Abstract
1,25-Dihydroxyvitamin D (1,25[OH]2D) regulates calcium (Ca), phosphate, and bone metabolism. Serum 1,25(OH)2D levels are reduced by low vitamin D status and high fibroblast growth factor 23 (FGF23) levels and increased by low Ca intake and high PTH levels. Natural genetic variation controls serum 25-hydroxyvitamin D (25[OH]D) levels, but it is unclear how it controls serum 1,25(OH)2D or the response of serum 1,25(OH)2D levels to dietary Ca restriction (RCR). Male mice from 11 inbred lines and from 51 BXD recombinant inbred lines were fed diets with either 0.5% (basal) or 0.25% Ca from 4 to 12 weeks of age (n = 8 per line per diet). Significant variation among the lines was found in basal serum 1,25(OH)2D and in the RCR as well as basal serum 25(OH)D and FGF23 levels. 1,25(OH)2D was not correlated to 25(OH)D but was negatively correlated to FGF23 (r = −0.5). Narrow sense heritability of 1,25(OH)2D was 0.67 on the 0.5% Ca diet, 0.66 on the 0.25% Ca diet, and 0.59 for the RCR, indicating a strong genetic control of serum 1,25(OH)2D. Genetic mapping revealed many loci controlling 1,25(OH)2D (seven loci) and the RCR (three loci) as well as 25(OH)D (four loci) and FGF23 (two loci); a locus on chromosome 18 controlled both 1,25(OH)2D and FGF23. Candidate genes underlying loci include the following: Ets1 (1,25[OH]2D), Elac1 (FGF23 and 1,25[OH]2D), Tbc1d15 (RCR), Plekha8 and Lyplal1 (25[OH]D), and Trim35 (FGF23). This report is the first to reveal that serum 1,25(OH)2D levels are controlled by multiple genetic factors and that some of these genetic loci interact with the dietary environment.
The classical role for vitamin D is to control calcium (Ca), phosphate (P), and bone homeostasis, but other research has revealed beneficial effects on other diseases including cancer, heart disease, and autoimmune disease (1). The regulation of vitamin D synthesis, metabolism, and action are complex processes. Vitamin D is acquired from the diet or by de novo synthesis in the skin, after which it is hydroxylated in the liver to 25-hydroxyvitamin D (25[OH]D) and released into the serum. Circulating 25(OH)D levels are used to assess vitamin D status. 25(OH)D is the inactive precursor to the hormonal metabolite, 1,25-dihydroxyvitamin D (1,25[OH]2D). 1,25(OH)2D acts through the vitamin D receptor to maintain mineral homeostasis by regulating gene expression in multiple tissues including the parathyroid gland, intestine, kidney, and bone (2). This method of regulation is sensitive to variations in dietary Ca and P levels. For example, habitually low dietary Ca intake stimulates production of PTH that induces an adaptive increase in the conversion of 25(OH)D to 1,25(OH)2D in the kidney by the enzyme CYP27b1 (2–4). Expression of the renal Cyp27b1 gene can also be suppressed by increased dietary P intake and by the P-regulating hormone fibroblast growth factor 23 (FGF23) (5).
Serum vitamin D metabolite levels are influenced by genetic and environmental factors (6, 7). The heritability of serum 25(OH)D in human populations ranges from 23% to 80% (7–12). However, to date, only a few genetic factors accounting for a small amount of variance (1%–14%) in serum 25(OH)D levels have been identified (6, 13). When combined, the known environmental and genetic factors have explained just 54% of total variance in serum 25(OH)D levels (13). These data suggest novel genetic factors influencing vitamin D status remain to be identified.
The genetic contribution to serum 1,25(OH)2D is less clear. Heritability of 1,25(OH)2D has been estimated at 16%–48% in human populations (11, 12), but studies examining the effect of genetic modifiers on serum 1,25(OH)2D level are limited and inconclusive. This is because 1,25(OH)2D metabolism is sensitive to environmental (eg, dietary Ca) and physiological cues (eg, serum Ca and P). Research studies on racial groups suggests that natural genetic variation can interact with the dietary Ca intake to influence 1,25(OH)2D-mediated Ca homeostasis (14–16). However, gene-by-diet interactions affecting serum 1,25(OH)2D levels have not been carefully studied.
Here we have used two genetically diverse populations of inbred mice to characterize the impact of natural genetic variation on basal serum 1,25(OH)2D levels on the response of serum 1,25(OH)2D levels to dietary Ca restriction and on the basal serum levels of 25(OH)D and FGF23. By using animal models, we were able to control both the impact of genetics and the environment on these phenotypes, thus overcoming a weakness that limits studies in free-living human populations. This is the first genetic mapping study to identify loci controlling serum 1,25(OH)2D levels, the 1,25(OH)2D response to dietary Ca restriction, or serum FGF23 levels.
Materials and Methods
Animal models
Study 1 examined a genetically diverse population of 11 inbred mouse lines: 129S1/SV1mJ, A/J, AKR/J, C3H/HeJ, C57BL/6J (B6), CAST/EiJ, CBA/J, DBA/2J (DBA), PWK/PhJ, SWR/J, and WSB/EiJ. Intestinal Ca absorption and bone-related phenotypes for this population have been reported previously (17). Study 2 was a forward genetic linkage mapping study to identify quantitative trait loci (QTLs) influencing serum vitamin D metabolites using 51 lines from the BXD recombinant inbred (RI) panel (18). BXD lines are each defined by a fixed recombination pattern of alleles from the B6 and DBA inbred mouse lines (18). Whereas individual F2 progeny from standard crosses are genetically unique, each BXD line is inbred and each individual within a line is genetically identical. This allows genetic experiments that include biological replicates and permits investigators to use interventions to test for the existence of gene-by-environment interactions. (18) A list of the specific lines used is provided in Supplemental Table 1.
Experimental design
Male mice were obtained at 4 weeks of age (The Jackson Labs). At arrival, an equal number of mice from each line was randomly assigned to either a 0.5% Ca (adequate) or 0.25% Ca (low) diet (AIN93G base with 200 IU/kg vitamin D3 diet; Research Diets) (n = 8 per diet per line). Dietary Ca levels were chosen to meet the rodent dietary Ca requirement (0.5% Ca) or elicit an adaptive increase in serum 1,25(OH)2D levels (0.25% Ca) (17). The low-Ca diet reflects the low Ca intake commonly seen in the United States (ie, 50% of the requirement [19, 20]), and it induces an adaptive increase in Ca absorption (+90%) in C57BL/6J mice (17). Mice were maintained in an UV-free environment (12 h light, 12 h dark) and given food and water ad libitum. At 12 weeks of age, mice were fasted overnight and then anesthetized with a ketamine-xylazine mix, euthanized by exsanguination, and the left kidney removed. Serum was prepared and both kidney and serum were frozen at −80°C. Commercial RIAs were used to measure the serum levels of 25(OH)D (intraassay percentage coefficient of variation [%CV], 5%, interassay % CV, 8.1% at 57 nmol/L) and 1,25(OH)2D (intraassay %CV, 8.8%, interassay %CV, 13% at 46 pmol/L) (IDS, Plc). Intact FGF23 was measured in the serum of mice fed the 0.5% Ca diet using a commercial ELISA according to the manufacturer's protocol (Kainos Laboratories Inc) as we have previously described (21) (interassay %CV, 2.1 at 42.4 pg/mL, intraassay %CV, 2.0 at 33.6 pg/mL). For study 1, mRNA was isolated from kidney and renal Cyp24 and Cyp27b1 mRNA was measured by a real-time PCR as previously described (17, 22). All animal experiments were approved by the Purdue University Animal Care and Use Committee.
