Abstract
Accurate detection of carbapenemase-producing Enterobacteriaceae (CPE) constitutes a major laboratory diagnostic challenge. We evaluated an electrochemical technique (the BYG Carba test) which allows detection of CPE in less than 35 min. The BYG Carba test was first validated in triplicate against 57 collection isolates with previously characterized β-lactam resistance mechanisms (OXA-48, n = 12; KPC, n = 8; NDM, n = 8; VIM, n = 8; IMP, n = 3; GIM, n = 1; GES-6, n = 1; no carbapenemase, n = 16) and against a panel of 10 isolates obtained from the United Kingdom National External Quality Assessment Service (NEQAS). The test was then evaluated prospectively against 324 isolates referred to the national reference center for suspicion of CPE. The BYG Carba test results were compared with those obtained with the Carba NP test using multiplex PCR sequencing as the gold standard. Of the 57 collection and the 10 NEQAS isolates, all but one GES-6-producing isolate were correctly identified by the Carba BYG test. Among the 324 consecutive Enterobacteriaceae isolates tested prospectively, 146 were confirmed as noncarbapenemase producers by PCR while 178 harbored a carbapenemase gene (OXA-48, n = 117; KPC, n = 25; NDM, n = 23; and VIM, n = 13). Prospectively, in comparison with PCR results, the BYG Carba test displayed 95% sensitivity and 100% specificity versus 89% and 100%, respectively, for the Carba NP test. The BYG Carba test is a novel, rapid, and efficient assay based on an electro-active polymer biosensing technology discriminating between CPE and non-CPE. The precise electrochemical signal (electrochemical impedance variations) allows the establishment of real-time objective measurement and interpretation criteria which should facilitate the accreditation process of this technology.
INTRODUCTION
The worldwide emergence and dissemination of carbapenemase-producing Gram-negative rods, in particular, carbapenemase-producing Enterobacteriaceae (CPE) that are resistant to carbapenems, is a major public health concern. Rapid detection and confirmation of CPE are essential for appropriate choice of antimicrobial therapy as well as for the implementation and/or maintenance of appropriate infection control measures (1). The transmissible carbapenemases are divided into three different classes, class A (serine carbapenemases, such as KPC), class B (metallo-β-lactamases [MBLs], such as VIM, IMP, and NDM), and class D (OXA carbapenemases, such as OXA-48) (1–3).
Various phenotypic screening and confirmatory tests for the detection of carbapenemases have been proposed, including inhibition tests of carbapenemase activity, for example, combined-disk tests using specific inhibitors such as EDTA and boronic acid, the modified Hodge test (MHT) (4), the carbapenem inactivation method (5), and detection of carbapenem hydrolysis by the Carba NP test (6) or by other closely related tests (7, 8). However, although allowing, in some formats, the differentiation between class A and B carbapenemases (9), these tests cannot specify types within each class of carbapenemases, (e.g., IMP, VIM, NDM, SIM, and GIM in class B), and they also are unable to confirm in a single test the occurrence of class D OXA-48 carbapenemase (10). To confirm the presence of OXA-48, Tsakris and colleagues (11) also recently proposed a confirmatory disk test (96.3% sensitivity and 97.7% specificity) based on the use of an imipenem disk, EDTA, and a combination of EDTA and phenyl boronic acid.
Molecular detection methods such as PCR and sequencing of carbapenemase genes are nevertheless more reliable for confirmation of carbapenemases (10), but they are only rarely implemented in most routine clinical microbiology laboratories because of their high cost and the skill level and the special equipment required.
However, rapid non-molecular-based confirmatory testing of the presence of a carbapenemase can be sufficient to implement clinical management strategies and infection control measures to limit the spread and cross-transmission of these organisms.
Various confirmatory tests rely on direct monitoring of β-lactam hydrolysis. For example, a spectrophotometric method (12) is reported as a very specific and sensitive gold standard but requires a spectrophotometer and is applied only in specialized laboratories.
In 2011, two different groups demonstrated that matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was able to detect by-products resulting from the hydrolysis of a carbapenem in the presence of a bacterial extract of CPE (13, 14). Since then, several investigators have improved the sensitivity of the method as well as the interpretation of the results (15–19).
Recently, Nordmann and colleagues developed a colorimetric method for the specific detection of carbapenemase activity, the Carba NP test (6). In this test the pH decrease concomitant with the hydrolysis of imipenem by a carbapenemase is monitored by phenol red, which acts as an acid-base colorimetric indicator. Phenol red color changes are estimated by the naked eye of the operator but are not easily traceable in the current laboratory information management systems (LIMS). The original method allows the detection of carbapenemase-producing organisms in a maximum of 2.5 h and is now commercialized by bioMérieux (Rapidec Carba NP).
In the present paper, we propose the BYG (Bogaerts-Yunus-Glupczynski) Carba test, a new and original electrochemical method for the rapid confirmation of carbapenemase-producing bacteria. The BYG Carba test detects the variations of conductivity of a polyaniline (an electro-sensing polymer)-coated electrode which is highly sensitive to the modifications of pH and of redox activity occurring during the imipenem enzymatic hydrolysis reaction (20–22). The modifications of the conductivity are monitored, analyzed, and reported in real time by the BYG Carba test and are indicative of the presence of a carbapenemase.
MATERIALS AND METHODS
Electrochemical instrumentation.
The portable potentiostat is a homemade electronic device (23), computer controlled via a USB serial port of a microcontroller board (Arduino uno interface) programmed as a control and acquisition card.
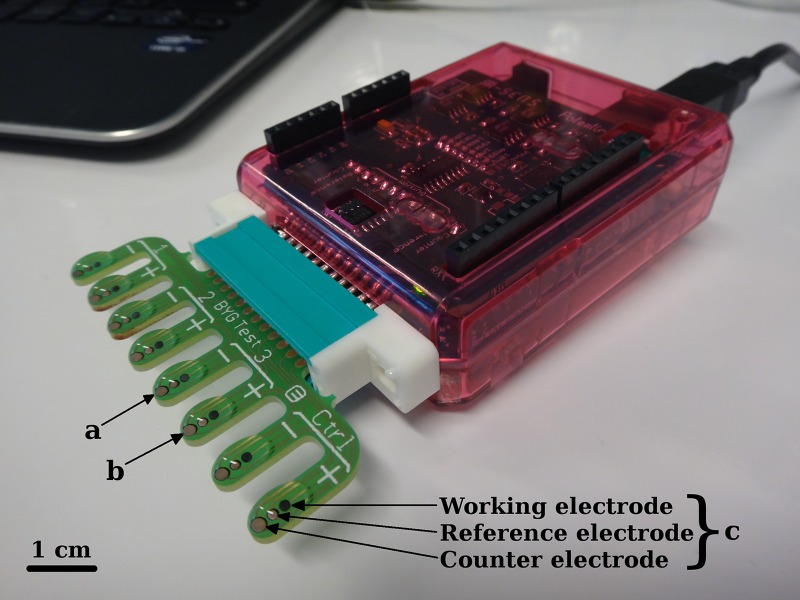
The potentiostat is multiplexed and analyzes eight different probes in parallel (Fig. 1). The instrument has a size of about 75 mm by 55 mm by 20 mm.
