of tamoxifen on MCF-7 and MCF-7/TAM cell proliferation. Cells were treated with various concentrations of tamoxifen for 24 h, and then cell viability was determined by the MTT assay. Data were expressed as means ± SEM of three independent experiments
Nrf2 pathway is activated in resistant breast cancer cells
Nrf2 is an essential transcription factor that binds to ARE sequences and regulates an aforementioned anti-oxidant protein expression [34, 35]. Upregulation of the Nrf2 signaling is thought to account for the drug resistance phenotype. Thus, we measured nuclear (activated) levels of Nrf2 in both MCF-7 and MCF-7/TAM cells. Our results showed that in MCF-7/TAM cells, the expression of total (A) and nuclear Nrf2 protein (B) and its downstream genes HO-1 and NQO1 was distinctly higher than that in MCF-7 cells (Fig 2a, b).
Fig. 2.
The expression of total (a) and nuclear (b) protein Nrf2 and its downstream gene HO-1 and NQO1 was assessed by Western blot in MCF-7 and MCF-7/TAM cells. Results represent mean values of three experiments ± SEM
Epicatechin gallate reduced the resistance of MCF-7/TAM cells to tamoxifen
To determine whether EGCG can reverse the resistance of MCF-7/TAM cells to tamoxifen, MTT assay was carried out and the RF values was then calculated. The RF values, indicating the potency of reversal, were obtained from fitting the IC50 of cytotoxic drug alone/IC50 of cytotoxic drug in the presence of the test drugs [32]. RF >1 indicates enhanced drug sensitivity in the presence of modulator, RF = 1 indicates no effect, and RF <1 indicates decreased drug sensitivity; the greater the RF magnitude, the more significant the reversal effect. As shown in Table 2, EGCG enhanced the sensitivity of MCF-7/TAM cells to tamoxifen by 1.24-, 1.72-, and 2.12-fold at 10, 50, and 100 μM of EGCG, respectively.
Table 2.
Effect of EGCG on tamoxifen cytotoxicity in MCF-7/TAM cells
| Group | IC50 of tamoxifen (μM) | RF |
|---|---|---|
| Control | 10.2 ± 0.9 | 1.00 |
| EGCG (10 μM) | 8.1 ± 0.5 | 1.24 |
| EGCG (50 μM) | 5.8 ± 0.4 | 1.72 |
| EGCG (100 μM) | 4.7 ± 0.6 | 2.12 |
Cells were treated with various concentrations of tamoxifen in the presence of 10, 50, and 100 μM EGCG for 24 h. IC50 values for tamoxifen were calculated and the reversal folds (RF) were evaluated. Data were expressed as means ± SDs of three independent experiments
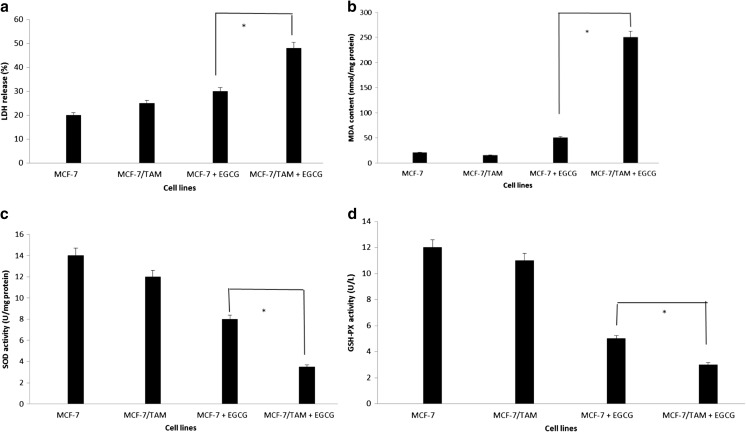
EGCG increased LDH release and MDA contents, downregulated SOD and GSH-PX activities in MCF-7/TAM cells
To investigate the effect of EGCG on the plasma membrane instability, we measured the LDH release assay. Our results showed that the LDH release of MCF-7/TAM cells (48.25 ± 2.14 %) was higher than that of MCF-7 cells (30.35 ± 3.12 %) after treatment with EGCG for 24 h (p < 0.05) (Fig. 3a). It demonstrated that EGCG could greatly impact the plasma membrane structure of MCF-7/TAM cells. To better investigate the change of oxidative stress caused by EGCG, we detected MDA contents and SOD and GSH-PX activities both in MCF-7 cells and MCF-7/TAM cells. Compared to control MCF-7 cells, the MDA contents in MCF-7/TAM cells increased significantly (p < 0.05) (Fig. 3b), while SOD and GSH-PX activities were decreased (p < 0.05) (Fig. 3c, d).
