Summary
MAPK signaling is important for T lymphocyte development, homeostasis, and effector responses. To better understand the role of Mekk1 (encoded by Map3k1) in T cells, we conditionally deleted Map3k1 in LckCre/+Map3k1f/f mice, and these display larger iNKT cell populations within the liver, spleen, and bone marrow. Mekk1 signaling controls splenic and liver iNKT cell expansion in response to glycolipid antigen. LckCre/+Map3k1f/f mice have enhanced liver damage in response to glycolipid antigen. Mekk1 regulates Jnk activation in iNKT cells and binds and transfers Lys63-linked poly-ubiquitin onto Carma1. Map3k1 is critical for the regulation of p27Kip1 (encoded by Cdkn1b).
Graphical Abstract
Highlights
-
•
iNKT cell expansion is aberrant in LckCre/+ Map3k1f/f mice
-
•
LckCre/+ Map3k1f/f mice have enhanced liver damage in response to glycolipids
-
•
Mekk1 regulates TCR-dependent Jnk activation
-
•
Mekk1 regulates p27Kip1 expression to regulate proliferation
Suddason et al. use a T-cell-specific deletion of Map3k1 to show that Mekk1 regulates TCR-dependent Jnk activation and Cdkn1b expression to drive proliferation in response to antigen.
Introduction
Mitogen-activated protein kinase (MAPK) kinase (MAP2K) kinases (MAP3Ks) are important regulators of IκB kinases (IKKs) and MAP2Ks (Dong et al., 2002, Ghosh and Karin, 2002, Kyriakis and Avruch, 2001, Raman et al., 2007, Suddason and Gallagher, 2015). Nineteen MAP3Ks are present in mammals, though their precise roles in regulating the immune system are not fully understood (Karin and Gallagher, 2005, Suddason and Gallagher, 2015). Mek kinase 1 (Mekk1) is unique in containing both a kinase domain and a plant homeodomain (PHD), that can bind transforming growth factor (Tgf)-β-activated kinase 1 (Tak1)-binding protein 1 (Tab1) and act as an E3 ubiquitin (Ub) ligase (Suddason and Gallagher, 2015, Charlaftis et al., 2014). Map3k1ΔKD B cells regulate Jnk and p38 signaling from tumor necrosis factor (TNF) receptor family members (TNFRs) (Matsuzawa et al., 2008, Gallagher et al., 2007, Karin and Gallagher, 2009). Analysis of Map3k1ΔKD T cells demonstrated that Mekk1 is an important regulator of T helper 2 (Th2) cytokine production by the Jnk-dependent activation of Itch (Enzler et al., 2009, Gallagher et al., 2006, Gao et al., 2004, Fang et al., 2002, Venuprasad et al., 2006). Moreover, an intact Mekk1 PHD motif is required for Itch phosphorylation following T cell receptor (TCR) signaling (Suddason and Gallagher, 2015, Charlaftis et al., 2014), though the means by which Mekk1 is recruited to the TCR remain to be clarified. Map3k1ΔKD CD8+ T cells display enhanced expansion in response to viruses, but the mechanism remains uncertain (Labuda et al., 2006). The analysis of the precise role of Mekk1 in T cells using Map3k1ΔKD mice has been complicated by both B lymphocyte defects and also the partial lethality of Map3k1ΔKD mice on the C57BL/6 background (Bonnesen et al., 2005, Gallagher et al., 2007).
T lymphocytes form a critical cellular component of the adaptive immune response and can be broadly subdivided into conventional and unconventional subtypes (Kronenberg and Gapin, 2002, von Boehmer, 1990). Of these, natural killer T (NKT) cells constitute a unique unconventional T cell population of the immune system (Kronenberg and Gapin, 2007). By contrast to conventional CD4+ and CD8+ T cells, which are reactive to major histocompatibility complex (MHC) class I- or II-associated peptides, NKT cells can recognize lipids in the context of CD1d molecules (Bendelac et al., 1995, Spada et al., 1998, Brossay et al., 1998). NKT cells may express a skewed range of TCR variable region genes and the natural killer (NK) cell marker NK1.1 (Sköld et al., 2000). NKT cells can be subdivided into three categories based on their reactivity to the glycolipid α-galactosylceramide (α-GalCer), TCR α chain diversity, and CD1d dependency. Type I invariant NKT (iNKT) cells have invariant Vα14-Jα18 TCR α chains and react to α-GalCer in a CD1d-dependent manner. Type II nonclassical NKT cells are unreactive to α-GalCer and have TCR α chain diversity but are CD1d dependent. NKT-like (or type III) cells are CD1d independent, unresponsive to α-GalCer, and possess diverse TCR α chains (Bendelac et al., 2007). Following TCR engagement by glycolipid presented by CD1d, iNKT cells undergo proliferative expansion and secrete cytokines (Kawano et al., 1997, Crowe et al., 2003, Parekh et al., 2005, Godfrey et al., 2010). Type I iNKT cells are abundant within the liver, where they are important regulators of inflammation and liver damage (Van Kaer et al., 2013).
Here, we investigate Map3k1 by T-cell-specific and germline ablation in mice. Map3k1 regulates iNKT cell proliferative expansion in response to glycolipid antigen. CARD-containing MAGUK protein 1 (Carma1), a TCR-associated scaffold protein, is a target for the Mekk1 PHD motif and provides a mechanism for Mekk1 recruitment to the TCR (Blonska and Lin, 2009, Rincón and Davis, 2007). Microarray gene profiling of Map3k1-deficient iNKT cells undergoing their clonal burst in response to glycolipid antigen identified Cdkn1b as a cell-cycle gene that is aberrantly expressed in Map3k1-deficient mice (Kiyokawa et al., 1996). The regulation of p27Kip1 by Mekk1 signaling provides a cell intrinsic molecular explanation for the altered proliferative expansion observed in both Map3k1ΔKD and LckCre/+ Map3k1f/f iNKT cells.