FIG 1.
Homemade potentiostat and eight-probe disposable electrode. Four isolates were analyzed in parallel (numbers 1 to 3 and one control [Ctrl]). Fifty-microliter drops of bacterial suspension in lysis buffer with (+) or without (−) imipenem were loaded on fingers corresponding to the isolates to be analyzed. Probe a was loaded with a drop of strain 3 suspended in buffer without imipenem; probe b was loaded with a drop of strain 3 suspended in buffer with imipenem; probe c was comprised of one working, one reference, and one counter electrode. The software subtracts the data obtained without imipenem from the data obtained with imipenem and generates a resulting real-time curve imaging the conductance of the polyaniline.
The electrodes.
The system uses disposable electrodes of eight probes that are produced by classical printed circuit board (PCB) realization techniques. The copper circuitry is protected by a solder mask varnish.
As polyaniline cannot be electro-synthesized on copper and as copper can be easily oxidized, all the electrode areas are coated with a screen-printed carbon layer having a resistance of approximately 14 to 20 Ω/square at a 25-μm dry film thickness.
Individual probes are composed of three electrode round spots (Fig. 1). The top spot has a diameter of 1 mm and constitutes the working electrode on which polyaniline is electro-synthesized. The middle electrode is the reference electrode; it has a diameter of 1 mm and is functionalized by applying a small spot of solid Ag/AgCl amalgam (Dupont 5874 silver/silver chloride composition; 4 h of curing at 80°C) on top of the carbon layer. This solid Ag/AgCl reference electrode has been checked for its stability, repeatability, and reliability in different measurement setups from pH 2 to 12. This reference displays an electrode potential 100 mV higher (+300 mV versus a standard hydrogen electrode [SHE]) than that of a commercial Ag/AgCl reference electrode (+197 mV versus SHE). The bottom electrode has a diameter of 1.5 mm and constitutes the counter electrode. It has a bigger surface and is also covered with the Ag/AgCl amalgam in order to prevent it from being the current limitation against the working electrode.
Each of the eight probes of the prepared electrodes is assignable by multiplexers present on the potentiostat card. The reference and counter electrodes are common between probes regarding the potentiostat electronic circuit. The electrode probes are about 4 mm.
Polyaniline electro-polymerization is performed by using the potentiostat in coulometry on the eight electrode probes placed in a row of a 96-multiwell platform. Each cell is filled with 300 μl of a 0.2 M aniline–2 M HCl aqueous solution. Electro-polymerization is performed up to 60 μC of charge on each working electrode at 890 mV against the solid Ag/AgCl reference electrode. After electro-synthesis, the electrode's probes are rinsed three times with distilled water, twice with 1 M aqueous ammonia, and finally three more times with distilled water. The electrodes are then dried using N2 and stored in common 96-multiwell platforms before any subsequent test. Aniline was distilled under reduced pressure prior to any experimentation. All chemicals were purchased from Sigma-Aldrich (Diegem, Belgium). Electro-synthesis and measurements were all performed at room temperature (20° to 25°C).
Conductometry.
The electrochemical equilibrium potential of the polyaniline electrode is first determined using the potentiostat and a method described previously (22, 23). The electrode is then brought to 10 mV above the equilibrium potential, and the current transient response is measured at a 34,487-Hz sampling rate during 11.6 ms (400 measured values). The electrode is then reset to its first equilibrium potential during 1 s. Finally, the electrode is brought to 10 mV below the equilibrium potential, and the current transient response is measured again at a 34,487-Hz sampling rate during 11.6 ms (400 measured values). Generally, the two measured current decays are mirror images, and their absolute values can be averaged. This decay is then integrated over the 400 measured values in order to obtain an arbitrary unit value that reflects the polyaniline electrochemical cell conductivity. This entire procedure is repeated over time whenever a stable equilibrium potential is attained during the hydrolysis measurement.
In a typical measurement, this type of signal, representing the conductance of the polyaniline and expressed in arbitrary units (AU), is recorded simultaneously for the four isolates that can be analyzed simultaneously on the electrode of the BYG test during 30 min. For each tested strain, two probes are used (Fig. 1, strains 1, 2, and 3 and a control strain, 4); one probe measures the signal with imipenem and the second probe measures the signal without imipenem (Fig. 1, + and −, respectively).
BYG Carba test and Carba NP test.