Statistical analysis
Statistical analyses were conducted using SAS Enterprise Guide 4.2 (SAS Institute Inc). In addition to the data obtained on each diet, a parameter reflecting the response of serum 1,25(OH)2D to dietary Ca restriction (RCR) was calculated as the percentage difference between the phenotype value for an individual (i) fed the 0.25% Ca diet (x) and the line (j) mean for the phenotype value from the 0.5% Ca diet (y), standardized to the line mean from the 0.5% Ca diet and multiplied by 100, ie, (17). All data were tested for normality using the Anderson-Darling test, and values were transformed when data were not normally distributed; study 1 required the following transformations: 1,25(OH)2D (y0.25) and Cyp24 and Cyp27b1 mRNA (natural log). No transformations were necessary for study 2. Data points with a Z-score in the extreme 2.5% of either end of a line/diet group distribution were removed as outliers.
Analysis of covariance was used to test for the main effects of genetic background (ie, line) and diet as well as a line-by-diet interaction while controlling for the effect of body weight (BW), femur length, or serum analysis kit as covariates. In study 1, 25(OH)D, 1,25(OH)2D, and Cyp27b1 mRNA required a correction for BW. In study 2, BW and the analysis kit were identified as confounding covariates for serum 1,25(OH)2 D. Data are expressed as analysis of covariance adjusted least square means (LSmeans) ± SEM.
In study 1, selected, a priori post hoc comparisons were made using Fisher's least significant differences (ie, significance compared with the reference B6 line). Relationships between phenotypes were determined using Pearson's correlation tests; covariate corrected individual values were used for study 1, whereas LSmeans for each line were used for study 2.
Heritability estimates and QTL mapping
The confounding effect of covariates was removed by linear regression, and residuals were used for linkage mapping (23). Narrow-sense heritability for each dietary group was calculated using the r2 of a one-way ANOVA (main effect is the line) for phenotype. Marker information and BXD genotypes were downloaded from The GeneNetwork (http://www.genenetwork.org/genotypes/BXD.geno), and the genetic location of each marker was updated using the Mouse Map Converter tool at the Jackson Labs Center for Genome Dynamics (http://cgd.jax.org/mousemapconverter/) (24). Markers with duplicate genetic locations or perfectly correlated genotypes in our panel of 51 BXD lines were removed. The final genetic map contained 1558 markers (the list is available upon request).
Composite interval mapping (CIM) was conducted using Windows QTL Cartographer version 2.5_011 (statgen.ncsu.edu/qtlcart/WQTLCart.htm) with RI line means (n = 51). Forward selection identified five significant background markers for each phenotype. CIM was carried out using a Haldane map function, 2 cM walking speed, and a 10-cM window. Each diet group and the response to dietary Ca restriction (RCR, 1,25[OH]2D only) were mapped separately. Five hundred permutations were used to determine the significance for each analysis (25).
Bioinformatic analysis
The QTL candidate region was defined in one of two ways. For significant loci, we used 1-log of the odds (LOD) support intervals (26), which approximate 95% confidence intervals (27), whereas for putative loci, we searched the region ± 5 Mb from the peak loci marker. Sex-averaged genetic locations of peaks (centimorgans) were converted to base pair positions (GRCm38) using the Mouse Map Converter tool (24). This application uses the New Standard Genetic Map for the Laboratory Mouse generated by Cox et al (28), and QTL candidate regions were populated with genome features including protein-coding genes, noncoding RNA genes, gene fragments, and unclassified genes from the Mouse Genome Informatics database (http://www.informatics.jax.org/) (29). Genes within loci were initially identified as the region from the first to last exon plus 5000 bp upstream of the gene coordinates for exon 1 (ie, the proximal promoter region). Regions within loci that were identical by decent (IBD) between B6 and DBA were identified using the Mouse Phylogeny Viewer (http://msub.csbio.unc.edu/) (30), and 100% of the IBD regions were removed. Genes that contained non-IBD sections and all of the remaining non-IBD regions in a loci were used to query for polymorphisms between B6 and DBA using the single-nucleotide polymorphism (SNP)/variation query tool at the Mouse Phenome Database (31). The Sanger1 and Sanger2 mouse SNP and genotypic variation data sets were merged to retrieve variation between B6 and DBA mice. Mouse Phenome Database annotations (dbSNP 138) were used to categorize polymorphisms by gene attribute: intronic, mRNA untranslated region (5′ and 3′ untranslated region), promoter region (5000 bp upstream), and exon-associated (ie, synonymous and nonsynonymous codons, stop codons, splice sites, or frameshift mutations).
Effects of nonsynonymous amino acid changes and insertion/deletions were examined for potential functional effects using PROVEAN version 1.1 with a cutoff set at −2.5 (32). Occasionally the PROVEAN analysis did not recognize a sequence; in this case the impact of polymorphisms on protein function was determined by manually entering sequence data into PolyPhen2 (33). Genome-wide deoxyribonuclease 1 (DNase1) hypersensitive site (HSS) data from the mouse ENCODE project (34) was used to identify potential regulatory regions within the QTL regions. Initially we used merged DNase1 HSS peak information for 10 mouse adult tissues (fat pad, genital fat pad, heart, kidney, large intestine, liver, lung, skeletal muscle, spleen, and brain) and identified polymorphisms present in the DNase1 HSS using the merging tool from the Galaxy project platform (http://galaxyproject.org/). For select expression QTL (eQTL) candidate genes, we extended the search for overlap between HSS and polymorphism to include the entire region between CTCF insulator sites that define the boundaries between gene transcriptional domains (35). For these searches we used DNase I HSS data from the tissues in which the eQTL was found to be significant. No DNase I HSS dataset exists for bone, so we used a merged data set of peaks from the kidney and large intestine.
Expression QTL mapping
The identification of local (cis) and distant (trans) eQTL was conducted in silico using publically available microarray data sets from the BXD panel that are available at the GeneNetwork (www.genenetwork.org/webqtl/main.py). These data sets include data sets from bone (Gene Network accession numbers GN410 [n = 30 total BXD lines per 29 BXD lines overlap with the lines in our study] and GN414 [n = 30/29]); kidney (accession numbers GN240 [n = 35/32], GN117 [n = 54/39] GN118 [n = 54/39]; and liver (accession numbers GN103 [n = 38/32] and GN432 [n = 39/1]). Searches were conducted on the NCBI37/mm9 location of the QTL in megabases (Mb) plus an inclusion buffer of 10 Mb and a likelihood ratio statistic score of 9 or greater (LOD ≥ 2.0) for cis-eQTL and likelihood ratio statistic of 15 or greater (LOD ≥ 3.3) for trans-eQTL. Associations between candidate gene mRNA levels and LSMeans for the phenotypes were determined by Pearson's correlations.
Annotation of potential candidate genes
For the analysis and interpretation of candidate genes within our QTL regions, we searched for functional annotations and gene attributes using Gene Ontology terms at Mouse Genome Informatics database (http://www.informatics.jax.org). Information regarding functional relationships between each candidate gene and the serum hormones, skeletal biology, or Ca metabolism was obtained using the Coremine Medical Explorer (http://www.coremine.com/). Finally, information on tissue gene expression patterns was obtained using the gene annotation portal, BioGPS (http://biogps.org/).