Results
Map3k1 Regulates Conventional T Cells
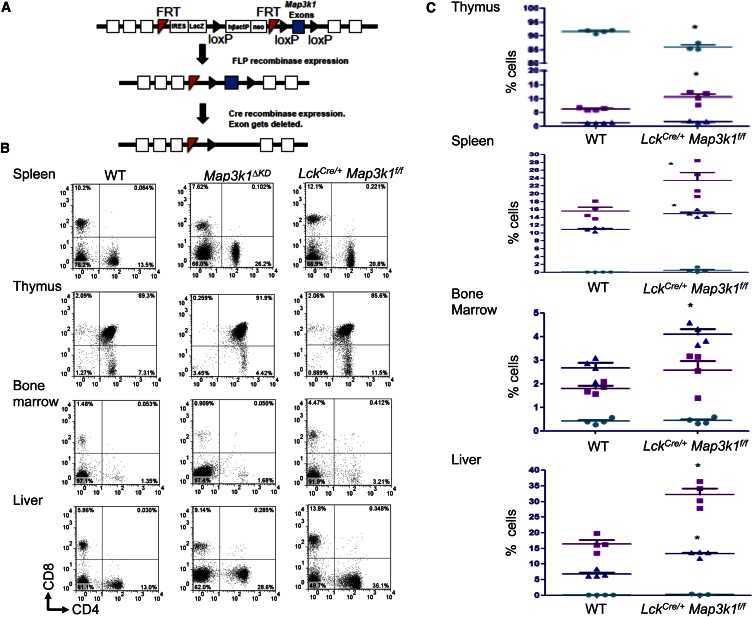
Because Map3k1ΔKD mice have both B cell defects and partial lethality on the C57BL/6 background, we generated LckCre/+ Map3k1f/f mice (Figures 1A, S1A, and S1B) to better understand the roles of Map3k1 in T cells (Gallagher et al., 2007, Bonnesen et al., 2005). Within the thymus of LckCre/+ Map3k1f/f mice, there is a minor development defect with significantly fewer CD4+CD8+ double-positive thymocytes than WT but a significantly larger than WT population of CD4+ single-positive thymocytes (Figures 1B and 1C; Chang et al., 2011, Charlaftis et al., 2014). However, the total number of thymocytes in Map3k1ΔKD and LckCre/+ Map3k1f/f mice is similar to WT (Table S1; Gao et al., 2004, Venuprasad et al., 2006, Labuda et al., 2006). Splenic CD4+ T cells isolated from LckCre/+ Map3k1f/f mice display an enhanced production of Il4 following TCR crosslinking with anti-CD3 and anti-CD28 antibodies (Figure S1C), the same Itch activation-dependent Th2 phenotype observed in Map3k1ΔKD CD4+ T cells (Gao et al., 2004, Venuprasad et al., 2006). By contrast, γδ T cells isolated from LckCre/+ Map3k1f/f or WT mice are not significantly different (Figure S1D; Maki et al., 1996). LckCre/+ Map3k1f/f mice display significantly more CD4+ and CD8+ T cells within the spleen and liver and a significantly larger CD8+ T cell population within the bone marrow (Figures 1B and 1C; Chang et al., 2011, Charlaftis et al., 2014). No significant difference was detected in T cells isolated from the thymus, spleen, liver, or bone marrow between WT and LckCre/+ mice (Figure S1E; data not shown).
Figure 1.
T Cell Development and Homeostasis within LckCre/+Map3k1f/f Mice
(A) A schematic diagram representing the construction of the Map3k1f/f allele.
(B) Thymocytes, splenocytes, bone marrow, and liver cells from WT, Map3k1ΔKD, and LckCre/+Map3k1f/f mice (all on the C57BL/6 background) were isolated, stained with anti-CD4 and anti-CD8 antibodies, and analyzed by flow cytometry as indicated. Data are representative of three independent experiments. Numbers in the profiles indicate the percentages of the gated populations.
(C) The average percentage (±SEM) of three cell populations CD4+CD8+, CD4+CD8−, and CD4−CD8+ cells from LckCre/+Map3k1f/f and WT mice from six independent experiments was statistically analyzed (green circle, CD4+CD8+; purple square, CD4+CD8−; blue triangle, CD4−CD8+), where appropriate, by two-tailed Student’s t test (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
Map3k1 Regulates iNKT Cells
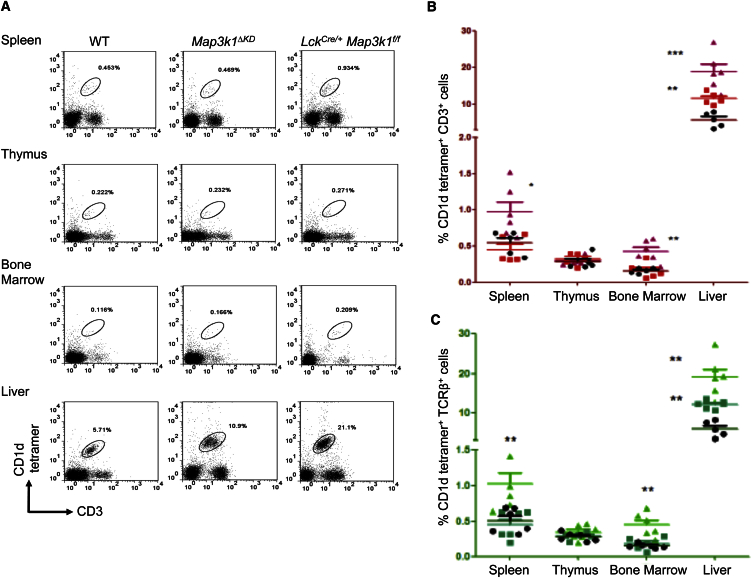
LckCre/+ Map3k1f/f mice have significantly higher numbers of iNKT cells (CD1d tetramer+CD3+, CD1d tetramer+TCRβ+, or CD1d tetramer+NK1.1+) within the liver, spleen, and bone marrow relative to WT or LckCre/+ mice (Figures 2A–2C, S2A, and S2B; data not shown; Ansari et al., 2010). Map3k1ΔKD mice, which display a germline deletion of the Map3k1 exons encoding the Mekk1 kinase domain (Gao et al., 2004), similarly displayed significantly higher numbers of iNKT cells (CD1d tetramer+CD3+, CD1d tetramer+TCRβ+, or CD1d tetramer+NK1.1+) in the liver (Figures 2B and 2C; data not shown). However, iNKT cell development in the thymus is normal for both Map3k1ΔKD and LckCre/+ Map3k1f/f mice (Figure S2C).
