Abstract
Microfluidics has been the focus of interest for the last two decades for all the advantages such as low chemical consumption, reduced analysis time, high throughput, better control of mass and heat transfer, downsizing a bench-top laboratory to a chip, i.e., lab-on-a-chip, and many others it has offered. Microfluidic technology quickly found applications in the pharmaceutical industry, which demands working with leading edge scientific and technological breakthroughs, as drug screening and commercialization are very long and expensive processes and require many tests due to unpredictable results. This review paper is on drug candidate screening methods with microfluidic technology and focuses specifically on fabrication techniques and materials for the microchip, types of flow such as continuous or discrete and their advantages, determination of kinetic parameters and their comparison with conventional systems, assessment of toxicities and cytotoxicities, concentration generations for high throughput, and the computational methods that were employed. An important conclusion of this review is that even though microfluidic technology has been in this field for around 20 years there is still room for research and development, as this cutting edge technology requires ingenuity to design and find solutions for each individual case. Recent extensions of these microsystems are microengineered organs-on-chips and organ arrays.
I. INTRODUCTION
Microfluidics is the science and technology that deals with devices and methods that control fluids in microchannels with a typical length scale of one hundred nanometers to several hundreds of micrometers. Microfluidic technology has been applied to biology, chemistry, and engineering and has been first used to analyze and carry out separations and detection.1 Microfabrication techniques allowed the improvement of the electronic industry with micro-electro-mechanical-systems (MEMS) and the biochemical industry with micro-total-analysis-systems (μTAS).2 These micro systems opened up the application of microfluidics to chemical and biochemical analysis, detection, and sensing. Lab-on-a-chip technology, being a subset of MEMS, was introduced later to perform single or multiple laboratory experiments and has been applied widely for non-analysis purposes. Working with very small volumes ranging from microliters to picoliters, shortening of the analysis time, facilitating the control of reactions and the transport of chemicals at the molecular level, and leaving very little chemical waste are the benefits causing the switching from conventional bench-top to lab-on-a-chip experiments. In recent years, these new microfluidic technologies have been transformed into successfully commercialized devices.3–6
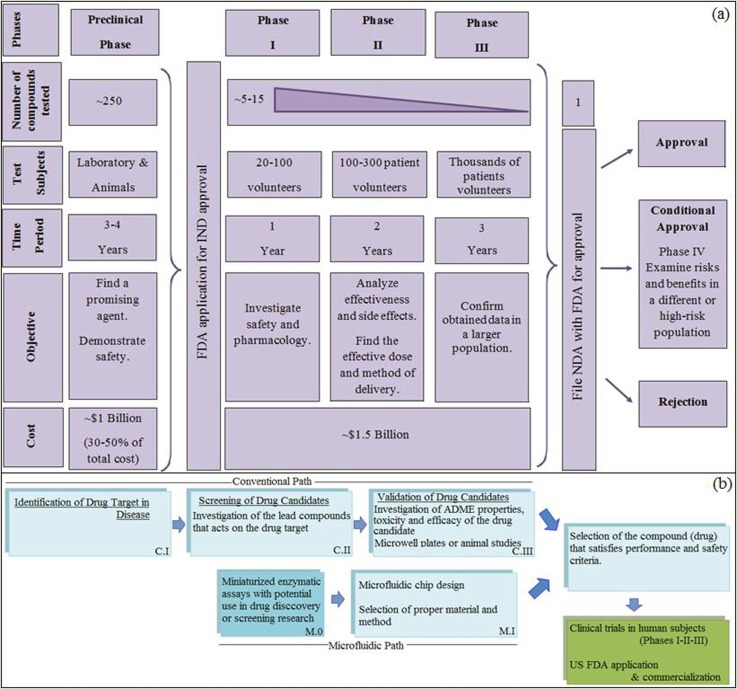
So far, various biological and biochemical processes have been successfully accomplished in microfluidic devices including polymerase chain reaction (PCR),7 drug screening,8 cell counting,9 DNA sequencing,10 and many enzymatic assays.11 Section II introduces an overview of microfluidic technologies. In the last decade, some review articles which were concentrated on different aspects of biological applications of lab-on-a-chip technology have been published between the mid 2000 s until present covering molecules/cells,12–15 drugs,16–22 enzymatic reactions,23 droplets,24 and organ-on-a-chip.25 In the present work, we have only focused on the drug-related studies, i.e., drug screening, drug toxicity, gradient based drug-dose response measurements, protein-drug (ligand) binding, and drug targeted enzyme inhibition (Sections III–V). Drug screening, which is performed before commercialization of a new drug in the pharmaceutical industry, is a long and expensive process where around $135 billion is spent each year. For an approved new compound, the costs are as high as 1 billion and 1.5 billion US dollars in the preclinical (pre-human) stage and clinical stages, respectively, reaching a total of 2.5 billion US dollars according to 2013 data (Figure 1(a)).26 On average, obtaining FDA approval and the rights to market a drug takes about 15 years.27 The steps for Drug Research and Approval are summarized in Figure 1(a). Conventionally, after basic research (experimentally and computationally) in laboratories, a preclinical (non-human) stage is necessary where tests on activity, drug-response, toxicity, ADME (absorption, distribution, metabolism, and elimination) properties, etc., are done. These lab tests are followed by in vivo animal tests to provide evidence that the drug in question might be used in humans. These steps are displayed in Figure 1(b), denoted as CI-III. Within the framework of this article, we reviewed the microfluidic systems, denoted by MI, which can replace CII and CIII stages. The stage M0 gives information about microfluidic enzyme inhibition assay platforms with potential use in drug discovery. With advances in microfluidic technology which have led to a reduction in analysis time and chemicals, there is a high possibility of decreasing these expenses in the pharmaceutical industry. Hence, recently, there has been an increasing interest in drug screening research to explore the opportunities offered by microfluidic technology, which deserves to be collected and reviewed. While there are no specific guidance on use of microfluidics in the drug discovery, the expectation would be to meet the International Conference on Harmonization (ICH) guidelines summarized in section Q2 (linearity, range, accuracy, repeatability, reproducibility, robustness, detection limit, and quantification limit) to qualify and validate the assay.28,29 Among the tested compounds, the one(s) satisfying the performance and safety criteria is (are) chosen for clinical trials in humans. Phase I-II-III trials are performed on human volunteers (Figure 1(a)) to determine the selected drug's real health benefits. Finally, the results (data from animal studies and human clinical trials) are submitted to New Drug Application (NDA) approval, which is required for commercialization since 1938. The Center for Drug Evaluation and Research (CDER) team reviews the NDA documentation submitted to FDA. Once the “drug's health benefits outweigh its known risks,” safe and effective in its proposed use, proposed labeling and manufacturing methods are appropriate, the candidate drug is ready for market.
FIG. 1.
Overview of drug discovery and development (a) steps from lab to market, (b) steps in preclinical phase: The stages of conventional path, C.II-III, can be replaced by microfluidic platforms, M1.
Drug discovery requires screening a high number of drug candidates for their efficacy, cytotoxicity, and possible side effects. In vivo animal models are common for drug screening, however; in recent years, due to ethical issues in animal testing, there has been an increasing research in mimicking in-vivo conditions with microfluidic systems such as tissue-on-a-chip and organ-on-a-chip.24 In order to perform pre-clinical drug screening using in vitro models before bringing a drug into the market, microfluidic chips with multiplexed cell culture chambers have emerged as a more efficient platform compared to high-density well plates, which are similarly used to perform cell viability assays, to determine IC50 values, to measure doubling times,30 or to find inhibitors against certain enzymes.31,32 Microfluidic chips for drug testing usually employ primary hepatocytes, which show many metabolizing enzyme activities, and differentiated functions, such as albumin synthesis, basal/induced cytochrome P450 1A1/2, and UDP-glucuronosyltransferase (UGT) activities, being more predictive of in vivo responses. The cytochrome P450 class of enzymes determines drug bioavailability and indicates a drug's potential to cause in vivo adverse drug interactions. In the case of microfluidic research, mainly the design and the fabrication of the device (Lab-on-a-Chip Platform) are important issues, and the experiments reported to have been performed in these microdevices can be altered/replaced and the researcher can apply his own system of interest. Therefore, we have also investigated the methods used for enzyme activity and inhibition, and the techniques that have the potential to be applied for further drug screening research are presented in Section IV. A summary of drug screening processes with microfluidic devices; their cell/enzyme/protein-ligand types and investigated parameters are shown in Table I. A detailed version of this table, Table S1 (including fabrication material and technique; detection method) can be found in the supplementary material (see DrugScreeningTable file).33
TABLE I.
Summary of drug screening processes with microfluidic devices.
| Cell/enzyme/protein-ligand type | Investigated parameters | |
|---|---|---|
| Enzyme inhibition by a drug candidate | ||
| Hadd et al.34 | Acetylcholinesterase (AChE) inhibitors | Michaelis constant, Km, inhibition constant, Ki |
| Cohen et al.35 | Protein kinase A (PKA) | Km, Ki |
| Puckett et al.36 | Calmodulin-Phenothiazine | Binding interaction |
| Wang et al.37 | Bovine carbonic anhydrous II (bCAII) | In situ click chemistry screening |
| Pihl et al.38 | hERG K+ channels, ligand-gated GABAA receptors | IC50, EC50 |
| Horiuchi39 | Kinase inhibitors | IC50 |
| Mangru40 | Botulinum neurotoxin serotype A (BoNT-A) | Inhibition curve |
| Iyer et al.41 | Thrombin & factor Xa (FXa) | Dose response, IC50 |
| Drug candidate screening by droplet microfluidics | ||
| Lombardi and Dittrich42 | Warfarin-Human serum albumin (HSA) | Association constant, KA |
| Miller et al.43 | Protein tyrosine phosphatase 1B (PTP1B) | Dose response, IC50 |
| Gu et al.44 | AChE inhibitors (carbaryl, chlorpyrifos, and tacrine) | Dose response, IC50 |
| Litten et al.45 | Cytochrome P450 | Enzyme activity, IC50 |
| Brouzes et al.46 | Monocytic (U937) cells | Dose response curve, IC50 |
| Toxicity assessment by a drug candidate | ||
| Viravaidya et al.47,48 | Rat hepatocytes (H4IIE), rat lung (L2), human hepatocytes (HepG2/C3A) cells | Toxicity level |
| Tirella et al.49 | Myoblast (C2C12) cells | Drug dose response |
| Toh et al.50 | Hepatocyte | IC50, LD50 |
| Yang et al.51 | Zebrafish embryo | Dose response |
| Kim et al.52 | Cardiomyocyte H9c2(2–1) cell line | Cardiotoxicity, IC50 |
| Su et al.53 | Human embryonic kidney (HEK) cells | Cardiotoxicity level |
| Cytotoxicity (cell death, apoptotic cell loss) | Cell/enzyme/protein-ligand type | Investigated parameters |
| Lee et al.54–56 | Breast cancer (MCF-7) cells | IC50 |
| Ye et al.57 | Liver carcinoma (HepG2) cells | Dose response |
| Kim et al.58,59 | HepG2 cells | Dose response, cell viability |
| Sung and Shuler60 | Colon cancer (HCT-116) & HepG2/C3A cells, myeloblasts (Kasumi-1) | Cell viability |
| Chao et al.61 | Hepatocyte | Hepatic clearance |
| Kim et al.62 | Chang liver cell line | Morphological change, cell apoptosis, dose response, EC50 |
| Lim et al.63 | Chang liver cell line | Morphological change, cell apoptosis, dose response, EC50 |
| Wang et al.64 | BALB/3T3, Cervical cancer (HeLa), bovine endothelial cells | Cell viability |
| Komen et al.65 | MCF-7 cells | Cell viability |
| Liu et al.66 | MCF-7 cells | Cell viability, GSH level |
| Jedrych et al.67 | Lung cancer (A549), colon cancer (H-29) cells | Cell viability % and IC50 |
| Sugiura et al.68 | HeLa cells | Cell growth inhibition |
| Liu et al.69 | Lung adenocarcinoma (A549/DD) cells | Cell viability |
| Cytotoxicity (cell death, apoptotic cell loss) | ||
| Ma et al.70 | HepG2 cells | Cell viability |
| Kim et al.71 | Prostate cancer (PC3) cells | Cell viability |
| Khanal et al.72 | PC3 and Ramos B cells | Hypoxia induced apoptosis |
| Seidi et al.73 | Dopaminergic nerve (PC12) cells | Cell viability |
| Hsiung et al.74 | MCF-7 cells | Cell viability, IC50 |
| Bogojevic et al.75 | HeLA cells | Apoptotic cell loss |
| Mao et al.76 | HepG2 cells | Cell viability |
| Song et al.77 | MCF-7 cells | Dose response |
| Enzyme assays with potential use for drug screening | ||
| Hadd et al.78 | β-galactosidase (β-gal) | Michaelis-Menten constants |
| Damean et al.79 | E. coli alkaline phosphatase | Michaelis-Menten constants |
| Garcia et al.80 | β-gal | IC50 |
| Clausell-Tormos et al.11 | β-gal | IC50 |
| Cai et al.81 | β-gal | IC50 |
| Sjostrom et al.82 | β-gal | Michaelis-Menten constants |
| Gielen et al.83 | β-glucosidase | Michaelis-Menten constants IC50 |
| Du et al.84 | β-gal | IC50 |
| Sun et al.85 | Leukemia cells | |
| Luk et al.86 | Trypsin and pepsin | Digestion level |
| Hughes and Herr87 | Calf intestinal alkaline phosphatase (CIP) & horseradish peroxidase (HRP) | Michaelis-Menten constants |
| Popovtzer et al.88 | Alkaline phosphatase | Enzymatic activity |
| Upadhyaya and Selvanagapathy89 | Bovine serum albumin (BSA) | |
| Werner et al.90 | β-secretase (BACE1) | IC50 |
II. AN OVERVIEW OF MICROFLUIDIC TECHNOLOGIES
Microfluidic devices offer many advantages over conventional systems, besides reducing the volume down to picoliters, such as fast heat and mass transfer due to the high surface area to volume ratio.91 The type of fluid flow in a microfluidic device is laminar with typical Reynolds number less than unity.92 Therefore, mixing in these microdevices is mainly limited to molecular diffusion,93 which gives precise control over the concentration profiles. On the other hand, the slow mixing is overcome by the integration of passive or active mixers.94 Multiple processes can be performed on a single microfluidic device, and high-throughput can be achieved by parallelization.95 Sample and medium are generally introduced into a microfluidic device through separate inlets and then delivered into sub-microchannel arrays creating a concentration gradient.49,51,57,62,66,67,74,96 Usually, serpentine (or zigzag) shaped channels are used as the channels need to be long for a complete mixing.96,97 A microfluidic concentration gradient generator (CGG) employs micron sized channels to manipulate the reagent volumes to generate a range of concentrations.98,99 The reader can find the details of concentration generator applications in Section III B.