Results
Variation in serum vitamin D metabolites, serum FGF23, and renal vitamin D-metabolizing enzyme mRNA levels across the 11 inbred line
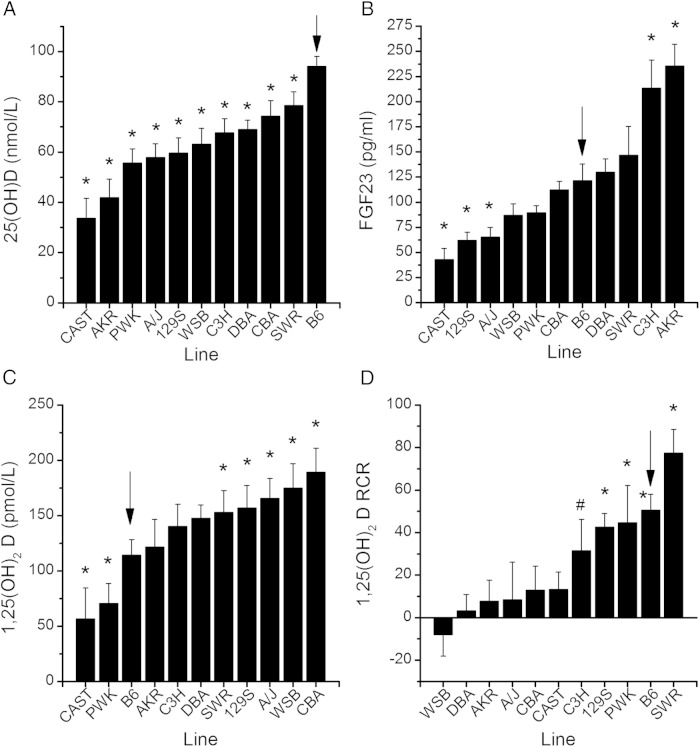
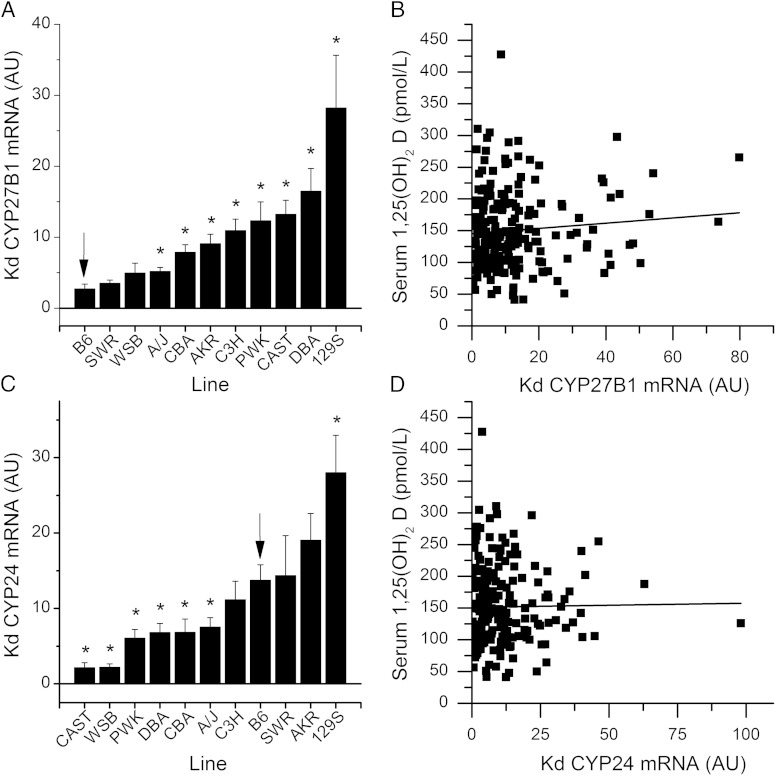
For study 1, line means for all phenotypes on each diet and their RCR are provided in Supplemental Table 2. Significant differences were found among the 11 lines for serum 1,25(OH)2D, 25(OH)D, and FGF23 in mice fed the 0.5% Ca diet (P < .0001, Figure 1, A–C). In addition to the significant line effect for serum 1,25(OH)2D, there was a diet main effect (P < .0001) and a trend for a gene-by-diet interaction observed (P = .09). As we previously observed (17), the serum 1,25(OH)2D RCR was significantly different across the lines (P < .0001, Figure 1D) but dietary Ca restriction increased serum 1,25(OH)2D in only five lines: B6, 129S1/SV1mJ, PWK/PhJ, SWR/J (P < .05), and C3H/HeJ (P < .1). There was no diet effect nor was there a line-by-diet interaction affecting serum 25(OH)D. Cyp27b1 mRNA levels in the kidney were significantly affected by line (P < .0001, 0.5% Ca diet values in Figure 2A) and diet (P = .0005) main effects, but no line-by-diet interaction was seen (P = .4). Renal Cyp24 mRNA levels were significantly different across the lines (P < .0001, Figure 2C), but no diet effect (P = .6) or line-by-diet interaction (P = .12) was observed. Serum 1,25(OH)2D levels were not significantly correlated to either renal Cyp27b1 (r = 0.08, P = .3) or Cyp24 (r = −0.03, P = .7) mRNA levels (Figure 2, B and D). This suggests that mechanisms other than the renal metabolism of vitamin D account for the variability we observed in serum 1,25(OH)2D levels among the 11 inbred lines.
Figure 1.
Serum vitamin D metabolite and FGF23 levels vary in a genetically diverse panel of 11 inbred mouse lines. Values for 25(OH)D (A), FGF23 (B), and 1,25(OH)2D (C) for lines fed the 0.5% Ca diet. D, The response of serum 1,25(OH)2D levels to RCR (dietary calcium restriction). Bars reflect the LSmeans ±SEM (n = 6–20 per diet per line) for covariate-corrected line means. *, In A–C, line mean differs significantly relative to the B6 reference line (P < .05). In D, the RCR significantly differs from 0 (*, P < .05; #, P < .1). Arrow, The reference B6 line. AKR, AKR/J; CAST, CAST/EiJ; CBA, CBA/J 13; C3H, C3H/HeJ; DBA, DBA/2Jl; PWK, PWK/PhJ; 129S1/SV1mJ; SWR, SWR/J; WSB, WSB/EiJ.
Figure 2.
Renal Cyp27b1 and Cyp24 mRNA levels vary in a genetically diverse panel of 11 inbred mouse lines. Cyp27b1 (A) and Cyp24 (C) mRNA levels for lines fed the 0.5% Ca diet (LSmeans ±SEM, n = 6–20 per diet per line) are shown. *, Line mean differs significantly relative to the B6 reference line (P < .05). The renal Cyp27b1 (B) or Cyp24 (D) mRNA relationship to serum 1,25(OH)2D levels is shown. Solid line, Regression line; dotted line, the 95% confidence interval (n = 202 individual observations). In panels A and B, the arrow is the reference B6 line. AKR, AKR/J; CAST, CAST/EiJ; CBA, CBA/J 13; C3H, C3H/HeJ; DBA, DBA/2Jl; Kd, kidney; PWK, PWK/PhJ; 129S1/SV1mJ; SWR, SWR/J; WSB, WSB/EiJ.