Figure 2.
iNKT Cell Development and Homeostasis in Map3k1ΔKD and LckCre/+Map3k1f/f Mice
(A) Cell suspensions were isolated from the spleen, thymus, bone marrow, and liver from WT, Map3k1ΔKD, and LckCre/+Map3k1f/f mice, stained with CD1d tetramer and anti-CD3 antibody, and analyzed by flow cytometry as indicated. Data are representative of five independent experiments. Numbers in the profiles indicate the percentages of the gated populations.
(B) Statistical analysis of iNKT populations (CD1d tetramer+CD3+) within the spleen, thymus, bone marrow, and liver from WT, Map3k1ΔKD, and LckCre/+Map3k1f/f mice. The average percentage (±SEM) of CD1d-tetramer and CD3-positive cells from five independent experiments is shown (black circle, WT; red square, Map3k1ΔKD; purple triangle, LckCre/+Map3k1f/f mice). Statistical differences were analyzed by two-tailed Student’s t test (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
(C) Statistical analysis of iNKT populations (CD1d tetramer+TCRβ+) within the spleen, thymus, bone marrow, and liver from WT, Map3k1ΔKD, and LckCre/+Map3k1f/f mice. The average percentage (±SEM) of CD1d-tetramer and TCRβ-positive cells from five independent experiments is shown (black circle, WT; green square, Map3k1ΔKD; green triangle, LckCre/+Map3k1f/f mice). Statistical differences were analyzed by two-tailed Student’s t test (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
Mekk1 Regulates TCR-Dependent Jnk Activation in iNKT Cells
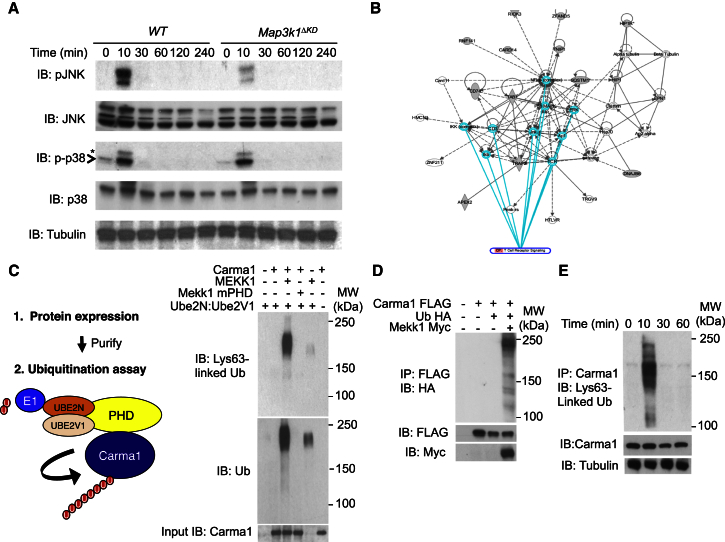
Map3k1ΔKD iNKT cells were isolated and stimulated in vitro by TCR crosslinking with antibodies (Figure 3A; Gao et al., 2004, Nagaleekar et al., 2011). WT iNKT cells display a transient Jnk activation at 10 min that is significantly reduced in Map3k1ΔKD iNKT cells following TCR crosslinking with antibodies (Figures 3A and S3A). Phosphorylation of c-Jun is similarly reduced in Map3k1ΔKD iNKT and conventional cells following TCR crosslinking with antibodies (data not shown), but there is no significant defect in p38 activation (Figure S3B; Gao et al., 2004). Because Mekk1 binds and ubiquitinates proteins by its PHD motif, we analyzed a comprehensive Mekk1 PHD protein array screen by ingenuity pathway analysis (IPA) bioinformatics to identify hits that are important for TCR signaling and identified Carma1 as a possible Mekk1 PHD substrate (Figure 3B; Suddason and Gallagher, 2015, Charlaftis et al., 2014). The Mekk1 PHD binds and, in association with Ub-conjugating enzyme E2N (Ube2N), transfers Lys63-linked Ub chains onto Carma1 (Figures 3C and 3D). Mekk1 and Carma1 transiently co-purify from iNKT cells 10 min following TCR crosslinking (Figure S3C), and endogenous Carma1 is transiently ubiquitinated following TCR crosslinking with antibodies (Figures 3E and S3D).
Figure 3.
Map3k1 Regulates Jnk Activation in iNKT Cells
(A) iNKT cells were isolated (four mice per experiment) and stimulated by TCR crosslinking with antibodies over a 240-min time course as indicated. Cell lysates were made and analyzed by IB with the indicated antibodies. Arrowhead indicates phospho-p38 and asterisk a non-specific band.
(B) IPA network diagram of TCR signaling to show the presence of the Mekk1 PHD substrate Carma within this pathway.
(C) In vitro ubiquitination assays using Mekk1 PHD, Mekk1 mPHD, Ube2N:Ube2V1, E1, Ub, and Carma1. Reactions were performed as indicated and analyzed by IB as indicated. A fraction of the ubiquitination reactions was taken pre-incubation, boiled, analyzed by IB as shown, and indicated as input.
(D) HEK293 cells were transfected with the indicated constructs. To detect in vivo ubiquitination, lysates were made under denaturing conditions for IP (Gallagher et al., 2007) and IB performed as indicated. Lysates were also made under non-denaturing conditions as a loading control and IB performed with the indicated antibodies.
(E) iNKT cells were isolated (four mice per experiment) and stimulated by TCR crosslinking with antibodies over a 60-min time course. To detect in vivo ubiquitination, lysates were made under denaturing conditions for IP (Gallagher et al., 2007) and IB performed as indicated. Lysates were also made under non-denaturing conditions as a loading control and IB performed with the indicated antibodies.