Fig. 3.
The effect of EGCG on release of LDH (a), MDA contents (b), SOD (c), and GSH-PX (d) activities in tamoxifen-resistant MCF-7 cells. Results represent mean values of three experiments ± SEM
EGCG inhibited the Nrf2 signaling pathway
Real-time quantitative PCR analysis revealed that EGCG suppressed the mRNA expression of HO-1 and NQO1 in a dose-dependent manner. EGCG at a concentration of 50 and 100 μM reduced the mRNA levels of these genes. Interestingly, Nrf2 mRNA was downregulated even more than its target genes. In MCF-7/TAM cells, Nrf2 mRNA level dropped more than 53 % in the presence of 50 μM EGCG (Fig. 4a). In contrast, Keap1 mRNA level was affected less than other ones, suggesting that EGCG-mediated inhibition is independent of Keap1. In addition, EGCG reduced protein levels of nuclear Nrf2 as well as Nrf2-target genes, including HO-1 and NQO1 in a dose-dependent manner, consistent with their mRNA expressions (Fig. 4b). Taken together, these results suggest that EGCG inhibits transcription of the ARE-driven genes in MCF-7/TAM cells.
Fig. 4.

Effects of EGCG on Nrf2 expression in MCF-7/TAM cells. MCF-7 and MCF-7/TAM cells were treated with indicated concentrations of EGCG for 24 h, and the change of mRNA (a) and protein expression (b) levels of total Nrf2 and its downstream genes NQO1 and HO-1 in MCF-7 and MCF-7/TAM cells were detected by qRT-PCR and immunoblot analysis, respectively. Results represent mean values of three experiments ± SEM
Genetic knockdown of Nrf2 sensitized MCF-7/TAM cells to tamoxifen
To assess the effects of Nrf2 on cancer cell proliferation and anticancer drug resistance, the expression of Nrf2 was knocked down using Nrf2 siRNA to reduce the endogenous Nrf2 expression in MCF-7/TAM cells. As shown in Fig. 5a, the expression of total and nuclear protein of Nrf2 was decreased significantly, accompanied with the downregulation of intracellular HO-1 and NQO1 in contrast to control siRNA group. Following 48-h siRNA transfection, cells were treated with tamoxifen (10 μM) for 24 h, and cell viability was assessed by MTT assay. Knockdown of Nrf2 with siRNA potently inhibited the proliferation of MCF-7/TAM cells (Fig. 5b). Co-treatment (Nrf2 siRNA and tamoxifen) enhanced tamoxifen-induced cell death (40.21 ± 1.24 % vs. 25.14 ± 0.48 %, p < 0.05). Furthermore, Annexin V/PI staining revealed that the apoptotic ratios (early and late stages) of Nrf2 siRNA group and co-treatment group were 45.12 ± 2.37 % vs. 64.22 ± 3.52 %, respectively (p < 0.01) (Fig. 5c). Our data showed that modulation of the Nrf2-mediated cellular defense response is an effective means to manipulate the sensitivity of MCF-7/TAM cells to tamoxifen.
Fig. 5.