Relationships between serum metabolites in the 51 BXD RI lines
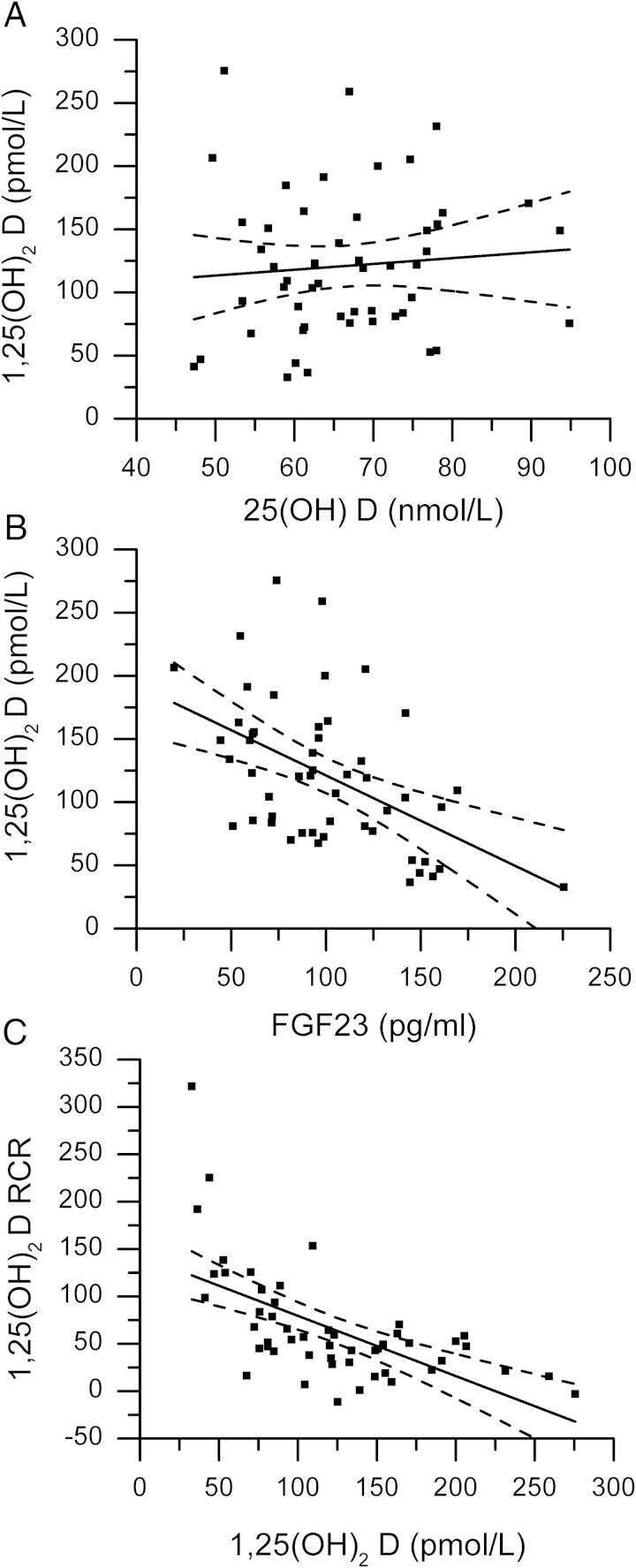
Line means for each phenotype are provided in Supplemental Table 3. There was no association between serum 25(OH)D and 1,25(OH)2D levels in mice fed either diet (Figure 3A for mice fed the 0.5% Ca diet). In mice fed the 0.5% Ca diet, serum 1,25(OH)2D was negatively associated with serum FGF23 (r = −0.501, P = .0002, Figure 3B). In addition, the serum 1,25(OH)2D RCR was negatively correlated to the basal serum 1,25(OH)2D level (r = −0.605, P < .0001, Figure 3C), indicating that lines with high basal 1,25(OH)2D levels were less likely to increase serum 1,25(OH)2D levels in response to low Ca intake.
Figure 3.
Correlations among serum vitamin D metabolites and FGF23 in 51 BXD RI lines. Covariate corrected LSMeans were used to assess relationships between the serum levels of (A) 25(OH) D and 1,25(OH)2 D (r = 0.08, NS); (B) FGF23 and 1,25(OH)2 D (r = –0.501, P = .002); and (C) 1,25(OH)2 D and the response of serum 1,25(OH)2 D to dietary Ca restriction (RCR, r = –0.61, P < .0001).
Genetic loci controlling serum 1,25(OH)2D and its response to dietary Ca restriction
Serum 1,25(OH)2D was significantly affected by line and diet main effects (P < .0001) as well as a line-by-diet interaction (P = .0002) in the BXD RI panel (see Z-scores in Supplemental Figure 1, C and D). Serum 1,25(OH)2D had a narrow-sense heritability (h2) of 0.66 and 0.65 in the 0.5% and 0.25% Ca diet groups, respectively. The serum 1,25(OH)2D RCR was significantly affected by line (P < .0001, Supplemental Figure 1D) and had a heritability of 0.59.
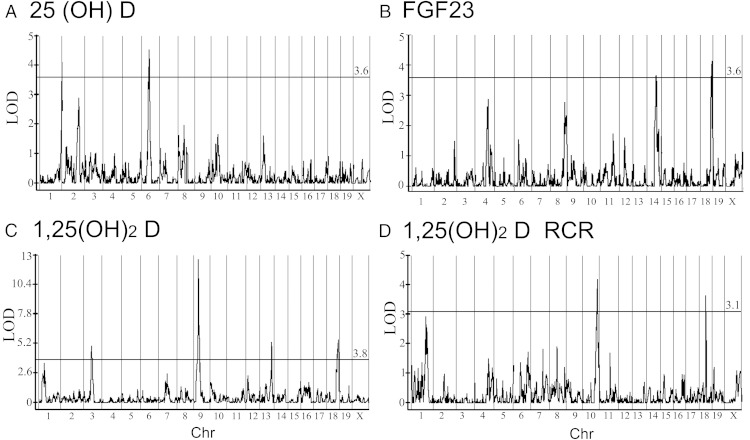
Nine QTLs for serum 1,25(OH)2D were significant in at least one diet or for the RCR (Table 1 and Figure 4, C and D). Four of these QTLs were identified as significant or putative within both diet groups (125D1a, 7, 9, and 18b). Because the two diet groups are comprised of unique individuals, this observation provides evidence of replication for several robust, diet-independent genetic effects on serum 1,25(OH)2D. The locus 125D9 (chromosome 9, peak at 33.1 Mb) accounted for the largest proportion of phenotype variance (34%). Two significant loci and one putative loci controlled the 1,25(OH)2D RCR: 125D1b (chromosome 1, 140 Mb), 125D10 (chromosome 10, 114.7 Mb), and 125D18a (chromosome 18, 47.1 Mb) (Table 1 and Figure 4D). None of the loci for RCR overlapped, with the loci controlling serum 1,25(OH)2D levels demonstrating that there are unique genetic controls on the basal and RCR phenotypes.
Table 1.
QTL Influencing Serum 25(OH)D, 1,25(OH)2D, and FGF23 Levels in Male BXD Mice
| Identification | Chr | Point Estimate, cM | Point Estimate, Mba,b | Parental Influence | Analysis | LODa | Variancee, %c |
|---|---|---|---|---|---|---|---|
| 1,25(OH)2D | |||||||
| 1,25D1a | 1 | 21.9/15.2 | 43.0/36.2 | B6 | Both | 3.5/2.2 | 6.2/3.3 |
| 1,25D1b | 1 | 61.6 | 140 | B6 | RCRd | 2.93 | 12.2 |
| 125D3 | 3 | 33.7 | 76.0 | B6 | 0.50% | 5.0 | 9.7 |
| 125D7 | 7 | 34.4/38.1 | 62.6/69.5 | B6 | Both | 2.5/3.7 | 3.5/5.9 |
| 125D9 | 9 | 18.1/18.1 | 33.1/33.1 | B6 | Both | 12.7/10.5 | 34.1/33.7 |
| 125D10 | 10 | 61.7 | 114.7 | DBA | RCR | 4.2 | 19.0 |
| 125D13 | 13 | 49.3 | 94.9 | B6 | 0.50% | 5.3 | 10.4 |
| 125D15 | 15 | 51.3 | 96.0 | DBA | 0.25% | 7.9 | 15.0 |
| 125D18a | 18 | 25.0 | 47.1 | DBA | RCR | 3.6 | 21.0 |
| 125D18b | 18 | 47.9/48.77 | 73.3/73.4 | B6 | Both | 5.6/6.4 | 11.0/11.7 |
| 25(OH)D | |||||||
| 25D1 | 1 | 95.4/95.4 | 189.9/189.9 | B6 | Both | 4.1/4.9 | 17.2/16.7 |
| 25D2 | 2 | 71.8 | 146.5 | DBA | 0.50% | 2.89 | 11.4 |
| 25D6 | 6 | 28.2/24.0 | 58.6/49.7 | B6 | Both | 4.5/2.1 | 19.2/5.2 |
| 25D9 | 9 | 56.62 | 104.8 | B6 | 0.25% | 2.6 | 7.9 |
| 25D13 | 13 | 14.4 | 36.4 | B6 | 0.25% | 6.3 | 23.4 |
| 25D15 | 15 | 37.57 | 78.7 | B6 | 0.25% | 4.1 | 13.7 |
| 25DX | X | 0.01 | 0.0 | B6 | 0.25% | 2.5 | 7.8 |
| FGF23 | |||||||
| FGF4 | 4 | 54.5 | 118.3 | B6 | 0.50% | 2.9 | 11.7 |
| FGF8 | 8 | 60.3 | 113.9 | B6 | 0.50% | 2.8 | 11.2 |
| FGF14 | 14 | 34.8 | 70.7 | DBA | 0.50% | 3.6 | 15.0 |
| FGF18 | 18 | 47.9 | 73.3 | DBA | 0.50% | 4.1 | 16.4 |
Abbreviation: Chr, chromosome. QTLs in bold were prioritized for candidate region characterization.
