Abstract
Familial Adenomatous Polyposis (FAP) is the second most common inherited predisposition to colorectal cancer (CRC) associated with the development of hundreds to thousands of adenomas in the colon and rectum. Mutations in APC are found in ~ 80% polyposis patients with FAP. In the remaining 20% no genetic diagnosis can be provided suggesting other genes or mechanisms that render APC inactive may be responsible. Copy number variants (CNVs) remain to be investigated in FAP and may account for disease in a proportion of polyposis patients. A cohort of 56 polyposis patients and 40 controls were screened for CNVs using the 2.7M microarray (Affymetrix) with data analysed using ChAS (Affymetrix). A total of 142 CNVs were identified unique to the polyposis cohort suggesting their involvement in CRC risk. We specifically identified CNVs in four unrelated polyposis patients among CRC susceptibility genes APC, DCC, MLH1 and CTNNB1 which are likely to have contributed to disease development in these patients. A recurrent deletion was observed at position 18p11.32 in 9% of the patients screened that was of particular interest. Further investigation is necessary to fully understand the role of these variants in CRC risk given the high prevalence among the patients screened.
Abbreviations: ALL, acute lymphoblastic leukaemia; BH, Bengamini and Hochberg; CHAS, Chromosome Analysis Suite; CN, copy number; CNV, copy number variation; COSMIC, Catalogue of Somatic Mutations in Cancer; CRC, colorectal cancer; DGV, Database of genomic variants; DNA, deoxyribose nucleic acid; FAP, familial adenomatous polyposis; HMDD, human microRNA disease database; Kb, kilobase; KEGG, Kyoto Encyclopaedia of Genes and Genomes; lncRNA, link RNA; LOH, loss of heterozygosity; mapd, median absolute pairwise difference; miR, microRNA; MLPA, multiplex ligation-dependant probe amplification; MMR, mismatch repair; ng, nanogram; NTC, no template control; QC, quality control; RNA, ribose nucleic acid; SNP, single nucleotide polymorphism; TAM, Tool for the annotation of microRNAs; TCGA, The Cancer Genome Atlas; UCSC, University of California, Santa Cruz
Keywords: Cancer, polyposis, CNV, long non-coding RNAs, diagnostic testing
Highlights
-
•
Catalogue of 139 CNVs unique to the polyposis patients which represent candidates for involvement in their disease
-
•
Identification of CNVs in four unrelated polyposis patients in APC, DCC, MLH1 and CTNNB1 which are likely to be associated with disease in affected individuals
-
•
A recurrent deletion at 18p11.32 was identified that affected 9% of the polyposis patients which harbours a lncRNA
1. Introduction
FAP is an autosomal dominant inherited disease, which affects nearly 1 in 12,000 individuals and accounts for approximately 0.5% of all CRCs. Typically, FAP is characterized by the early development of hundreds to thousands of adenomas in the colon and rectum. The development of adenomas commences during early childhood and adolescence and commonly becomes malignant if untreated, with an average age of cancer onset of 35-36 years1. A less severe form of FAP termed attenuated FAP is characterized by fewer adenomas and a later age of disease onset(Lynch and de la Chapelle, 2003, Rustgi, 2007).
Mutations in the APC gene were found to be the genetic basis of FAP and since its discovery over 1500 pathogenic mutations have been reported(Fokkema et al., 2011, Stenson et al., 2009). Up to 20% of polyposis patients do not have a family history of disease but do harbour germline APC mutations. Mutations in the APC gene account for the majority of patients diagnosed with FAP and more recently, mutations in the base excision repair gene MUTYH have been shown to be associated with a recessive form of colorectal polyposis(Pezzi et al., 2009). Up to 20% polyposis patients that are clinically tested for mutations in these genes do not have a germline mutation and no genetic diagnosis for their disease.
CNVs represent a form of structural genetic variation associated with a gain or loss of genomic material. CNVs have been shown to contribute to the development of disease directly through the disruption of functional gene sequences; via promoter region inactivation; or as a result of more cryptic changes such as alterations in epigenetic marks, changes to microRNA controlling species, transcription read through, unmasking recessive alleles and via disruption of non-coding gene sequences(Stella et al., 2007, Morak et al., 2011, Clendenning et al., 2011, Chan et al., 2001, Ligtenberg et al., 2009, Hochstenbach et al., 2012).
Furthermore, while CNVs which are commonly observed in the population may contain cancer related genes, it is the rare CNVs (low population frequencies) which are proposed to harbour genes or other regulatory elements that are likely to be disease susceptibility factors(Shlien and Malkin, 2010). Several studies have recently investigated the contribution of rare CNVs in cancer; one study identified 26 rare CNVs which they proposed to contribute to breast cancer susceptibility, while another has reported the enrichment of disrupted genes that affect the maintenance of genomic integrity i.e. DNA double-strand break repair also in familial breast cancer(Krepischi et al., 2012, Pylkas et al., 2012).
In this study we have focused on the role of CNVs in the genomes of patients diagnosed with polyposis that do not harbour germline mutations in APC or MUTYH as assessed by direct DNA sequencing and multiplex ligation probe amplification (MLPA). High throughput microarray technology has continuously improved since its introduction such that now continually smaller CNVs can be detected in ever larger patient cohorts. We used the Affymetrix Cyto2.7M microarray, which at the time of this study provided the highest genomic coverage of any commercially available microarray; containing 400,000 SNP probes and > 2.1 million CNV probes with an average spacing of 1395 base pairs (bp). CNV analysis was conducted on DNA derived from 56 polyposis patients (APC/MUTYH mutation negative) and compared to 40 controls and the Database of Genomic Variants (DGV) with the aim of identifying CNVs, which may be involved in the pathogenesis of the observed disease.