Transient knockdown of Nrf2 by siRNA sensitizes MCF-7/TAM cells to tamoxifen. a The Nrf2, NQO1, and HO-1 protein levels as well as the nuclear Nrf2 levels were compared in MCF-7/TAM cells transfected with Nrf2 siRNA or control siRNA. b MCF-7/TAM cells, transfected with Nrf2 siRNA or control siRNA for 48 h, were treated with the indicated doses of tamoxifen for 24 h, and cell viability was determined by the MTT assay. c Apoptosis analysis by flow cytometry of MCF-7/TAM cells, transfected with Nrf2 siRNA and treated with the indicated doses of tamoxifen. The data are presented as means ± SEM
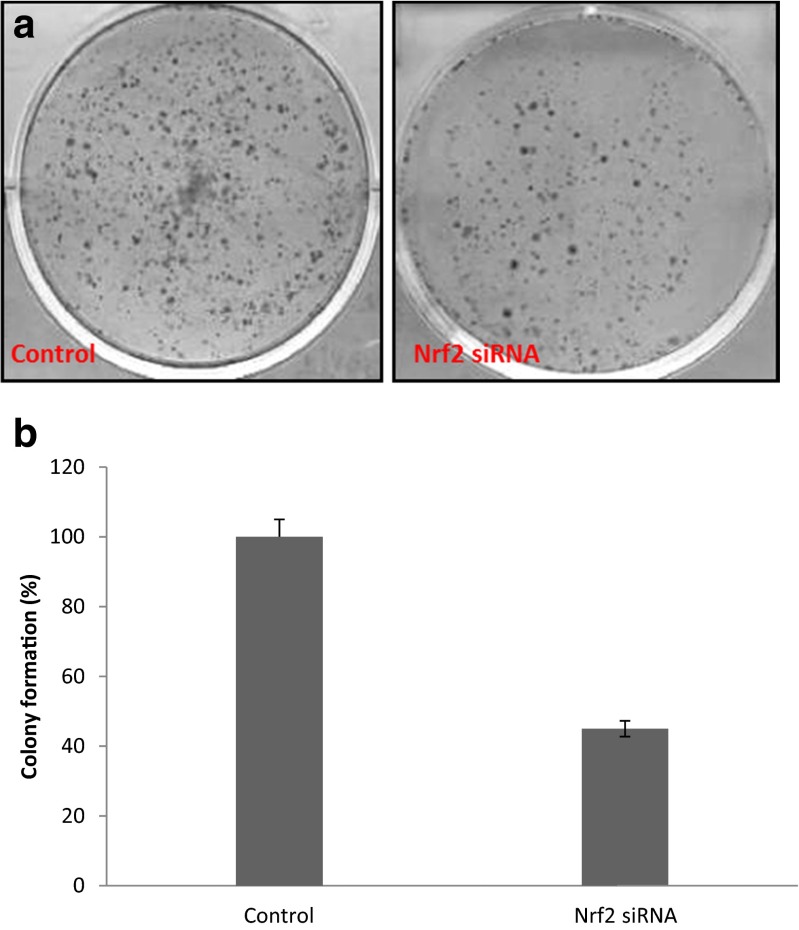
Decrease in Nrf2 protein level inhibits colony formation
We then tested whether RNAi-mediated reductions in Nrf2 levels could influence the ability of MCF-7/TAM cells to form colonies in soft agar. Transfected MCF-7/TAM cells with Nrf2siRNA were placed into medium with soft agar, and colonies were counted after 2 weeks. RNAi directed against Nrf2 resulted in a significant decrease (about 55 %) in colony formation in MCF-7/TAM cells (Fig. 6) (p < 0.001). These results showed that the reduction in Nrf2 protein level decreased the ability of MCF-7/TAM cells to form colonies in soft agar.
Fig. 6.
Knockdown of Nrf2 by RNAi reduces colony formation in soft agar. MCF-7/TAM cells were transfected with Nrf2 siRNA and seeded in 0.3 % agarose containing Dulbecco’s modified Eagle’s medium with 10 % fetal bovine serum. The colony numbers were counted 10 days later. The numbers of colonies of treated cells were standardized against the control cells (set at 100 %). The data shown are means and SD from two independent triplicate experiments. The difference between treatments is statistically significant (p < 0.001)
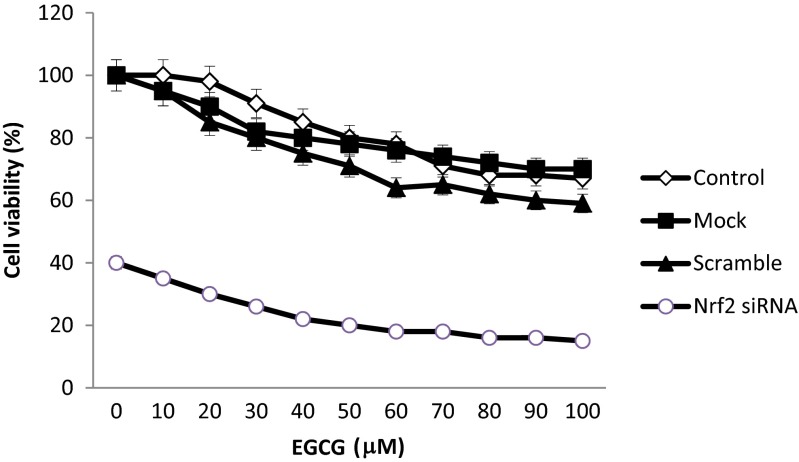
Nrf2 siRNA and EGCG increase cytotoxicity in a synergic manner
The MTT values for MCF-7/TAM cells after transfection with Nrf2siRNA, in the presence or absence of 50 and 100 μM EGCG, are presented in Fig. 7. The results showed that Nrf2siRNA and EGCG induce a decrease in number of live cells by 20 and 15 %, respectively, after 24 h of treatment, without any noticeable cytotoxicity associated with delivery system. Next, to determine whether Nrf2 siRNA and EGCG can reverse the resistance of MCF-7/TAM cells to tamoxifen, the RF values was calculated. Based on data obtained from Table 3, co-treatment of MCF-7/TAM with Nrf2 siRNA and EGCG with RF >1 enhanced the sensitivity of tamoxifen-resistant breast cancer cells to tamoxifen by 2.61- and 3.3-fold at 50 and 100 μM EGCG, respectively.