When loci is present in both diet groups, data are presented as 0.5% Ca diet value/0.25% Ca diet value.
Location is from mouse genome build GRC38/mm10.
Proportion of the phenotype variance that is explained by the loci within a diet group.
RCR is the response of serum 1,25(OH)2D levels to dietary Ca restriction.
Figure 4.
Results from Composite Interval Mapping (CIM) across all chromosomes. A, Serum 25(OH)D levels on the basal, 0.5% Ca diet. B, Serum FGF23 on the 0.5% Ca diet. C, Serum 1,25(OH)2D on the 0.5% Ca diet. D, The response of serum 1,25(OH)2D to RCR (dietary calcium restriction). Significance was determined separately for each data set by permutation (n = 500). The LOD cutoff is shown as a solid horizontal line, and the score is provided to the right on top of the line. Chr, chromosome.
The two significant RCR QTLs and three of the loci controlling basal serum 1,25(OH)2D levels (125D9, 125D15, and 125D18b) were selected for in-depth, bioinformatic analyses. The number of genome features, genes, and functional polymorphisms at each locus is given in Supplemental Table 4. Of the protein coding genes within the five high-priority loci we examined, only Sema6a (125D18a, LP:204) and Tph2 (125D10, PR447) have polymorphisms predicted to alter protein coding (Table 2 and Supplemental Table 5). Tph2 is expressed almost exclusively in the retina, whereas Sema6a is semaphorin that regulates microtubule dynamics, angiogenesis, and neural cell biology. A number of cis eQTLs were found within our high-priority loci, but just four of these mRNAs were significantly correlated (P < .05) to our phenotypes and were considered to be candidate genes underlying our loci: Ets1, Fli1, Tbc1d15, and Elac1 (Table 2 and Supplemental Tables 6 and 8). Many DNase I HSSs were found within the regulatory regions of these genes, and for each candidate gene, these HSSs contain more than 40 polymorphisms that might alter gene regulation (Supplemental Table 7).
Table 2.
Summary of Potential Candidate Genes Within Prioritized QTL
| Type | QTL |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 125D9 | 125D10 | 125D15 | 125D18a | 125D18b | 25D1 | 25D6 | FGF14 | FGF18 | |
| Cna | Kcnj1 (2) | Caps2 | Slc38a (2) | Pggtr1b | LAS2 (4) | Cenpf (3) | Ggct | Adam7 | LAS2 (4) |
| Tbc1d15 | 4833403l15Rik (3) | Stard6 | Ptpn | Gars | Adamdec1 | Stard6 | |||
| Tph2 (PR:447)b | Commd10 | Poli (2) | Lyplal1 (IT:58)b | Crhr2 | Adam28 | Poli (2) | |||
| Sema6a (LP:204)b | Mbd2 Dcc |
Ush2a (2) (PS:1484)b | Fam188b | Npm2 (GC:36)b | Mbd2 | ||||
| Rd3 (ML:1, HL:48)b | Pnma2 (RW:277)b Nuggc (PK:45, AG:805)b |
Dcc | |||||||
| cis eQTLc | Ets1 (−0.39/6.8/K)d | Tbc1d15 (−0.53, 4.8/K,L)d | Ano6 (−0.28/6.7/K)e | None | Elac1 (0.42/14.6/B)d | Mfsd7b (0.43/3.8/B)d | Immt (−0.38/6.1/B)d | Bnip3l (−0.39/6.9/L)d | Elac1 (−0.41/14.6/B)d |
| Fli1 (0.44/11.7/K)d | Lyplal1 (−0.31/5.3/L)# | Nod1 (0.45/6.0/L)e | Rgcc (−0.43/7.1/B)d | ||||||
| Tatdn3 (0.43/3.1/B)d | PlekHa8 (0.44/5.5/L)e | Trim35 (−0.41/2.8/B)d | |||||||
| Trans eQTL | None | None | Depdc2 (0.44/3.4/B)d | None | Poc7b (−0.42/4.1/B)d | Olfr2 (−0.36/3.7/L)d | None | Elf4 (0.36/3.7/B)d | Nvl (0.43/4.0/B)d |
| Sun1 (−0.49/4.7/B)d | Brox (0.53/3.5/L)d | Hnf6a (−0.44/3.9/B)d | |||||||
| Poc7b (0.52/4.1/B)d | |||||||||
| Sun1 (0.40/4.7/B)d | |||||||||
Cn means that the polymorphisms cause nonsynonymous amino acid substitutions (if different from 1).
Potentially deleterious nonsynonymous change is based on bioinformatics (amino acid substitution-position).
genes with eQTLs that map to the same loci (Pearson's r/LOD/tissue with the eQTL). B, bone; K, kidney; L, liver.
mRNA level are significantly correlated to phenotype (P < .05).
mRNA level is significantly correlated to phenotype (P < .10).
Genetic loci controlling serum 25(OH)D
A significant line effect influencing serum 25(OH)D was seen in the BXD RI panel (P < .0001). The main effect of diet was not significant (P = .2), but a significant line-by-diet interaction (P < .0001) was observed (see Supplemental Figure 1A for Z-scores). The narrow sense heritability (h2) of 25(OH)D was 0.40 and 0.45 in the 0.5% and 0.25% Ca diet groups, respectively. When QTL mapping results across the two diets were considered, four significant and three putative QTLs were identified (Figure 4A and Table 1). Two 25(OH)D QTLs were significant in both diet groups; these were selected for in-depth bioinformatics analysis: 25D1 (chromosome 1, 189.9 Mb) and 25D6 (chromosome 6, 58.6 Mb). Fourteen and 50 genome features remained in 25D1 and 25D6, respectively, after IBD filtering. Detailed classification of functional polymorphisms at each locus is given in Supplemental Table 4. In 25D1 there were polymorphisms that cause potentially disruptive nonsynonymous amino acid substitutions in Lyplal1, a palmitoyl-protein hydrolase, and Ush2a, a protein highly expressed in granulocytes (Supplemental Table 5). Of the cis eQTLs identified within the 25(OH)D loci, three mRNAs were significantly (P < .05) associated with serum 25(OH)D (ie, Myl5, Tatdn3 in 25D1, Immt in 25D6), and a trend (P < .10) was observed for three others (ie, Lyplal1 in 25D1, PlekHa8, Nod1 in 25D6) (Table 2 and Supplemental Tables 6 and 8). In the HSSs found within the regulatory regions of each candidate gene, we found between 8 and 181 polymorphisms that might alter gene regulation (Supplemental Table 7).