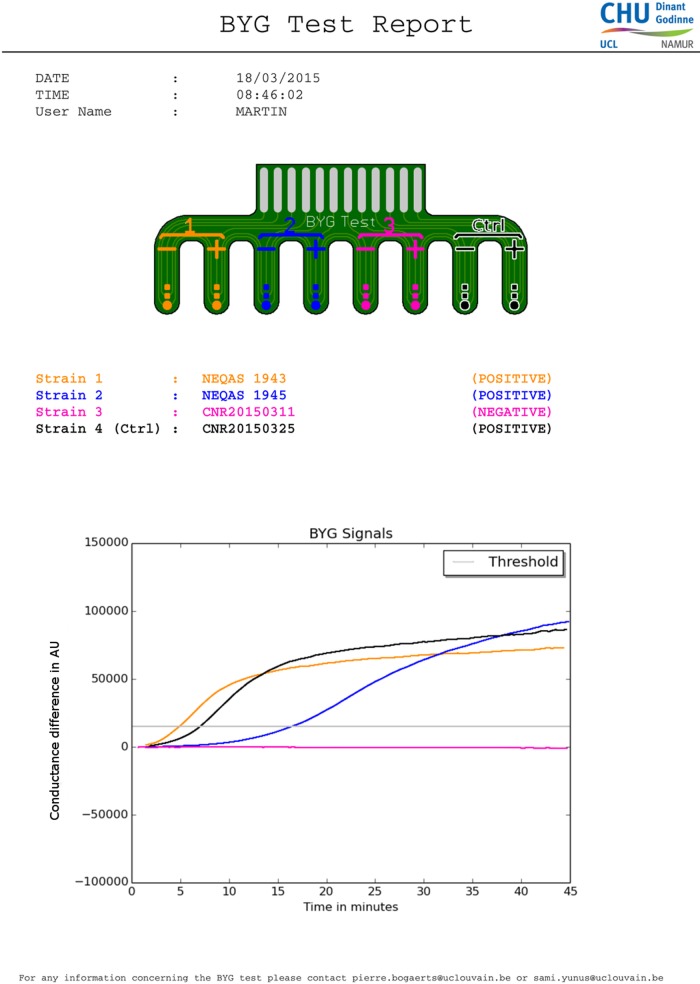
For the BYG Carba test, a full 10-μl loop of bacteria recovered from a fresh 18- to 24-h culture on tryptic soy agar (TSA)-sheep blood agar (Becton Dickinson, Erembodegem, Belgium) is suspended in 100 μl of an in-house prepared lysis buffer. Thirty microliters of this bacterial suspension is mixed with 100 μl of a 0.1 mM ZnSO4 solution with or without 3 mg/ml imipenem (6 mg/ml consisting of 3 mg/ml imipenem monohydrate plus 3 mg/ml cilastatin sodium [Tienam]; MSD France, Courbevoie, France). A 50-μl aliquot of this suspension is transferred on the probes (Fig. 1). A signal cutoff of 3.5 (arbitrary units) was chosen for the discrimination between carbapenemase and non-carbapenemase producers The results of the BYG Carba test are visualized as a curve appearing in real time. One curve is obtained for the signal (conductance) detected with imipenem, and another curve is obtained for the signal without imipenem (background curve). The software then subtracts the background from the signal obtained with imipenem (data not shown). Once the resulting curve crosses the cutoff, the corresponding isolate is reported as positive. At the end of the run, the software generates a report (Fig. 2). A preliminary control without bacterial extract was performed to confirm the stability of the imipenem in the solution during the experiment (data not shown).
FIG 2.
Real-time curve obtained with the BYG Carba test. A report of the results automatically generated by the software is shown. The gray horizontal line represents the cutoff line. The colored lines represent the real-time curve generated during the analysis. The orange (K. pneumoniae OXA-48 NEQAS 1943), blue (K. pneumoniae VIM-1 NEQAS 1945), and black (K. pneumoniae OXA-48 control strain CNR20150325) curves correspond to positive strains, and the pink flat curve corresponds to a negative Enterobacter aerogenes strain (CNR 20150311). The y axis is linked to the conductance of polyaniline, and values are expressed in arbitrary units.
For comparison, the Carba NP test was performed in parallel to the BYG Carba test on the same bacterial culture isolates according to the procedure initially published by Nordmann et al. (6).
Bacterial isolates.
The BYG Carba test was initially validated against a collection of 57 Enterobacteriaceae isolates previously characterized for their resistance mechanisms to β-lactam agents (24–26). These isolates comprised 41 carbapenemase producers and 16 noncarbapenemase producers (Table 1). The BYG Carba test was subsequently challenged against an external quality control (EQA) proficiency testing panel consisting of 10 putative CPE isolates (EQA exercise 2013, European survey on CPE [EuSCAPE] project; University Medical Center Groningen [UMCG] in collaboration with the United Kingdom National External Quality Assessment Service [NEQAS], September 2013) (Table 2). Imipenem, meropenem, and ertapenem MICs were determined by Etest (bioMérieux, Marcy l'Etoile, France) according to Clinical and Laboratory Standards Institute (CLSI) guidelines (27).
TABLE 1.
Validation of the BYG Carba test on 57 collection isolates
| Carbapenemase and isolate (n)a | MIC (μg/ml) of: |
BYG Carba resultb | BYG signal (AU)c | SD | Time to result (min) | ||
|---|---|---|---|---|---|---|---|
| Imipenem | Meropenem | Ertapenem | |||||
| OXA-48-like (12) | |||||||
| OXA-48 (11) | |||||||
| Klebsiella pneumoniae (5) | 0.19 to 1 | 0.38 to 3 | 0.75 to 2 | POS | 22.2 to 30.8 | 1.5 to 5.5 | <5 to 15 |
| Escherichia coli (2) | 0.75, 4 | 1.5, 8 | 1.5, 8 | POS | 33.2 to 35.5 | 4.8, 4.9 | <5 |
| Citrobacter freundii (1) | 1.5 | 0.75 | 3 | POS | 35.8 | 4.6 | <5 |
| Enterobacter cloacae (2) | 0.75, 1.5 | 0.38, 0.75 | 4, 12 | POS | 27.7, 31.2 | 1.2, 3.3 | <5 |
| Klebsiella oxytoca (1) | 0.5 | 3 | 1.5 | POS | 31.2 | 0.9 | >5 to 10 |
| OXA-162 (1) | |||||||
| Klebsiella pneumoniae (1) | 0.75 | 0.38 | 24 | POS | 23.5 | 4.5 | >5 to 10 |
| KPC (8) | |||||||
| KPC-2 (8) | |||||||
| Klebsiella pneumoniae (8) | 8 to >32 | 6 to >32 | 8 to >32 | POS | 34.5 to 53.9 | 0.9 to 9.9 | >5 to 10 |
| NDM (8) | |||||||
| NDM-1 (7) | |||||||
| Enterobacter cloacae (1) | 3 | 4 | 4 | POS | 35.6 | 2.8 | <5 |
| Escherichia coli (2) | 4 | 0.75 to >32 | 3 to >32 | POS | 32.2 and 51.4 | 2.7 and 6.3 | <5 |
| Klebsiella pneumoniae (3) | 2 to >32 | 4 to >32 | 12 to >32 | POS | 50.6 to 65.6 | 2.1 to 6.0 | <5 |
| Morganella morganii (1) | 24 | 6 | 1 | POS | 35.6 | 2.8 | <5 |
| NDM-5 (1) | |||||||
| Escherichia coli (1) | 16 | >32 | >32 | POS | 32.2 | 2 | <5 |
| VIM (8) | |||||||
| VIM-1 (4) | |||||||