Fig. 7.
The inhibitory effect of EGCG on Nrf2 siRNA transfected MCF-7/TAM cell proliferation. Cells were treated with various concentrations of EGCG 24 h, and cell viability was determined by the MTT assay
Table 3.
Effect of co-treatment of Nrf2 siRNA and EGCG on tamoxifen cytotoxicity in MCF-7/TAM cells
| Group | IC50 of tamoxifen (μM) | RF |
|---|---|---|
| Control | 10.2 ± 0.9 | 1.00 |
| Nrf2 siRNA | 4.1 ± 0.5 | 2.50 |
| Nrf2 siRNA + EGCG (50 μM) | 3.9 ± 0.2 | 2.61 |
| Nrf2 siRNA + EGCG (100 μM) | 3.1 ± 0.4 | 3.30 |
Transfected cells were treated with 50 and 100 μM EGCG for 24 h. IC50 values for tamoxifen were calculated and the reversal folds (RF) were evaluated. Data were expressed as means ± SDs of three independent experiments
EGCG and Nrf2 siRNA can induce apoptosis of tamoxifen-resistant breast cancer cells
To further determine if EGCG and/or Nrf2 siRNA treatment also induce cellular apoptosis, percentages of apoptotic cells in MCF-7/TAM cells before and after treatment were analyzed by flow cytometry following staining with Annexin V-FITC and PI. Apoptotic cells were significantly increased in both the late phase (upper right panel) and the early phase (lower right panel). Co-treatment of MCF-7/TAM cells with Nrf2 siRNA and EGCG resulted in a marked induction of apoptosis at both the early and late stages of apoptosis (Fig. 8a). We further compared the protein expressing level of caspase-3, caspase-9, caspase-8, Bcl2, and Bax in MCF-7/TAM cells co-treated with EGCG and Nrf2 siRNA. As expected, co-treatment of tamoxifen-resistant MCF-7 cells with EGCG and Nrf2 siRNA increased the protein expressions of caspase-3, caspase-9, caspase-8, and Bax, but decreased Bcl2 protein expression in MCF-7/TAM compared to control (Fig. 8b). These observations suggest that Nrf2 siRNA induces cell apoptosis in MCF7/TAM cells, and EGCG can significantly potentiate the apoptosis induction.
Fig. 8.
a Apoptosis analysis by flow cytometry of MCF-7/TAM cells, transfected with Nrf2 siRNA and co-treated with the indicated doses of EGCG. b Western blots analysis of apoptotic factors in MCF-7/TAM transfected with Nrf2 siRNA and co-treated with EGCG. The data are presented as means ± SEM
Discussion
Breast cancer is the leading cause of cancer death in women worldwide. Despite advances in detection and chemotherapy, many women with breast cancer continue to die of this malignancy [36]. Therefore, an understanding of the molecular mechanisms involved in breast cancer formation and progression should be helpful in developing more effective treatments for breast cancer. Drug resistance during chemotherapy is the major obstacle to the successful treatment of many cancers. Tamoxifen remains a commonly prescribed drug for the treatment and prevention of ER-positive breast cancer, as the drug increases survival and helps maintain disease-free status [37]. Tamoxifen works principally through ER antagonism, but also involves ER-independent pathways. Tamoxifen induces apoptosis by increasing oxidative stress, which may mediate its anti-tumor effect [38, 39].