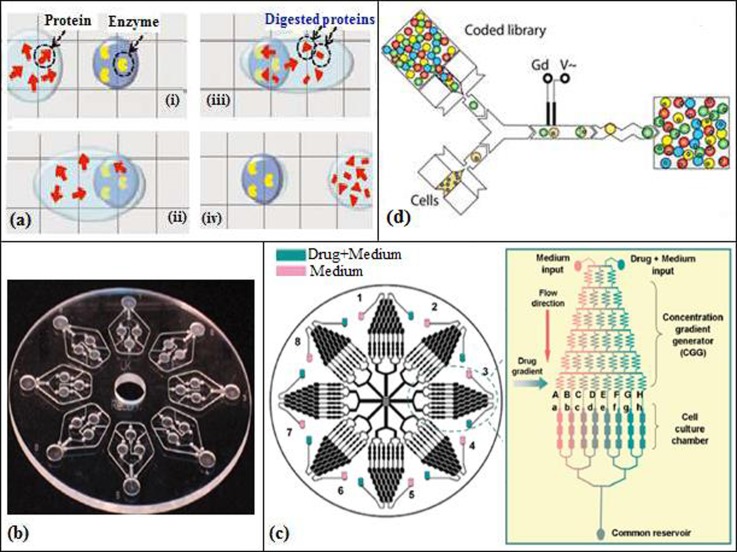
Flow in microfluidic devices can be divided into two categories, continuous and discrete. In the first one, the liquid flows continuously in the microchannel by external actuators such as a mechanical pump60,61,69 or by capillary forces and electrokinetic mechanisms.16,34,35,78 Continuous flow is easy to implement in microfluidic systems, and it is a common method employed in biological applications of microfluidic devices. However, due to the parabolic flow in the microchannel, residence time distribution occurs, and since analytes are in close contact with the channel walls, precipitation, coagulation, or contamination might also occur. To avoid sedimentation, surfactants or PEG (polyethylene glycol) is added to the buffer solution.100 In discrete flow, droplets are employed to separate the liquids, and this flow can be further divided into two categories, segmented flow and digital microfluidics (DMF). Microdroplets offer high throughput with reduced cost and reagent consumption where cross-contamination of the reagents is prevented and mixing is enhanced due to the short diffusion distance and chaotic mixing within the microdroplet (Section III C). In segmented flow, an aqueous phase is injected into an immiscible carrier phase creating a droplet; this type of flow prevents the problem of residence time distribution that occurs in continuous flow.11,79,81–83,85 In digital microfluidics, droplets are separately manipulated on an array of electrodes, and not in a microchannel (Figure 2(a)). These droplets can be moved, merged, mixed, split, or dispersed in the presence of a potential source.86,101
FIG. 2.
(a) Digital microfluidic device (DMF) containing hydrogel matrix. Reprinted with permission from Luk et al., Proteomics 12, 1310 (2012). Copyright 2012 John Wiley and Sons. (b) PMMA disc chip with a diameter of 120 mm containing eight identical structures. Reprinted with permission from Puckett et al., Anal. Chem. 76, 7263 (2004). Copyright 2004 American Chemical Society. (c) Serpentine-shaped concentration gradient generator. Reprinted with permission from Ye et al., Lab Chip 7, 1696 (2007). Copyright 2012 The Royal Society of Chemistry. (d) Droplet-based microfluidics for cell screening. Reprinted with permission from Brouzes et al., Proc. Natl. Acad. Sci. U.S.A. 106, 14195 (2009). Copyright 2009 National Academy of Sciences.
Various materials are used for the fabrication of microfluidic devices including poly(dimethylsiloxane) (PDMS),38,49,59,60,63,64,68–71,76,79,89,96,102 poly(methyl methacrylate) (PMMA),36 and glass.51,66,87 Oxygen and the water permeability of PDMS make it more advantageous in biological applications, whereas its hydrophobic nature should be used with care in applications such as screening of hydrophobic drugs, to prevent the absorption of the drug molecule into PDMS.103 Hybrid microdevices can also be made by combining PDMS and glass.65,67,82 The most favored fabrication techniques are soft-lithography,82 photo-lithography,86,89 and wet-etching.66,67 Materials and fabrication methods are chosen considering their compatibilities with the application.104
III. DRUG CANDIDATE SCREENING APPLICATIONS: INHIBITION OF TARGET ENZYME BY CANDIDATE DRUG AND DETERMINATION OF DYNAMIC PARAMETERS
A. Screening by microfluidic channels
Separation and diagnostics by a microchip are among the first applications of bio-microfluidics. Hadd et al.34 performed enzyme (AChE) inhibition assays in a microfluidic device, i.e., they designed a microchip on a microscope slide and fabricated it using photolithography, wet etching, and thermal bonding. The chip consisted of six wells of 70–140 μl volumes along with the injection ports for the enzyme (AChE), the substrate (acetylthiocholine chloride), the fluorophore (coumarinylphenylmaleimide, CPM), the inhibitor (tacrine, carbofuran, eserine), sample waste, a separation channel, a mixing region for the substrate, a reaction channel, a channel for mixing CPM to make the product fluorescent, and lastly a detection channel. Fluid flow and reagent mixing were achieved using electrokinetics (electroosmosis and electrophoresis), and the detection of CPM-thiocholine was done with a laser-induced fluorescence signal. Michaelis-Menten constants including the inhibitor equilibrium dissociation constant Ki and, consequently, the enzyme reaction rates were determined. For the inhibitor tacrine (used in dementia and Alzheimer's disease), this constant was determined as 1.5 ± 0.2 nM, being comparable to standard cuvette assays.
Hadd et al.34 started using microfluidics and miniaturizing enzymatic assays to overcome the limitations in conventional methods including the evaporation of reagents, the fixed concentration reagents, and not having a parallel reaction or a detection system. A related microchip assay was performed by Cohen et al.35 on a soda lime glass microchip (70 μm × 15 μm × 525 μm) fabricated using photolithography and thermal bonding similar to the microchips prepared by Hadd et al.34 to determine the competitive inhibition of protein kinase A (PKA) by H-89. As in Ref. 34, enzyme and substrate solutions were placed in wells and electrokinetics was used for transport and mixing. They ran their experiments in two steps. Using a simple design microchip, they optimized the substrate (Kemptide) and the enzyme (PKA) concentrations and the incubation time (around 75 s). Then, on a more complex chip, they analyzed the effect of the inhibitor H-89, which was diluted on the microchip, in contrast to Hadd et al.34 where the dilution was not done on the microchip but mixed with fixed concentrations of PKA and Kemptide. The product and substrate were separated using electrophoresis, and the separation occurred in 65 s. The detection was done with fluorescence. The inhibition constant, Ki, and the Michaelis constant, KM, were determined. Cohen et al.35 showed that these values were in good agreement with the ones obtained with conventional experiments. Protein kinases play an important role in the regulation of cell signaling by phosphorylation. The abnormal phosphorylation of proteins is the cause of many diseases such as cancer, diabetes, arthritis, Alzheimer, and hypertension. Therefore, the investigation of the inhibitors of protein kinase is necessary to inhibit this abnormal phosphorylation and prevent the emergence of these diseases.105 Horiuchi39 developed a microarray method to investigate the chemical-kinase interactome. The inhibitions of the kinase proteins were detected by the immunofluorescence staining, and their IC50 values were determined. This method included 10 microarrays each containing 6000 reactions and also managed to decrease the reagent consumption. High-throughput was achieved by aerosol deposition of the reagents into nanodroplets that are placed on the microarrays containing chemical compounds.
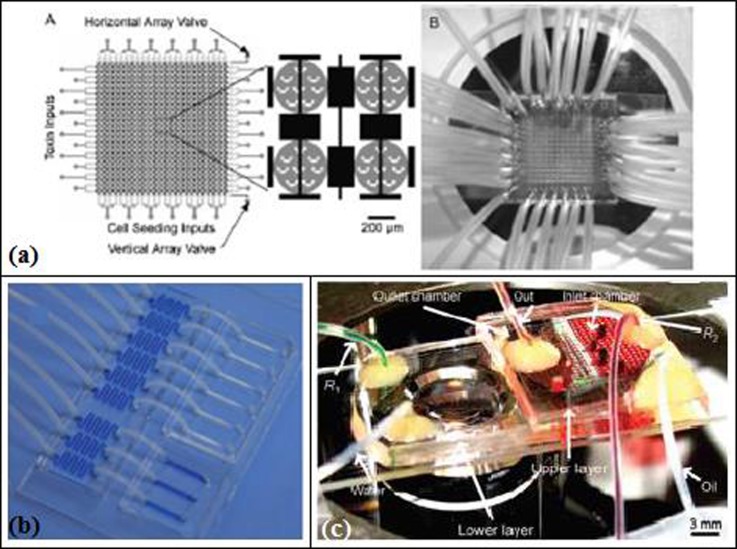
With the development of μTAS, fluid manipulation became one of the most intriguing research areas and has been mostly addressed via electrokinetic flow. In this method, the flow is generated with the use of a potential and electrostatic double layer. Due to difficulties like dependency of the flow on the pH of the solution, Puckett et al.36 incorporated a centrifugal pumping system into a μTAS system (Figure 2(b)) and applied it to protein–ligand binding assays. A servo motor supplied a rotational speed of up to 1050 rpm. They fabricated their microfluidics system, consisting of eight identical microstructures for parallel analysis with CNC machined 3-mm-thick PMMA of dimensions similar to a compact disc. The cover was made of a transparent tape. These microstructures, comprised four reservoirs of around 12 μl volume (water, analyte, dried protein, and detection) filled by pipetting, were connected by channels (635 μm wide × 635 μm deep and 127 μm wide × 63.5 μm deep). Puckett et al.36 evaluated the effectiveness of their microfluidic system by using fluorescence detection and characterizing the binding interaction between phenothiazine antidepressants and calmodulin. The analysis time was around 5–7 min, which was claimed to be a significant improvement compared to a microtiter plate analysis lasting 15 min.
A PDMS microdevice was developed by Mangru40 for inhibition assay of BoNT-A enzyme, which can cause fatal paralysis in human. The microdevice was fabricated by photolithography and consisted of pinch valves to control the fluid flow. Peptide inhibitor, Ac-CRATKML-amide, was tested against BoNT-A enzyme, and inhibition curves were obtained using fluorescent resonance energy transfer (FRET). The assay was repeated in macroscale 96-well plate, and the results of micro and macroscale systems were found to be in good agreement. The reagent consumption was 1000 fold less in microscale (40 nl) compared to the macroscale (45 μl).
The target-guided synthetic process of in situ click chemistry is known to be an effective approach for the synthesis of biligand enzymatic inhibitors. Wang et al.37 constructed integrated microfluidic chip platforms for parallel synthesis and screening of 32 (1st generation) and 1024 (2nd generation) in situ click chemistry reactions for the target bCAII. With the improved design, the time required for preparing a single click reaction was reduced to 17 s/cycle (2nd-generation), totaling to 290 min for 1024 reactions. Using a total reaction volume of 400 nl, containing approximately 12.4 pmol (360 ng) of enzyme, 120 pmol of both acetylene and azide and the complementary ligands, all click chemistry reactions between the selected acetylenes and the azides were screened under 4 reaction conditions, and consequently, 39 hit reactions were identified successfully.
In the pharmaceutical industry, the structure-activity relationship (SAR) has an important role, since it allows designing a drug with high potency and less side effects. Werner et al.90 obtained SAR data for two different aniline building blocks that were targeted to inhibit BACE1. For this research, they used a glass capillary (1.5 mm diameter, 1.5 m length) with 3 inlets and an outlet. The sample was injected into the capillary from the first inlet creating a concentration gradient by Taylor dispersion. Substrate and BACE1 were separately introduced into the capillary from the other inlets. Thus, a concentration gradient of 6 orders of magnitude was generated. IC50 values were determined and found to be in agreement with conventional methods.
B. Screening by using concentration gradient generators
In the last decade, microfluidic chips have been used to create gradients of drugs to investigate orders of magnitude of concentrations both in space and in time. Generating a concentration gradient is an important subject in biological applications, and the use of microfluidic devices to create a concentration gradient for drug screening purposes is preferable due to the high ability to control the systems and to have reduced reagent consumption. A concentration gradient is generated by adjusting the initial sample concentration, flow rates, and the shape of the microchannel network. In biological applications, it is not always possible to work at high flow rates to enhance the concentration gradient. High flow rates can increase the shear force applied to the cells and cause damage in them or flush away the adherent cells from the microfluidic device, and that can skew the results of the experiments and decrease the high-throughput.
Therefore, the design and integration of a CGG that can create a high range of a concentration gradients while reducing the reagent consumption for drug discovery studies have become outstanding.21,106 A serpentine-shaped CGG used for drug discovery experiments is shown in Figure 2(c).57
This gradient-based microchip method allows dose-response measurements for a large range and number of concentrations, which would be otherwise impossible. Pihl et al.38 built a microchip of PDMS with channel widths of 45 μm and heights of 61 μm using photolithography, electron beam, and plasma etching to investigate the gradient-based method. They used a deep reactive ion etching of silicon on an insulator (SOI) wafer for the master fabrication for a very high resolution of around ±0.5 μm. Mixing was achieved with pure diffusion in around 3–4 s. They investigated the technique for pharmacological screening of voltage-gated human hERG K+ channels and ligand-gated GABAA receptors and obtained dose-response curves at 23 different concentrations and 5 orders of magnitude in less than 30 min. They also reported the resulting IC50 and EC50values. In a work of Iyer et al.,41 inhibitors of thrombin and factor Xa (FXa), which are the enzymes of blood coagulation cascade, were identified using a microfluidic chip integrated with nano-liquid chromatography (nano-LC) in parallel with a mass spectrometer. The microfluidic chip (45 mm × 15 mm × 2.2 mm) consisted of 3 inlets (each for nano-LC, substrate, and enzyme) and had an open tubular microchannel (125 μm wide × 70 μm deep) which was bound to two microreactors, where an online inhibition assay was performed. Thus, dose response and IC50 data of inhibitors were successfully obtained.