| Citrobacter braakii (1) | 3 | 1.5 | 2 | POS | 14.2 | 5.1 | <5 |
| Klebsiella pneumoniae (1) | 32 | 24 | 16 | POS | 29.2 | 0.7 | >5 to 10 |
| Klebsiella oxytoca (1) | 24 | 2 | 8 | POS | 35 | 3.4 | <5 |
| Providencia vermicola (1) | >32 | >32 | 2 | POS | 30.8 | 8.4 | >5 to 10 |
| VIM-4 (2) | |||||||
| Serratia marcescens (1) | 32 | >32 | 32 | POS | 30.4 | 1.7 | >5 to 10 |
| Aeromonas caviae (1) | 0.75 | 0.064 | 0.34 | POS | 50.6 | 0.8 | <5 |
| VIM-27 (1) | |||||||
| Klebsiella pneumoniae (1) | >32 | >32 | >32 | POS | 35.3 | 5.9 | <5 |
| VIM-31 (1) | |||||||
| Enterobacter cloacae (1) | 1.5 | 1.5 | 0.5 | POS | 41.7 | 9.9 | <5 |
| IMP (3) | |||||||
| IMP-4 (1) | |||||||
| Klebsiella pneumoniae (1) | 1 | 1.5 | 1.5 | POS | 42.7 | 3.3 | <5 |
| IMP-8 (1) | |||||||
| Enterobacter cloacae (1) | 12 | 3 | 3 | POS | 39.3 | 3.8 | <5 |
| IMP-11 (1) | |||||||
| Serratia marcescens (1) | 3 | 2 | 3 | POS | 44.3 | 6.2 | <5 |
| GIM (1) | |||||||
| GIM-1 (1) | |||||||
| Enterobacter cloacae (1) | 0.38 | 1.5 | 3 | POS | 61.1 | 4.5 | <5 |
| GES (1) | |||||||
| GES-6 (1) | |||||||
| Citrobacter braakii (1) | 0.5 | 0.094 | 0.064 | NEG | −1.60 | 0.62 | |
| No carbapenemase (16)d | |||||||
| Citrobacter amalonaticus (1) | 0.19 | 0.047 | 0.016 | NEG | −3.3 | 1.8 | 30 |
| Citrobacter freundii (1) | 0.25 | 0.19 | 0.38 | NEG | −0.1 | 0.5 | 30 |
| Enterobacter aerogenes (1) | 0.25 | 0.064 | 0.25 | NEG | −0.8 | 0.6 | 30 |
| Enterobacter asburiae (1) | 0.19 | 0.125 | 0.19 | NEG | 0.2 | 0.7 | 30 |
| Escherichia coli (4) | 0.125 to 0.19 | 0.032 to 0.23 | 0.012 to 0.032 | NEG | −15.3 to 0.2 | 0.4 to 10.8 | 30 |
| Klebsiella pneumoniae (4) | 0.125 to 0.25 | 0.047 to 2 | 0.016 to 0.75 | NEG | −1.6 to 0.1 | 0.2 to 1.9 | 30 |
| Proteus mirabilis (1) | 1.5 | 0.19 | 0.125 | NEG | −0.1 | 0.9 | 30 |
| Providencia stuartii (1) | 0.5 | 0.125 | 0.023 | NEG | −4.6 | 1.9 | 30 |
| Salmonella sp. (1) | 0.25 | 0.094 | 0.047 | NEG | −1,5 | 1.5 | 30 |
| Serratia marcescens (1) | 0.25 | 0.094 | 0.125 | NEG | −0.7 | 0.4 | 30 |
n, number of isolates.
POS, positive; NEG, negative.
AU, arbitrary units.
Including the following: TEM-1 (n = 9), TEM-24 (n = 1), TEM-30 (n = 1), SHV-1 (n = 1), SHV-2a (n = 2), SHV-11 (n = 1), SHV-12 (n = 2), SHV-28 (n = 1), SHV-76 (n = 1), CTX-M-2 (n = 1), CTX-M-3 (n = 1), CTX-M-14 (n = 1), CTX-M-15 (n = 3), GES-7 (n = 1), OXA-1 (n = 1), OXA-10 (n = 1), OXA-163 (1), ACC-1 (n = 1), DHA-1 (n = 1), DHA-7 (n = 1), and extended-spectrum AmpC (n = 1).
TABLE 2.
Results of the BYG Carba Test on the carbapenemase-producing Enterobacteriaceae panel NEQAS 2013
| Specimen no. | Species | Carbapenemase | MIC (μg/ml) of: |
BYG Carba resulta | Time to result (min) | BYG signal (AU)b | ||
|---|---|---|---|---|---|---|---|---|
| Imipenem | Meropenem | Ertapenem | ||||||
| NEQAS 1940 | K. pneumoniae | KPC | >32 | >32 | >32 | POS | <5 | 64.3 |
| NEQAS 1941 | E. cloacae | NDM-1 | >32 | >32 | >32 | POS | <5 | 50.9 |
| NEQAS 1942 | K. pneumoniae | KPC-3 | >32 | >32 | >32 | POS | <5 | 62.8 |
| NEQAS 1943 | K. pneumoniae | OXA-48 | 1 | 1.5 | 4 | POS | <5 | 38.2 |
| NEQAS 1944 | K. pneumoniae | KPC-2 | >32 | >32 | 24 | POS | <5 | 54.9 |
| NEQAS 1945 | K. pneumoniae | VIM-1 | >32 | >32 | >32 | POS | >5–10 | 32.7 |
| NEQAS 1946 | K. pneumoniae | NDM-1 | >32 | >32 | >32 | POS | <5 | 50.3 |
| NEQAS 1947 | K. pneumoniae | IMP-1 | 12 | 3 | 8 | POS | <5 | 61.3 |
| NEQAS 1948 | K. pneumoniae | NDM-1 | >32 | >32 | >32 | POS | <5 | 58.3 |
| NEQAS 1949 | E. aerogenes | None | 6 | 3 | 32 | NEG | 30 | 1.0 |
S, positive; NEG, negative.
Arbitrary unit.
The performance of the test was also assessed prospectively on 324 consecutive Enterobacteriaceae isolates referred to the National Reference Laboratory (NRL) by Belgian laboratories in 2014 for nonsusceptibility to at least one carbapenem (ertapenem, imipenem, or meropenem). Nonsusceptibility to carbapenems was assessed by the referring laboratories according to CLSI (27) or to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (http://www.eucast.org), which are the two sets of guidelines used by most microbiologists in Belgium. Bacterial confirmation of identification to the species level was carried out centrally at the NRL using matrix-assisted laser desorption–ionization time of flight (MALDI-TOF) mass spectrometry on a Microflex LT (Bruker Daltonics GmbH, Bremen, Germany), and antimicrobial resistance profiles were analyzed phenotypically by a disk diffusion method according to CLSI guidelines (27). Meropenem and ertapenem MICs were determined using broth microdilution panels (Sensititre, Thermo Fisher Scientific, Cleveland, IL, USA) in the prospective study.