Genetic loci controlling serum FGF23
A significant line effect influencing serum FGF23 was seen in the BXD RI panel (P < .0001, see Supplemental Figure 1B for Z-scores). The narrow sense heritability (h2) of FGF23 in mice fed the 0.5% Ca diet was 0.37. QTL mapping identified two significant and two putative loci (Figure 4B and Table 1). The significant locus FGF18 (chromosome 18, 73.4 Mb) overlapped with 125D18b. This locus and FGF14 (chromosome 14, 70.7 Mb) were selected for in-depth bioinformatics analysis. Detailed classification of functional polymorphisms at each locus is given in Supplemental Table 4. In FGF14 and FGF18, only one polymorphism in the nuclear GTPase gene Npm2 caused a potentially damaging effect on protein function (Table 2 and Supplemental Table 5). In addition, four mRNAs were significantly, negatively (P < .05) correlated with serum FGF23 including Elac1 from FGF18 (also positively correlated to 1,25[OH]2 D in 125D18b) as well as Bnip31, Rgcc, and Trim35 in FGF14 (Table 2 and Supplemental Tables 6 and 8).
Discussion
Our findings are novel in that they are the first to carefully characterize the genetic factors controlling serum 1,25(OH)2D levels as well as their increase in RCR. Others have previously reported that the heritability of serum 1,25(OH)2D is variable across human subgroups, ie, 16%–20% in Hispanic Americans, 48% in African Americans, and 30% in European populations (11, 12). Engelman et al (12) previously reported that polymorphisms in the vitamin D binding protein (GC) gene were associated with 1,25(OH)2D levels in Hispanic Americans and African Americans, but Wjst et al (11) found no significant loci affecting 1,25(OH)2D levels in a linkage mapping analysis of German families. Collectively these findings suggest that the genetic determinants of 1,25(OH)2D are difficult to identify in a mixed, free-living population because environmental factors can modulate serum 1,25(OH)2D levels, eg, vitamin D status, dietary Ca, and phosphorus intake (36). In contrast, the BXD RI panel is ideal for the study of phenotypes like serum 1,25(OH)2D whose levels are influenced by both genetics and environment (18). By using a controlled, well-defined diet in our study, we eliminated these confounding influences and found that the heritability of serum 1,25(OH)2D in the BXD RI panel was 65%–66%, reflecting a strong genetic contribution to the control of this phenotype. An additional, novel aspect of our approach is that by using two diets, we were also able to show that heritability for the serum 1,25(OH)2D RCR was also high (h2 = 59%, ie, the gene by diet interaction).
We found that serum 1,25(OH)2D levels mapped to multiple genomic locations in the BXD RI panel, and only one locus overlapped with loci controlling either 25(OH)D or FGF23 (125D18a and FGF18). Four of the loci affecting serum 1,25(OH)2D were found in both diet groups. Thus, in addition to being robust and unaffected by diet, this replication provides independent validation of the four loci. A final novelty of our findings on 1,25(OH)2D is that the three loci controlling the 1,25(OH)2D RCR were distinct from those that control basal serum 1,25(OH)2D, thereby demonstrating independent genetic controls on these two phenotypes. However, it is likely that the genetic effects on the basal and RCR phenotypes interact to have a complex impact on serum 1,25(OH)2D levels. For example, we found that BXD lines with low basal serum 1,25(OH)2D levels were more likely to increase 1,25(OH)2D when challenged with a low-Ca diet than lines with high basal serum 1,25(OH)2D levels. The functional impact of this gene-by-diet interaction remains to be determined.
Our results also have novelty for the genetics of serum FGF23 and 25(OH)D levels. Whereas variants near the FGF23 gene have been implicated in the control of serum P levels (37), no genome-wide association studies, twin studies, or heritability estimates for serum FGF23 have been reported in humans or in preclinical models. Thus, our mapping study extends the limited information we have on the impact of genetics on the control of serum FGF23 level. Our data also clarify some issues regarding the genetic controls of serum 25(OH)D. Others have reported heritability values for 25(OH)D between 23% and 80% in human populations; this range is likely due to the confounding influence of variable UV exposure (7–12). In BXD RI lines, all vitamin D was obtained from the diet and the heritability of serum 25(OH)D was 40%–45%. Interestingly, the loci we identified as controlling serum 25(OH)D in BXD mice do not contain genes that are known to participate in vitamin D metabolism (eg, cubilin, megalin) or genes whose polymorphisms have been associated with human vitamin D status in either candidate gene studies, (ie, vitamin D binding protein [GC], 25-hydroxylase [CYP2R1] [12, 38]) or genome-wide association studies (ie, GC, 7-dehydrocholesterol reductase [DHCR7], acyl-coenzyme A dehydrogenase [ACADSB], and CYP2R1) (39). Cubilin and Cyp2r1 are located in regions that are IBD between B6 and DBA, so our QTL analysis would not identify these loci as influencing serum 25(OH)D. In addition, both Dhcr7 and Acadsb participate in UVB-induced synthesis of vitamin D from cholesterol (39, 40), but our mice were shielded from UVB radiation, thereby eliminating the any potential contribution of variants in these genes to serum 25(OH)D levels in BXD mice.
Our identification of new loci controlling serum vitamin D metabolite or FGF23 levels offers us an opportunity to gain new insight into the regulation of these hormones. However, genetic mapping in the BXD RI panel identifies large regions that contain multiple genes and many polymorphisms. This makes it difficult to definitively identify the causal variant and gene whose function controls these traits. Fortunately, bioinformatics analysis can inform candidate gene identification (41). In the following paragraphs, we will discuss candidate genes that we identified after the bioinformatics analysis of several loci.
The QTL 125D9 was identified as a significant locus in both diet groups. It accounts for 34% of the variation in serum 1,25(OH)2D levels in the BXD panel. Within 125D9 is the gene encoding the transcription factor E26 avian leukemia oncogene 1 (Ets1). Previous studies from our group and others (42, 43) have identified Ets1 as a transcription factor necessary for maximal 1,25(OH)2D-induced transcription of the CYP24 gene. In the BXD panel, the Ets1 gene has no potentially deleterious coding level polymorphisms but contains 167 polymorphisms that fall within DNase1 HSS in the kidney genome. As a result, variation in the Ets1 regulatory regions may influence Ets1 gene expression and the 1,25(OH)2D level through the regulation of Cyp24 transcription, ie, increased Ets1 expression would enhance the vitamin D-induced degradation of 1,25(OH)2D mediated through elevated CYP24 levels. Consistent with our hypothesis, renal Ets1 mRNA levels were significantly, negatively correlated with serum 1,25(OH)2D levels in the BXD panel. Future studies will be necessary to determine the relationship among Ets1 gene expression, CYP24 activity, and serum 1,25(OH)2D levels as well as to identify how specific regulatory region polymorphisms affect Ets1 gene expression.
QTLs controlling serum 1,25(OH)2D (125D18b) and FGF23 (FGF18) levels mapped to identical locations on chromosome 18. We and others have demonstrated that these hormones are inversely related to one another (Figure 3 and reference 44). As such, our result suggests that the variation at this locus either influences a common regulator of the two hormones or that it is having a direct impact on one hormone that then indirectly affects the other. Our data do not permit us to definitively differentiate between these two options. Within 125D18/FGF18 there were no disruptive, coding level polymorphisms, and only the cis eQTL for Elac1 from bone was significantly correlated to both serum 1,25(OH)2D (r = −0.42) and FGF23 (r = 0.42) levels. Elac1 is an endonuclease involved in tRNA maturation. Consistent with the fact that the cis eQTL was found using the bone transcriptome, data from BioGPS shows that Elac1 is well expressed in osteoblasts (where FGF23 is produced [45]) but poorly expressed in kidney (where the bulk of serum 1,25[OH]2D is made) (ie, 110 vs 15 AU). Although it is not clear how tRNA processing could specifically influence serum FGF23 levels, the links between Elac1 expression and bone cells suggests that the primary effect of the locus is through the control of FGF23 production. Osteoblastic bone formation is a potent inducer of FGF23 production and release into the circulation (46), so future studies should evaluate whether differences in Elac1 expression can influence osteoblast biology leading to increased serum FGF23 levels. The QTL FGF14 may also link to osteoblast function through the bone cis eQTL for Trim35 (aka Mair) mRNA. BioGPS shows that Trim35 mRNA (probe identification 145621_at) is highly expressed in osteoblasts. Trim35 promotes apoptosis (47), and this could explain why its bone mRNA level is negatively correlated with serum FGF23 levels.