RNA interference (RNAi) is a remarkable endogenous regulatory pathway that can bring about sequence-specific gene silencing. If harnessed effectively, RNAi could result in a potent targeted therapeutic modality with applications ranging from viral diseases to cancer [40]. The discovery of siRNAs—constructs that can be designed to specifically and efficiently silence genes of interest—has stirred considerable excitement. Most exciting is the potential therapeutic application of this technology. Though a number of challenges stand in the way of realizing this potential, the biggest bottleneck in siRNA delivery, over a decade of innovative engineering, has resulted in solutions to a number of these challenges, laying down a foundation for continuing headway toward making widespread therapeutic siRNA a reality hopefully in the not too distant future [41, 42]. Currently, several potential siRNA candidates are undergoing clinical trials for treatment of macular degeneration, respiratory diseases, and cancers. It has also been observed that blocking the expression of the Nrf2 gene or anti-apoptotic genes, such as Bcl2, survivin, XIAP, and RhoGDI, increases the susceptibility of tumor cells to chemotherapy-induced apoptosis [43].
Much evidence confirms the positive role of Nrf2 in resistance of cancer cell to chemotherapy agents [44, 45]. Here, we found a notably high level of Nrf2 and its target genes such as HO-1 and NQO1 in tamoxifen-resistant human breast carcinoma (MCF-7) cells compared to control MCF-7 cells (Fig. 2). These findings confirmed that the increase of Nrf2 was one of the reasons that led to resistance to tamoxifen in MCF-7 cells. Transfection of MCF-7/TAM with Nrf2 siRNA attenuated the Nrf2-mediated signaling, resulting in the increased sensitivity of MCF-7 cells to tamoxifen. Based on these observations, our results explained that activation of Nrf2-mediated signaling accounts for, at least in part, tamoxifen resistance in breast carcinoma cells.
Flavonoids are polyphenolic compounds that occur ubiquitously in food plants and vegetables. Flavonoids are generally safe and are associated with low toxicity, making them ideal candidates for cancer chemopreventive agents. Several flavonoid compounds have been reported to be potent Nrf2 inhibitors, such as epigallocatechin 3-gallate, luteolin, and brusatol [28, 46, 47]. The chemopreventive effects of green tea have been attributed to polyphenolic ingredients that have potent antioxidant properties. Among many polyphenolic compounds isolated from green tea, (−)-epigallocatechin gallate (EGCG) is recognized as a key active constituent in terms of cancer chemopreventive potential. It is reported that EGCG can significantly inhibit the growth of acute myeloblastic leukemia cells and induce apoptosis in human cancer cells [48, 49]. In one study done by Farabegoli and colleagues, they showed that EGCG could compete with 17-beta-estradiol for binding to estrogen receptor alpha (ERalpha), and functionally active ERalpha may have a role in EGCG cytotoxicity, increasing the sensitivity to the drug [50]. EGCG significantly enhanced the growth inhibition and apoptosis in both doxorubicin (DOX)-sensitive and -resistant MCF-7 cells induced by curcumin. The mechanism may be related to the further activation of caspase-dependent apoptotic signaling pathways and the enhanced cellular incorporation of curcumin by inhibiting P-glycoprotein (P-gp) pump function. Moreover, curcumin and EGCG in combination could enhance the toxicity of DOX and increase the intracellular level of DOX in resistant MCF-7 cells [51]. Current studies showed that EGCG downregulates Pg-P and BCRP in a tamoxifen-resistant MCF-7 cell line. These three plasma membrane proteins—Multidrug Resistance Protein (MRP1), P-glycoprotein (P-gp), and Breast Cancer Resistance Protein (BCRP)—are involved in the mechanism of drug resistance in MCF-7 cells [52]. These prompted us to determine whether EGCG in combination with Nrf2 siRNA exerts a synergic pro-apoptotic effect that are greater than those of each agent taken individually and can sensitize cancer cells to anticancer drugs through inhibiting Nrf2 signaling pathway. Based on these, the objective of this study, in part, was to use natural phytochemicals to enhance the efficacy of siRNA therapy. Our results showed that the EGCG in combination with Nrf2 iRNA has synergic apoptotic effects, which are greater than each compound taken individually. EGCG suppressed nuclear Nrf2 expression as well as the expression of its downstream antioxidant enzymes in MCF-7/TAM cells. This study addresses an important issue of enhancing the efficacy of chemotherapy by targeting Nrf2 signaling, based on the findings that overexpression of Nrf2 in cancer cells provides growth advantages and results in drug resistance. On the other hand, it seems worthy to mention that not all Nrf2 activators would be bad for cancer, depending on the stages of cancer, as well as whether the Nrf2 activators will also inhibit other signaling pathways, such as the NF-kB pathway, and induce cell cycle arrest and apoptosis. In conclusion, the present study demonstrates that MCF-7/TAM cells acquiring resistance to chemotherapeutic agents due to Nrf2 overexpression become susceptible by the drug treatment combined with EGCG. Because of the complexity of cancer, combination therapy is becoming increasingly important to overcome multidrug resistance in cancer and to enhance apoptosis. The present study provides evidence that Nrf2 siRNA has synergic effects when combined with EGCG in the induction of apoptosis and reducing drug resistance, as shown by MTT and PCR-array data. The advantage of combining EGCG with Nrf2-targeted RNAi therapy is that it leads to the activation of alternative apoptosis-inducing pathways and suggests promising new therapies for cancer inhibition. Further studies in animal models as well as in human clinical trials are necessary to test the efficacy of co-treatment of EGCG as well as other Nrf2 inhibitors during chemotherapy.