C. Screening by droplet microfluidics
Miniaturized systems based on droplet microfluidics (droplets down to picoliter volumes functioning as independent microreactors) are designed to carry out experiments in continuous or segmented flow, performing all required steps in a single device, thereby reducing sample consumption, manipulating very small volumes with flexible control of composition, enhancing reaction speed and efficiency by rapid mixing, and increasing reliability and reproducibility.
Droplets can be created in nano and pico scales using passive (T-junction, flow focusing, and co-flowing) and active (electric field, magnetic field, optical field, etc.) methods. A microdroplet is generated by the dispersion of an oil phase into an aqueous phase. The created droplet arrays can be mobile79 where reagents were introduced either before79 or after droplet formation. In mobile microdroplets, the assay is monitored continuously, making this process difficult. On the other hand, multiple static microdroplets can be monitored simultaneously since they do not move with the flow and are stored at predefined locations.
Brouzes et al.46 developed a PDMS microfluidic device to assess the cytotoxicity of mitomycin C on human monocytic (U937) cells using droplet based microfluidic technology (Figure 2(d)). Cells were encapsulated into the droplets (700 pl) through a flow focusing nozzle. 80% of the cells managed to stay viable in the droplets. Then, these droplets with cells were merged with other droplets (200 pl) containing drug and fluorescence label. Dose response curve and IC50values were obtained by creating droplets at different drug concentrations. High throughput and high reproducibility were achieved by a high droplet production rate (100 Hz) and low standard deviation, respectively. Lombardi and Dittrich42 developed droplet microreactors to determine binding mechanisms and kinetics of a ligand (drug) to a protein. They studied the binding of the anticoagulant drug warfarin to HSA to determine its affinity constant. Each individual droplet microreactor (around 1.25 nl) contained magnetic beads (200–700 beads at a concentration of 1–3.3 mg/ml and 1–2.8 μm diameter) coated with the target protein, HSA, and surrounded by a hydrophobic carrier solution, and mineral oil for preventing non-specific interactions with the surfaces of the microfluidic device. KA determined by this droplet microfluidics approach agreed well with that of the bulk assay, confirming its application in pharmacokinetic studies. Performance reduction was observed in high flow rates (above 5 μl/min) and very high bead concentrations (50 mg/ml). Because of the high reliability and reproducibility of micro-droplet reactors, Lombardi and Dittrich42 used fluorinated or silicone oil micro-droplets, depending on the lipophilicity of the drug, with magnetic beads of different surface groups for drug discovery and testing studies.
Using the droplet-based microfluidics approach, controllable concentration gradient generation methods with very small reagent and sample volumes have been developed measuring dose-response curves, estimating IC50 values and performing online detection such as the fluorescence or electrochemical method.43,44 Miller et al.43 generated a PDMS droplet based microfluidic device for screening a chemical library (more than 700 drugs/inhibitors with molecular weights between 113 and 1882 Da) for inhibition of a target enzyme (PTP1B, a target for type 2 diabetes mellitus, obesity, and cancer). For that purpose, various concentrations were injected with a sampler, and a compartment of the drug was formed using Taylor-Aris dispersion. Then, the drug with the enzyme and the substrate were encapsulated in a droplet of around 140 pl volume, which was formed in continuous phase (oil). Around 10 000 droplets were formed at each run, i.e., per compound. This number is 1000 times more than those obtained in conventional systems, resulting in extremely precise measurements of dose–response relationships using minimal quantities of reagents (17.5 μm—a 25 000 fold reduction in reagent consumption per dose-response data point). Throughput was one compound every 157 s, and the entire screen, which lasted 4.2 h, was performed without manual intervention using a single microfluidic device. Initial reaction rates and IC50 values determined were in good agreement with those obtained in conventional microplate assays at a much higher precision (approximately 26-fold). The microfluidic system can operate effectively over concentration ranges exceeding 3 orders of magnitude (dose-response curves covering wide concentration ranges) and can be adapted for cell based assays (CBAs) like intracellular calcium flux assays for identifying agonists and antagonists, in addition to focused or iterative drug screening. Similarly, Gu et al.44 designed a microfluidic chip integrated with microelectrodes for droplet generation and electrochemical detection, to measure dose response curves and to determine IC50 values of three types of inhibitors for AChE. The effects of oil flow rate and surfactant on electrochemical sensing were investigated where current response decreased slightly as the oil flow rate increased, and the droplet size decreased. Compared with traditional enzyme inhibition assay methods, the sample consumption in the droplet-based system was reduced by 1000-fold, the whole procedure of enzyme inhibition assay was achieved within 6 min on a single microchip, and the total consumption of reagents was less than 5 μl. In droplet microfluidics, there occurs artefacts like partitioning of reagents from droplet to the carrier fluid and interaction of the biological reagents between water and oil interfaces. To understand these problems, Litten et al. developed a droplet microfluidics assay, where the partitioning of drug-like molecules in different oils, protein binding at the water-oil interface, and its effect on cytochrome P450 enzyme inhibition (on IC50 values) were investigated within the droplets of 200 μm diameter (4.2 nl).45
Microdroplets can also be created by the droplet-on-demand method (DOD). In flow-focusing droplet generators, the droplet production frequency (>kHz level107) is relatively higher than the production rate obtained in DOD method. Enhanced droplet generation can be useful to achieve high-throughput in drug screening assays. However, if a smaller library is needed for screening or a higher control over the droplet generation process is required, DOD technique can be adopted safely. Gielen et al. developed a fully automated DOD platform combined with an absorbance detection unit and investigated enzyme kinetics and inhibition. In this platform, samples were sequentially and continuously sucked into a polytetrafluoroethylene (PTFE) tubing (I.D. 200 μm) from a sample loading carousel by a computer controlled unit. A linear concentration gradient in droplets was achieved by producing microdroplet pairs at different sizes and keeping the total volume of droplet pairs at 60 nl. Then, the droplet pairs were merged by increasing their flow rates. The highest dilution ratio achieved in the experiments was 1–5 where the volume of the droplet pairs was 10–50 nl. For enzyme kinetics, the hydrolysis of the chromogenic substrate 4-nitrophenyl glucopyranoside by sweet almond β-glucosidase was investigated. Michaelis-Menten parameters were determined, consuming a total enzyme solution of 720 nl and a substrate of 360 nl. For inhibition assay, 1-deoxynojirimycin was used and its IC50 value was calculated. The same enzyme kinetic assays with inhibitor were also tested in 96-well plate system using 240 μl samples.
IV. ASSESSMENT OF TOXICITY—SIDE EFFECTS INDUCED BY DRUG CANDIDATE ON TISSUES AND CELL CULTURES
The investigation of the ADME properties of drug candidates is an important research area for the pharmaceutical industry since it involves a long and expensive process. In addition to ADME properties, the toxicity of a potential drug must be assessed before its release into the market due to safety reasons in therapeutic applications. At this point, the use of microfluidic devices to test the toxic effects of a new drug is reasonable in order to reduce the time and the chemical amount consumed. Also, a new opportunity for the application of microfluidics in preclinical drug discovery is the organ-on-chip technology where the functional units of human organs are built.22,25,108,109
A. Cytotoxicity
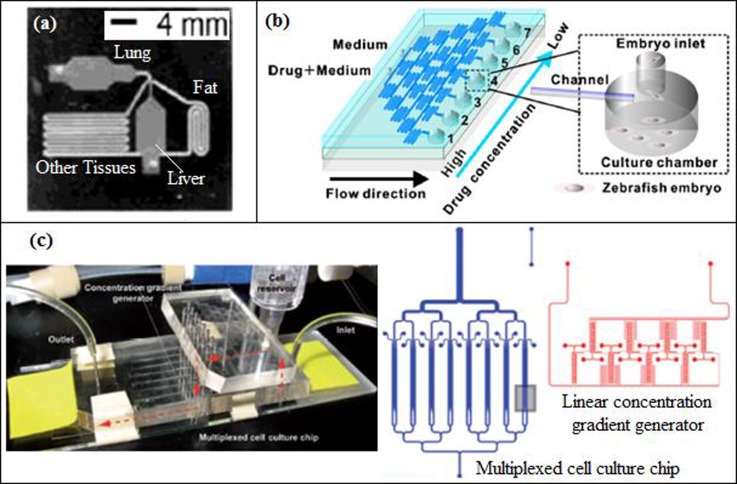
Viravaidya et al.48 and Viravaidya and Shuler47 designed a silicon microfluidic device (Figure 3(a)) with four chambers based on the PBPK (physiologically based pharmacokinetic) model of a 220 g rat, using lithography method. These chambers consisted of the lung, liver, fat, and other tissues that can exactly mimic the organs and even the flow split between the organs. Viravaidya et al.48 and Viravaidya and Shuler47 studied the toxicity of naphthalene and naphthoquinone in H4IIE, L2 (Type II epithelial cells), and HepG2/C3A cell cultures. Glutathione (GSH) is a substantial antioxidant that prevents damage to the cells caused by the reactive oxygen species (ROS), and it (GSH) can be depleted in the lung and liver cells in the presence of naphthalene metabolites. Naphthalene and naphthoquinone induced GSH toxicity level was measured using fluorescence image analysis in both 3T3-L1 adipocyte present and absent media.
FIG. 3.
(a) Micro cell culture analog (μCCA) device containing fat, lung, liver, and other tissue chambers. Reprinted with permission from Viravaidya et al., Biotechnol. Prog. 20, 316 (2004). Copyright 2004 John Wiley and Sons. (b) Microfluidic chip for zebrafish embryo with a concentration gradient generator. Reprinted with permission from Biomicrofluidics 5, 1 (2011). Copyright 2011 AIP Publishing LLC. (c) 3D Hepatox chip design with a concentration gradient generator. Reprinted with permission from Toh et al., Lab Chip 9, 2026 (2009). Copyright 2009 The Royal Society of Chemistry.
CGGs are used to measure the cytotoxic effects of a drug in a few orders of magnitude of its concentration using small sample and reagent volumes. Tirella et al.49 examined drug toxicity using a CGG and found this method more advantageous compared to single shot testing, especially at low drug concentrations. In this work, a PDMS microfluidic CGG, which was obtained using a serpentine shaped channel, was designed and manufactured using soft-lithography method. The toxicological effects of two anesthetic drugs, bupivacaine and lidocaine, on C2C12cell cultures were investigated at a constant flow rate of 170 μl/min. The control of the flow rate was found to be just as an important issue since the flow rate used to create the required concentration range can have a damaging effect on the shear sensitive C2C12 cells. In addition to the experimental work, Tirella et al.49 modeled the network of the experiments using COMSOL Multiphysics.
Similar to the morphology based cytotoxicity assessment, phenotype based evaluation can be performed for the toxicity measurements of drug candidates. For the phenotype based toxicity measurements, zebrafish embryo can be chosen as the target model organism because the zebrafish morphology and physiology are similar to those of mammals. The zebrafish is also preferred due to its transparency, easy accessibility, lack of ethical issues, and its perception of pain. The drawbacks of using zebrafish, on the other hand, are listed as the evaporation of the solution and the nonselective absorption that causes a change in the concentration and pH of the solution. Yang et al.51 worked with zebrafish embryos to investigate the toxic effects of doxorubicin, 5-flurouracil (5-FU), cisplatin, and vitamin C on phenotype characteristics. With the proposed microfluidic system of Yang et al.,51 the reduction of evaporation due to open surface design and nonselective absorption was achieved by supplying enough flow, at a velocity of 3–5 μl/min. The microfluidics glass device (Figure 3(b)) was manufactured using photolithography and wet-etching methods and included a concentration gradient, by serpentine shaped microchannels. Toxicities of the drugs were evaluated according to observations such as heart rate, body motion and shape, development of notochord, tail and fin morphology, and pigmentation. Dose response curves for doxorubicin were obtained for zebrafish embryos at different developmental ages, and it was found that, for certain developmental ages, even the low concentrations of the drug had irreversible and fatal side effects.
B. Hepatoxicity
In drug testing, it is important to eliminate the false leading drug candidates using in vitro models since the design cost of a new drug is extremely high. The drugs can be tested in commercially available microfluidic chips during their pre-clinical trial period. However, these chips are not cell-based and the microfluidic chips should include cell cultures like hepatocytes to better predict the in vivo drug metabolism and toxicity for drug discovery. Toh et al.50 developed a multiplexed microfluidic hepatocyte culture system (3D HepaToxChip, Figure 3(c)) containing 8 parallel cell culture channels (each cell culture channel having dimensions of 1 cm (length), 600 μm (width), 100 μm (height), and having an array of 30 μm × 50 μm elliptical micropillars to separate the cell culture channel into 3 compartments) and integrated with a linear CGG. 5 model hepatotoxic drugs (acetaminophen (AP), diclofenac, quinidine, rifampin, and ketoconazole) were used to assess hepatotoxicity and to estimate IC50 values from the dose-response curves. Although the IC50 values obtained by 3D HepaToxChip using cell necrosis as the cytotoxic marker were slightly higher than the IC50 values reported in the literature, they were well correlated to the reported in vivo lethality dose, LD50 values, which were used to predict the in vivo acute toxic potential of a drug (the LD50 value of a drug is the concentration that results in 50% mortality in animals). They also showed that LD50 values can be estimated in an in vitro environment to determine the required initial drug dose.