More than 35% of the variation in basal serum 25(OH)D levels in the BXD panel is controlled by 25D1 and 25D6. Two genes are likely candidates account for the influence of these loci: Lyplal1 in 25D1 and PlekHa8 in 25D6. A cis eQTL and a potentially deleterious, nonsynonymous coding variant were found for Lyplal1 (Table 2), a lysophospholipase with palmitoyl-hydrolase activity. The B6 allele at this locus is associated with elevated serum 25(OH)D levels, and Lyplal1 mRNA levels are low in B6 mouse liver compared with other mouse lines in the diversity outbred cross (cgd.jax.org/apps/eqtlviewer-beta/). Consistent with an inhibitory effect of Lyplal1 on serum 25(OH)D, we found that liver Lyplal1 mRNA levels are negatively correlated with serum 25(OH)D in the BXD population. In humans, variations in this gene have been closely linked to obesity and nonalcoholic fatty liver (48, 49). Whereas serum 25(OH)D levels are negatively correlated to body mass in humans (50), our analysis was conducted after correction for body size as a covariate, so it is unlikely that this loci simply reflects an indirect effect of obesity. However, palmitoylation is a regulatory process that alters subcellular localization of proteins and influences protein-protein interactions (51), and this could influence vitamin D metabolism in ways that have yet to be explored.
Plekha8 (also called Fapp2) was identified as a cis eQTL in 25D6 whose liver mRNA levels are positively correlated to serum 25(OH)D levels. Liver Plekha8 mRNA levels are higher in B6 mice compared with other lines in the diversity outbred cross, consistent with the hypothesis that high Plekha8 expression supports elevated serum 25(OH)D levels. Plekha8 is a cargo transport protein that mediates apical transport from the Golgi apparatus, and it plays a role in the synthesis of complex glycoshingolipids (52). In addition to being expressed in the liver, Plekha8 mRNA levels are high in the kidney (from BioGPS) where it has been reported to control apical membrane trafficking in polarized kidney epithelial cells and the structural conformation of apical tubule carriers (53, 54). Megalin and cubilin are cell surface receptors that bind and internalize GC-bound 25(OH)D during renal reabsorption of vitamin D (55). We hypothesize that the variation in Plekha8 influences membrane placement or recycling of megalin and cubilin, thereby influencing serum 25(OH)D levels by influencing urinary 25(OH)D loss. Between B6 and DBA mice, Plekha8 has no nonsynonymous coding polymorphisms but does contain polymorphisms within DNase1 HSS. This suggests that the regulation of Plekha8 gene expression, rather than protein function, is altered in the BXD RI panel. Future studies are needed to identify whether renal Plekha8 mRNA or urinary 25(OH)D levels differ across the BXD panel.
In summary, using a classical, forward genetics approach we have identified considerable genetic diversity controlling serum vitamin D metabolite and FGF23 levels in the mouse. Our study is novel because it was done in a large recombinant inbred mouse population under strict environmental control, and it is the first to examine the genetics controlling serum 1,25(OH)2D, the response of serum 1,25(OH)2D to dietary Ca restriction, and serum FGF23 levels. Another important finding from our study is that distinct loci control serum 1,25(OH)2D and the adaptation of 1,25(OH)2D to low Ca intake. Although we have not been able to definitively identify the causal variants affecting serum vitamin D metabolite levels, our bioinformatic approach points to altered gene regulation of several plausible candidate genes as areas for future study. The genetic loci identified in this study serve as a starting point to identify novel pathways or genes controlling vitamin D metabolism, and this has the potential to expand our knowledge of vitamin D physiology.
Acknowledgments
This work was supported by National Institutes of Health Grants R21-ES019103 (to J.C.F.), R01-DK063934 and R01-DK95784 (to K.E.W.), and F32-AR065389 (to E.L.C.). P.R.-F. was partially supported by a scholarship from Consejo Nacional de Ciencia y Technologia (Mexico). R.A.R. was supported by a fellowship from the National Institute of Food and Agriculture (Grant 2012–67011-19963).
Disclosure Summary: J.C.F. is on the Scientific Advisory Board for Innophos, Inc. K.E.W. received royalties from Kyowa Hakko Kirin Co, Ltd for the licensing of the FGF23 gene and for anti-FGF23 monoclonal antibody trials. The other authors have nothing to disclose.
Footnotes
- B6
- C57BL/6J
- BW
- body weight
- CIM
- composite interval mapping
- DBA
- DBA/2J
- Ca
- calcium
- DNase1
- deoxyribonuclease 1
- eQTL
- expression QTL
- FGF23
- fibroblast growth factor 23
- HSS
- hypersensitive site
- IBD
- identical by decent
- LOD
- log of the odds
- LSmeans
- least square means
- 25(OH)D
- 25-hydroxyvitamin D
- 1,25(OH)2D
- 1,25-dihydroxyvitamin D
- P
- phosphate
- QTL
- quantitative trait locus
- RCR
- response to dietary calcium restriction
- RI
- recombinant inbred
- SNP
- single-nucleotide polymorphism.
References
- 1. Christakos S, DeLuca HF. Minireview: vitamin D: is there a role in extraskeletal health? Endocrinology. 2011;152:2930–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fleet JC, Schoch RD. Molecular mechanisms for regulation of intestinal calcium absorption by vitamin D and other factors. Crit Rev Clin Lab Sci. 2010;47:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centeno V, de Barboza GD, Marchionatti A, Rodriguez V, Tolosa de Talamoni N. Molecular mechanisms triggered by low-calcium diets. Nutr Res Rev. 2009;22:163–174. [DOI] [PubMed] [Google Scholar]
- 4. Murayama A, Takeyama K, Kitanaka S, Kodera Y, Hosoya T, Kato S. The promoter of the human 25-hydroxyvitamin D3 1α-hydroxylase gene confers positive and negative responsiveness to PTH, calcitonin, and 1α,25(OH)2D3. Biochem Biophys Res Commun. 1998;249:11–16. [DOI] [PubMed] [Google Scholar]
- 5. Perwad F, Portale AA. Vitamin D metabolism in the kidney: regulation by phosphorus and fibroblast growth factor 23. Mol Cell Endocrinol. 2011;347:17–24. [DOI] [PubMed] [Google Scholar]
- 6. Wang TJ, Zhang F, Richards JB, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karohl C, Su S, Kumari M, et al. Heritability and seasonal variability of vitamin D concentrations in male twins. Am J Clin Nutr. 2010;92:1393–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hunter D, de Lange M, Snieder H, et al. Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res. 2001;16:371–378. [DOI] [PubMed] [Google Scholar]
- 9. Orton SM, Morris AP, Herrera BM, et al. Evidence for genetic regulation of vitamin D status in twins with multiple sclerosis. Am J Clin Nutr. 2008;88:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shea MK, Benjamin EJ, Dupuis J, et al. Genetic and non-genetic correlates of vitamins K and D. Eur J Clin Nutr. 2009;63:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wjst M, Altmuller J, Braig C, Bahnweg M, Andre E. A genome-wide linkage scan for 25-OH-D(3) and 1,25-(OH)2-D3 serum levels in asthma families. J Steroid Biochem Mol Biol. 2007;103:799–802. [DOI] [PubMed] [Google Scholar]
- 12. Engelman CD, Fingerlin TE, Langefeld CD, et al. Genetic and environmental determinants of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D levels in Hispanic and African Americans. J Clin Endocrinol Metab. 2008;93:3381–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lucas RM, Ponsonby AL, Dear K, et al. Vitamin D status: multifactorial contribution of environment, genes and other factors in healthy Australian adults across a latitude gradient. J Steroid Biochem Mol Biology. 2013;136:300–308. [DOI] [PubMed] [Google Scholar]