Acknowledgment
This work was financially supported by Iran National Science Foundation (INSF, grant number 92031220).
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Sharma GN, Dave R, Sanadya J, Sharma P, Sharma KK. Various types and management of breast cancer: an overview. J Adv Pharm Technol Res. 2010;1:109–126. [PMC free article] [PubMed] [Google Scholar]
- 2.Park SH, Ito K, Olcott W, Katsyv I, Halstead-Nussloch G, Irie HY. PTK6 inhibition promotes apoptosis of Lapatinib-resistant Her2+ breast cancer cells by inducing Bim. Breast Cancer Res. 2015;17:86. doi: 10.1186/s13058-015-0594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose C, Thorpe SM, Andersen KW, Pedersen BV, Mouridsen HT, Blichert-Toft M, Rasmussen BB. Beneficial effect of adjuvant tamoxifen therapy in primary breast cancer patients with high oestrogen receptor values. Lancet. 1985;1:16–19. doi: 10.1016/S0140-6736(85)90966-3. [DOI] [PubMed] [Google Scholar]
- 4.Clemons M, Danson S, Howell A. Tamoxifen (‘Nolvadex’): a review. Cancer Treat Rev. 2002;28:165–180. doi: 10.1016/S0305-7372(02)00036-1. [DOI] [PubMed] [Google Scholar]
- 5.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osburn WO, Wakabayashi N, Misra V, et al. Nrf2 regulates an adaptive response protecting against oxidative damage following diquat-mediated formation of superoxide anion. Arch Biochem Biophys. 2006;454:7–15. doi: 10.1016/j.abb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh K, Tong KI, Yamamoto M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med. 2004;36:1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 8.Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 10.Lee JM, Johnson JA. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J Biochem Mol Biol. 2004;37:139–143. doi: 10.5483/BMBRep.2004.37.2.139. [DOI] [PubMed] [Google Scholar]
- 11.Baird L, Dinkova-Kostova AT. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol. 2011;85:241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 12.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:79757984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P, Singh A, Yegnasubramanian S, Esopi D, et al. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, Zhang DD. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 2010;70:5486–5496. doi: 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akhdar H, Loyer P, Rauch C, Corlu A, Guillouzo A, Morel F. Involvement of Nrf2 activation in resistance to 5-fluorouracil in human colon cancer HT-29 cells. Eur J Cancer. 2009;45:2219–2227. doi: 10.1016/j.ejca.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Kim SK, Yang JW, Kim MR, et al. Increased expression of Nrf2/ARE-dependent anti-oxidant proteins in tamoxifen-resistant breast cancer cells. Free Radic Biol Med. 2008;45:537–546. doi: 10.1016/j.freeradbiomed.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Boumendjel A, Di Pietro A, Dumontet C, Barron D. Recent advances in the discovery of flavonoids and analogs with high-affinity binding to P-glycoprotein responsible for cancer cell multidrug resistance. Med Res Rev. 2002;22:512–529. doi: 10.1002/med.10015. [DOI] [PubMed] [Google Scholar]
- 18.Kandaswami C, Lee LT, Lee PP, et al. The antitumor activities of flavonoids. In Vivo. 2005;19:895–909. [PubMed] [Google Scholar]
- 19.Yang CS, Prabhu S, Land J. Prevention of carcinogenesis by tea polyphenols. Drug Metab Rev. 2001;33:237–253. doi: 10.1081/DMR-120000651. [DOI] [PubMed] [Google Scholar]
- 20.Inoue M, Tajima K, Hirose K, Hamajima N, Takezaki T, Kuroishi T, Tominaga S. Tea and coffee consumption and the risk of digestive tract cancers: data from a comparative case-referent study in Japan. Cancer Causes Control. 1998;9:209–216. doi: 10.1023/A:1008890529261. [DOI] [PubMed] [Google Scholar]