All isolates were verified for the presence of carbapenemase by in-house multiplex PCR targeting blaVIM, blaIMP, blaNDM, blaKPC, and blaOXA-48 (26) and other β-lactamase genes (24, 25). Results of the molecular tests were considered the gold standard for the presence of a carbapenemase. In the case of discrepancies observed between hydrolysis tests and PCR results, the resistance genes detected by PCR were sequenced using an external service company (Macrogen, Inc., Seoul, South Korea). The sequence obtained was compared with the genes present in GenBank and aligned with the reference gene referred at the Lahey clinic (http://www.lahey.org/Studies/).
RESULTS
Validation of collection isolates and external quality assessment.
Of the 57 collection isolates tested in triplicate (Table 1), the BYG Carba test correctly and repeatedly detected 40 of the carbapenemase producers but missed one single GES-6-producing isolate, a very weak and rarely reported class A serine carbapenemase (28). On the other hand, all noncarbapenemase producers yielded a negative BYG Carba test result. Based on these results, the BYG Carba test was found to display a sensitivity of 97.6% and a specificity of 100%. All 40 CPE isolates were detected as positive by the BYG Carba test within 15 min, with 34 CPE isolates being detected in less than 5 min (including 9 out of 12 OXA-48-like producers). No correlation between the intensity of the signal and MICs could be observed (Table 1). The maximum observed signal value after 30 min of reaction was 70.2 (arbitrary units [AU]) for an NDM-1-producing K. pneumoniae isolate, and the minimum was 9.9 AU for a VIM-1-producing Citrobacter braakii (data not shown).
The BYG Carba test was further validated with an external quality control provided by the NEQAS which distributed a panel of 10 suspected CPE isolates during September 2013 (Table 2). The agreement with expected results was 100%. The BYG Carba test correctly identified the 9 CPE isolates and the single non-carbapenemase-producing Enterobacter aerogenes. Eight out of the nine CPE isolates were detected in less than 5 min, including one OXA-48-producing K. pneumoniae isolate (NEQAS 1943), while the VIM-1-producing K. pneumoniae isolate (NEQAS 1945) was detected between 5 and 10 min after the start of the test. The maximum value (64.3) was observed for a KPC producer (NEQAS 1940), and the lowest (32.7) was obtained for the VIM-1 producer (NEQAS 1945). The value at 30 min for the non-carbapenemase-producing E. aerogenes isolate (NEQAS 1949) was 1.0.
Prospective evaluation.
The BYG Carba test was then evaluated prospectively on consecutive strains referred to the NRL for the detection of CPE (Table 3).
TABLE 3.
Prospective evaluation of the BYG Carba test on 324 isolates
| Carbapenemase, isolate, and prospective result (n)a | Meropenem MIC (μg/ml) | Ertapenem MIC (μg/ml) | BYG Carba resultb | BYG signal (AU)c | Time to result (min) | Carba NP result (no. of isolates and appearance) |
|---|---|---|---|---|---|---|
| OXA-48 (117) | ||||||
| BYG positive (109) | ||||||
| Klebsiella pneumoniae (65) | <0.25 to >32 | <0.25 to >32 | POS | 3.6 to 42 | <5 to 30 | 52 POS (50 orange, 2 yellow), 12 NEG, 1 noninterpretable |
| Escherichia coli (15) | <0.25 to 32 | <0.25 to >32 | POS | 13.4 to 43.8 | <5 to 20 | 12 POS (orange), 3 NEG |
| Citrobacter freundii (14) | 0.5 to 8 | 0.5 to 2 | POS | 27 to 46 | <5 | 13 POS (12 orange, 1 yellow), 1 mucoid |
| Enterobacter cloacae (9) | <0.25 to 4 | 2 to 16 | POS | 25.7 to 36.9 | <5 to 10 | 9 POS (7 orange, 2 yellow) |
| Klebsiella oxytoca (5) | <0.25 to 2 | 0.5 to 4 | POS | 19.3 to 31.3 | <5 to 15 | 5 POS (3 orange, 2 yellow) |
| Serratia marcescens (1) | >32 | >32 | POS | 26.3 | <10 | 1 POS (orange) |
| BYG false negative (8) | ||||||
| Klebsiella pneumoniae (7) | <0.25 to >32 | 2 to >32 | NEG | −1 to 2.2 | 30 | 3 POS (orange), 4 NEG |
| Escherichia coli (1) | <0.25 | <0.25 | NEG | 0.4 | 30 | 1 NEG |
| KPC (25) | ||||||
| BYG positive (25) | ||||||
| Klebsiella pneumoniae (25) | 2 to >32 | 8 to >32 | POS | 40.2 to 76.8 | <5 | 25 POS (23 yellow, 2 orange) |
| NDM (23) | ||||||
| BYG positive (23) | ||||||
| Enterobacter cloacae (13) | 4 to >32 | 8 to >32 | POS | 35.9 to 59.0 | <5 | 13 POS (11 yellow, 2 orange) |
| Escherichia coli (4) | 2 to 16 | 4 to 16 | POS | 45.9 to 56.6 | <5 to 10 | 4 POS (3 yellow, 1 orange) |
| Klebsiella pneumoniae (3) | 8 to >32 | 16 to >32 | POS | 43.5 to 63.7 | <5 | 3 POS (1 yellow, 2 orange) |
| Aeromonas caviae (1) | 2 | POS | 49.7 | <10 | 1 POS (orange) | |
| Citrobacter freundii (1) | 4 | 8 | POS | 56.2 | <5 | 1 POS (yellow) |
| Klebsiella oxytoca (1) | 32 | 32 | POS | 53.8 | <5 | 1 POS (yellow) |
| VIM (13) | ||||||
| BYG positive (12) | ||||||
| Enterobacter cloacae (4) | 4 to 32 | 8 to 32 | POS | 38.4 to 58.3 | <5 to 10 | 4 POS (3 yellow, 1 orange) |
| Klebsiella pneumoniae (3) | <0.25 to 1 | <0.25 to 2 | POS | 10.3 to 32.6 | >5 to 15 | 3 POS (3 yellow) |
| Escherichia coli (2) | <0.25 and 1 | <0.25 and 1 | POS | 10.6 and 19.1 | 20 and 25 | 2 POS (2 yellow) |
| Citrobacter amalonaticus (1) | 16 | 8 | POS | 38.8 | <5 | 1 POS (yellow) |
| Citrobacter freundii (1) | 8 | 8 | POS | 32.9 | <5 | 1 POS (orange) |
| Klebsiella oxytoca (1) | >32 | 32 | POS | 22.1 | <5 | 1 POS (yellow) |
| BYG false negative (1) | ||||||
| Klebsiella oxytoca (1) | 32 | >32 | NEG | 1.6 | 30 | 1 POS (yellow) |
| No carbapenemase (146)d | ||||||
| BYG negative (146) | ||||||
| Klebsiella pneumoniae (51) | <0.25 to 32 | <0.25 to >32 | NEG | −19.4 to 3.2 | 30 | 47 NEG, 3 noninterpretable, 1 mucoid |
| Enterobacter cloacae (34) | <0.25 to 2 | <0.25 to 16 | NEG | −20.2 to 1.3 | 30 | 32 NEG, 2 mucoid |
| Citrobacter freundii (3) | <0.25 to 4 | 1 to 16 | NEG | −29.9 to −0.2 | 30 | 2 NEG, 1 mucoid |
| Enterobacter aerogenes (21) | <0.25 to 16 | <0.25 to >32 | NEG | −1.07 to 1.8 | 30 | 3 NEG |
| Escherichia coli (24) | <0.25 to 4 | <0.25 to 32 | NEG | −17.2 to 0.9 | 30 | 23 NEG, 1 mucoid |
| Klebsiella oxytoca (6) | <0.25 to 8 | 1 to 16 | NEG | −1.4 to 2.8 | 30 | 5 NEG, 1 mucoid |
| Morganella morganii (2) | <0.25 | <0.25 | NEG | −12.25 and 1.8 | 30 | 2 NEG |
| Proteus mirabilis (2) | <0.25 and 1 | <0.25 | NEG | −2.9 and −0.5 | 30 | 2 NEG |
| Serratia marcescens (2) | <0.25 and 2 | 1 and 16 | NEG | 0.2 | 30 | 2 NEG |
| Citrobacter braakii (1) | <0.25 | 1 | NEG | −0.2 | 30 | 1 NEG |
n, number of isolates.