2. Methods
2.1. Samples
The study including patient recruitment and all experimental protocols were approved by the Hunter New England Human Research Ethics Committee and the University of Newcastle Human Research Ethics Committee. The methods employed in this study were carried out in accordance with the approved guidelines of the University of Newcastle. Genomic DNAs were obtained from polyposis patients who had given informed consent for their DNA to be used for studies into their disease and control DNA samples from the Hunter Community Study was used in the current study(McEvoy et al., 2010). DNA was extracted from whole blood by the salt precipitation method(Miller et al., 1988).
The inclusion criteria for this study was a patient diagnosed with adenomatous polyposis or and who did not have a detectable APC or MUTYH mutation as assessed by complete Sanger sequencing and MLPA analysis. A cohort of 56 clinically histologically confirmed polyposis patients was used in this study. All patients were unrelated and were diagnosed after colorectal resection who then sought genetic testing for their condition. The average age of diagnosis was 51 years (range 10 - 74), 32 of the probands had a family history of colonic polyposis or CRC, 21 had no family history and for 3 patients no information on family history was available (see Table 1 in Masson et al., submitted for publication). Polyp counts ranged from 5 through to over 1000, however most patients had less than 100 polyps suggesting that the majority of patients presented with an attenuated form of polyposis. From the 55 patients examined for CNVs in this study 8 were diagnosed with typical polyposis and the remaining 47 were diagnosed with attenuated form of the disease (see Table 1 in Masson et al., submitted for publication).
The control patients were all over 55 years of age and healthy at the time of phlebotomy. None had reported any change in bowel habits prior to blood collection. A total of 96 samples were included in the study.
2.2. Genomic array preparation and data processing
The genomic DNA was processed on the Affymetrix Cyto2.7M array according to manufacturer’s protocols. Affymetrix Chromosome Analysis Suite (ChAS) (Version CytoB-N1.2.0.232; r4280) was used to analyse the array data (NetAffx Build 30.2 (Hg18) annotation).
A training set of 20 randomly selected samples was used to further optimize a series of quality control (QC) parameters reduce the number of false-positive CNVs being included in the analysis as many of these QC thresholds were more stringent than default settings alone.
The Cyto2.7M array is comprised of both CNV probes and SNP probes. For the confident detection of CNVs samples were required to have a minimum quality threshold of: mapdQC < 0.27 (Median Absolute Pair-wise Difference; QC of CN probes compared to a reference model); snpQC > 1.1 (SNP probe QC measuring distances between the distribution of alleles AA, AB and BB alleles in which larger differences in allele distribution is associated with an increased ability to call a given genotype); and wavinessSd < 0.1 (measure of standard deviation in data waviness; the GC content across the genome correlates with average probe intensities).
For the data that fulfilled the array performance QC, CNV calling QC was then undertaken to minimise the inclusion of false positive or negative CNV calls being incorporated into the analysis. This included evaluation of CNV calls with respect to having > 90% confidence, a CNV having to be of autosomal origin (not located on either sex chromosome), and CNVs had to have a minimum of 24 probes used to call the CNV region. Visual inspection was used to confirm all CNV calls, verify the suggested CN state, and to further identify regions associated with low marker coverage and excluded across all samples (i.e. centromeric and telomeric regions; see Table 2 in Masson et al., submitted for publication). The smallest CNV detected with confidence was 6.03 Kilobases (Kb) across all samples.
2.3. CNV and statistical analysis
CNVs were subject to a series of analyses which included: (Galiatsatos and Foulkes, 2006) Identification of abundant genomic regions and genes affected by a CNV in patients; (Lynch and de la Chapelle, 2003) Statistical assessment of the distribution of CNVs across the genome of patients compared to controls; and (Rustgi, 2007) interrogation of CNV data for CN gains and losses residing in or ± 100 Kb of 77 genes comprising the WNT signalling and mismatch repair (MMR) pathways as well as other reported CRC susceptibility genes, likely to be associated with polyposis (see Table 3 in Masson et al., submitted for publication)(Pezzi et al., 2009, Molatore et al., 2010). Associations (e.g. numbers and sizes of CNVs) were statistically compared between patients and controls using a two tailed un-paired t-test Graphpad Prism (Version 6; available http://www.graphpad.com/quickcalcs/ttest1/). The derived p-values were corrected for multiple testing using Bonferroni correction (Alpha = 0.05, R = 22). The Bonferroni's adjustment resulted in the confidence level being < 0.0023.
2.4. Validation of CNV results
CN gains and losses were subject to validation using pre-designed TaqMan Copy Number Assays (Applied Biosystems). Where possible, two CN assays (test assays) located within and two located just outside (control assay) the CNV of interest were utilized (assay information summarized in Table 4 in Masson et al., submitted for publication). The sample(s) of interest were tested along with a no template control (NTC) and several calibrator samples (of known CN for the region assessed). All samples were assayed in triplicate and real-time PCR was conducted according to manufacturer’s instructions using 10 ng of DNA sample in a final reaction volume of 20 μL. Using the real-time PCR (Applied Biosystems 7500; SDS software Version v1.4) the assay was run according manufacturer’s protocols. CopyCaller v2.0 software (Applied Biosystems) was used to analyse the results.
Several CNVs were further validated using this independent method (see Table 5 in Masson et al., submitted for publication). These CNVs included both CN gains and CN losses in the genes DCC and APC as well as in the genomic region 18p11.32. As we observed high concordance between array data and all CNVs were validated using an independent method, it was considered unnecessary to confirm every CNV identified as analysis parameters were consistent across all samples.
For the variant identified in the promoter region of the APC gene family studies were undertaken and the result was confirmed in a number of affected relatives. Unfortunately, we were unable to extend this aspect of the study for other potentially causative CNVs due to either there being no living affected relative or any other person diagnosed with disease in the respective family.