- 21.Ji BT, Chow WH, Hsing AW, McLaughlin JK, Dai Q, Gao YT, Blot WJ, Fraumeni JF., Jr Reduced risk of esophageal cancer associated with green tea consumption. Int J Cancer. 1997;70:255–258. doi: 10.1002/(SICI)1097-0215(19970127)70:3<255::AID-IJC1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 22.Nakachi K, Suemasu K, Suga K, Takeo T, Imai K, Higashi Y. Influence of drinking green tea on breast cancer malignancy among Japanese patients. Jpn J Cancer Res. 1998;89:254–261. doi: 10.1111/j.1349-7006.1998.tb00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhtar H, Ahmad N. Mechanism of cancer chemopreventive activity of green tea. Proc Soc Exp Biol Med. 1999;220:234–238. doi: 10.3181/00379727-220-44372. [DOI] [PubMed] [Google Scholar]
- 24.Katiyar SK, Agarwal R, Zaim MT, Mukhtar H. Protection against N-nitrosodiethylamine and benzo[a]pyrene-induced forestomach and lung tumorigenesis in A/J mice by green tea. Carcinogenesis. 1993;14:849–855. doi: 10.1093/carcin/14.5.849. [DOI] [PubMed] [Google Scholar]
- 25.Yamane T, Hagiwara N, Tateishi M, Akachi S, Kim M, Okuzumi J, Kitao Y, Inagake M, Kuwata K, Takahashi T. Inhibition of azoxymethane-induced colon carcinogenesis in rat by green tea polyphenol fraction. Jpn J Cancer Res. 1991;82:1336–1339. doi: 10.1111/j.1349-7006.1991.tb01801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuart EC, Scandlyn MJ, Rosengren RJ. Role of epigallocatechin gallate (EGCG) in the treatment of breast and prostate cancer. Life Sci. 2006;79:232923–232936. doi: 10.1016/j.lfs.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 27.Katiyar SK, Mukhtar H. Inhibition of phorbol ester tumor promoter 12-O-tetradecanoylphorbol-13-acetate-caused inflammatory responses in SENCAR mouse skin by black tea polyphenols. Carcinogenesis. 1997;18:1911–1916. doi: 10.1093/carcin/18.10.1911. [DOI] [PubMed] [Google Scholar]
- 28.Kweon MH, Adhami VM, Lee JS, Mukhtar H. Constitutive overexpression of Nrf2-dependent heme oxygenase-1 in A549 cells contributes to resistance to apoptosis induced by epigallocatechin 3-gallate. J Biol Chem. 2006;281:33761–33772. doi: 10.1074/jbc.M604748200. [DOI] [PubMed] [Google Scholar]
- 29.Gottardis MM, Jordan VC. Development of tamoxifen-stimulated growth of MCF-7 tumors in athymic mice after long-term antiestrogen administration. Cancer Res. 1988;48:5183–5187. [PubMed] [Google Scholar]
- 30.Choi HK, Yang JW, Roh SH, Han CY, Kang KW. Induction of multidrug resistance associated protein 2 in tamoxifen-resistant breast cancer cells. Endocr Relat Cancer. 2007;14:293–303. doi: 10.1677/ERC-06-0016. [DOI] [PubMed] [Google Scholar]
- 31.Paglia DE, Valentine WN. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J Lab Clin Med. 1967;70:158–169. [PubMed] [Google Scholar]
- 32.Kakkar P, Das B, Viswanathan PN. A modified spectrophotometric assay of superoxide dismutase. Indian J Biochem Biophys. 1984;21:130–132. [PubMed] [Google Scholar]
- 33.Draper HH, Hadley M. Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 1990;186:421–431. doi: 10.1016/0076-6879(90)86135-I. [DOI] [PubMed] [Google Scholar]