C. Cardiotoxicity
Drug-induced cardiac dysfunction leads to morbidity and high mortality. The change in ion channel activity is one of the major causes of drug-induced cardiotoxicity, which is associated with an unintended hERG blockage and has to be checked for successful drug development, i.e., screen identified or developed compounds for hERG blockage (hERG is a major target in drug safety programs). Kim et al.52 investigated drug-induced changes in ion channel activity such as the permeability of K+ channel and intracellular Na+ concentration and discrimination of simultaneous cellular events (apoptosis and necrosis triggered by anticancer drugs) in the microfluidic platform (single cell multicolor imaging system of 6 channels, channel dimension: 3.8 mm × 17 mm × 0.4 mm) using fluorescence detection. The responses of cardiac cells to cisapride, known as a hERG channel blocker, and to digoxin, known as Na+/K+ pump blocker, were examined for K+ ion channel permeability and intracellular ion level (Na+) where the cardiomyocyte H9c2(2–1) cell lines on the microchannels were treated with different concentrations of each drug. Moreover, two anticancer drugs, an already known drug (camptothecin) and a potential one (SH-03), were also screened for cardiotoxicity and induction of apoptosis/necrosis by direct visualization, and monitoring a series of cellular responses at a particular cellular state (affinity of the drug to the ion channel is cellular state dependent). Compared to conventional 96 or 12 wells, the microfluidic platform having single cell multicolor imaging system gave satisfactory IC50 values, and it is very advantageous for the achievement of reduced sample consumption and measurement time as well as producing less cell contamination. Ion channel activity exists in many cell types and is related to many processes, such as heartbeat, muscle contraction, hormone secretion, and pain perception as well as ion transportation, regulation of potential difference in plasma membrane, cell signaling, etc. Hence, the usage of such a microfluidic platform for the investigation of drug-induced toxicity by monitoring ion channel activity has considerable potential in the application to cells in other organs. In another work, Su et al.53 studied a hERG screening assay in polystyrene (PS), cyclo-olefin polymer (COP), and PDMS microchannels to assess drug-related hERG membrane trafficking. A 16 × 12 array of straight microchannels (5.25 mm × 10 mm × 140 μm, channel volume 750 nl) existed in PS and COP microdevices, where PDMS microchannels were elliptical, had a volume of 3.2 μl, and were fabricated by soft-lithography. To study the cardiotoxicity of the drugs (fluoxetine, aspirin, acetaminophen, ivermectin, and arsenic trioxide (ATO)), the growth and culturing properties of human embryonic kidney (HEK) cells transfected with hERG channel protein were investigated. The results showed that PS and COP are more applicable than PDMS as a fabrication material for the drug-related research, since hydrophobic drug molecules can be absorbed by PDMS, and this may skew the results of toxicity assays.
V. ASSESSMENT OF CYTOTOXICITY (CELL DEATH, APOPTOTIC CELL LOSS) BY CELL BASED MICROFLUIDIC ASSAYS
Cell-on-chip technology allows the combination of microfluidic techniques with cell culture technology and enables in vitro toxicology assays to be performed precisely at a microscale resolution. In the designing of microfluidic devices that are used for cell based assays in drug discovery research, the aim is to fabricate a device that can exactly mimic the in vivo environment under in vitro conditions. A third dimension in cell cultures has been shown to increase the validity of assays, since a 3D cell culture can mimic the original cell culture more accurately than a 2D cell culture where the cells adhere to the surface.110 However, manufacturing 3D microfludic channels brings additional complexity to the design and manufacturing methods.
A. 3D hydrogel matrices
Lee et al.54,55 developed a 3D cell culture array (DataChip) with dimensions of 25 × 75 mm combined with a metabolizing enzyme toxicology assay chip (MetaChip) for high-throughput toxicology analysis of drug candidates and their cytochrome P450 generated metabolites. MCF-7 cell lines were encapsulated in different 3D hydrogel matrices, i.e., collagen and alginate, and then the dose response cytotoxicity of model drugs (doxorubicin, 5-FU, and tamoxifen) on MCF-7 cells was measured with the fluorescent live-cell staining method. Thus, the IC50 values of the drugs were determined, and the results obtained using DataChip were in good agreement with the results of the conventional 96-well plate. The DataChip, consisted of 1080 cell culture chambers, each with a volume of 20 nl, was miniaturized 2000-fold compared to a 96-well plate device and decreased the stamping procedure (adherence of the cell monolayer on solgel on MetaChip) from a period of 1-to-7 days to only 6 h. In a later work of Lee et al.,56 DataChip and MetaChip were constructed on a micropillar chip (DataChip 2.0) and a microwell chip (MetaChip 2.0), respectively. The metabolism and the cytotoxicity of a therapeutic molecule, ajone, on human hematoma cells (Hep3B) were investigated, and dose response profiles and their corresponding IC50 values were obtained for MetaChip2.0 and DataChip2.0.
In pharmaceutical technology, high content screening (HCS) is used for drug discovery. HCS allows multiple readouts; however, it provides low assay throughput and requires additional parts for liquid handling. These disadvantages, or handicaps, can be overcome using a lab-on-a-chip device that can increase the assay throughput and reduce the cost and amount of equipment used in an experiment. Ye et al.57 employed the cell-based HCS method with concentration gradients of drugs on a lab-on-a-chip device to detect the apoptotic effects of different drugs on HepG2 cells. There existed eight uniform units where each unit consisted of a set of channels acting as a CGG upstream and parallel cell culture chambers downstream, i.e., CGGs were connected to the arrays of parallel cell culture chambers (1000 μm long × 400 μm wide) with micro channels of 2000 μm long × 75 μm wide. Thus, eight concentrations of eight different drugs were tested simultaneously. Dose response curves of anticancer drugs were plotted against mitochondrial membrane potential (MMP) nuclear size, plasma membrane permeability, and fluorescent intensities of ROS and GSH. The changes in nuclear morphology and membrane permeability displayed signs of late stage and irreversible apoptosis, where ROS and GSH were parameters of cell oxidative stress and regulators of apoptosis. Using this microfluidic device, HCS was performed using both less amount of sample and time, while high throughput was achieved by the usage of a concentration gradient.
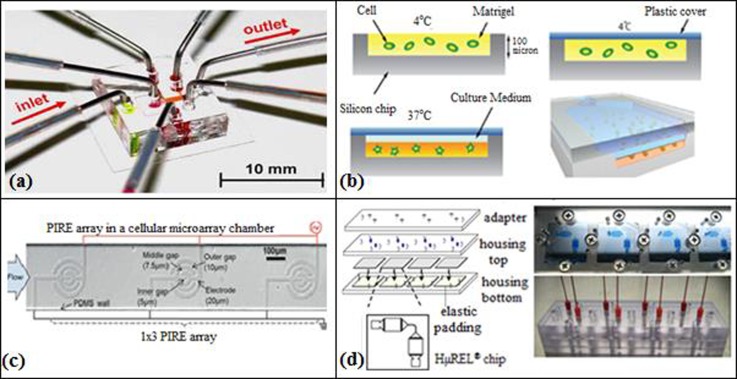
Microfluidics offers a well-confined microenvironment and precise manipulation of fluids. Considering the importance of mimicking the original cell culture, Kim et al.58 designed a microfluidic device (Figure 4(a)) for in situ cell based assays with a linear concentration gradient in a peptide scaffold. However, cells were not distributed uniformly in this microfluidic device and the peptide scaffold was seriously damaged due to the pressure difference caused by tube removal, which occurs only in 3D cell cultures, but not in 2D cell culture. In a later work, Kim et al.,59 using multilayer soft lithography method, fabricated a CBA on a 3D microfluidic platform with microvalves (microvalve-assisted patterning, MAP) for higher functionality that is necessary for biomedical research. Kim et al.59 performed CBAs for cellular dynamics. The designed microchannel, made of PDMS, consisted of main valves, side valves, solution valves, and layers of fluidic channel, PDMS membrane, and a control channel. Microvalves were used for bubble removal and the exchange of solutions without breaking the cell-matrix culture, as well as for 3D cell patterning in a microchannel, by opening and closing the channels temporarily with the PDMS membrane. The detection area for the cell based assays was located in the main channel with dimensions of 800 μm × 1400 μm. They hydrodynamically focused hydrogel (Puramatrix), in the middle of a main channel, and simultaneously immobilized HepG2 cells which were then cultured in scaffolds inside the device. A linear concentration gradient of drugs was generated in the detection area using side valves. The hepatoxicity of HepG2 cells was measured at different drug concentrations, and cell viability was followed using fluorescence imaging technique. It was demonstrated that the morphology of HepG2 cells cultured in the designed scaffold was improved compared with that of conventional cell culture (96-well plate) by reducing the drug amount 10 fold, and the experiment time 3 fold. In addition, the culturing time for colorimetric microtiter (MTT) assay and MAP took 12 h and 3 h, respectively, and a time delay occurred for LIVE/DEAD® image analysis in MTT assays.
FIG. 4.
(a) Microfluidic device for 3D-cell culture. Reprinted with permission from Kim et al., Biomed. Microdevices 9, 25 (2007). Copyright 2007 Springer. (b) Cell-embedded Matrigel (hydrogel) in a μCCA device. Reprinted with permission from Sung and Shuler, Lab Chip 9, 1385 (2009). Copyright 2009 The Royal Society of Chemistry. (c) 1 × 3 PIRE designed to uniformly pattern cells via DCP. Reprinted with permission from Hsiung et al., Lab Chip 11, 2333 (2011). Copyright 2011 The Royal Society of Chemistry. (d) HμREL biochip. Reprinted with permission from Chao et al., Biochem. Pharmacol. 78, 625 (2009). Copyright 2009 Elsevier.
Like Kim et al.58 and Kim et al.,59 Sung and Shuler60 designed a microfluidic device with 3D hydrogel cell cultures that can produce multi-organ interactions to assess cytotoxic effects of anticancer drugs (Figure 4(b)). The design of the micro cell culture analog (μCCA) device consisted of separate sections for the liver, colon cancer, and bone marrow cells. These sections were connected to each other with microchannels which were designed to mimic the residence time of blood in the corresponding organs in the human body. The PDMS microchip was fabricated using the photolithography method. Cells were encapsulated in 3D hydrogel with a depth of 100 μm in silicon chip. The top of the silicon chip was covered with a plastic cover, and then the culture medium was distributed through the chip between the hydrogel matrix and the plastic cover. Tubing was connected to the microchip to be used as inlets and outlets for the culture medium. To prevent bubble formation in the system, micro sized bubble traps were attached to the tubing. The bubble traps consisted of two 50 μm PDMS layers where the top layer acted as a barrier to capture bubbles within a specific range and the bottom layer worked as a bypass channel to discharge the captured air bubble. After working for three days without any problem, air bubbles then started being trapped in the system due to air leakage between the tubing and the μCCA. The cytotoxicity effects of the drugs (tegafur and 5-FU) on tumor cells, liver cells, and myeloblasts were tested by cell viability, hepatoxicity, and hematological toxicity assays. Cell viability was measured by staining the cells with a fluorescent LIVE/DEAD staining solution. The same experiments were also performed using a conventional 96-well plate MTT assay. μCCA was found to be more expensive, less user friendly, and less adaptable to high throughput applications but provided a physiologically more realistic environment than the conventional MTT systems.
The process of disease progression as well as drug inhibition or activation on a target protein is influenced by the communication between the target cell/protein and the surrounding cells or interacting proteins. Hence, the development of three-dimensional microfluidic cell arrays enables the reconstruction of an in vivo cell/tissue microenvironment (physiological or pathological environment) and can provide fast and reliable screening of drugs (antimicrobial, anticancer, etc.) and better prediction of drug responses. Moreover, multilayer devices can imitate the process of drug delivery in micro-tissue arrays with blood circulation. These microengineered systems need to be scaled up for high throughput drug screening in the pharmaceutical industry.
B. Dielectrophoresis and digital microfluidics based systems
To achieve uniform MCF-7 cell arrays, Hsiung et al.74 designed a dielectrophoresis-based cellular microarray and investigated the effects of anticancer drugs (cisplatin and docetaxel) on MCF-7 cells. Dielectrophoresis force was assessed by the simulation conducted on COMSOL Multiphysics 3.5a. The design of Hsiung et al.74 consisted of a serpentine shaped CGG with antipulse valves (APVs) and anticrosstalk valves (ACVs). MCF-7 cells were first suspended in a dielectrophoretic cell patterning (DCP) buffer, which was used to achieve uniform and viable cellular arrays, and then the cells were introduced into the microarray chambers where an AC signal of 5 Vpp (pp: peak to peak) at 10 MHz was applied to the cells to pattern them on collagen-coated planar interdigitated ring electrode (PIRE) arrays uniformly using dielectrophoresis forces (Figure 4(c)). To prevent the convective-diffusive crosstalks in the manifold channels, pressure-driven ACVs were put into the microarray. Hydraulic pulses, which could disturb the patterned cellular microarray, were prevented using APV. Cell viability was assessed by Calcein-AM and propidium iodide (PI) staining method where live and dead cells were measured, respectively, using fluorescence detection. IC50 values of the anticancer drugs were calculated. A comparison with conventional 96-well plate method showed that only one third of the cell amount was required to perform the cell viability test in the microarray.
One of the alternative methods to lab-on-a-chip systems is DMF. DMF allows control over discrete droplets on an array of electrodes.111 In these individual droplets, biological processes can be conducted; also samples and reagents can be moved, merged, or dispensed from reservoirs. Bogojevic et al.75 implemented the first cell based assay on a multiplexed DMF device and investigated Caspase-3 activity in HeLa cells in response to staurosporine (antibiotic), that is to say the effect of drug concentration on HeLa cells was assessed. The detection of Caspase-3 activity was done by NucView staining and using fluorescence detection method; the apoptotic cell loss was observed and dose-response profiles were obtained. In conventional systems, one of the drawbacks is the loss of HeLa cells after the washing process (HeLa cells can become detached due to the changes occurring in their morphological characteristics during apoptosis). Therefore, apoptotic cell loss cannot be measured properly. The use of a DMF device proved more efficient over a 96-well plate (conventional), as it prevented the loss of apoptotic cells when the system was washed (38% apoptotic cell loss in 96-well plate vs negligible apoptotic cell loss in DMF device). The shear stress, applied to the HeLa cells causing them to detach, was low or negligible in DMF devices due to flow recirculation occurring in the droplets. Other advantages of the DMF device over 96-well plate were a 33-fold reduction in reagent consumption and decreased standard of error.