POS, positive; NEG, negative.
AU, arbitrary units.
Including the following: TEM (n = 28), SHV (n = 30), OXA-1 group (n = 29), CTX-M of group 1 (n = 22), CTX-M of group 2 (n = 1), CTX-M of group 9 (n = 1), and DHA (n = 9).
Over a 4-month period during the first half of 2014, 324 suspected CPE isolates collected in 66 Belgian laboratories were received by the NRL. All isolates were analyzed for carbapenem susceptibility testing by the Carba NP test, by the BYG Carba test, and by multiplex PCR for the identification of carbapenemase genes according to the procedures described in Materials and Methods. All 324 Enterobacteriaceae isolates could be analyzed and yielded an interpretable result with the BYG Carba test while the Carba NP test gave definite results for 313 isolates (96%) and no result for 11 (3.4%) strains (7 mucoid isolates could not be sampled by pipetting because of hyperviscosity after the lysis step of the Carba NP test and 4 isolates had noninterpretable results, with the control without imipenem yielding an orange color not distinguishable from that of the strain with imipenem). Among the 11 isolates with an indeterminate Carba NP test result (K. pneumoniae, n = 5; Enterobacter cloacae, n = 2; Citrobacter freundii, n = 2; Escherichia coli and Klebsiella oxytoca, n = 1 each), two isolates produced an OXA-48 and were positive with the BYG test, and nine were noncarbapenemase producers with a negative BYG test result.
In comparison with results from PCR, considered the reference gold standard, the BYG Carba test gave 315/324 (97.2%) interpretable and correct results after 30 min, and the Carba NP test yielded 294/324 (90.7%) interpretable and correct results after 2.5 h (including lysis). Neither the BYG Carba nor Carba NP test presented false-positive results (100% specificity for each of the tests).
Nine strains out of 178 carbapenemase producers (5.1%) yielded a false-negative result with the BYG Carba test (OXA-48, n = 7; OXA-232, n = 1; and VIM-2, n = 1), while 19 false-negative results (10.7%) occurred with the Carba NP test (OXA-48, n = 16, and OXA-232, n = 3). Four OXA-48-like producing K. pneumoniae isolates (OXA-48, n = 3, and OXA-232, n = 1) were not detected by any of the tests (Table 4).
TABLE 4.
False-negative and discrepant results between the BYG Carba and the Carba NP tests
| OXA-48-like discrepant result and isolate (n)a | Meropenem MIC (μg/ml) | Ertapenem MIC (μg/ml) | BYG Carba resultb | BYG signal (AU)c | Time to result (min) | Carba NP result (color) |
|---|---|---|---|---|---|---|
| False-negative Carba NP only (15) | ||||||
| K. pneumoniae OXA-48 (10) | <0.25 to 1 | 1 to 8 | POS | 5.4 to 34.4 | 30 | NEG |
| K. pneumoniae OXA-232 (2) | 16 and >32 | >32 | POS | 4.1 and 4.4 | 30 | NEG |
| E. coli OXA-48 (3) | <0.25 to 32 | <0.25 to >32 | POS | 22.8 to 27.5 | <5 to 15 | NEG |
| False-negative BYG Carba only (5) | ||||||
| K. pneumoniae OXA-48 (3) | 1 to >32 | 4 to >32 | NEG | 1.5 to 2.1 | 30 | POS (orange) |
| E. coli (1) | <0.25 | <0.25 | NEG | 0.4 | 30 | POS (orange) |
| K. oxytoca VIM-2 (1) | 32 | >32 | NEG | 1.6 | 30 | POS (yellow) |
| False-negative BYG Carba and Carba NP (4) | ||||||
| K. pneumoniae OXA-48 (3) | <0.25 to 4 | 2 to 32 | NEG | −1.0 to 0.5 | 30 | NEG |
| K. pneumoniae OXA-232 (1) | >32 | >32 | NEG | 2.2 | 30 | NEG |
n, number of isolates. Total number of isolates, 23.
POS, positive; NEG, negative.
AU, arbitrary units.
Overall, the BYG Carba test yielded sensitivity, specificity, positive predictive values (PPV), and negative predictive values (NPP) of 95, 100, 100, and 94%, respectively, and the values obtained with the Carba NP test were 89, 100, 100, and 88%, respectively, not taking into account the 11 noninterpretable results.
One definite advantage of the BYG Carba test is related to the fact that once a cutoff limit is set, the interpretation becomes objective by reporting of a number. With the current method of analysis, for the 169 isolates (out of 178 CPE isolates) with a positive BYG Carba test result, the values obtained ranged between 3.63 and 76.83 AU (mean, 32.08 AU; median, 31.34 AU; standard deviation [SD], 16.50 AU). For the 155 BYG-negative strains including 146 non-CPE isolates, the values of the BYG signal ranged between −29.91 and 3.17 AU (mean, −1.19 AU; median, −0.20 AU; SD, 4.1 AU), with a negative value indicating that, after 30 min, the sample without imipenem presented a higher value than the sample with imipenem. The highest values were observed for KPC producers (mean, 52.25 AU; SD, 12.07 AU) followed by NDM producers (mean, 50.25 AU; SD, 7.45 AU), VIM producers (mean, 30.78 AU; SD, 10.35 AU), and OXA-48 producers (mean, 22.16 AU; SD, 12.08 AU) (Table 3).
