Abstract
We evaluated an immunochromatographic lateral flow assay to detect OXA-48-like carbapenemases (OXA-48 K-SeT) in Enterobacteriaceae (n = 82). One hundred percent sensitivity and specificity were observed using bacteria recovered from both solid medium and spiked blood culture bottles, and the results were obtained in <10 min.
TEXT
The identification, treatment, and control of multidrug-resistant (MDR) bacterial infections are global health priorities. Enterobacteriaceae with plasmids carrying genes encoding class A (Klebsiella pneumoniae carbapenemases [KPC]), B (IMP, VIM, and NDM), and D (OXA) carbapenemases (carbapenemase-producing Enterobacteriaceae [CPE]) are one of the most important groups of pathogens due to the burden of disease, lack of any new treatments, and potential for dissemination (1). Rapid and effective diagnostics therefore underpin any strategy aimed at tackling the problem of CPE. Considerable effort has been made to develop novel assays using both genotypic and phenotypic approaches. These approaches include the genetic detection of resistance gene profiles (PCR, loop-mediated isothermal amplification [LAMP], microarrays, and genome sequencing) (2), selective culture medium (chromogenic or supplemented) (3), combination disk testing (4), and direct or indirect detection of carbapenem-hydrolyzing enzymes (matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS], acidometric Carba NP, and Blue-Carba) (5–7). These methods require varied levels of technical skill, investment into equipment, and quality assurance optimization. Strains producing OXA-48-like carbapenem-hydrolyzing class D enzymes (CHDL) have proven to be particularly difficult to detect in clinical laboratories. This is due in part to relatively low MICs, conflicting interpretive rules associated with automated systems (8), and a lack of suitable inhibitor compounds for use in confirmatory tests. A novel means of detecting OXA-48-like enzymes using an antibody-mediated approach was recently developed (9). The OXA-48 K-SeT assay relies on the immunological capture of two epitopes specific to the OXA-48 enzyme using colloidal gold nanoparticles bound to a nitrocellulose membrane within a lateral flow device. Capture and detection antibodies were designed to bind all current CHDL OXA-48-like variants (OXA-48, -181, -204, -232, and -244).
In this study, we assessed the performance of the Coris OXA-48 K-SeT assay for detecting OXA-48-like-mediated carbapenem resistance in a large collection of carbapenem-resistant Enterobacteriaceae and also determined whether it could provide robust results when working directly with organisms recovered from blood culture bottles.
Eighty-two enterobacterial isolates were used in the evaluation. Seventy-eight were clinical isolates (K. pneumoniae, n = 60; Escherichia coli, n = 11; Enterobacter cloacae, n = 6; Enterobacter aerogenes, n = 1) with resistance to one or more carbapenems (ertapenem, imipenem, and/or meropenem) along with a susceptible type strain as a representative control for each bacterial species (K. pneumoniae NCTC 9633, E. coli NCTC 12241, E. cloacae 13380, and E. aerogenes NCTC 9375). Resistance or reduced susceptibility to carbapenems in clinical isolates was identified by disk diffusion and/or Etest (bioMérieux, Marcy L'Etoile, France) and confirmed by broth microtiter dilution (ertapenem MIC, ≥1 μg/ml), according to Clinical and Laboratory Standards Institute (CLSI) methodology. Multiplex PCRs were used to detect the presence of genes encoding class A, B, C, and D β-lactamases (10). PCR detection and Sanger sequencing of entire blaOXA-48 alleles were undertaken and used as the standard for all isolates designated OXA-48-like-positive strains (3, 11). OXA-48 K-SeT assay kits (Coris BioConcept, Gembloux, Belgium) were obtained commercially through BioConnections (Knypersley, United Kingdom) at the catalogue price in August 2015 (£5/$7.6 per cassette).
Isolates were grown and maintained on Mueller-Hinton II cation-adjusted agar plates (Sigma-Aldrich, Gillingham, United Kingdom). The detection of CHDL-producing isolates from solid medium was undertaken, according to the manufacturer's instructions. Briefly, a single colony was resuspended in 10 drops of LY-A buffer (Tris-HCl, NaN3 [pH 7.5]), and 3 drops of the homogenized solution was then applied to the sample well. The tests were read by eye within 15 min (Fig. 1). The lower limit of detection (in CFU per milliliter) of the OXA-48 K-SeT device was determined using K. pneumoniae NCTC 9633 and KP41 (blaOXA-48) grown in Trypticase soy broth (TSB) (Sigma). Cells from 1 ml of overnight cultures were harvested by centrifugation, resuspended in phosphate-buffered saline (PBS), plated in serial dilutions (10−1 to 10−9) onto MH agar, and viable counts (in CFU per milliliter) were recorded after 18 h of incubation. Ten drops of LY-A buffer were added to the cells pelleted from each dilution and used to inoculate OXA-48 K-SeT assay cassettes, as described above.