C. Miscellaneous microdevices
Cell death process was originally characterized by morphological categorization, i.e., circularity and the shrinkage of adherent cells. The methods used to measure cell death, such as fluorescence based microscopy and flow cytometry, are widely applied in high-throughput screening because of their high sensitivity and the ease of the application. However, these methods are not based on morphological classification (rounding, shrinkage, etc.) due to their deficiency regarding imaging skills when the cell death processes are investigated. Therefore, Kim et al.62 investigated the Cd2+ induced apoptosis of a Chang liver cell line where a morphology based measurement method was applied using a microfluidic image cytometry (μFIC). The morphological properties of the adherent cells, like circularity and shrinkage, were observed when a toxin concentration gradient was created by laminar flow and diffusional mixing. Kim et al.62 estimated the dose response curve of the effective concentration, EC50 by the relationship between the Cd2+ concentration and cell rounding. The estimated EC50 was in an agreement with the values reported in the literature. When the results of the microfluidic assay were compared with the conventional 96-well colorimetric MTT test, a large difference between two methods was observed. The reason for this disagreement was explained by the difference between the monitored parameters, meaning mitochondrial enzyme activity in MTT and morphological changes in μFIC. It was suggested that the μFIC is a more sensitive method than the conventional MTT assay, and the cell rounding is an early sign of cell apoptosis. In the experiments, only 0.3 μl sample volume and 2000 adherent cells were used, whereas the conventional MTT method required 2000 times more sample volume and cell numbers.
In a continued work of this group, Lim et al.63 combined the morphology based μFIC method with the colorimetric MTT absorbance based image cytometry assay to quantify the cell death process. This modified microfluidic device was made of PDMS using the soft lithography technique; the dimensions and the volume of each cell culture compartment were 654 μm × 213 μm and 72 nl, respectively, and also the incubation time required for the MTT reagent was decreased from 4 h to 80 min. The cell apoptosis was estimated at two endpoints, the mitochondrial activity and the circularity of the cell. The EC50 value obtained from this modified technique63 was much higher than that obtained by the work of Kim et al.62 because the mode of the toxin exposure was changed from the flow mode to the static mode using a pneumatic valve system.
Similar to Sung and Shuler,60 Chao et al.61 designed a microfluidic device called HμREL® chip that can assess pharmacodynamic and pharmacokinetic properties of drugs in in vitro systems. HμREL system consisted of three parts, biochips (four biochips were placed between top and bottom housings), a fluid reservoir, and a peristaltic pump (Figure 4(d)). In HμREL device, the cell viability was measured using fluorescent LIVE/DEAD staining solution. Hepatic clearance data of different drugs obtained by HμREL device were compared with those of in vivo systems. It was reported that HμREL device can also be used to investigate drug-protein binding, drug-drug interaction, and interactions between different organs.
Developing a cheap and less labor-intensive microfluidic cell array providing a high-throughput system is very important for preclinical cytotoxicity assays of drug candidates. Microfluidic devices are used for uniform cell seeding and drug loading to the cytotoxicity assay array. If the cells are not uniformly loaded, the fluid flow can be perturbed, which may dislocate the seeded cells and cause cell clumping and local nutrient losses that can stress the cell cultures.
To overcome this problem, Wang et al.64 developed a PDMS microfluidic array (Figure 5(a)) using soft lithography method for the cytotoxicity analysis of five toxins (digitonin, saponin, CoCl2, NiCl2, and acrolein) on BALB/3T3 (mouse embryonic fibroblast cell line), HeLa, and bovine (embryonic stem cells) cells. This microfluidic device was equipped with two orthogonal microchannels (40 μm × 100 μm) for toxin exposure with a circular chamber of 400 μm diameter at each channel intersection. Pneumatically actuated valves were integrated vertically and horizontally to seed the cell lines into the desired column or row in a controlled manner. Each circular chamber included 8 U-shaped cell sieves (8 μm × 20 μm × 40 μm) where each sieve had two apertures with a diameter of 8 μm. The geometry and the locations of the cell sieves were determined using computational fluid dynamics (CFD) simulations (CFD tool, STAR-CD 3.15a). The viability of the cells against the toxins was measured by fluorescent LIVE/DEAD staining method. The results of the cell cytotoxicity assay in the microfluidic array and in the 96-well plate were found to be in good agreement. Song et al.77 designed a PDMS-glass microfluidic device consisting of 16 × 8 chambers. Similar to the work of Wang et al.,64 they used 8 cell sieves in each chamber to solve the cell seeding problem. Apoptotic and proliferation inhibitory effects of mitomycin C and tamoxifen on MCF-7 cells were detected by fluorescence staining, and the results provided by this microfluidic device were found to be in good agreement with the data obtained from conventional 96-well plate technique. Like Wang et al.64 and Song et al.,77 Komen et al.65 designed and manufactured a PDMS microfluidic device with cell traps to improve the cell loading in experiments investigating apoptosis. This microdevice consisted of a single microchannel (1 cm length × 200 μm width × 50 μm depth) with a volume of 0.1 μl. The microchannel included vertical columns (17 μm length × 10 μm width × 40 μm depth) which were separated from each other by a distance of 5 μm. The PDMS chip was placed onto a Pyrex glass (1 cm × 2 cm) due to the easy attachment of cells onto Pyrex. Cells were loaded into the microchannel by pipetting and using hydrostatic forces, and then they were trapped in the cell traps. However, pipetting was not very efficient for this design due to the sudden strong flow which occurred in the microchannel when a large volume of liquid was pipetted into a relatively smaller volume. A sudden flow resulted in cell detachments and aggregations, which can change the results of cell viability assays. Therefore, a second microdevice was designed to overcome this problem. In this design, the main channel was connected to a broader cell culture chamber with a volume of 4.4 μl (10 mm length × 5 mm width × 44 μm depth) that included an array of pillars to increase stability. Cell loading was improved by increasing the volume of the cell chamber. Flow was introduced into the microchamber in a controlled manner to prevent the cells from detaching and to reduce the shear stress applied to the cells. Finally, in this second microdevice, chemosensitivity of MCF-7 cells to staurosporine was investigated and cell viability was detected by Calcein-AM for live cells and PI dead cells.
FIG. 5.
(a) Microfluidic chip with cell sieves for cell trapping. Reprinted with permission from Wang et al., Lab Chip 7, 740 (2007). Copyright 2007 The Royal Society of Chemistry. (b) Microfluidic device for drug metabolism and cytotoxicity assay. Reprinted with permission from Mao et al., Lab Chip 12, 219 (2012). Copyright 2012 The Royal Society of Chemistry. (c) Microfluidic chip used for parallel microdroplets (PμD) technology. Reprinted with permission from Damean et al., Lab Chip 9, 1707 (2009). Copyright 2009 The Royal Society of Chemistry.
For the investigation of the chemotherapy sensitivity of MCF-7 cells, Liu et al.66 designed a glass microchip with a concentration gradient using wet etching technique. In most of the studies reported in the literature, microdevices were made of PDMS using soft-lithography method due to low cost and short turnaround time. However, PDMS may be very easily deformed112 and has a low reproducibility, which decreases the control over the concentration gradient profile. Therefore, Liu et al.66 preferred glass as a design material for its rigidity, stability, and ability to be etched at multiple depths. As Wang et al.,64 Liu et al.66 also focused on solving cell positioning and seeding problems. In both works, dam and weir structures were used for cell positioning and seeding in the microchip design, respectively. The microchip had five pyramidal branches where each branch worked as a concentration gradient generating module and induced a downstream cell culture module. At each branch, six different concentrations were generated at the terminals of the two inlets. At the end of each branch, there was a cell culture chamber (2000 μm × 1000 μm × 30 μm) and to prevent the migration of the cells to the up-stream channel, dam structures were located at the intersections of these cell culture chambers and the CGGs. Finally, arc shaped weir structures (270 μm in long axis and 100 μm in semi-short axis) were placed in the cell chambers to ease cell seeding by increasing surface area and decreasing the flow speed. To investigate the effect of GSH modulators, N-acetyl cysteine-NAC and ATO on the chemotherapy sensitivity of MCF-7 cells, cell viability, and GSH level were measured and dose dependent relationships of GSH with NAC and ATO treatments were obtained using fluorescent LIVE/DEAD morphological test.
As mentioned before, cytotoxicity tests of chemotherapeutic agents require supplying different concentrations of drugs into the microfluidic device. For this purpose, Jedrych et al.67 designed a hybrid (PDMS/glass) microfluidic device with a concentration gradient using photolithography and wet etching methods. The microfluidic device consisted of a matrix of 5 × 5 cell culture microchambers (diameter 1 mm, depth 30 μm) connected to a set of serpentine shaped microchannels (width 100 μm × depth 50 μm), which, in turn, included two inlets and five outlets to produce five different drug concentrations acting as a CGG. Using this hybrid microfluidic device, the cytotoxic effects of 5-FU drug on A549 and H-29 cells were investigated. The same experiments were also performed with a conventional 96-well plate device. The percentage of cell viability and IC50 values were obtained for both micro and macro (conventional) systems. Cell viabilities for A549 and HT-29 for both micro and macro systems were almost the same.
For delivering cell culture media into the microfluidic devices, syringe pumps have been widely used. However, the delivery of multiple types of drugs can be problematic and difficult due to the requirement of many tube connections. The usage of microvalves could be another option; for example, Wang et al.64 preferred microvalves in their microfluidic chip design and achieved drug perfusion successfully, but the fabrication and operation of the microvalves can be challenging. Sugiura et al.68 overcame this problem by using a pressure-driven liquid delivery system. Their PDMS microchip of three layers was fabricated by multilayer photolithography method and consisted of 8 × 5 arrays of 40 microchambers. Culture media were added into the macroscopic medium-stock chambers (6 μm diameter and 8 μm depth) by a micropipette and then were delivered from these chambers into the cell culture microchambers by using a single pressure source. Each cell culture microchamber (180 μm depth) was connected to a medium-inlet branch channel (4 μm depth) and surrounded with terrace structure (65 μm depth) that helps remove air and prevent bubble formation in the microchambers while cell loading. The hydrodynamic simulation of velocity profile in the microchambers was done in COMSOL Multiphysics, and a design for a more uniform velocity profile was determined. Cytotoxicity tests of seven anticancer drugs on HeLa cells were performed and inhibition of cell growth was followed by Calcein-AM staining and fluorometric quantification. The results were compared with those of conventional 96-well plate and were found to be in good agreement. So, the integration of a microfluidic gradient generator to the chip enabled the achievement of a high-throughput, and also the cell loading can be managed easily and uniformly without dealing with the problem of bubble formation.
Using photolithography, Liu et al.69 developed a multilayer PDMS microdevice integrated with a ladder shaped CGG that is capable of assessing apoptosis of A549/DD cells due to Cisplatin (drug). The first layer of the microdevice consisted of pear-shaped microchambers in order to culture A549/DD cells and micropillars to prevent these cells from flowing through the outlet. In microdevices, mostly circular113 and rectangular114,115 microchambers were used. However, Liu et al.69 showed that a pear-shaped microchamber provided a more uniform fluid velocity which, in turn, resulted in uniform cell distribution. The fluid velocity was high at the center of the pear-shaped microchamber, and micropillars helped keep the cells at the center. Five cascaded mixing steps were found in the second layer. The flow rates of the merging solutions were controlled by changing the length of the micro channels which led to a concentration gradient generation in the microdevice. In the third layer, microvalves controlled the fluid (drug or medium) that should be introduced into the cell microchambers. The air inlets were located on the fourth layer and distributed the incoming air to the microvalves, but that air flow caused a deformation on the microvalve and plugged the microchambers. Each microchamber was loaded with cells by a syringe pump. Finally, a drug solution was loaded into the culture chamber after the microvalves were turned off. The cell viability was measured at different drug concentrations. Cell apoptosis was found to be weak in the microdevice due to the resistance of cells to the drug Cisplatin and absorption of drug molecules by PDMS. Nevertheless, the microdevice provided stable conditions where high throughput could be achieved, with less time and sample consumption.
In addition to the cytotoxicity assays of drugs, it is also important to characterize the drug metabolites to prevent undesirable biological results. The characterization of drug metabolites was reported in the previous studies;54 however, it was not possible to perform drug metabolite characterization and cytotoxicity simultaneously, and hence, they had a lesser high-throughput, which slowed down the drug discovery process. Mao et al.76 developed a microfluidic device (Figure 5(b)) that brought the metabolite characterization and cytotoxicity assay together. They designed and fabricated a PDMS microfluidic device with three layers, i.e., a 1 mm quartz layer embedded with separation microchannels and a three-microwell array using photolithography. The three-microwell array having human liver microsome (HLM) encapsulated in sol-gel used to produce drug metabolites where cell culture microchambers (diameter 1 mm and depth 40 μm) each having 9 pillars (diameter 150 μm) were used for cytotoxicity assays. As mentioned earlier, cell distribution is an important problem in the experiments performed in microfluidic devices. However, as in the work of Wang et al.,64 the cell distribution problem was prevented by placing pillars into the microchambers. Using this microdevice, UGT metabolism of AP and the metabolism based drug-drug interaction between AP and phenytoin (PH) were characterized where their cytotoxic effects on HepG2 cells were investigated using fluorescence LIVE/DEAD staining and following the cell viability. Ma et al.70 was able to bring the cytotoxicity assay and metabolite detection together by developing a microfluidic device integrated with ESI-Q-TOF mass spectroscopy. Drug metabolism, bioreactor, cytotoxicity assay, and integrated solid-phase extraction (SPE) columns were combined in a single microdevice. UGT metabolism of AP and the cytotoxicity of AP and acetaminophen-glucuronides (APG) on HepG2 cells were investigated. This microdevice provided less reagent consumption (4 μg HLM protein), high throughput, and fast analysis time (30 min).
Kim et al.71 managed to design a fully automatic and programmable microdevice for the purpose of investigating the cell viability of PC3 cells under the effect of a drug combination (doxorubicin and mitoxantrone) with tumor necrosis factor (TNF)-related-apoptosis-inducing ligand (TRAIL). The device was fabricated using soft-lithography, and it consisted of an array of 64 individual cell culture chambers and a concentration generator module. The concentration gradient was created using the method of diffusive mixing. The cell viability was detected by Calcein-AM LIVE/DEAD staining method, and the results were compared with conventional method 96-well plates. It was found that the cell viability was lower than those obtained in 96-well plates. This was explained as follows: (i) the media was refreshed and the waste of the process was removed continuously in the microdevice, whereas the media was introduced only at the beginning of the experiment and could not be removed, and, consequently, accumulated in the well plates. Therefore, the cell viability was not high in the microdevice as was the case in the conventional method. (ii) The cells were seeded into the microdevice more uniformly compared to the 96-well plates which led to higher cell growth rate and lower drug efficacy in the microdevice. (iii) Finally, the drug might have been absorbed into the PDMS, as this is one of the disadvantages of PDMS.