Map3k1 Regulates Splenic and Liver iNKT Cell Expansion
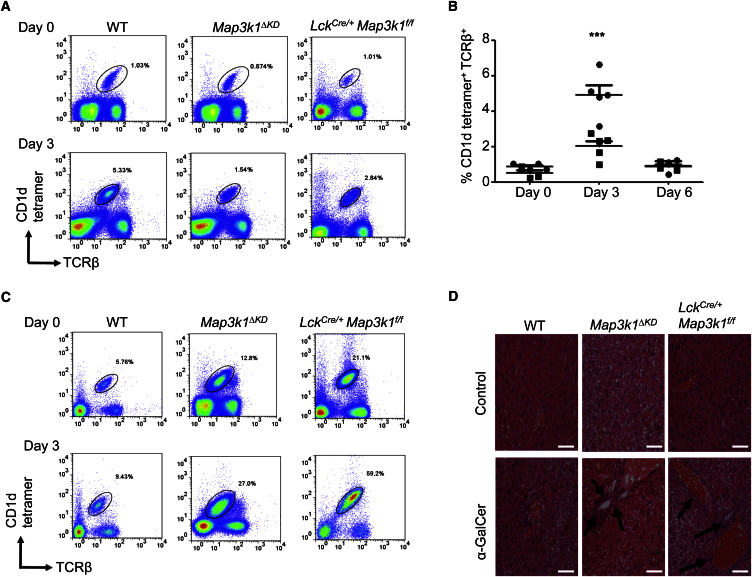
To assess the role of Mekk1 signaling in iNKT cell responses to antigen, WT, Map3k1ΔKD, and LckCre/+ Map3k1f/f mice were immunized with the iNKT cell TCR agonist α-GalCer (Figures 4 and S4). Short-term stimulation of Map3k1ΔKD and LckCre/+ Map3k1f/f mice with α-GalCer lead to normal iNKT activation and cytokine production (Figure S4A; data not shown). By contrast, splenic iNKT cells from Map3k1ΔKD and LckCre/+ Map3k1f/f mice display significantly reduced long-term proliferative expansion following immunization with α-GalCer (Figures 4A, 4B, and S4B; data not shown). Conversely, liver iNKT cells from Map3k1ΔKD and LckCre/+ Map3k1f/f mice showed significantly enhanced long-term proliferative expansion following immunization with α-GalCer (Figures 4C, S4B, and S4C). Analysis of the livers from Map3k1ΔKD and LckCre/+ Map3k1f/f mice revealed significantly enhanced lymphocyte infiltration and liver damage following long-term immunization with α-GalCer relative to control mice (Figures 4D and S4D).
Figure 4.
iNKT Cell Expansion in Map3k1-Deficient Mice
(A) WT, Map3k1ΔKD, and LckCre/+Map3k1f/f mice were i.p. injected with α-GalCer for 3 days. Splenocytes were harvested at days 0 and 3, stained with anti-TCRβ antibody and CD1d tetramer, and analyzed by flow cytometry as indicated. Data are representative of three independent experiments. Numbers in the profiles indicate the percentages of the gated populations.
(B) Statistical analysis of α-GalCer-dependent iNKT expansion at days 0, 3, and 6 in Map3k1ΔKD mice. The average percentage (±SEM) of PBS-57-loaded CD1d tetramer+ TCRβ + cells from five independent experiments is shown (black circle, WT; black square, Map3k1ΔKD mice). Differences were analyzed by two-tailed Student’s t test (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
(C) Liver cells were harvested at days 0 and 3 from WT, Map3k1ΔKD, and LckCre/+Map3k1f/f mice following i.p. immunization with α-GalCer. Liver cells were stained with CD1d tetramer and anti-TCRβ antibodies and analyzed by flow cytometry as indicated. Data are representative of three independent experiments. Numbers in the profiles indicate the percentages of the gated populations.
(D) Representative H&E-stained liver sections were prepared from unstimulated (upper panels) and 3-day α-GalCer stimulated (lower panels) WT, Map3k1ΔKD, and LckCre/+Map3k1f/f mice (original magnification ×40; scale bar, 10 μM). Arrows indicate lymphocyte infiltration. Data are representative of three independent experiments (two mice per experiment).
Mekk1 Controls iNKT Cell Proliferative Expansion by the Regulation of Jnk-Dependent p27Kip1 Expression
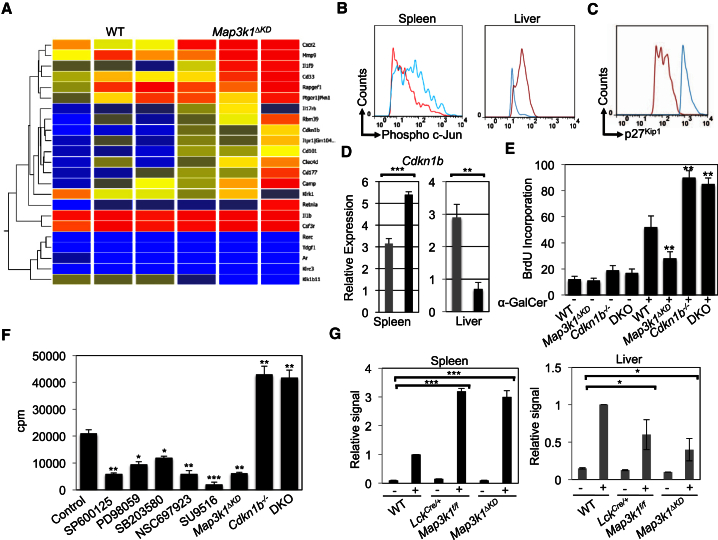
To understand the molecular basis underpinning the aberrant proliferative expansion of both LckCre/+ Map3k1f/f and Map3k1ΔKD iNKT cells, we analyzed global gene expression patterns following the long-term immunization of Map3k1ΔKD mice with α-GalCer (Figure 5A; Table S2). Bioinformatics analysis of the screened hits identified Cdkn1b (encoding p27Kip1) as a Map3k1-dependent cell-cycle regulator (Figure S5A). LckCre/+ Map3k1f/f mice have enhanced long-term phospho-c-Jun in splenic iNKT cells but reduced phospho-c-Jun in liver iNKT cells, relative to WT following immunization with α-GalCer (Figure 5B). Cdkn1b and p27Kip1 expression is significantly enhanced in splenic Map3k1ΔKD and LckCre/+ Map3k1f/f iNKT cells, but Cdkn1b expression is significantly reduced in liver Map3k1ΔKD and LckCre/+ Map3k1f/f iNKT cells following long-term stimulation by α-GalCer (Figures 5C and 5D; data not shown). Similarly, other screened hits (including Rorc, Il1β, Il1f9, and Cxcr2) identified by global gene expression analysis were verified by real-time PCR in splenic (Figure S5B; data not shown) and liver tissues (Figure S5C; data not shown). Integrin gene expression (Itgb7, Itgb21, and Itgb1) is equivalent between WT and Map3k1ΔKD iNKT cells (Figure S5D). Whereas splenic iNKT cells from Map3k1ΔKD and LckCre/+ Map3k1f/f mice hypoproliferate, splenic Cdkn1b−/− iNKT cells hyperproliferate following long-term stimulation by α-GalCer (Figure 5E). iNKT cell proliferation in response to TCR crosslinking with antibodies is significantly reduced by chemical inhibition of Jnk, Ube2N, or cyclin-dependent kinases (CDKs) (Figure 5F). Splenic, in contrast to liver, Map3k1ΔKD and LckCre/+ Map3k1f/f iNKT cells display greater phosphorylation of c-Jun at the Cdkn1b promoter activator protein-1 (AP-1)-binding site following long-term stimulation by α-GalCer (Figure 5G; Khattar and Kumar, 2010).