2.5. Pathway analysis and annotation
In silico analysis conducted in this study involved the analysis of 49 of the 148 genes unique to the patients that were also considered rare as they have not been reported in the DGV).
Pathway analysis was performed using WebGestalt software (Version 2013)(Zhang et al., 2005). This software was used to assess gene lists derived from the refined CNV results obtained from ChAS according to Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways, cytoband enrichment analysis, and miR targets. Analysis was performed using hypergeometric statistical method, Benjamini and Hochberg (BH) correction for multiple testing and a biological significance threshold of < 0.05 with a minimum of two genes per category required to assess any enrichment.
TAM (Tool for Annotations of miRs) (Version 2)(Lu et al., 2010) software was used to annotate miRs according to miR family, cluster, function, Human miR associated disease database (HMDD) and tissue specificity. Annotations were performed using the following parameters: all miRs in the TAM database were used as a background; to identify meaningful categories we looked at miR over-representation in all categories and analysis was limited to at least one miR in a given category. Enrichment analysis for miRs categories was conducted using hypergeometric testing and p-values were corrected according to Bonferroni correction for multiple testing.
3. Results
3.1. Array resolution and CNV detection
A total of 278 CNVs were identified in the 96 participants involved in this study (Table 1). CNVs ranged in size from 6.03 Kb to 1435.95 Kb. The average number of CNVs identified per sample did not differ significantly between patients and controls (p = 0.4383) nor did the average CNV burden (p = 0.5173) or average CNV size (p = 0.1664).
Table 1.
Summary of CNV results obtained from the Cyto2.7M array analysed in ChAS.
| CNV Count |
CNV Size (Kb) |
|||||
|---|---|---|---|---|---|---|
| Median CNVs per sample | Mean CNVs per sample | Total CNV affected genome per group | Mean total CNV affected genome per sample | Mean size of a CNV | ||
| Patients | 56 | 2 | 3.11 | 14,018.83 | 250.34 | 82.18 |
| Controls | 40 | 2 | 2.6 | 11,820.75 | 295.52 | 106.57 |
| p | - | - | 0.4383 | - | 0.5173 | 0.1664 |
*statistically significant
3.2. Abundant genomic regions and genes associated with CNVs
Analysis of the control population revealed a total of 104 CNVs of which 12 genomic regions were disrupted by a CNV in more than one individual. Eight of the genomic regions (2p16.1, 4p15.31, 4q13.1, 5p13.3, 5q21.2, 7p14.1, 8q12.1 and 8q24.23) were disrupted by a CNV in two unrelated individuals; three genomic regions (4q32.2, 6q22.31 and 12p13.31) were disrupted by a CNV in three control participants; and one genomic region (3q26.31) was found to be affected in five individuals (see Table 6 in Masson et al., submitted for publication). In total 66 of the 104 CNVs (63.46%) disrupted 96 genes. The 96 genes disrupted by CNVs were screened against the current cancer genome census list (COSMIC database available: http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) to determine whether any were likely to be associated with cancer. None of these genes were in common with those known to be associated with a cancer predisposition. Three CNVs were identified to disrupt MLL3, PBX1 and PLAG1, respectively, that have been observed in medulloblastomas, pre-BALL/myoepitheliomas and salivary adenomas.
Of the 174 CNVs identified in the polyposis patients, 15 genomic regions disrupted by a CNV (comprising 32 CNVs detected in 28 unrelated patients) were common to regions also disrupted by a CNV in the controls (see Table 7 in Masson et al., submitted for publication). These CNVs were not included in further analysis as they were considered most likely to be neutral. Of the remaining 142 CNVs unique to the patients, 6 genomic regions contained CNVs which were common to multiple patients (Table 2). The genomic region 2q32.3, was disrupted by a CNV in two patients, as were the CNVs that encompassed regions located at 2q34, 3q26.1 and 4q12; one genomic region (3q26.32) was disrupted by a CNV in three patients; and another genomic region (18p11.32) was disrupted by a CNV in five patients.
Table 2.
Recurrent CNVs unique to polyposis patients. Note location of recurrent CNV, the type of CNV identified in each patient, description of the CNV (Chr, Start, End, size) and patient ID.