Normally, effects of drugs on cancer cells are tested in an environment at normal oxygen level. However, it was found that hypoxia (condition of oxygen deprivation) in cancer cells causes a resistance towards therapeutics. Therefore, the efficacy of drug candidates may give misleading results under normal oxygen concentrations. Considering this, Khanal et al.72 investigated the effect of a lack of oxygen on the apoptosis of PC3 and Ramos B cells in response to the drug staurosporine. Apoptosis assay was performed in a microfluidic chip that was fabricated by soft lithography. The chip consisted of 256 cell culture chambers each having a volume of 5.7 nl and could contain 10–50 cells. Apoptosis was successfully detected by Mitotracker and Annexin-V stained fluorescent method.
Apart from testing of candidate drugs, microdevices are also preferred in the usage of mimicking the environment of a disease in in vitro conditions. As an example of this case, Seidi et al.73 designed a microdevice to model Parkinson's disease in vitro. Parkinson's disease is caused by the loss of PC12 cells due to apoptosis. To selectively damage the dopaminergic neurons, a synthetic compound, 6-hydroxydopamine (6-OHDA), is used in research. Thus, new drug candidates and treatments can be developed for in vitro modeled cell apoptosis. Seidi et al.73 developed a CGG in a microchannel of dimensions 100 μm (height) × 30 mm (length) × 2 mm (width). To create a concentration gradient, the microchannel was first filled with a water solution or a culture medium, and then a second solution (6-OHDA) was introduced from the inlet, while the outlet was emptied by a micropipette, creating a forward flow. Finally, the forward flow was reversed by placing a drop of the first solution into the outlet and emptying 10 μl of fluid from the inlet. The cell viability was detected by LIVE/DEAD cell staining kit. Neurons can be damaged either by apoptosis or by necrosis depending on the concentration level of 6-OHDA. To assess the concentration ranges of these cell death processes in the microdevice, Annexin (apoptosis) and PI (necrosis) stainings were used, and the ratio of apoptosis/necrosis rates of neurons were obtained. The microdevice of Seidi et al.73 gave similar results to the conventional well plate method but managed to create and stabilize a concentration gradient in less than 30 s and provided the usual benefits of microdevices, i.e., low cost, high throughput, and simple operation.
VI. MINIATURIZED ENZYMATIC ASSAYS WITH POTENTIAL USE IN DRUG DISCOVERY/SCREENING RESEARCH
Microdevices offer a great opportunity to develop new drugs and to investigate drug efficacy and cytotoxicity. Moreover, microfluidic devices can also be used for enzyme assays.116 Enzyme assays are measurements of the rate of reactions and may be applied to drug screening research to test the effect of inhibitors on the enzyme. The enzyme inhibition assays have been generally performed using the model system of β-gal enzyme, its fluorogenic substrate, resorufin-β-D-galactopyranoside (RBG), and the inhibitors like phenylethyl β-D-thiogalactoside (PETG), lactose, galactose, etc. The pioneers in that area, Hadd et al.,78 developed the first microchip device for enzyme assays, which had a potential use in drug discovery, disease diagnostics, and biochemical detection. As early as 1997 (before the chip of 1999 mentioned in Section III A), a microchip with dimensions of 9 × 35 μm (depth × width) was fabricated using photolithography and wet chemical etching, where the model β-gal enzyme system and inhibitor, PETG, were controlled using electrokinetic flow. The microchip includes mixing and reaction channels with 3 inlets for substrate, enzyme and -inhibitor. A potential was applied to channel terminuses to dilute and mix reagents at required concentrations. For reaction kinetics, resorufin, the hydrolysis product of RBG, was detected using laser-induced fluorescence, and Michaelis-Menten constants were obtained in the presence and absence of the inhibitor PETG. The results were compared with those of a conventional enzyme assay and found to be in agreement. Using a microchip, enzyme assay could be performed in 20 min, and reagent consumption was decreased by 4 orders of magnitude (120 pg enzyme and 75 ng substrate) compared to the conventional method.
A. Enzyme inhibition assays using droplet based microfluidics
In recent years, the use of microdroplets has become important for biological and chemical purposes.117
1. Mobile droplet arrays
Damean et al.79 developed a PDMS based microfluidic device that could generate mobile microdroplets in parallel (Figure 5(c)). At the beginning of the bottom layer of this device, a set of serpentine shaped microchannels (50 μm deep × 50 μm wide) were used to obtain a concentration gradient (4 different concentrations). The fluid at different concentrations was mixed with the second fluid by a passive micromixer. Then, strings of microdroplets with identical volumes (5–60 pl) were generated by flow focusing method. The desired droplet size was obtained by manipulating the flow rates. A chaotic advection was achieved in the microdroplets to enhance the mixing of the reagents, with the help of serpentine shaped microchannels. Multiple reactions can be performed simultaneously with this microfluidic device, and the gradient of concentration profile can be increased. To test the device, Damean et al.79 performed an enzymatic assay, hydrolysis reaction of fluorescein diphosphate by E. coli, and Michaelis-Menten constants were obtained and found to be in agreement with the results of 96-well plate method.
Garcia et al.80 developed a continuous flow microfluidic method of assaying enzyme inhibition and estimating IC50 values. This assay requires feeding a fluorogenic or colorimetric substrate and the same concentration of substrate and an inhibitor through two inlets of a Y-shaped microchannel. The speed of the solution in the main channel was 74 μm/s. The inhibitor was given 7 mm length for the molecular diffusion to occur and to generate a concentration gradient of the inhibitor after which the reaction of the enzyme with the substrate took place. The enzyme was deposited as microdroplets of 100 μm diameter on the glass which was already precoated with silanes terminated with aldehyde groups. Using the model system of β-gal enzyme and the competitive inhibitors, lactose and galactose, the microfluidic inhibition assay took 2 min, and the IC50 results agreed perfectly with those of conventional 96-well plate microtiter plate method.
Clausell-Tormos et al.11 also investigated an enzymatic inhibition reaction using droplet-based microfluidics. They generated 5 μl of plugs by injecting samples into a tubing with 300 μm inner diameter. Each plug was split into eight equal plugs (625 nl), and then they were met with 188 nl of enzyme and substrate in the chamber resulting in an 813 nl plug. Inhibition of β-gal enzyme was investigated with this device, and IC50 value of PETG (inhibitor) was obtained. This method can create a minimal plug volume of 150 nl at low cost and can decrease the assay volume 1000-fold compared to 96-well plate. Cai et al.81 also preferred mobile microdroplets with a concentration gradient and used this method to investigate enzyme inhibition. They combined flow injection gradient (FIG) technique with droplet-based microfluidics. In conventional FIG applications, a sample in microliter scale is needed, which can be high for biological assays like drug screening. However, Cai et al.81 managed to decrease the sample consumption 1000-fold by applying FIG technique to droplet microfluidics. A microfluidic device with a microchannel (depth 30 μm and width 150 μm) was fabricated by photolithography and wet etching. The inhibitor was injected into the carrier fluid creating a concentration gradient. This solution was merged with enzyme and substrate at the first channel junction and then dispersed into oil resulting in microdroplets at the second channel junction. Inhibition of β-gal by PETG and diethylene-triaminepentaacedic acid (DTPA) inhibitors was performed in 2–5 min by consuming a 16 nl sample and creating a concentration gradient with 3–4 orders of magnitude using FIG technique. IC50 values of inhibitors, PETG and DTPA, were determined by fluorescent detection technique. Sjostrom et al.82 used droplet picoinjectors to investigate enzyme inhibition on a PDMS-glass microfluidic chip. The microchip was fabricated by two-layer soft-lithography, and it consisted of two circuits. The first circuit was used to create 23 pl droplets at a speed of 1200 droplets/s. The droplets included enzyme (β-gal) and different concentrations of inhibitor (PETG). At the second circuit, fluorogenic substrate (RBG) was injected into the droplets with the help of electrically controlled picoinjector. Using this microchip, enzyme kinetics of β-gal and its inhibition were studied, and Michaelis-Mentens parameters were determined. This microchip was found to be a promising device for drug screening purposes due to its ability to achieve high-throughput and monitor different conditions (different inhibitors and concentrations) simultaneously using a low sample amount.
2. Static droplet systems
Du et al.84 used droplet-based microfluidics to investigate enzyme inhibition without fabricating microchips or the microchannel network. A 6 cm long capillary with tapered tip (25–30 μm inner diameter) was used to create micro-to-pico scaled static droplets at different concentrations, and the generated droplets were stored in a nanowell plate. Du et al.84 were able to generate 400 droplets with a minimal volume of 20 pl at a speed of 4.5 s/droplet. Inhibition of β-gal by DTPA (inhibitor) was performed in 2 nl droplets, and IC50 values were obtained for different reagent compositions. This method was not capable of producing droplets at a higher speed. However, the system was simple and did not require any microfabrication techniques. On the other hand, the DOD method adopted by Gielen et al.83 (see Section III C) is fully unsupervised and gives precise control over the experiments. Gielen et al.83 obtained more data points with a 1000-fold sample reduction and performed pipetting using 20 times fewer steps, since the liquid handling was automated during the experiments. The concentration gradient generated in mobile droplets (2 orders of magnitude) was not as high as the gradient achieved in static microdroplets. Sun et al.85 also worked with static droplets using a concentration gradient. They developed a microfluidic device with a microchannel network. There existed repeated loops in this network where each loop included a bypass channel connected to a microfluidic trap with a higher hydrodynamic resistance (i.e., bypass channel had a lower resistance than the trap). First, a sample plug was aspirated into a cartridge. Then, the sample plug was introduced into the microfluidic device. When all of the loops were filled with the sample plug, a diluting sample (water) was introduced into the device. The plug diluted the first drop of the loop and the next drops were diluted with diluting solution, which had an increasing amount of sample material, as the solution moved down the microchannel network. Thus, a desired concentration gradient was achieved by changing only the concentration of the first sample plug. Using four diluting plugs, 100 000-fold dilution of the first sample plug compared to the last drop was achieved. By this method, it was also possible to add or remove a reagent from a droplet. Sun et al.85 managed to achieve a five orders of magnitude greater concentration gradient in 1–5 min using static droplets; Dose-response investigations and cytotoxicity assays can be performed with this microfluidic device, as a result of its ability to get a high range of concentration gradient in a short period of time and to consume less amount of sample (60 different concentrations in 20 nl droplets, total sample consumption 2 μl).
Luk et al.86 investigated proteomic applications by digital microfluidics (Figure 2(a)). They combined digital microfluidics with hydrogel. The digital microfluidics device was fabricated by photolithography and etching and had 68 square actuation electrodes (2 mm × 2 mm). Enzymes (trypsin and pepsin) were immobilized in agarose gel discs (1 mm diameter and 140 μm height). Gel discs were placed between the top and bottom plates, and the droplets containing protein were actuated between these plates by a potential of 200–250 Vpp. As the sample droplet was actuated, the droplet was moved onto the gel disc and the digestion occurred. The digestion level of each enzyme was quantified by mass spectroscopy. By combining hydrogel with digital microfluidics, the droplets can be easily manipulated and sent to the desired location on the hydrogel. In addition, since the droplets can be split into daughter droplets, the droplets can be filled with multiple enzymes and delivered to multiple locations for parallel digestion assays.
B. Miscellaneous systems
Microfluidics can also be used in an automated platform for the determination of an enzyme's molecular weight, amount, and activity. It is usually difficult to measure enzyme activities due to their low abundance. Using techniques like pore-limit electrophoresis (PLE) allows sensitivities at exceptionally low levels, reaching zeptomoles. Hughes and Herr87 employed a single microchannel chip to use a two-step gradient-gel zymography for measuring calf intestinal alkaline phosphatase (CIP) (500 pM stock solutions) and horseradish peroxidase (HRP). They fabricated glass chips using wet-etching, and four straight channels with a depth of 10 μm and a width of 70 μm per chip were etched. Channels were cleaned, and then functional hydrogels were patterned. Once the channels were ready, first, molecule sizing and pseudoimmobilization of the enzymes were achieved thanks to polyacrylamide gel density gradients and PLE; then, the substrates were loaded on the same channel, electrophoretically transported along the separation channel with immobilized enzymes and products were formed and detected due to the fluorescence of the product. The assay took around 40 min, and the detection limit was 5 zmol (∼3000 molecules) of CIP. The catalytic activities of the enzymes were increased greatly as the enzymes were not chemically immobilized.
Cancer can be treated by removing or killing tumor cells. However, the drugs used for treatments are toxic, and further investigation is required before their application as a medical treatment. One of the methods to treat cancer is differentiation therapy (a treatment method by differentiating cancer cells and restraining cell growth). For this purpose, Popovtzer et al.88 used butyric acid (a differentiation agent) to modify HT-29 cells, and enzymatic activity of alkaline phosphatase was measured to understand the efficacy of the differentiation agent. They designed a microchip that consisted of an array of eight electrochemical cells. Each cell having a volume of 10 nl was equipped with three electrodes. A potential was applied to working and reference electrodes to quantify the enzyme activity, and the current was measured when the product of enzymatic reaction, p-aminophenylphosphate (PAP), was oxidized on the working electrode. With this electrochemical microchip, high throughput was achieved by performing multiple experiments in parallel. The diffusion distance of PAP molecules became shorter due to the high surface-area-to-volume ratio of the microchip. Thus, the response time was decreased when the sensitivity was increased in the experiments.
Conventional microwell plates are frequently used for dose dependent drug research. Microwell plates provide high throughput, but application of microwell plates can be expensive and difficult to miniaturize. Considering this situation, Upadhyaya and Selvaganapathy89 designed and fabricated a PDMS microfluidic array by photolithography that consisted of three layers: agar gel on the top layer, nanoporous interface membrane (8 μm thickness) in the middle, and two microchannels (width 0.5 mm, spacing 2 mm) at the bottom layer of each drug spot. Platinum wires were placed in the reservoirs of one of the microchannels, and an electric potential (Ex. 30 V for 30 s) was applied through the wires to pump the desired dose of drug from the microchannel to the agar gel. This microfluidic device gave a precise control on drug dose with an accuracy of 50 μg. A density of 156 drug spots/cm2 (15 000 spots/plate) was achieved which is higher than the densities of conventional 384-well and 1536-well plates. This device has a potential use for drug discovery research with fast response time regardless of drug characteristics.