FIG 1.
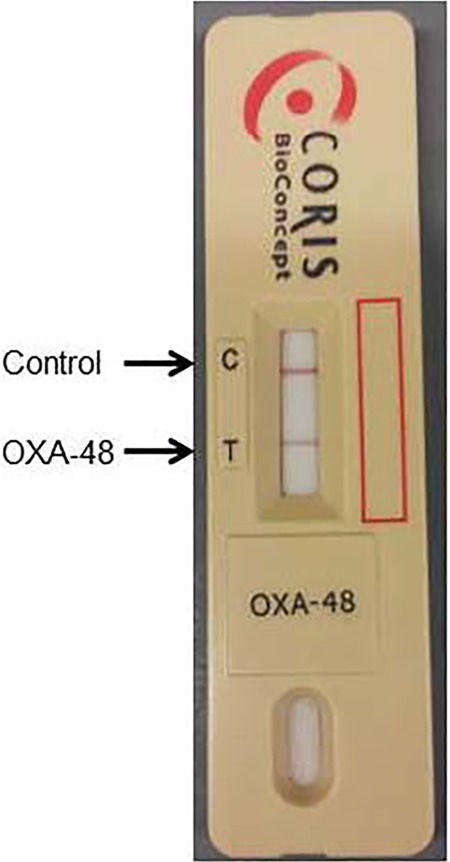
Detection of OXA-48-producing K. pneumoniae using OXA-48 K-SeT assay cassettes. The device was inoculated with 2 × 106 CFU/ml of OXA-48-producing KP41 and read after 5 min.
The ability of the OXA-48 K-SeT system to directly detect OXA-48-producing isolates recovered using commercial microbial blood culture medium was performed using the BacT/Alert system (bioMérieux, Durham, NC). Aerobic FA Plus 30-ml bottles were supplemented with 10 ml of heparinized horse blood (Oxoid, Basingstoke, United Kingdom) before inoculation with 102 CFU of each of the test organisms. The bottles were incubated at 37°C for up to 18 h before 0.1-ml aliquots were harvested, lysed, and analyzed directly without further dilution. Adequate growth of each isolate under simulated blood culture conditions was confirmed by subculture on MH II agar.
Molecular analysis of the isolates used in the analysis confirmed that 53 isolates produced an OXA-48-like carbapenemase, including 2 K. pneumoniae isolates with the OXA-181 and OXA-232 variants. Twenty-five isolates were carbapenem resistant due to the production of either KPC, VIM, or NDM (n = 13) β-lactamases or had hyper-ampC and/or permeability lesions. The OXA-48 K-SeT assay gave a positive result for all 53 OXA-48-producing strains and was negative for all of the other isolates tested, including 5 isolates that produced OXA-1. A correlation of the viable colony counts with the OXA-48 K-SeT assay results obtained with K. pneumoniae NCTC 9633 and KP41 assessed the lower limit of detection of the assay to be 2.41 × 106 CFU/ml. The results were identical whether the cassettes were inoculated with lysates prepared from MH II plates or from BacT/Alert blood culture bottles. In this evaluation, complete agreement between the molecular and OXA-48 K-SeT assay results was demonstrated (Table 1), and the sensitivity and specificity of the assay for detecting OXA-48-producing isolates were both calculated to be 100% (95% confidence interval, 91.9 to 100% and 84.2 to 100%, respectively). Positive and negative results were clearly differentiated within 10 min. Furthermore, the reads were not obscured by the presence of red cells when using lysates from blood culture sets.
TABLE 1.