| Location | CNV Type | Chr | Start (bp) | End (bp) | Size (Kb) | Patient IDs |
|---|---|---|---|---|---|---|
| 2q34 | Gain | 2 | 209,751,678 | 209,923,081 | 171.4 | FAP6 |
| Gain* | 2 | 209,793,755 | 209,821,161 | 27.41 | FAP23 | |
| 2q32.3 | Loss | 2 | 194,626,162 | 194,695,204 | 69.04 | FAP19 |
| Loss | 2 | 194,626,162 | 194,696,112 | 69.95 | FAP20 | |
| 3q26.1 | Loss | 3 | 166,523,809 | 166,565,186 | 41.38 | FAP4 |
| Loss | 3 | 166,523,809 | 166,565,186 | 41.38 | FAP17 | |
| 4q12 | Gain | 4 | 57,745,642 | 57,794,798 | 49.16 | FAP24 |
| Gain | 4 | 57,745,642 | 57,794,798 | 49.16 | FAP4 | |
| 3q26.32 | Loss | 3 | 177,370,126 | 177,396,832 | 26.71 | FAP21 |
| Loss | 3 | 177,370,126 | 177,396,832 | 26.71 | FAP5 | |
| Loss | 3 | 177,370,126 | 177,396,832 | 26.71 | FAP22 | |
| 18p11.32 | Loss | 18 | 1,891,809 | 1,974,284 | 82.48 | FAP4 |
| Loss | 18 | 1,894,368 | 1,974,284 | 79.92 | FAP1 | |
| Loss | 18 | 1,894,368 | 1,974,284 | 79.92 | FAP2 | |
| Loss | 18 | 1,894,368 | 1,974,284 | 79.92 | FAP3 | |
| Loss | 18 | 1,964,144 | 2,015,983 | 51.84 | FAP5 |
indicates the CNVs not reported in the DGV
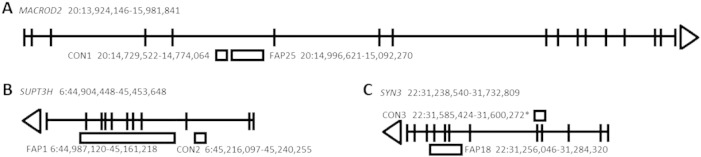
In three patients CNVs located 6p12.3, 20p12.1 and 22q12, respectively, harboured large deletions encompassing part of the genes SUPT3H, MACROD2 and SYN3 and were subject to special consideration as the three genes were also affected by CNV changes in 3 control subjects (see Fig. 1). Two of the deletions occurring in the patients encompassed coding regions of SUPT3H (exons 4-10) and SYN3 (exons 7-9) suggesting a loss of gene function in both instances. The SUPT3H CNV in another control subject occurred in intron 2 and appears to be less likely to affect function. The SYN3 CNV in a control subject was a duplication that included exons 4 and 5 and which did not alter the reading frame of SYN3 and may have affected gene function. The CNV affecting MACROD2 in intron 5 of a polyposis patient appeared not to alter exonic structure of the gene. Similarly, a control subject was also found to harbour a CNV in intron 5 of MACROD2 but residing 5' to that identified in the patient. Neither CNV encompassed an exon. Since two adjacent deletions in intron 5 of MACROD2 were observed that did not appear to alter the exonic structure of the gene they were not considered to be disruptive.
Fig. 1.
Genes within which non-overlapping CNVs were identified in patients and controls: (A) MACROD2, (B) SUPT3H and (C) SYN3 respectively.
Note the location of the CNVs (duplication above and deletions below) with respect to the gene identifying exons and introns and direction of transcription (direction of arrow). Representation only, not to scale.
Of the remaining 139 CNVs (see Table 8 in Masson et al., submitted for publication), 85 (61.15%) disrupted a total of 148 genes and these were considered candidate genes for disease development in these patients. Furthermore a subgroup of 10 genes: EVI2B, EVI2A, SMAP2, BOD1L, NAMPT, NF1, HSD11B1, G0S2, DOCK4 and A2BP1 were found to be affected by a CNV in more than one patient (Table 3) and therefore were considered to have a higher probability of being associated with disease warranting further investigation.
Table 3.
The 10 genes associated with CNVs unique to polyposis patients (identified as FAP11-16). Note the disrupted gene (its symbol and description), if the gene is expressed in the colon (www.proteinatlas.org), the CN type observed in the current dataset and a general column outlining the predicted interpretation of the effect different types of CNVs have on disease development.
| Gene | Description | Expression in colon | CN type | Region of gene disputed | Interpretation (predicted) |
|---|---|---|---|---|---|
| EVI2B | ecotropic viral integration site 2B | medium | Gains | FAP11: whole gene | Whole gene duplication:increased expression/amplification of gene function. |
| FAP12: part gene | |||||
| EVI2A | ecotropic viral integration site 2A | medium | Gains | FAP11: whole gene | |
| FAP12: whole gene | |||||
| SMAP2 | small ArfGAP2 | medium | Gains | FAP12: part gene, exon and intron 1 | Partial duplication or deletion involving introns and exons:disruption of gene/loss of gene function. |
| FAP13: part gene , exon and intron 1 | |||||
| BOD1L | biorientation of chromosomes in cell division 1-like 1 | medium | Gains | FAP12: part gene, introns and exons | |
| FAP11: part gene, introns and exons | |||||
| NAMPT | nicotinamide phosphoribosyltransferase | medium | Gains | FAP12: part gene, most of it from start of gene | Partial duplication involving promoter:transcription of aberrant transcript leading to non-functional protein product/loss of gene function. |
| FAP13: whole gene | |||||
| NF1 | neurofibromin 1 | medium | Gains | FAP11: part gene, intronic | |
| FAP12: part gene, intronic | |||||
| HSD11B1 | hydroxysteroid (11-beta) dehydrogenase 1 | low | Gains | FAP12: part gene, upstream into exon 1 | Partial duplication involving exons:possible addition of duplicated exons into gene transcript creating a non-functional protein product/loss of gene function. |
| FAP11: part gene, upstream into exon 1 | |||||
| G0S2 | G0/G1 switch 2 | medium | Gains | FAP12: whole gene | |
| FAP11: whole gene | |||||
| DOCK4 | dedicator of cytokinesis 4 | low | Both | FAP11: part gene dup, intronic | Partial duplication or deletion involving introns:development of cryptic splice sites or the formation of pseudo exons leading to disruption in gene expression/loss of gene function. |
| FAP4: part gene del, intronic | |||||
| A2BP1 | RNA binding protein, fox-1 homolog (C. elegans) 1 (alias RBFOX1) | low | Both | FAP15: part gene dup, introns and exon | |
| FAP16: part gene del, introns and exons |
3.3. Distribution of CNVs across the genome in patients
The distribution of CNVs across chromosomes was compared between patients and controls revealing no statistically significant differences in CNV distribution. We did observe a trend in the over-representation of CNVs in patients (CNVs = 9) compared to controls (CNVs = 0) using Fisher’s exact test for chromosome 18 (p = 0.009) which did not remain statistically significant after correction for multiple testing (i.e. p > 0.0022).