Regarding the time to result, 90 out of 178 (50.6%) of the carbapenemase producers were detected in less than 5 min (including 40 OXA-48 producers), and more than 80% of the CPE isolates (144/178) were detected in less than 15 min (data not shown). No correlation could be observed between meropenem or ertapenem diameter or MIC and the intensity of the signal.
DISCUSSION
The detection of CPE is challenging for diagnostic microbiological laboratories (10) but is of utmost importance for prevention and outbreak control in clinical settings (29). Rapid, sensitive, and specific laboratory detection techniques based on the detection of carbapenem hydrolysis have been recently proposed in the literature (10). The most widespread colorimetric test, the Carba NP test, was reported by several laboratories as lacking some degree of sensitivity, especially for the detection of OXA-48 and for some metallo-β-lactamase producers (30–34). Several groups or companies subsequently developed slightly modified tests based on the principle of the Carba NP test. For example, Pires et al. proposed and evaluated a Carba NP-like test using bromothymol blue instead of the phenol red as a pH indicator with 93.3 to 100% sensitivity and 100% specificity (7, 35). The Rosco Diagnostica company (Taastrup, Denmark) developed two distinct commercial tests (Rosco Rapid Carb screen kits), also based on the hydrolysis of imipenem using either phenol red or bromothymol blue as the indicator. The published evaluation of this test revealed some difficulty in obtaining readings, resulting in up to 12% of noninterpretable results and poor specificity (83%) (8, 36). A recent evaluation of Dortet and colleagues further confirmed the poor specificity of this test (71%) (37).
Very recently, bioMérieux proposed a commercial ready-to-use version of the Carba NP test, the Rapidec Carba NP. The test still requires an incubation time of 30 min before incubation of the plastic tray at 37°C and interpretation of the results after 30, 60, and 120 min. Two evaluations of the test by the inventors revealed sensitivity and specificity varying between 96 and 99% and between 96 and 100%, respectively (37, 38). All of these tests have in common the fact that they are interpreted by naked eye (color modification), and hence readings may be subjective and variable between observers, especially when color changes are very faint.
In this article, we propose the BYG Carba test, a novel original electrochemical diagnostic tool for the rapid detection of CPE from bacterial suspensions.
The BYG Carba test indeed offers several advantages in comparison to colorimetrically based methods. First, the BYG technology reduces the time to results to about 30 min, and the test can be performed at room temperature without incubation at 37°C. Second, the BYG method takes its advantages from the fact that, in addition to the acidification of the medium, the oxydo-reduction process also participates in the signal detected by the system (20, 21), suggesting that the BYG test could be a better sensor for the hydrolysis of carbapenemases than only a colorimetric pH indicator or iodine indicators (39). In our hands, the test leads to a significant improvement for the detection OXA-48 producers compared to results with the Carba NP test (8 false-negative results for the BYG Carba test versus 19 with the Carba NP test; P < 0.02). However, this could be, at least partially, the consequence of the subjective analysis of the Carba NP test as Papagiannitis and colleagues recently showed that interpretation of the Carba NP test by an enzyme-linked immunosorbent assay (ELISA) reader improves the sensitivity of the test from 76 to 81% (18). In our hands the sensitivity for OXA-48-like detection by the Carba NP test was 83.8%, and with the BYG Carba test it was 93.2%.
Third, an electrochemical test permits the real-time objective measurement and the traceability of the signal, with the results being visualized as a real-time curve (Fig. 2). In the scope of the accreditation process for the clinical laboratory, this element may represent a substantial advantage. During the prospective evaluation, definitive positive results (i.e., production of carbapenemase) could be confirmed in less than 5 min in more than 50% of the cases. Moreover, in the current format, the BYG Carba test can analyze up to four strains in parallel, but the technology also allows parallel arrangement of the electrodes, allowing testing of up to 192 strains (48 electrodes). The electrodes are currently produced in the laboratory at about $2/electrode and $0.50/strain. The reader is also produced in-house (about $100), and a software program automatically interprets the data and generates a report (pdf format). In conclusion, the BYG Carba test is a novel electrochemical assay that was developed for the detection of CPE. This method is rapid (detection within 30 min), traceable, objective, sensitive, and specific, with slightly improved performance compared to that of the Carba NP test. Further evaluation of this test by independent investigator groups is currently ongoing.