Figure 5.
Mekk1 Signaling Controls p27Kip1 Expression to Regulate iNKT Cell Proliferation
(A) WT and Map3k1ΔKD mice were i.p. injected with α-GalCer for 3 days. RNA was isolated from WT and Map3k1ΔKD splenic iNKT cells, processed, and hybridized onto Affymetrix arrays. Bioinformatics analysis was performed, and a heatmap comparing gene hits between WT and Map3k1ΔKD iNKT cell microarray screens was constructed. The data are from three independent experiments (four mice per experiment).
(B) Splenic and liver iNKT cells were isolated from 3-day α-GalCer-immunized LckCre/+Map3k1f/f, WT mice stained with anti-phospho c-Jun antibody, and flow cytometry performed as indicated (red line, WT; blue line, LckCre/+Map3k1f/f). Data were representative of three independent experiments. Histograms show the phospho c-Jun present in the gated iNKT cell population.
(C) Splenic iNKT cells from 3-day α-GalCer-immunized LckCre/+Map3k1f/f and WT mice were isolated and stained with anti-p27Kip1 antibody and flow cytometry performed as indicated (red line, WT; blue line, LckCre/+Map3k1f/f). Data were representative of three independent experiments. Histogram shows the p27Kip1 present in the gated iNKT cell population.
(D) iNKT cells from the spleen and liver of WT and LckCre/+Map3k1f/f mice were isolated 3 days post-i.p. injection with α-GalCer and their RNA analyzed by real-time PCR as indicated (gray square, WT; black square, LckCre/+Map3k1f/f). The average relative expression (±SEM) of genes from three independent experiments was statistically analyzed, where appropriate, by two-tailed Student’s t test (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
(E) WT, Map3k1ΔKD, Cdkn1b−/−, or Map3k1ΔKD/Cdkn1b−/− (DKO) mice were treated with water containing BrdU and i.p. immunized with α-GalCer (day 3) or left unstimulated (day 0). Splenocytes were extracted and analyzed as indicated. Representative results (±SEM) from three quantitated iNKT proliferation experiments were statistically analyzed, where appropriate, by two-tailed Student’s t test (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
(F) WT, Cdkn1b−/−, Map3k1ΔKD, or Map3k1ΔKD/Cdkn1b−/− (DKO) iNKT cells were isolated and incubated in [3H] thymidine containing media for 24 hr in the presence of DMSO (control), SP600125, PD98059, SB203580, NSC697923, or SU9516. The average cpm (±SEM) from three independent experiments was statistically analyzed, where appropriate, by two-tailed Student’s t test (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
(G) WT, Map3k1ΔKD, and LckCre/+Map3k1f/f mice were immunized with α-GalCer for 3 days, splenic or liver iNKT cells isolated, and Cdkn1b ChIP performed with anti-phospho c-Jun antibody as indicated. The average relative signal (±SEM) from three independent experiments was statistically analyzed, where appropriate, by two-tailed Student’s t test (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
Discussion
We have shown, using LckCre/+ Map3k1f/f mice, that Mekk1 has important roles in T cells. Thymic development is moderately skewed in LckCre/+ Map3k1f/f mice, with reduced numbers of CD4+CD8+ double-positive thymocytes and enhanced numbers of CD4+ single-positive thymocytes. Our findings differ from LckCre/+ Map3k7f/f mice that display reduced numbers of CD4+ and CD8+ single-positive thymocytes (Wan et al., 2006) and LckCre/+ Map3k2−/− Map3k3 f/f mice that display normal thymic development (Chang et al., 2011). The CD4+ and CD8+ T cell populations are also larger within the spleen, bone marrow, and liver tissues of LckCre/+ Map3k1f/f mice. Map3k1ΔKD mice have elevated numbers of T cells in the liver, and analysis of the iNKT cell population revealed that these are significantly expanded in the liver of both Map3k1ΔKD and LckCre/+ Map3k1f/f mice. Two factors complicating the analysis of T cells using Map3k1ΔKD mice have been the germline kinase domain mutation impacting B cells and partial embryonic lethality (Gao et al., 2004, Gallagher et al., 2007, Bonnesen et al., 2005). Bone marrow chimeras and in vitro assays using cells from Map3k1ΔKD mice have previously demonstrated that B and T cell Mekk1 signaling defects are intrinsic (Gao et al., 2004, Venuprasad et al., 2006, Labuda et al., 2006, Gallagher et al., 2007). As such, conditional deletion of Map3k1 in T cells using LckCre/+ Map3k1f/f mice represents a significant refinement of the analysis of Map3k1 in T cells.