VII. FUTURE STRATEGIES
Microfluidic technology is still a new field for drug discovery applications. The focus of these applications has been to develop a microfluidic device that can be fabricated at a low cost and to enable high-throughput in drug screening, cytotoxicity analysis, drug-dose response measurements, protein-drug (ligand) binding, and drug targeted enzyme inhibition.
The challenges such as creating an extracellular in vivo environment exactly in a microfluidic device and correlating extracellular in vitro results with those of intracellular in vivo systems are expected to enhance research with better outcomes. Furthermore, due to ethical issues regarding animal testing, there is a trend towards in vitro microfluidic systems such as tissue-on-a-chip and organ-on-a-chip. The ultimate aim is to connect these microengineered systems to form human-on-a-chip. The NIH and the FDA have invested more than 100 × 106 US dollars to create the tissue chips for drug screening.118,119 The goal of replacing animal studies with in vitro systems leads to the development of SEURAT-15 and the Body-on-a-Chip6 in Europe. In order to first reduce and then to replace animal studies, 3D microengineered tissue systems mimicking the human physiological environment and the pharmacological response will take over in the near future. Microphysiological Systems (MPS) technology, an improved approach for more predictive preclinical drug discovery via a highly integrated experimental/computational paradigm, is thus another challenge in microfluidic systems. Griffith lab developed Human Physiome on a Chip, where they merged tissue engineering and systems pharmacology.120
In addition to ethical issues, in vitro microfluidic drug screening platforms allow for high-throughput and reproducible screening with little to no batch-to-batch variation at a relatively lower cost compared to in vivo animal models.8 3D cultures in organ-on-a-chip systems are capable of mimicking native tissues and can better represent in vivo cellular responses to drug treatment. These cell-based high throughput screening (HTS) platforms provide more relevant in vivo biological information than biochemical assay, help reduce the number of animal tests, and accelerate the drug discovery process.121 The drugs, Eltrombopag (Promacta/Revolade; GlaxoSmithKline), a TPO receptor against BMS-790052 hepatitis C virus (HCV) NS5A (Bristol-Myers Squibb), and Bortezomib, used in cancer chemotherapy are some of the successful commercial examples developed by cell-based HTS.121 Furthermore, Janssen Biotech, Inc., and Emulate are in a collaboration to develop a Thrombosis-on-Chip platform where human response will be modeled in vitro environment and drug candidates causing thrombosis will be tested. The drug candidates that are obtained from Thrombosis-on-Chip assays can be used in human clinical trials and increase the success of them.122
Another aspect of the use of 3D systems is patients' tumor heterogeneity, which necessitates tailoring chemotherapies to individual patients. Hence biopsies, which preserve the tumor microenvironment, may be used in microfluidic platforms for drug screening and can provide quick solutions for personalized therapies.
The key to improving the lives of millions of people worldwide is stated to be automation, i.e., robot systems, reducing the time and expenses involved in screening. One such example is Eve, the first AI-robot scientist, that has been used in the identification of new drug candidates for infectious (e.g., malaria, Chagas disease) and tropical diseases (e.g., African sleeping sickness). By means of artificial intelligence, the robot Eve has learned from early successes in her screens and selected compounds that had a high probability of being active against a chosen drug target. Mankind, which is both exposed and prone to severe or life-threatening illnesses, will reap significant benefits from automation, which could speed up new drug discoveries and potentially improve the lives of millions of people worldwide.123
The future focus is to stabilize and automate current microfluidic devices and integrate them into the pharmaceutical industry and also to develop new application areas beyond diagnosis, analysis, and therapy. Eventually, these microfluidic devices may speed up the cumbersome and time consuming drug discovery processes for lethal and rare diseases and reduce the cost of treatment.
ACKNOWLEDGMENTS
This work was financially supported by the Bogazici University Research Grants 9701 R (KOU) and 8085 (AKU).
References
- 1. Whitesides G. M., Nature 442, 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- 2. Manz A., Graber N., and Widmer H. M., Sens. Actuators, B 1, 244 (1990). 10.1016/0925-4005(90)80209-I [DOI] [Google Scholar]
- 3. Volpatti L. R. and Yetisen A. K., Trends Biotechnol. 32, 347 (2014). 10.1016/j.tibtech.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 4. Yetisen A. K. and Volpatti L. R., Lab Chip 14, 2217 (2014). 10.1039/c4lc00399c [DOI] [PubMed] [Google Scholar]
- 5.See http://www.seurat-1.eu/ for SEURAT-1-Towards the Replacement of in vivo Repeated Dose Systemic Toxicity Testing (2015).
- 6.See http://cordis.europa.eu/project/rcn/104027_en.html for European Commission CORDIS. Projects & Results Service: The Body-on-a-Chip (BoC) (2015).
- 7. Zhang C., Xu J., Ma W., and Zheng W., Biotechnol. Adv. 24, 243 (2006). 10.1016/j.biotechadv.2005.10.002 [DOI] [PubMed] [Google Scholar]
- 8. Tsui J. H., Lee W., Pun S. H., Kim J., and Kim D. H., Adv. Drug Delivery Rev. 65, 1575 (2013). 10.1016/j.addr.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hassan U. and Bashir R., Biomed. Microdevices 16, 697 (2014). 10.1007/s10544-014-9874-0 [DOI] [PubMed] [Google Scholar]
- 10. Abate A. R., Hung T., Sperling R. A., Mary P., Rotem A., Agresti J. J., Weiner M., and Weitz D., Lab Chip 13, 4864 (2013). 10.1039/c3lc50905b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clausell-Tormos J., Griffiths A. D., and Merten C. A., Lab Chip 10, 1302 (2010). 10.1039/b921754a [DOI] [PubMed] [Google Scholar]
- 12. El-Ali J., Sorger P. K., and Jensen K. F., Nature 442, 403 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- 13. Craighead H., Nature 442, 387 (2006). 10.1038/nature05061 [DOI] [PubMed] [Google Scholar]
- 14. Wu M.-H., Huang S.-B., and Lee G.-B., Lab Chip 10, 939 (2010). 10.1039/b921695b [DOI] [PubMed] [Google Scholar]
- 15. Duncombe T. A., Tentori A. M., and Herr A. E., Nat. Rev. Mol. Cell Biol. 16, 554 (2015). 10.1038/nrm4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nguyen N.-T., Shaegh S. A. M., Kashaninejad N., and Phan D.-T., Adv. Drug Delivery Rev. 65, 1403 (2013). 10.1016/j.addr.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 17. Esch M. B., King T. L., and Shuler M. L., Annu. Rev. Biomed. Eng. 13, 55 (2011). 10.1146/annurev-bioeng-071910-124629 [DOI] [PubMed] [Google Scholar]
- 18. Xu F., Wu J., Wang S., Durmus N. G., Gurkan U. A., and Demirci U., Biofabrication 3, 034101 (2011). 10.1088/1758-5082/3/3/034101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neuži P., Giselbrecht S., Länge K., Huang T. J., and Manz A., Nat. Rev. Drug Discovery 11, 620 (2012). 10.1038/nrd3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vladisavljević G. T., Khalid N., Neves M. A., Kuroiwa T., Nakajima M., Uemura K., Ichikawa S., and Kobayashi I., Adv. Drug Delivery Rev. 65, 1626 (2013). 10.1016/j.addr.2013.07.017 [DOI] [PubMed] [Google Scholar]
- 21. Zheng X. T., Yu L., Li P., Dong H., Wang Y., Liu Y., and Li C. M., Adv. Drug Delivery Rev. 65, 1556 (2013). 10.1016/j.addr.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 22. Usta O. B., McCarty W. J., Bale S., Hegde M., Jindal R., Bhushan A., Golberg I., and Yarmush M. L., Technology 3, 1 (2015). 10.1142/S2339547815300012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matosevic S., Szita N., and Baganz F., J. Chem. Technol. Biotechnol. 86, 325 (2011). 10.1002/jctb.2564 [DOI] [Google Scholar]
- 24. Kintses B., van Vliet L. D., Devenish S. R., and Hollfelder F., Curr. Opin. Chem. Biol. 14, 548 (2010). 10.1016/j.cbpa.2010.08.013 [DOI] [PubMed] [Google Scholar]
- 25. Bhatia S. N. and Ingber D. E., Nat. Biotechnol. 32, 760 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 26. DiMasi J., see http://csdd.tufts.edu/files/uploads/Tufts_CSDD_briefing_on_RD_cost_study_-_Nov_18,_2014.pdf for Cost of Developing a New Drug (2014).
- 27. Petrova E., in Innovation and Marketing in the Pharmaceutical Industry, edited by Ding S., Eliashberg M., and Stremersch J. ( Springer, New York, 2014), pp. 19–81. [Google Scholar]
- 28.See http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM073381.pdf for FDA Q2A Text on Validation of Analytical (2015).
- 29.See http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM073384.pdf for FDA Q2B Validation of Analytical Procedures: Methodology (2015).
- 30. Lee D. W., Lee M.-Y., Ku B., and Nam D.-H., J. Biomol. Screening 20, 1178 (2015). 10.1177/1087057115597635 [DOI] [PubMed] [Google Scholar]
- 31. Perrin D., Fremaux C., Besson D., Sauer W. H., and Scheer A., J. Biomol. Screening 11, 996 (2006). 10.1177/1087057106294094 [DOI] [PubMed] [Google Scholar]
- 32. Wright B. D., Simpson C., Stashko M., Kireev D., Hull-Ryde E. A., Zylka M. J., and Janzen W. P., J. Biomol. Screening 20, 655 (2015). 10.1177/1087057114564057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.See supplementary material at http://dx.doi.org/10.1063/1.4940886E-BIOMGB-10-013601 for Table S1 giving detailed information on drug screening processes and the list of abbreviations.
- 34. Hadd A. G., Jacobson S. C., Ramsey J. M., and Chem H. M. A., Anal. Chem. 71, 5206 (1999). 10.1021/ac990591f [DOI] [Google Scholar]
- 35. Cohen C. B., Chin-Dixon E., Jeong S., and Nikiforov T. T., Anal. Biochem. 273, 89 (1999). 10.1006/abio.1999.4204 [DOI] [PubMed] [Google Scholar]
- 36. Puckett L. G., Dikici E., Lai S., Madou M., Bachas L. G., and Daunert S., Anal. Chem. 76, 7263 (2004). 10.1021/ac049758h [DOI] [PubMed] [Google Scholar]
- 37. Wang Y., Lin W.-Y., Liu K., Lin R. J., Selke M., Kolb H. C., Zhang N., Zhao X.-Z., Phelps M. E., Shen C. K. F., Faull K. F., and Tseng H.-R., Lab Chip 9, 2281 (2009). 10.1039/b907430a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pihl J., Sinclair J., Sahlin E., Karlsson M., Petterson F., Olofsson J., and Orwar O., Anal. Chem. 77, 3897 (2005). 10.1021/ac050218+ [DOI] [PubMed] [Google Scholar]
- 39. Horiuchi K. Y., J. Biomol. Screening 11, 48 (2005). 10.1177/1087057105282097 [DOI] [PubMed] [Google Scholar]
- 40. Mangru S., J. Biomol. Screening 10, 788 (2005). 10.1177/1087057105278927 [DOI] [PubMed] [Google Scholar]
- 41. Iyer J. K., Otvos R. A., Kool J., and Kini R. M., J. Biomol. Screening 21, 212 (2016) 10.1177/1087057115602648. [DOI] [PubMed] [Google Scholar]
- 42. Lombardi D. and Dittrich P. S., Anal. Bioanal. Chem. 399, 347 (2011). 10.1007/s00216-010-4302-7 [DOI] [PubMed] [Google Scholar]
- 43. Miller O. J., El A., Mangeat T., Baret J., Frenz L., El B., Mayot E., Samuels M. L., Rooney E. K., Dieu P., Galvan M., Link D. R., and Griffiths A. D., Proc. Natl. Acad. Sci. U.S.A. 109, 378 (2012). 10.1073/pnas.1113324109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gu S., Lu Y., Ding Y., Li L., Zhang F., and Wu Q., Anal. Chim. Acta 796, 68 (2013). 10.1016/j.aca.2013.08.016 [DOI] [PubMed] [Google Scholar]
- 45. Litten B., Blackett C., Wigglesworth M., Goddard N., and Fielden P., Biomicrofluidics 9, 052607 (2015). 10.1063/1.4927324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brouzes E., Medkova M., Savenelli N., Marran D., Twardowski M., Hutchison J. B., Rothberg J. M., Link D. R., Perrimon N., and Samuels M. L., Proc. Natl. Acad. Sci. U.S.A. 106, 14195 (2009). 10.1073/pnas.0903542106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Viravaidya K. and Shuler M. L., Biotechnol. Prog. 20, 590 (2004). 10.1021/bp034238d [DOI] [PubMed] [Google Scholar]