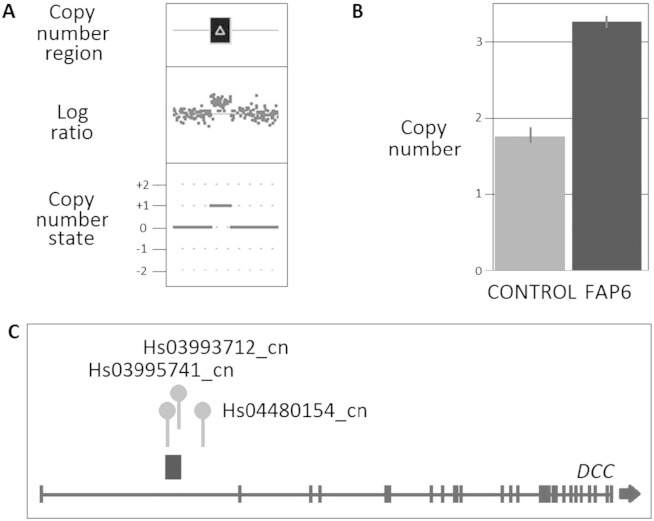
Among the CNVs located on chromosome 18, a CNV gain was detected in the first intron of the DCC gene (18:48,381,778-48,412,417; 30.6 Kb; 93% confidence, detected by 52 probes), which was subsequently confirmed by TaqMan CN assays (see Fig. 2).
Fig. 2.
CNV results for the duplication in the DCC gene in an FAP patient (FAP6). (A) CNV profile from Cyto2.7M array data using ChAS noting the defined CN region (dark box above gene representing the CN deletion) in relation to the log ratio plot (relative fluorescence of each probe, dot, on the array showing a decrease in fluorescence indicating a loss in genomic material) and the CN state (0 = normal two copies present, + 1 = one extra copy, + 2 = two extra copies, -1 = one less copy and -2 = two less copies); (B) Validation using TaqMan CN assay showing results for assay Hs03995741_cn noting the normal two copies of this region identified in the control (CON1), confirmation of the aberrant three copy in the affected FAP patient and the error bars associated with the three technical repeats for each sample; and (C) Location of CN duplication with respect to the gene and the TaqMan CN assays used in validating the variants.
A CNV gain was also identified in one patient which encompassed exon 1 and extending into the first intron of the USP14 gene located on chromosome 18 (position 18:50,739-154,914, size 104 Kb; 90% confidence and detected by 34 probes). This CNV encompassing part of the irritable bowel disease (IBD) locus(Hetzenecker et al., 2012) is intriguing and suggests there may be a relationship between IBD and colorectal malignancy.
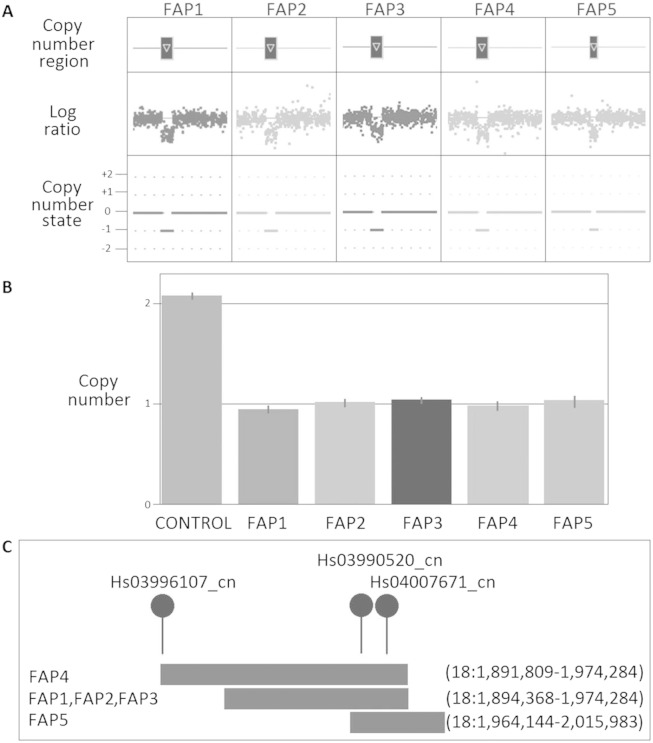
Of note, five patients harboured the same CNV loss at the 18p11.32 locus (Fig. 3). The predicted size of the largest CNV identified a loss of 82.48 Kb (93% confidence and detected by 65 probes); three other unrelated patients all harboured a similar sized CNV loss of 79.92 Kb (all > 91% confidence and detected by 63 probes); and one patient was found to have a CNV loss of 51.84 Kb (94% confidence and detected by 59 probes). All CNVs located in the 18p11.32 region overlapped each other by 10.14 Kb and this was confirmed by TaqMan CN assay to be lost in all five patients. This region of loss is proposed to contain the long non-coding RNA (lnc-RNA) TCONS_00026231 (18:1,963,908-1,972,876 Hg19; UCSC Genome Browser).
Fig. 3.
CNV results for 18p11.32 deletion in the FAP patients (FAP1, FAP2, FAP3, FAP4 and FAP5). (A) CNV profile from Cyto2.7M array data using ChAS noting the defined CN region (dark box above gene representing the CN deletion) in relation to the log ratio plot (relative fluorescence of each probe, dot, on the array showing a decrease in fluorescence indicating a loss in genomic material) and the CN state (0 = normal two copies present, + 1 = one extra copy, + 2 = two extra copies, -1 = one less copy and -2 = two less copies); (B) Validation using TaqMan CN assay showing results for assay Hs03990520_cn noting the normal two copies of this region identified in the control (CON2), confirmation of the aberrant one copy in all affected FAP patients and the error bars associated with the three technical repeats for each sample; and (C) Location of CN deletions with respect to each other and the TaqMan CN assays used in validating the variants.