TCR signaling leads to the rapid activation of MAPK and the phosphorylation of its downstream targets (e.g., c-Jun), and these can then initiate T cell effector responses (Su et al., 1994, Dong et al., 2002). More recently, E3 Ub ligase Itch was identified as a downstream Mekk1-signaling target that is phosphorylated by Jnk1 to induce a conformational change within the protein leading to Itch activation and canonical ubiquitination of Jun transcription factors (Gallagher et al., 2006, Gao et al., 2004, Fang et al., 2002, Venuprasad et al., 2006). As with conventional CD4+ T cells, TCR signal transduction rapidly activates Jnk in iNKT cells and this is significantly reduced in Map3k1-deficent iNKT cells (Gao et al., 2004). Mekk1 transiently binds and ubiquitinates Carma1, a scaffold known to regulate Jnk activation, following TCR engagement, and this provides a mechanism of recruitment for Mekk1 to the TCR that differs from TNFRs or Tgf-β receptors (Suddason and Gallagher, 2015, Charlaftis et al., 2014, Gaide et al., 2002).
Our work identifies a role for Map3k1 in the regulation of the iNKT cell proliferative expansion in response to glycolipid antigen. In order to identify the mechanism underpinning Map3k1-dependent iNKT cell expansion, we analyzed their global gene expression profile to identify Cdkn1b as a regulated target. The modulation of p27Kip1 expression in T cells by Mekk1 signaling represents a molecular mechanism that regulates T cell proliferation. We have identified increased iNKT cell infiltration into the liver and a higher degree of liver damage in Map3k1ΔKD and LckCre/+ Map3k1f/f mice. The aberrant iNKT cell expansion within the spleen and liver of Map3k1-deficient mice can be explained by altered c-Jun-dependent Cdkn1b expression. Our results reinforce the importance of Mekk1 signaling in T cells.
Experimental Procedures
Gene Targeting of Map3k1
Map3k1 was targeted by insertion of a FRT site followed by a LacZ sequence and a loxP site into chromosome 13 upstream of the exons of the Map3k1 gene (to generate the Map3k1f allele; Skarnes et al., 2011). The first loxP site was followed by neo under the control of the human B-actin promoter, SV40 poly-A, a second FRT site, and a second loxP site. A third loxP site was inserted downstream of the Map3k1 exons. Map3k1+/f ESCs (C57BL/6) were generated by standard procedures (Gossler et al., 1986) and genotyped by Southern blotting or genomic PCR (Ledermann, 2000, Gao et al., 2004, Charlaftis et al., 2014). Four independently generated Map3k+/f ESC clones were injected into blastocysts, and the resulting transgenics were genotyped by PCR (Gao et al., 2004, Charlaftis et al., 2014). Genomic PCR was carried out on mice biopsies using primers to detect the Map3k1-specific WT allele, the shorter mutant allele, and the recombinant allele (5′ to 3′ primers: TCGTGGTATCGTTATGCGCC; AATAGGCCACACGTTGACTGG; and CAACCCACGAAAGGAGGTTC; Charlaftis et al., 2014). Map3k1+/f mice were crossed with ACTFLPe mice (Jackson Laboratory) to initiate recombination at the FRT sites, deleting the lacZ and neo and resulting in offspring that contain a Map3k1f/f allele.
Cell Line and Cell Culture Conditions
HEK293 cells were maintained in DMEM (22320; Invitrogen) supplemented with 10% FBS (SH3007003; Thermo Scientific) and antibiotics in a humidified atmosphere at 37°C. Cells were passaged every 2 or 3 days when approaching full confluence (Charlaftis et al., 2014).
Transfection
HEK293 cells were plated in 6-well plates at a density of 1 × 106 cells per well. The following day, cells were transfected with Lipofectamine 2000 (11668-019; Invitrogen) or Jet Prime (114-07; Polyplus) transfection reagents according to the manufacturer’s instructions. Cells were collected and lysed 48 hr later.
Tissue Preparation
Spleen and perfused liver tissues were mashed through a 70-μm and a 100-μm strainer, respectively, and resuspended in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS (ThermoScientific). Following low-speed centrifugation, the splenic pellet was treated with red blood cell (RBC) lysis buffer (Sigma), washed, and resuspended in medium. The liver cell pellet was resuspended in 38% Percoll (GE Healthcare) and then centrifuged at 500 g for 20 min at room temperature. Cell pellets were treated with RBC lysis buffer, washed, and resuspended in medium.
Isolation of iNKT Cells
Mouse iNKT cells isolation from spleen or liver tissues was performed using PE-conjugated and α-GalCer-loaded CD1d tetramer (Nagaleekar et al., 2011). Tetrameric CD1d:α-GalCer cell complexes were purified using anti-PE MicroBeads (Miltenyi Biotec). Residual B cells were depleted prior to iNKT cell enrichment using a CD45R (B220) MicroBead kit (Miltenyi Biotec), and the final iNKT cell purity obtained was greater than 95%.
Flow Cytometry
Cell staining with CD1d tetramer was followed by intracellular staining performed using a Fix/Perm kit (BD PharMingen). For intracellular staining, the cells were incubated with 50 μg/ml PMA (Sigma-Aldrich), 1 μM ionomycin (Sigma-Aldrich), and 10 μg/ml brefeldin A (Sigma-Aldrich) for 2 hr before processing. For 5-bromo-2-deoxyuridine (BrdU) labeling, mice were fed with BrdU (0.8 mg/ml) in drinking water supplemented with 5% (weight/volume) glucose 1 day prior to α-GalCer (2 μg) i.p. injection. Mice were treated with BrdU in drinking water for 3 days to study proliferation. Cells were surface stained, and BrdU staining was performed according to the manufacturer protocol (BD PharMingen BrdU Flow Kit). Cells were analyzed on a Cyan ADP (DakoCytomation) flow cytometer and further analyzed on a workstation using FlowJo software (TreeStar).