- 34.Chan JY, Kwong M. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim Biophys Acta. 2000;1517:19–26. doi: 10.1016/S0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 35.Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O'Connor T, Harada T, Yamamoto M. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE regulated drug metabolizing enzymes and antioxidant genes. Toxicol Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 36.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 37.Fisher B, Redmond C, Legault-Poisson S, Dimitrov NV, Brown AM, Wickerham DL, et al. Postoperative chemotherapy and tamoxifen compared with tamoxifen alone in the treatment of positive-node breast cancer patients aged 50 years and older with tumors responsive to tamoxifen: results from the National Surgical Adjuvant Breast and Bowel Project B-16. J Clin Oncol. 1990;8:1005–1018. doi: 10.1200/JCO.1990.8.6.1005. [DOI] [PubMed] [Google Scholar]
- 38.Duthie SJ, Melvin WT, Burke MD. Drug toxicity mechanisms in human hepatoma HepG2 cells: cyclosporin A and tamoxifen. Xenobiotica. 1995;25:1151–1164. doi: 10.3109/00498259509061915. [DOI] [PubMed] [Google Scholar]
- 39.Gundimeda U, Chen ZH, Gopalakrishna R. Tamoxifen modulates protein kinase C via oxidative stress in estrogen receptor-negative breast cancer cells. J Biol Chem. 1996;271:13504–13514. doi: 10.1074/jbc.271.23.13504. [DOI] [PubMed] [Google Scholar]
- 40.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Targeting multiple signaling pathways by green tea polyphenol (−)-epigallocatechin-3-gallate. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 41.Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 42.Gavrilov K, Saltzman WM. Therapeutic siRNA: principles, challenges, and strategies. Yale J Biol Med. 2012;85:187–200. [PMC free article] [PubMed] [Google Scholar]
- 43.Berindan-Neagoe I, Cornelia B, Alexandru I. Combining the chemotherapeutic effects of epigallocatechin 3-gallate with siRNA-mediated p53 knock-down results in synergic pro-apoptotic effects. Int J Nanomed. 2012;7:6035–6047. doi: 10.2147/IJN.S36523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang XJ, Sun Z, Villeneuve NF, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohnuma T, Matsumoto T, Itoi A, Kawana A, Nishiyama T, Ogura K, Hiratsuka A. Enhanced sensitivity of A549 cells to the cytotoxic action of anticancer drugs via suppression of Nrf2 by procyanidins from Cinnamomi Cortex extract. Biochem Biophys Res Commun. 2011;413:623–629. doi: 10.1016/j.bbrc.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 46.Tang X, Wang H, Fan L, Wu X, Xin A, Ren H, Wang XJ. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic Biol Med. 2011;50:1599–1609. doi: 10.1016/j.freeradbiomed.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hibasami H, Achiwa Y, Fujikawa T, Komiya T. Induction of programmed cell death (apoptosis) in human lymphoid leukemia cells by catechin compounds. Anticancer Res. 1996;16:1943–1946. [PubMed] [Google Scholar]
- 49.Paschka AG, Butler R, Young CY. Induction of apoptosis in prostate cancer cell lines by the green tea component, (−)-epigallocatechin-3-gallate. Cancer Lett. 1998;130:1–7. doi: 10.1016/S0304-3835(98)00084-6. [DOI] [PubMed] [Google Scholar]
- 50.Farabegoli F, Barbi C, Lambertini E, Piva R. Epigallocatechin-3-gallate downregulates estrogen receptor alpha function in MCF-7 breast carcinoma cells. Cancer Detect Prev. 2007;31:499–504. doi: 10.1016/j.cdp.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 51.Wang S, Chen R, Zhong Z, Shi Z, Chen M, Wang Y. Epigallocatechin-3-gallate potentiates the effect of curcumin in inducing growth inhibition and apoptosis of resistant breast cancer cells. Am J Chin Med. 2014;42:1279–1300. doi: 10.1142/S0192415X14500803. [DOI] [PubMed] [Google Scholar]
- 52.Farabegoli F, Papi A, Bartolini G, Ostan R, Orlandi M. Epigallocatechin-3-gallate downregulates Pg-P and BCRP in a tamoxifen resistant MCF-7 cell line. Phytomedicine. 2010;17:356–362. doi: 10.1016/j.phymed.2010.01.001. [DOI] [PubMed] [Google Scholar]