- 48. Viravaidya K., Sin A., and Shuler M. L., Biotechnol. Prog. 20, 316 (2004). 10.1021/bp0341996 [DOI] [PubMed] [Google Scholar]
- 49. Tirella A., Marano M., Vozzi F., and Ahluwalia A., Toxicol. In Vitro 22, 1957 (2008). 10.1016/j.tiv.2008.09.016 [DOI] [PubMed] [Google Scholar]
- 50. Toh Y.-C., Lim T. C., Tai D., Xiao G., van Noort D., and Yu H., Lab Chip 9, 2026 (2009). 10.1039/b900912d [DOI] [PubMed] [Google Scholar]
- 51. Yang F., Chen Z., Pan J., Li X., Feng J., and Yang H., Biomicrofluidics 5, 024115 (2011) 10.1063/1.3605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim M. J., Lee S. C., Pal S., Han E., and Song J. M., Lab Chip 11, 104 (2011). 10.1039/C0LC00110D [DOI] [PubMed] [Google Scholar]
- 53. Su X., Young E. W. K., Underkofler H. A. S., Kamp T. J., January C. T., and Beebe D. J., J. Biomol. Screening 16, 101 (2011). 10.1177/1087057110386218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee M.-Y., Park C. B., Dordick J. S., and Clark D. S., Proc. Natl. Acad. Sci. U.S.A. 102, 983 (2005). 10.1073/pnas.0406755102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee M.-Y., Kumar R. A., Sukumaran S. M., Hogg M. G., Clark D. S., and Dordick J. S., Proc. Natl. Acad. Sci. U.S.A. 105, 59 (2008). 10.1073/pnas.0708756105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee D. W., Lee M. Y., Ku B., Yi S. H., Ryu J. H., Jeon R., and Yang M., Arch. Toxicol. 88, 283 (2014). 10.1007/s00204-013-1102-9 [DOI] [PubMed] [Google Scholar]
- 57. Ye N., Qin J., Shi W., Liu X., and Lin B., Lab Chip 7, 1696 (2007). 10.1039/b711513j [DOI] [PubMed] [Google Scholar]
- 58. Kim M. S., Yeon J. H., and Park J. K., Biomed. Microdevices 9, 25 (2007). 10.1007/s10544-006-9016-4 [DOI] [PubMed] [Google Scholar]
- 59. Kim M. S., Lee W., Kim Y. C., and Park J.-K., Biotechnol. Bioeng. 101, 1005 (2008). 10.1002/bit.21962 [DOI] [PubMed] [Google Scholar]
- 60. Sung J. H. and Shuler M. L., Lab Chip 9, 1385 (2009). 10.1039/b901377f [DOI] [PubMed] [Google Scholar]
- 61. Chao P., Maguire T., Novik E., Cheng K. C., and Yarmush M. L., Biochem. Pharmacol. 78, 625 (2009). 10.1016/j.bcp.2009.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim M. J., Lim K. H., Yoo H. J., Rhee S. W., and Yoon T. H., Lab Chip 10, 415 (2010). 10.1039/b920890a [DOI] [PubMed] [Google Scholar]
- 63. Lim K. H., Park J., Rhee S. W., and Yoon T. H., Cytometry, Part A 81, 691 (2012). 10.1002/cyto.a.22079 [DOI] [PubMed] [Google Scholar]
- 64. Wang Z., Kim M.-C., Marquez M., and Thorsen T., Lab Chip 7, 740 (2007). 10.1039/b618734j [DOI] [PubMed] [Google Scholar]
- 65. Komen J., Wolbers F., Franke H. R., Andersson H., Vermes I., and vam den Berg A., Biomed. Microdevices 10, 727 (2008). 10.1007/s10544-008-9184-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu D., Wang L., Zhong R., Li B., Ye N., Liu X., and Lin B., J. Biotechnol. 131, 286 (2007). 10.1016/j.jbiotec.2007.06.014 [DOI] [PubMed] [Google Scholar]
- 67. Jedrych E., Flis S., Sofinska K., Jastrzebski Z., Chudy M., Dybko A., and Brzozka Z., Sens. Actuators, B 160, 1544 (2011). 10.1016/j.snb.2011.08.074 [DOI] [Google Scholar]
- 68. Sugiura S., Edahiro J. I., Kikuchi K., Sumaru K., and Kanamori T., Biotechnol. Bioeng. 100, 1156 (2008). 10.1002/bit.21836 [DOI] [PubMed] [Google Scholar]
- 69. Liu C., Wang L., Xu Z., Li J., Ding X., Wang Q., and Chunyu L., J. Micromech. Microeng. 22, 065008 (2012). 10.1088/0960-1317/22/6/065008 [DOI] [Google Scholar]
- 70. Ma B., Zhang G., Qin J., and Lin B., Lab Chip 9, 232 (2009). 10.1039/B809117J [DOI] [PubMed] [Google Scholar]
- 71. Kim J., Taylor D., Agrawal N., Wang H., Kim H., Han A., Rege K., and Jayaraman A., Lab Chip 12, 1813 (2012). 10.1039/c2lc21202a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Khanal G., Hiemstra S., and Pappas D., Analyst 139, 3274 (2014). 10.1039/C3AN02324A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Seidi A., Kaji H., Annabi N., Ostrovidov S., Ramalingam M., and Khademhosseini A., Biomicrofluidics 5, 022214 (2011). 10.1063/1.3580756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hsiung L.-C., Chiang C.-L., Wang C.-H., Huang Y.-H., Kuo C.-T., Cheng J.-Y., Lin C.-H., Wu V., Chou H.-Y., Jong D.-S., Lee H., and Wo A. M., Lab Chip 11, 2333 (2011). 10.1039/c1lc20147f [DOI] [PubMed] [Google Scholar]
- 75. Bogojevic D., Chamberlain M. D., Barbulovic-Nad I., and Wheeler A. R., Lab Chip 12, 627 (2012). 10.1039/C2LC20893H [DOI] [PubMed] [Google Scholar]
- 76. Mao S., Gao D., Liu W., Wei H., and Lin J.-M., Lab Chip 12, 219 (2012). 10.1039/C1LC20678H [DOI] [PubMed] [Google Scholar]
- 77. Song H., Chen T., Zhang B., Ma Y., and Wang Z., Biomicrofluidics 4, 44104 (2010). 10.1063/1.3497376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hadd A. G., Raymond D. E., Halliwell J. W., Jacobson S. C., and Ramsey J. M., Anal. Chem. 69, 3407 (1997). 10.1021/ac970192p [DOI] [PubMed] [Google Scholar]
- 79. Damean N., Olguin L. F., Hollfelder F., Abell C., and Huck W. T. S., Lab Chip 9, 1707 (2009). 10.1039/b821021g [DOI] [PubMed] [Google Scholar]
- 80. Garcia E., Hasenbank M. S., Finlayson B., and Yager P., Lab Chip 7, 249 (2007). 10.1039/B608789B [DOI] [PubMed] [Google Scholar]
- 81. Cai L. F., Zhu Y., Du G. S., and Fang Q., Anal. Chem. 84, 446 (2012). 10.1021/ac2029198 [DOI] [PubMed] [Google Scholar]
- 82. Sjostrom S. L., Joensson H. N., and Svahn H. A., Lab Chip 13, 1754 (2013). 10.1039/c3lc41398e [DOI] [PubMed] [Google Scholar]
- 83. Gielen F., Van Vliet L., Koprowski B. T., Devenish S. R., Fischlechner M., Edel J. B., Niu X., Demello A. J., and Hollfelder F., Anal. Chem. 85, 4761 (2013). 10.1021/ac400480z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bin Du W., Sun M., Gu S. Q., Zhu Y., and Fang Q., Anal. Chem. 82, 9941 (2010). 10.1021/ac1020479 [DOI] [PubMed] [Google Scholar]
- 85. Sun M., Bithi S. S., and Vanapalli S. A., Lab Chip 11, 3949 (2011). 10.1039/c1lc20709a [DOI] [PubMed] [Google Scholar]
- 86. Luk V. N., Fiddes L. K., Luk V. M., Kumacheva E., and Wheeler A. R., Proteomics 12, 1310 (2012). 10.1002/pmic.201100608 [DOI] [PubMed] [Google Scholar]
- 87. Hughes A. J. and Herr A. E., Anal. Chem. 82, 3803 (2010). 10.1021/ac100201z [DOI] [PubMed] [Google Scholar]
- 88. Popovtzer R., Neufeld T., Popovtzer A., Rivkin I., Margalit R., Engel D., Nudelman A., Rephaeli A., Rishpon J., and Shacham-Diamand Y., Nanomed. Nanotechnol., Biol. Med. 4, 121 (2008). 10.1016/j.nano.2008.03.002 [DOI] [PubMed] [Google Scholar]
- 89. Upadhyaya S. and Selvaganapathy P. R., Lab Chip 10, 341 (2010). 10.1039/B918291H [DOI] [PubMed] [Google Scholar]
- 90. Werner M., Kuratli C., Martin R. E., Hochstrasser R., Wechsler D., Enderle T., Alanine A. I., and Vogel H., Angew. Chem., Int. Ed. 53, 1704 (2014). 10.1002/anie.201309301 [DOI] [PubMed] [Google Scholar]
- 91. Pennathur S., Meinhart C. D., and Soh H. T., Lab Chip 8, 20 (2008). 10.1039/B717986N [DOI] [PubMed] [Google Scholar]
- 92. Whitesides G. M., Ostuni E., Jiang X., and Ingber D. E., Annu. Rev. Biomed. Eng. 3, 335 (2001). 10.1146/annurev.bioeng.3.1.335 [DOI] [PubMed] [Google Scholar]
- 93. Capretto L., Cheng W., Hill M., and Zhang X., Top. Curr. Chem. 11, 27 (2011). 10.1007/128_2011_150 [DOI] [PubMed] [Google Scholar]
- 94. DeMello A. J., Nature 442, 394 (2006). 10.1038/nature05062 [DOI] [PubMed] [Google Scholar]
- 95. Dittrich P. S. and Manz A., Nat. Rev. Drug Discovery 5, 210 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- 96. Toh A. G. G., Wang Z. P., Yang C., and Nguyen N. T., Microfluid. Nanofluid. 16, 1 (2014). 10.1007/s10404-013-1236-3 [DOI] [Google Scholar]
- 97. Dertinger S. K. W., Chiu D. T., Jeon N. L., and Whitesides G. M., Anal. Chem. 73, 1240 (2001). 10.1021/ac001132d [DOI] [Google Scholar]
- 98. Irimia D., Geba D. A., and Toner M., Anal. Chem. 78, 3472 (2006). 10.1021/ac0518710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhou B., Xu W., and Wang C., Microfluid. Nanofluid. 18, 175 (2015). 10.1007/s10404-014-1432-9 [DOI] [Google Scholar]
- 100. de Gennes P. G., Adv. Colloid Interface Sci. 27, 189 (1987). 10.1016/0001-8686(87)85003-0 [DOI] [Google Scholar]
- 101. Pollack M. G., Pamula V. K., Srinivasan V., and Eckhardt A. E., Expert Rev. Mol. Diagn. 11, 393 (2011). 10.1586/erm.11.22 [DOI] [PubMed] [Google Scholar]
- 102. Lee P. J., Ghorashian N., Gaige T. A., and Hung P. J., JALA 12, 363 (2007) 10.1016/j.jala.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Becker H., Lab Chip 10, 3197 (2010). 10.1039/c0lc90065f [DOI] [PubMed] [Google Scholar]
- 104. Madou M. J., Fundamentals of Microfabrication and Nanotechnology, 3rd ed. ( CRC Press, Boca Raton, FL, 2012). [Google Scholar]
- 105. Cohen P., Nat. Rev. Drug Discovery 1, 309 (2002). 10.1038/nrd773 [DOI] [PubMed] [Google Scholar]
- 106. Zhou Y. and Lin Q., Sens. Actuators, B 190, 334 (2014). 10.1016/j.snb.2013.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bardin D., Kendall M. R., Dayton P. A., and Lee A. P., Biomicrofluidics 7, 034112 (2013). 10.1063/1.4811276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Esch E. W., Bahinski A., and Huh D., Nat. Rev. Drug Discovery 14, 248 (2015). 10.1038/nrd4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Prodanov L., Jindal R., Bale S. S., Hegde M., McCarty W. J., Golberg I., Bhushan A., Yarmush M. L., and Usta O. B., Biotechnol. Bioeng. 113, 241 (2016) 10.1002/bit.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Edmondson R., Broglie J. J., Adcock A. F., and Yang L., Assay Drug Dev. Technol. 12, 207 (2014). 10.1089/adt.2014.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Berthier J. and Silberzan P., Microfluidics for Biotechnology, 2nd ed. ( Artech House, Norwood, MA, 2010). [Google Scholar]
- 112. Dangla R., Gallaire F., and Baroud C. N., Lab Chip 10, 2972 (2010). 10.1039/c003504a [DOI] [PubMed] [Google Scholar]
- 113. Lee P. J., Hung P. J., Rao V. M., and Lee L. P., Biotechnol. Bioeng. 94, 5 (2006). 10.1002/bit.20745 [DOI] [PubMed] [Google Scholar]
- 114. Joon Sung Y., Young Hwan Kim J., Wan Bong K., and Jun Sim S., Analyst 141, 989 (2016). 10.1039/C5AN02211H [DOI] [PubMed] [Google Scholar]
- 115. Siyan W., Feng Y., Lichuan Z., Jiarui W., Yingyan W., Li J., Bingcheng L., and Qi W., J. Pharm. Biomed. Anal. 49, 806 (2009). 10.1016/j.jpba.2008.12.021 [DOI] [PubMed] [Google Scholar]
- 116.Biological Applications of Microfluidics, edited by Gomez F. ( Wiley-Interscience, Hoboken, NJ, 2008). [Google Scholar]
- 117. Theberge A. B., Whyte G., Frenzel M., Fidalgo L. M., Wootton R. C. R., and Huck W. T. S., Chem. Commun. 2009(41), 6225. 10.1039/B911594C [DOI] [PubMed] [Google Scholar]
- 118.See http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-11-022.html for U.S. NIH Integrated Microphysiological Systems for Drug Efficacy and Toxicity Testing in Human Health and Disease (UH/UH3) (2015).
- 119.See http://news.mit.edu/2012/human-Body-on-a-Chip-Research-Funding-0724 for DARPA and NIH to find “human body on a chip” research (2015).
- 120. Yu J., Cilfone N., Large E., Sarkar U., Wishnok J., Tannenbaum S., Hughes D., Lauffenburger D., Griffith L., Stokes C., and Cirit M., CPT: Pharmacometrics Syst. Pharmacol. 4, 585 (2015) 10.1002/psp4.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zang R., Li D., Tang I.-C., Wang J., and Yang S.-T., Int. J. Biotechnol. Wellness Ind. 1, 31 (2012) 10.6000/1927-3037.2012.01.01.02. [DOI] [Google Scholar]
- 122.See http://wyss.harvard.edu/viewpressrelease/204/emulate-Announces-Strategic-Collaboration-with-Johnson-Johnson-Innovation-to-Use-Organsonchips-Platform-to-Better-Predict-Human-Response-in-Drug-Development-Process for Wyss Institute (2015).
- 123. Williams K., Bilsland E., Sparkes A., Aubrey W., Young M., Soldatova L. N., De Grave K., Ramon J., de Clare M., Sirawaraporn W., Oliver S. G., and King R. D., J. R. Soc., Interface 12, 1 (2015) 10.1098/rsif.2014.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4940886E-BIOMGB-10-013601 for Table S1 giving detailed information on drug screening processes and the list of abbreviations.