Immunoblotting, Immunoprecipitation, Real-Time PCR, and Chromatin Immunoprecipitation
iNKT cells were isolated by magnetic selection using MACS LS columns according to the manufacturers protocols (Miltenyi Biotech). Immunoblotting (IB) and immunoprecipitation (IP) were carried out as previously described (Gao et al., 2004). iNKT cell RNA was prepared with the RNeasy kit (QIAGEN), and total RNA (500 μg) was converted to cDNA using the High Capacity cDNA RT-Kit (Applied Biosystems). Real-time PCR was performed in triplicate with the appropriate gene primers (Invitrogen; Table S3) using SYBR Green (Applied Biosystems) and an ABI Prism 7700 Sequence Detector (Applied Biosystems). β-actin was used for normalization of results. Chromatin IP (ChIP) was performed using an EpiTect ChIP qPCR primer assay for mouse Cdkn1b kit (QIAGEN) according to the manufacturer instructions.
Ubiquitination Assays
Carma1 cDNA was overexpressed in HEK293 cells and IP performed with anti-FLAG antibody, washed extensively, and protein eluted. Subsequently, Carma1 was incubated for 1 hr at 37°C with the ubiquitination assay enzymes E1 (100 nM), Ube2N:Ub-conjugating enzyme E2 variant 1 (Ube2V1; 0.36 μM), Ub, and ATP, with or without WT Mekk1 PHD (100 ng) or Mekk1 mutant PHD (mPHD; 100 ng; Charlaftis et al., 2014). All ubiquitination assay reagents were from Boston Biochem.
Microarray and Bioinformatics Analysis
Total RNA from iNKT cells was reverse transcribed into biotinylated cRNA with an RNA amplification kit according to the manufacturer’s instructions (Ambion). RNA quality was verified using a 2100 Bioanalyzer (Agilent Technologies). Samples were hybridized to Mouse Gene 1.0 ST arrays (Affymetrix; Charlaftis et al., 2014). Partek software was used according to the vendor protocols for data analysis, quality control, and for creating gene lists and scatterplots. GeneSpringX software was used according to the vendor protocols to generate heatmaps. Probes with a fold change of less than two were discarded. Probes were quantile normalized among all microarray data. Gene lists were uploaded into the IPA program (Ingenuity Systems) to generate relevant signaling networks and gene wheels according to the vendor instructions.
Liver Damage Assay
Mice were injected i.v. with 2 μg KRN7000. After 3 days, the livers were harvested, fixed in 4% paraformaldehyde, processed, and paraffin embedded. H&E staining was carried out on liver sections (4 μm). Slides were analyzed using an Olympus light microscope, and pictures were taken using Image Pro-Software at 40× magnification.
Statistical Analysis
Data were expressed as SEM. Statistical significance was determined by two-tailed Student’s t test. All analyses were performed using GraphPad Prism 5 software (GraphPad).
Author Contributions
T.S., S.A., N.C., and E.G. performed experiments, analyzed data, and wrote the manuscript.
Acknowledgments
Research was supported by grants from the Wellcome Trust (WT090939MA) and Cancer Research UK (C26616/A12679). We would like to thank Xin Lin (University of Texas, MD Anderson Cancer Center) for Carma1 constructs, Michael Karin (University of California at San Diego) for Map3k1ΔKD mice, A. Wahid Ansari (Imperial College London) for collegiate FACS advice, and Shashi Prajapati (Biogen) for thoughts on the manuscript.
Published: January 7, 2016
Footnotes
This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, five figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.12.047.
Contributor Information
Tesha Suddason, Email: teshas7@yahoo.com.
Ewen Gallagher, Email: ewengallagher@outlook.com.
Accession Numbers
The accession number for the dataset reported in this paper is ArrayExpress: E-MTAB-1561.
Supplemental Information
References
- Ansari A.W., Temblay J.N., Alyahya S.H., Ashton-Rickardt P.G. Serine protease inhibitor 6 protects iNKT cells from self-inflicted damage. J. Immunol. 2010;185:877–883. doi: 10.4049/jimmunol.1000651. [DOI] [PubMed] [Google Scholar]
- Bendelac A., Lantz O., Quimby M.E., Yewdell J.W., Bennink J.R., Brutkiewicz R.R. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- Bendelac A., Savage P.B., Teyton L. The biology of NKT cells. Annu. Rev. Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Blonska M., Lin X. CARMA1-mediated NF-kappaB and JNK activation in lymphocytes. Immunol. Rev. 2009;228:199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnesen B., Orskov C., Rasmussen S., Holst P.J., Christensen J.P., Eriksen K.W., Qvortrup K., Odum N., Labuda T. MEK kinase 1 activity is required for definitive erythropoiesis in the mouse fetal liver. Blood. 2005;106:3396–3404. doi: 10.1182/blood-2005-04-1739. [DOI] [PubMed] [Google Scholar]
- Brossay L., Chioda M., Burdin N., Koezuka Y., Casorati G., Dellabona P., Kronenberg M. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J. Exp. Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X., Liu F., Wang X., Lin A., Zhao H., Su B. The kinases MEKK2 and MEKK3 regulate transforming growth factor-β-mediated helper T cell differentiation. Immunity. 2011;34:201–212. doi: 10.1016/j.immuni.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlaftis N., Suddason T., Wu X., Anwar S., Karin M., Gallagher E. The MEKK1 PHD ubiquitinates TAB1 to activate MAPKs in response to cytokines. EMBO J. 2014;33:2581–2596. doi: 10.15252/embj.201488351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe N.Y., Uldrich A.P., Kyparissoudis K., Hammond K.J., Hayakawa Y., Sidobre S., Keating R., Kronenberg M., Smyth M.J., Godfrey D.I. Glycolipid antigen drives rapid expansion and sustained cytokine production by NK T cells. J. Immunol. 2003;171:4020–4027. doi: 10.4049/jimmunol.171.8.4020. [DOI] [PubMed] [Google Scholar]
- Dong C., Davis R.J., Flavell R.A. MAP kinases in the immune response. Annu. Rev. Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- Enzler T., Chang X., Facchinetti V., Melino G., Karin M., Su B., Gallagher E. MEKK1 binds HECT E3 ligase Itch by its amino-terminal RING motif to regulate Th2 cytokine gene expression. J. Immunol. 2009;183:3831–3838. doi: 10.4049/jimmunol.0803412. [DOI] [PubMed] [Google Scholar]