3.4. CRC susceptibility gene interrogation
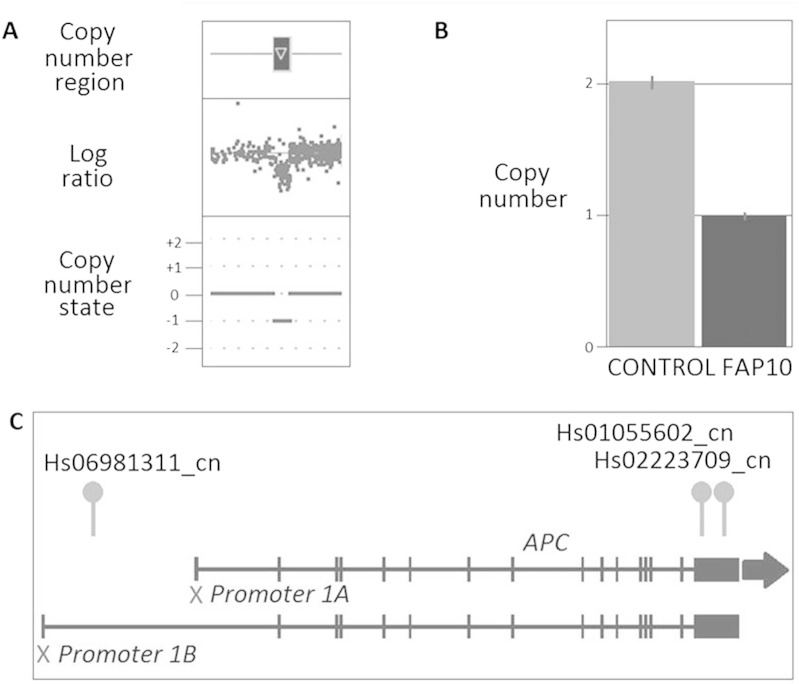
We determined whether CNVs within 100 Kb either side of 77 genes involved in pathways known to be associated with CRC risk, including members of the WNT signalling and MMR pathways (see Table 3 in Masson et al., submitted for publication), could potentially contribute to CRC development. Two unrelated polyposis patients harboured a CNV in the vicinity of the MLH1 and CTNNB1 genes, respectively. One patient harboured a CNV loss 18 Kb upstream MLH1 located 3:36,925,248-36,991,856 (66.6 Kb, 95% confidence and detected by 35 probes) while the other harboured a CNV loss extending upstream and into the promoter region of CTNNB1 located 3:41,068,578-41,119,502 (50.9 Kb, 94% confidence and detected by 24 probes). A third polyposis patient was identified harbouring a CNV loss located directly within the promoter 1B region of the APC tumour suppressor gene (5:112,065,033-112,096,002, 31 Kb; 91% confidence and detected by 56 probes) (Fig. 4). TaqMan CN assays confirmed the loss of this region in the affected patient and her two affected sons that were also recruited into the current study indicating this CN loss has been transmitted from one generation to the next. The loss of this region is likely to have contributed to disease development in all affected individuals.
Fig. 4.
CNV results for the APC promoter 1B deletion in the FAP patient (FAP10). (A) CNV profile from Cyto2.7M array data using ChAS noting the defined CN region (dark box above gene representing the CN deletion) in relation to the log ratio plot (relative fluorescence of each probe, dot, on the array showing a decrease in fluorescence indicating a loss in genomic material) and the CN state (0 = normal two copies present, + 1 = one extra copy, + 2 = two extra copies, -1 = one less copy and -2 = two less copies); (B) Validation using TaqMan CN assay showing results for assay Hs06981311_cn, noting the normal two copies of this region identified in the control (CON2), confirmation of the aberrant one copy in all affected FAP patient and the error bars associated with the three technical repeats for each sample; and (C) Location of CN duplication with respect to the gene and the TaqMan CN assays used in validating the variants.
3.5. Rare CNV events
The CNV dataset was also compared against the DGV. CNVs that were rare (not identified in the DGV and herein termed rare CNVs) corresponded to 31.29% (87 of 278) of the total CNVs identified in both patients and controls. In the control cohort 28.85% (30 of 104) of CNVs detected in 19 of the 40 controls (32.56%) were classified as rare whereas 32.76% (57 of 174) of CNVs detected in 23 of the 56 patients (41.07%) were rare. No significant difference was detected in the number of rare CNVs between patients and controls (p = 0.57). In total, 49 genes were associated with the 57 rare CNVs identified in the polyposis cohort representing genes most likely to be associated with disease (see Table 4). With the exception of ANKFN1, FAM184B, and HCN1 (CNV loss) and CCDC19 (CNV gain) which were not expressed in normal colon tissue and four other genes (SNORD12, SNORD12B, SNORD12C, C20orf199) where no information was available, 41 genes have been reported to be expressed in the colon and rectum (www.proteinatlas.org).
Table 4.