- 14. Bryant RJ, Wastney ME, Martin BR, et al. Racial differences in bone turnover and calcium metabolism in adolescent females. J Clin Endocrinol Metab. 2003;88:1043–1047. [DOI] [PubMed] [Google Scholar]
- 15. Braun M, Palacios C, Wigertz K, et al. Racial differences in skeletal calcium retention in adolescent girls with varied controlled calcium intakes. Am J Clin Nutr. 2007;85:1657–1663. [DOI] [PubMed] [Google Scholar]
- 16. Weaver CM, McCabe LD, McCabe GP, et al. Vitamin D status and calcium metabolism in adolescent black and white girls on a range of controlled calcium intakes. J Clin Endocrinol Metab. 2008;93:3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Replogle RA, Li Q, Wang L, Zhang M, Fleet JC. Gene-by-diet interactions influence calcium absorption and bone density in mice. J Bone Miner Res. 2014;29:657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heaney RP. Nutritional factors in osteoporosis. Ann Rev Nutr. 1993;13:287–316. [DOI] [PubMed] [Google Scholar]
- 20. Wallace TC, Reider C, Fulgoni VL., 3rd Calcium and vitamin D disparities are related to gender, age, race, household income level, and weight classification but not vegetarian status in the United States: analysis of the NHANES 2001–2008 data set. J Am Coll Nutr. 2013;32:321–330. [DOI] [PubMed] [Google Scholar]
- 21. Farrow EG, Yu X, Summers LJ, et al. Iron deficiency drives an autosomal dominant hypophosphatemic rickets (ADHR) phenotype in fibroblast growth factor-23 (Fgf23) knock-in mice. Proc Natl Acad Sci USA. 2011;108:E1146–E1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song Y, Fleet JC. Intestinal Resistance to 1,25 Dihydroxyvitamin D in mice heterozygous for the vitamin D receptor knockout allele. Endocrinology. 2007;148:1396–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lang DH, Sharkey NA, Lionikas A, et al. Adjusting data to body size: a comparison of methods as applied to quantitative trait loci analysis of musculoskeletal phenotypes. J Bone Miner Res. 2005;20:748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ackert-Bicknell CL, Karasik D, Li Q, et al. Mouse BMD quantitative trait loci show improved concordance with human genome-wide association loci when recalculated on a new, common mouse genetic map. J Bone Miner Res. 2010;25:1808–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Ooijen JW. Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet. 1992;84:803–811. [DOI] [PubMed] [Google Scholar]
- 27. Dupuis J, Siegmund D. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics. 1999;151:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cox A, Ackert-Bicknell CL, Dumont BL, et al. A new standard genetic map for the laboratory mouse. Genetics. 2009;182:1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang JR, de Villena FP, McMillan L. Comparative analysis and visualization of multiple collinear genomes. BMC Bioinform. 2012;3(suppl 13):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grubb SC, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2014;42:D825–D834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yue F, Cheng Y, Breschi A, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim S, Yu NK, Kaang BK. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med. 2015;47:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DeLuca HF. The control of calcium and phosphorus metabolism by the vitamin D endocrine system. Ann NY Acad Sci. 1980;355:1–17. [DOI] [PubMed] [Google Scholar]
- 37. Kestenbaum B, Glazer NL, Kottgen A, et al. Common genetic variants associate with serum phosphorus concentration. J Am Soc Nephrol. 2010;21:1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bu FX, Armas L, Lappe J, et al. Comprehensive association analysis of nine candidate genes with serum 25-hydroxyvitamin D levels among healthy Caucasian subjects. Hum Genet. 2010;128:549–556. [DOI] [PubMed] [Google Scholar]
- 39. Ahn J, Yu K, Stolzenberg-Solomon R, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19:2739–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waterham HR, Wanders RJ. Biochemical and genetic aspects of 7-dehydrocholesterol reductase and Smith-Lemli-Opitz syndrome. Biochim Biophys Acta. 2000;1529:340–356. [DOI] [PubMed] [Google Scholar]
- 41. DiPetrillo K, Wang X, Stylianou IM, Paigen B. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 2005;21:683–692. [DOI] [PubMed] [Google Scholar]
- 42. Dwivedi PP, Omdahl JL, Kola I, Hume DK, May BK. Regulation of rat cytochrome P450C24 (CYP24) gene expression—evidence for functional cooperation of Ras-activated Ets transcription factors with the vitamin D receptor in 1,25-dihydroxyvitamin D-3-mediated induction. J Biol Chem. 2000;275:47–55. [DOI] [PubMed] [Google Scholar]
- 43. Cui M, Zhao Y, Hance KW, Shao A, Wood RJ, Fleet JC. Effects of MAPK signaling on 1,25-dihydroxyvitamin D-mediated CYP24 gene expression in the enterocyte-like cell line, Caco-2. J Cell Physiol. 2009;219:132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dai B, David V, Alshayeb HM, et al. Assessment of 24,25(OH)2D levels does not support FGF23-mediated catabolism of vitamin D metabolites. Kidney Int. 2012;82:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kolek OI, Hines ER, Jones MD, et al. 1α,25-Dihydroxyvitamin D3 upregulates FGF23 gene expression in bone: the final link in a renal-gastrointestinal-skeletal axis that controls phosphate transport. Am J Physiol Gastrointest Liver Physiol. 2005;289:G1036–G1042. [DOI] [PubMed] [Google Scholar]
- 46. Samadfam R, Richard C, Nguyen-Yamamoto L, Bolivar I, Goltzman D. Bone formation regulates circulating concentrations of fibroblast growth factor 23. Endocrinology. 2009;150:4835–4845. [DOI] [PubMed] [Google Scholar]
- 47. Kimura F, Suzu S, Nakamura Y, et al. Cloning and characterization of a novel RING-B-box-coiled-coil protein with apoptotic function. J Biol Chem. 2003;278:25046–25054. [DOI] [PubMed] [Google Scholar]
- 48. Nettleton JA, Follis JL, Ngwa JS, et al. Gene × dietary pattern interactions in obesity: analysis of up to 68 317 adults of European ancestry. Hum Mol Genet. 2015;24:4728–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sharma M, Mitnala S, Vishnubhotla RK, Mukherjee R, Reddy DN, Rao PN. The riddle of nonalcoholic fatty liver disease: progression from nonalcoholic fatty liver to nonalcoholic steatohepatitis. J Clin Exp Hepatol. 2015;5:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wortsman J, Matsuoka LY, Chen TC, Lu Z, Holick MF. Decreased bioavailability of vitamin D in obesity. Am J Clin Nutr. 2000;72:690–693. [DOI] [PubMed] [Google Scholar]
- 51. Aicart-Ramos C, Valero RA, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta. 2011;1808:2981–2994. [DOI] [PubMed] [Google Scholar]
- 52. D'Angelo G, Rega LR, De Matteis MA. Connecting vesicular transport with lipid synthesis: FAPP2. Biochim Biophys Acta. 2012;1821:1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vieira OV, Verkade P, Manninen A, Simons K. FAPP2 is involved in the transport of apical cargo in polarized MDCK cells. J Cell Biol. 2005;170:521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cao X, Coskun U, Rossle M, et al. Golgi protein FAPP2 tubulates membranes. Proc Natl Acad Sci USA. 2009;106:21121–21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Christensen EI, Birn H, Storm T, Weyer K, Nielsen R. Endocytic receptors in the renal proximal tubule. Physiology (Bethesda). 2012;27:223–236. [DOI] [PubMed] [Google Scholar]