Detection of OXA-48 production in carbapenem-resistant Enterobacteriaceae using OXA-48 K-SeT cassettes
| OXA-48 K-SeT result (n) by organism | Type (no.) of carbapenem-resistant Enterobacteriaceae (n = 78) by type |
||
|---|---|---|---|
| OXA-48-like carbapenemase | Other carbapenemase | No carbapenemasea | |
| Positive (53) | |||
| K. pneumoniae | 46; OXA-181 (1), OXA-232 (1) | 0 | 0 |
| E. coli | 5 | 0 | 0 |
| E. cloacae | 2 | 0 | 0 |
| Negative (25) | |||
| K. pneumoniae | 0 | 8; KPC (2), NDM (3), VIM (2), KPC+NDM (1) | 6 |
| E. coli | 0 | 2; KPC (1), NDM (1) | 4 |
| E. cloacae | 0 | 3; NDM (2), VIM (1) | 1 |
| E. aerogenes | 0 | 0 | 1 |
Porin loss ± hyper-ampC production.
The OXA-48 K-SeT assay required minimal hands-on analytical time and no investment in any special equipment outside that available in a routine microbiology laboratory. The assay was highly sensitive and specific and was able to either rule in or rule out the presence of an OXA-48-producing strain within minutes. With a limit of detection of 106 CFU/ml, it might also be useful for the direct detection of OXA-48-producing strains in urinary samples or other biological fluids. Although we did not undertake a formal cost/benefit analysis, this assay has clear potential for development as a simple rapid local point-of-care test able to identify and guide the treatment and control of carbapenem-resistant infections due to OXA-48-producing Enterobacteriaceae.
ACKNOWLEDGMENTS
No specific funding was used to undertake this study, as it was performed as part of our routine activities.
We declare no conflicts of interest and have no association with Coris BioConcept.
REFERENCES
- 1.Doi Y, Paterson DL. 2015. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med 36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osei Sekyere J, Govinden U, Essack SY. 2015 Review of established and innovative detection methods for carbapenemase-producing Gram-negative bacteria. J Appl Microbiol 119:1219–1233. doi: 10.1111/jam.12918. [DOI] [PubMed] [Google Scholar]
- 3.Hornsey M, Phee L, Woodford N, Turton J, Meunier D, Thomas C, Wareham DW. 2013. Evaluation of three selective chromogenic media, CHROMagar ESBL, CHROMagar CTX-M and CHROMagar KPC, for the detection of Klebsiella pneumoniae producing OXA-48 carbapenemase. 66:348–350. doi: 10.1136/jclinpath-2012-201234. [DOI] [PubMed] [Google Scholar]
- 4.van Dijk K, Voets GM, Scharringa J, Voskuil S, Fluit AC, Rottier WC, Leverstein-Van Hall MA, Cohen Stuart JW. 2014. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin Microbiol Infect 20: 345–9. doi: 10.1111/1469-0691.12322. [DOI] [PubMed] [Google Scholar]
- 5.Dortet L, Agathine A, Naas T, Cuzon G, Poirel L, Nordmann P. 2015. Evaluation of the RAPIDEC CARBA NP, the Rapid CARB Screen and the Carba NP test for biochemical detection of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 70:3014–3022. doi: 10.1093/jac/dkv213. [DOI] [PubMed] [Google Scholar]
- 6.Mirande C, Canard I, Buffet Croix Blanche S, Charrier JP, van Belkum A, Welker M, Chatellier S. 2015. Rapid detection of carbapenemase activity: benefits and weaknesses of MALDI-TOF MS. Eur J Clin Microbiol Infect Dis 34:2225–2234. doi: 10.1007/s10096-015-2473-z. [DOI] [PubMed] [Google Scholar]
- 7.Pires J, Novais A, Peixe L. 2013. Blue-Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol 51:4281–4283. doi: 10.1128/JCM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodford N, Eastaway AT, Ford M, Leanord A, Keane C, Quayle RM, Steer JA, Zhang J, Livermore DM. 2010. Comparison of BD Phoenix, Vitek 2, and MicroScan automated systems for detection and inference of mechanisms responsible for carbapenem resistance in Enterobacteriaceae. J Clin Microbiol 48:2999–3002. doi: 10.1128/JCM.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ote I, Bogaerts P, Denome L, Borlon C, Thunissen C, Glupczynski Y, Leclipteux T, Mertens P. 2015. Development of a novel immunochromatographic confirmatory test for the detection of OXA-48 carbapenemase in Enterobacteriaceae. Abstr; 25th Eur Cong Clin Microbiol Infect Dis, 25 to 28 April 2015, Copenhagen, Denmark. [Google Scholar]
- 10.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 11.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