List of 49 genes which may be implicated in disease (unique to the polyposis patients and not observed in the DGV). Note the gene symbol, description and if the gene is normally expressed in the colon (www.proteinatlas.org).
| Gene | Description | Expression |
|---|---|---|
| ADD3 | adducin 3 (gamma) | High |
| AIM1 | absent in melanoma 1 | Medium |
| AMICA1 | adhesion molecule, interacts with CXADR antigen 1 | Medium |
| ANKFN1 | ankyrin-repeat and fibronectin type III domain containing 1 | Not Detected |
| APC | adenomatous polyposis coli | Low |
| ARHGAP25 | Rho GTPase activating protein 25 | Low |
| ARHGAP26 | Rho GTPase activating protein 26 | Medium |
| ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | High |
| BCL2A1 | BCL2-related protein A1 | Low |
| BOD1L | biorientation of chromosomes in cell division 1-like 1 | Medium |
| C17orf95 | methyltransferase like 23 | Medium |
| C20orf199 | ZNFX1 antisense RNA 1 | Unknown |
| C5orf56 | chromosome 5 open reading frame 56 | Low |
| CCDC19 | coiled-coil domain containing 19 | Not Detected |
| CDH11 | cadherin 11, type 2, OB-cadherin (osteoblast) | Medium |
| CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | High |
| DDX10 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 | Medium |
| ETV6 | ets variant 6 | Medium |
| FAM184B | family with sequence similarity 184, member B | Not Detected |
| FKBP1A | FK506 binding protein 1A, 12kDa | High |
| G0S2 | G0/G1switch 2 | Medium |
| GALC | galactosylceramidase | Medium |
| GAS7 | growth arrest-specific 7 | Medium |
| GPR65 | G protein-coupled receptor 65 | Low |
| HCN1 | hyperpolarization activated cyclic nucleotide-gated potassium channel 1 | Not Detected |
| HSD11B1 | hydroxysteroid (11-beta) dehydrogenase 1 | Low |
| JMJD6 | jumonji domain containing 6 | Medium |
| LAMB3 | laminin, beta 3 | Medium |
| LCA5 | Leber congenital amaurosis 5 | Low |
| MFSD11 | major facilitator superfamily domain containing 11 | Medium |
| MPZL3 | myelin protein zero-like 3 | Low |
| MXRA7 | matrix-remodelling associated 7 | Medium |
| NAMPT | nicotinamide phosphoribosyltransferase | Medium |
| NUMA1 | nuclear mitotic apparatus protein 1 | High |
| PLEK | pleckstrin | Medium |
| QKI | QKI, KH domain containing, RNA binding | Medium |
| RFT1 | RFT1 homolog (S. cerevisiae) | Medium |
| SELL | selectin L | Low |
| SFRS2 | arginine/serine rich splicing factor 2 | High |
| SH3BGRL2 | SH3 domain binding glutamic acid-rich protein like 2 | High |
| SMAP2 | small ArfGAP2 | Medium |
| SMC3 | structural maintenance of chromosomes 3 | Medium |
| SNORD12 | small nucleolar RNA, C/D box 12 | Unknown |
| SNORD12B | small nucleolar RNA, C/D box 12B | Unknown |
| SNORD12C | small nucleolar RNA, C/D box 12C | Unknown |
| STK17B | serine/threonine kinase 17b | Medium |
| STX8 | syntaxin 8 | High |
| TAGLN2 | transgelin 2 | High |
| ZNFX1 | zinc finger, NFX1-type containing 1 | Medium |
3.6. Pathway analysis and miR annotation of rare genes in polyposis patients
The 45 of the 49 rare genes were successfully mapped and investigated further using WebGestalt pathway analysis(Zhang et al., 2005) for enrichment among KEGG pathways, cytogenetic band regions and 3’UTR regions of genes (i.e. miR targets).
KEGG analysis revealed no significant pathways however, enrichment among cytogenetic bands identified 11 cytobands (20q13, 17q25, 6q14, 20q, 10q25, 6q, 2p13, 11q23, 1q32, 17q and 5q31; all with p < 0.0443; see Table 9 in Masson et al., submitted for publication) that were associated with 22 of the 49 rare genes (see Table 10 in Masson et al., submitted for publication). Enrichment analysis for the targets of miRs identified 26 significant regions (all with p < 0.0421) within the 3’UTR of 19 of the 49 rare genes for which 42 miRs were suggested to target (see Tables 8 and 9 in (Masson et al.)).
TAM21 of the 42 miRs subsequently identified a total of 161 miR categories: 10 families, 13 clusters, 34 functional categories, 102 Human miR associated disease database (HMDD) and 2 tissue specificity categories. Specifically, miRs were significantly over-represented in the family category miR-15 (miR-15a, miR-195, miR-15b and miR-16; p = 0.002) and the HMDD categories for glioblastoma (miR-15a, miR-195, miR-16, miR-181a-c, miR-18a and miR-32; p = 0.0205), breast neoplasia (miR-15a, miR-520a, miR-320, miR-200a, miR-181b, miR-135b, miR-135b, miR-497, miR-133a, miR-27a, miR-302c, miR18a, miR-195, miR-30a, miR-18b and miR-519c; p = 0.002) and leukaemia (mir-15a, miR-181a, miR-181b and miR-16; p = 0.008).
4. Discussion
CNVs have yet to be intensively investigated for their involvement in polyposis and consequent contribution to disease development. Here we have presented a comparison between 56 unrelated APC and MUTYH mutation negative patients all diagnosed with polyposis and 40 healthy controls. We have furthermore compared our results the COSMIC database and DGV and have assessed our data in terms of CNV abundance, size and distribution using a whole genome approach in search of genes and genomic regions that could be associated with polyposis.
An increased CNV burden has been suggested to be associated with an increased risk of disease development, while variation in CNV burden is associated with phenotypic variation(Girirajan and Eichler, 2010). We did not identify any significant differences in the number or size of CNVs between polyposis patients and controls. This finding suggests that the numerical burden of CNVs (> 6.03 Kb) does not appear to contribute to an increased disease risk. However, since our analysis was limited to the detection of CNVs greater than 6.03 Kb we cannot rule out the involvement of smaller CNVs in the aetiology of this disease.