- Fang D., Elly C., Gao B., Fang N., Altman Y., Joazeiro C., Hunter T., Copeland N., Jenkins N., Liu Y.C. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 2002;3:281–287. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- Gaide O., Favier B., Legler D.F., Bonnet D., Brissoni B., Valitutti S., Bron C., Tschopp J., Thome M. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat. Immunol. 2002;3:836–843. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- Gallagher E., Gao M., Liu Y.C., Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl. Acad. Sci. USA. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher E., Enzler T., Matsuzawa A., Anzelon-Mills A., Otero D., Holzer R., Janssen E., Gao M., Karin M. Kinase MEKK1 is required for CD40-dependent activation of the kinases Jnk and p38, germinal center formation, B cell proliferation and antibody production. Nat. Immunol. 2007;8:57–63. doi: 10.1038/ni1421. [DOI] [PubMed] [Google Scholar]
- Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y.C., Karin M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Stankovic S., Baxter A.G. Raising the NKT cell family. Nat. Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- Gossler A., Doetschman T., Korn R., Serfling E., Kemler R. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc. Natl. Acad. Sci. USA. 1986;83:9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Gallagher E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life. 2005;57:283–295. doi: 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- Karin M., Gallagher E. TNFR signaling: ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunol. Rev. 2009;228:225–240. doi: 10.1111/j.1600-065X.2008.00755.x. [DOI] [PubMed] [Google Scholar]
- Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- Khattar E., Kumar V. Mitogenic regulation of p27(Kip1) gene is mediated by AP-1 transcription factors. J. Biol. Chem. 2010;285:4554–4561. doi: 10.1074/jbc.M109.029280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa H., Kineman R.D., Manova-Todorova K.O., Soares V.C., Hoffman E.S., Ono M., Khanam D., Hayday A.C., Frohman L.A., Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- Kronenberg M., Gapin L. The unconventional lifestyle of NKT cells. Nat. Rev. Immunol. 2002;2:557–568. doi: 10.1038/nri854. [DOI] [PubMed] [Google Scholar]
- Kronenberg M., Gapin L. Natural killer T cells: know thyself. Proc. Natl. Acad. Sci. USA. 2007;104:5713–5714. doi: 10.1073/pnas.0701493104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis J.M., Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- Labuda T., Christensen J.P., Rasmussen S., Bonnesen B., Karin M., Thomsen A.R., Odum N. MEK kinase 1 is a negative regulator of virus-specific CD8(+) T cells. Eur. J. Immunol. 2006;36:2076–2084. doi: 10.1002/eji.200535163. [DOI] [PubMed] [Google Scholar]
- Ledermann B. Embryonic stem cells and gene targeting. Exp. Physiol. 2000;85:603–613. [PubMed] [Google Scholar]
- Maki K., Sunaga S., Komagata Y., Kodaira Y., Mabuchi A., Karasuyama H., Yokomuro K., Miyazaki J.I., Ikuta K. Interleukin 7 receptor-deficient mice lack gammadelta T cells. Proc. Natl. Acad. Sci. USA. 1996;93:7172–7177. doi: 10.1073/pnas.93.14.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa A., Tseng P.H., Vallabhapurapu S., Luo J.L., Zhang W., Wang H., Vignali D.A., Gallagher E., Karin M. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science. 2008;321:663–668. doi: 10.1126/science.1157340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaleekar V.K., Sabio G., Aktan I., Chant A., Howe I.W., Thornton T.M., Benoit P.J., Davis R.J., Rincon M., Boyson J.E. Translational control of NKT cell cytokine production by p38 MAPK. J. Immunol. 2011;186:4140–4146. doi: 10.4049/jimmunol.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh V.V., Wilson M.T., Olivares-Villagómez D., Singh A.K., Wu L., Wang C.R., Joyce S., Van Kaer L. Glycolipid antigen induces long-term natural killer T cell anergy in mice. J. Clin. Invest. 2005;115:2572–2583. doi: 10.1172/JCI24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M., Chen W., Cobb M.H. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- Rincón M., Davis R.J. Choreography of MAGUKs during T cell activation. Nat. Immunol. 2007;8:126–127. doi: 10.1038/ni0207-126. [DOI] [PubMed] [Google Scholar]
- Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sköld M., Faizunnessa N.N., Wang C.R., Cardell S. CD1d-specific NK1.1+ T cells with a transgenic variant TCR. J. Immunol. 2000;165:168–174. doi: 10.4049/jimmunol.165.1.168. [DOI] [PubMed] [Google Scholar]
- Spada F.M., Koezuka Y., Porcelli S.A. CD1d-restricted recognition of synthetic glycolipid antigens by human natural killer T cells. J. Exp. Med. 1998;188:1529–1534. doi: 10.1084/jem.188.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B., Jacinto E., Hibi M., Kallunki T., Karin M., Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Suddason T., Gallagher E. A RING to rule them all? Insights into the Map3k1 PHD motif provide a new mechanistic understanding into the diverse roles of Map3k1. Cell Death Differ. 2015;22:540–548. doi: 10.1038/cdd.2014.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kaer L., Parekh V.V., Wu L. Invariant natural killer T cells as sensors and managers of inflammation. Trends Immunol. 2013;34:50–58. doi: 10.1016/j.it.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad K., Elly C., Gao M., Salek-Ardakani S., Harada Y., Luo J.L., Yang C., Croft M., Inoue K., Karin M., Liu Y.C. Convergence of Itch-induced ubiquitination with MEKK1-JNK signaling in Th2 tolerance and airway inflammation. J. Clin. Invest. 2006;116:1117–1126. doi: 10.1172/JCI26858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H. Developmental biology of T cells in T cell-receptor transgenic mice. Annu. Rev. Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- Wan Y.Y., Chi H., Xie M., Schneider M.D., Flavell R.A. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat. Immunol. 2006;7:851–858. doi: 10.1038/ni1355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.