ACKNOWLEDGMENTS
This work was partially supported by the Belgian Walloon region (First Spin-off E-Sens and POC Project, VALBYG Convention 1117301).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that we have no conflicts of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers in Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 2.Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen Ø, Seifert H, Woodford N, Nordmann P, European Network on Carbapenemases . 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect 18:413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. [DOI] [PubMed] [Google Scholar]
- 4.Doyle D, Peirano G, Lascols C, Lloyd T, Chuirch DL, Pitout JD. 2012. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol 50:3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. 2015. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pires J, Novais A, Peixe L. 2013. Blue-Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol 51:4281–4283. doi: 10.1128/JCM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang TD, Berhin C, Bogaerts P, Glupczynski Y. 2014. Comparative evaluation of two chromogenic tests for rapid detection of carbapenemase in Enterobacteriaceae and in Pseudomonas aeruginosa isolates. J Clin Microbiol 52:3060–3063. doi: 10.1128/JCM.00643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrabák J, Chudáčková E, Papagiannitsis CC. 2014. Detection of carbapenemases in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin Microbiol Infect 20:839–853. doi: 10.1111/1469-0691.12678. [DOI] [PubMed] [Google Scholar]
- 11.Tsakris A, Poulou A, Bogaerts P, Dimitroulia E, Pournaras S, Glupczynski Y. 2015. Evaluation of a new phenotypic OXA-48 disk test for differentiation of OXA-48 carbapenemase-producing Enterobacteriaceae clinical isolates. J Clin Microbiol 53:1245–1251. doi: 10.1128/JCM.03318-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernabeu S, Poirel L, Nordmann P. 2012. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn Microbiol Infect Dis 74:88–90. doi: 10.1016/j.diagmicrobio.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 49:3222–3227. doi: 10.1128/JCM.00984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol 49:3321–3324. doi: 10.1128/JCM.00287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauget M, Cabrolier N, Manzoni M, Bertrand X, Hocquet D. 2014. Rapid, sensitive and specific detection of OXA-48-like-producing Enterobacteriaceae by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Microbiol Methods 105:88–91. doi: 10.1016/j.mimet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Johansson Å, Nagy E, Sóki J. 2014. Instant screening and verification of carbapenemase activity in Bacteroides fragilis in positive blood culture, using matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Med Microbiol 63:1105–1110. doi: 10.1099/jmm.0.075465-0. [DOI] [PubMed] [Google Scholar]
- 17.Knox J, Jadhav S, Sevior D, Agyekum A, Whipp M, Waring L, Iredell J, Palombo E. 2014. Phenotypic detection of carbapenemase-producing Enterobacteriaceae by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and the Carba NP test. J Clin Microbiol 52:4075–4077. doi: 10.1128/JCM.02121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papagiannitsis CC, Študentová V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, Hrabák J. 2015. Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol 53:1731–1735. doi: 10.1128/JCM.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasserre C, De Saint Martin L, Cuzon G, Bogaerts P, Lamar E, Glupczynski Y, Naas T, Tandé D. 2015. Efficient detection of carbapenemase activity in Enterobacteriaceae by matrix-assisted laser desorption ionization-time of flight mass spectrometry in less than 30 minutes. J Clin Microbiol 53:2163–2171. doi: 10.1128/JCM.03467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang JC, MacDiarmid AG. 1986. Polyaniline: protonic acid doping of the emeraldine form to the metallic regime. Synth Met 13:193–205. doi: 10.1016/0379-6779(86)90070-6. [DOI] [Google Scholar]
- 21.MacDiarmid AG, Chiang JC, Richter AF, Epstein AJ. 1987. Polyaniline: a new concept in conducting polymers. Synth Met 18:285–290. doi: 10.1016/0379-6779(87)90893-9. [DOI] [Google Scholar]
- 22.Yunus S, Attout A, Vanlancker G, Bertrand P, Ruth N, Galleni M. 2011. A method to probe electrochemically active material state in portable sensor applications. Sens Actuators B Chem 156:35–42. doi: 10.1016/j.snb.2011.03.070. [DOI] [Google Scholar]
- 23.Yunus S, Bertrand P, Attout A. July 2011. Smart sensor system using an electroactive polymer. International patent WO2011082837.
- 24.Bogaerts P, Hujer AM, Naas T, de Castr Rezende R, Endimiani A, Nordmann P, Glupczynski Y, Bonomo RA. 2011. Multicenter evaluation of a new DNA microarray for rapid detection of clinically relevant bla genes from β-lactam-resistant gram-negative bacteria. Antimicrob Agents Chemother 55:4457–4460. doi: 10.1128/AAC.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Nordmann P. 2012. Evaluation of a DNA microarray for the rapid detection of extended-spectrum β-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM). J Antimicrob Chemother 67:1865–1869. doi: 10.1093/jac/dks156. [DOI] [PubMed] [Google Scholar]
- 26.Bogaerts P, Rezende de Castro R, de Mendonça R, Huang TD, Denis O, Glupczynski Y. 2013. Validation of carbapenemase and extended-spectrum β-lactamase multiplex endpoint PCR assays according to ISO 15189. J Antimicrob Chemother 68:1576–1582. doi: 10.1093/jac/dkt065. [DOI] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement update. Document M100-S24 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 28.Vourli S, Giakkoupi P, Miriagou V, Tzelepi E, Vatopoulos AC, Tzouvelekis LS. 2004. Novel GES/IBC extended-spectrum beta-lactamase variants with carbapenemase activity in clinical Enterobacteria. FEMS Microbiol Lett 234:209–213. doi: 10.1111/j.1574-6968.2004.tb09535.x. [DOI] [PubMed] [Google Scholar]
- 29.Schwaber MJ, Carmeli Y. 2014. An ongoing national intervention to contain the spread of carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 58:697–703. doi: 10.1093/cid/cit795. [DOI] [PubMed] [Google Scholar]
- 30.Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. 2013. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osterblad M, Hakanen AJ, Jalava J. 2014. Evaluation of the Carba NP test for carbapenemase detection. Antimicrob Agents Chemother 58:7553–7556. doi: 10.1128/AAC.02761-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chong PM, McCorrister SJ, Unger MS, Boyd DA, Mulvey MR, Westmacott GR. 2015. MALDI-TOF MS detection of carbapenemase activity in clinical isolates of Enterobacteriaceae spp., Pseudomonas aeruginosa, and Acinetobacter baumannii compared against the Carba-NP assay. J Microbiol Methods 111:21–23. doi: 10.1016/j.mimet.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 33.Maurer FP, Castelberg C, Quiblier C, Bloemberg GV, Hombach M. 2015. Evaluation of carbapenemase screening and confirmation tests with Enterobacteriaceae and development of a practical diagnostic algorithm. J Clin Microbiol 53:95–104. doi: 10.1128/JCM.01692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dortet L, Bréchard L, Poirel L, Nordmann P. 2014. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the Carba NP test. J Med Microbiol 63:772–776. doi: 10.1099/jmm.0.071340-0. [DOI] [PubMed] [Google Scholar]
- 35.Novais Â, Brilhante M, Pires J, Peixe L. 2015. Evaluation of the recently launched rapid Carb Blue kit for detection of carbapenemase-producing gram-negative bacteria. J Clin Microbiol 53:3105–3107. doi: 10.1128/JCM.01170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simner PJ, Gilmour MW, DeGagne P, Nichol K, Karlowsky JA. 2015. Evaluation of five chromogenic agar media and the Rosco Rapid Carb screen kit for detection and confirmation of carbapenemase production in gram-negative bacilli. J Clin Microbiol 53:105–112. doi: 10.1128/JCM.02068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dortet L, Agathine A, Naas T, Cuzon G, Poirel L, Nordmann P. 2015. Evaluation of the RAPIDEC® CARBA NP, the Rapid CARB Screen® and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 70:3014–3022. doi: 10.1093/jac/dkv213. [DOI] [PubMed] [Google Scholar]
- 38.Poirel L, Nordmann P. 2015. Rapidec Carba NP test for rapid detection of carbapenemase producers. J Clin Microbiol 53:3003–3008. doi: 10.1128/JCM.00977-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawai T, Takahashi I, Yamagishi S. 1978. Iodometric assay method for beta-lactamase with various beta-lactam antibiotics as substrates. Antimicrob Agents Chemother 13:910–913. doi: 10.1128/AAC.13.6.910. [DOI] [PMC free article] [PubMed] [Google Scholar]