This study revealed several CNVs affecting recurrent loci that included genes known to be associated with CRC, in multiple patients: DOCK4 variants have been reported to give rise to various cancers including ovarian, prostate, glioma and CRC(Kuo et al., 2009); DOCK4 is also involved with the regulation of β-catenin in the WNT signalling pathway(Upadhyay et al., 2008), which has been directly implicated in FAP development; NAMPT, which is involved in the metabolism and proliferation of cells(Zhang et al., 2012), has been identified to be over-expressed in CRC(Hufton et al., 1999); NAMPT is also a target of mir-26b (a putative tumour suppressor-miR) which binds to the 3’ UTR of NAMPT(Zhang et al., 2013); while duplications in the NF1 gene have also been observed in CRCs(Cacev et al., 2005) suggesting that CNVs associated with these genes contribute to disease.
Furthermore, two of the recurrent CNV deletions observed in the current study also fell into regions containing recurrent deletions peaks reported by the TCGA for colon adenocarcinoma tumour data (located 16p13.3 and 7q 31.3), one of which is reported to harbour the disease candidate gene A2BP1(Cancer Genome Atlas, 2012). Several other CNV regions identified among patients in the present study were also recurrent in the TCGA dataset (located 11q22.3, 15q21.1, 1p33, 20p12.1, 5q22.2, 7q31.3 and 5p12) containing several candidate disease genes including APC, B2M, AGBL4, MACROD2 and HCN130. Overall the results from the current study are consistent with previous reports on CNV burden in CRC, however our data suggests several additional genomic regions may contribute to disease in these polyposis patients.
The distribution of CNVs on individual chromosomes was also compared between patients and controls, which failed to reveal any significant difference in the frequency of total CNVs between the two groups. The frequency of CNVs on each chromosome could not be shown to be significantly different, but a trend was observed indicating a greater number of CNVs on chromosome 18 compared to the controls. Among these CNVs was a gain in the DCC gene (identified in one patient). DCC is reported as a tumour suppressor and is frequently observed to be down-regulated in CRC (~ 70% of patients) which has been attributed to the loss of genomic material in the 18q21 region in which DCC resides(Fearon et al., 1990, Thiagalingam et al., 1996). Here we report the possible loss of DCC gene function as a result of an intronic CN gain. It has been revealed by others that some deep intronic variants have been shown to contribute to CRC via the formation of pseudoexons, the activation of cryptic splice sites and the expression of aberrant mRNA transcripts(Spier et al., 2012). Validation studies using TaqMan CN assays confirmed the CN gain in the affected patient, however further studies will be required to understand the role of DCC in CRC.
Intriguingly CN losses rather than CN gains were shown to be statistically enriched on chromosome 18 in polyposis patients. We observed a region of CN loss at 18p11.32 that affects nearly 9% of the polyposis patients in our study. GWAS and meta-analysis studies have previously recognized 18p11.32 as susceptibility loci for bipolar disease, childhood acute lymphoblastic leukemia (ALL) and leisure time exercise behaviour(De Moor et al., 2009, Ferreira et al., 2008, Trevino et al., 2009). More recently, loss of heterozygosity (LOH) at 18p11.32 has been reported in CRC adenomas (but not normal mucosa) and is suggested to be involved in CRC tumour development(Costi et al., 2011); and a second study reporting genomic losses at 18p11.32 in CRCs are suggesting that this region is associated with adenoma-carcinoma progression(Shi et al., 2012). Of particular note was the occurrence of the recently reported lncRNA (TCONS_00026231) residing in this region of loss. LncRNAs (non-coding nucleotides, 200-100,000 bp in size) are proposed to be master regulators whose functions include post-transcriptional regulation of gene expression, regulation of epigenetic marks, gene activation in cis and they have been shown to influence processes such as pluripotency(Loewer et al., 2010, Nagano et al., 2008, Orom et al., 2010). Validation studies using TaqMan CN assays confirmed the CN losses in all affected patients. The frequency of this variant in a series of polyposis patients suggests that it may be associated with a predisposition of CRC in a proportion of APC/MUTYH mutation negative patients. Further studies are required to ascertain the precise involvement of this lncRNA in the genesis of CRC and more specifically whether it is involved in controlling WNT signalling and therefore adenomatous polyposis development.
Investigation of CNVs residing in or in the proximity of known cancer genes or pathways may expand our understanding of their contribution to disease risk in polyposis. Herein we interrogated the CNV data in search for variants associated with genes in the WNT signalling and MMR pathways focusing on APC and MUTYH(Pezzi et al., 2009, Molatore et al., 2010). CNVs arising in or in the proximity of any of these genes may contribute to disease directly or via more cryptic means. Two unrelated polyposis patients harboured CNVs near MLH1 and CTNNB1. Germline variants arising in MLH1 are typically associated with Lynch syndrome(Rustgi, 2007); whereas mutations associated with CTNNB1 occur in sporadic CRC and other malignancies(Hirata et al., 2012, Sygut et al., 2012); furthermore mutations in CTNNB1 are reported to be enriched in desmoid disease(Le Guellec et al., 2012), the second major cause of mortality in FAP. Interestingly the TCGA results on colon adenocarcinoma also reports CTNNB1 as one of the most significantly mutated genes in (5%) non-hypermutated colon tumours(Cancer Genome Atlas, 2012). The involvement of these two genes is concordant with both Lynch syndrome and FAP, respectively(Jasperson et al., 2010).
We identified a genomic loss located directly within the promoter 1B region of APC. It has been estimated that up to 2% of the mutations identified in FAP cases are large deletions, including deletions that extend from the promoter into the coding region(Gismondi et al., 1998). Of APCs two promoter regions, promoter 1A and 1B, the latter of these is suggested to only play a minor role in APC gene regulation(Tsuchiya et al., 2000). The first report attempting to characterize promoter-specific deletions in FAP was described in 2008(Charames et al., 2008), while (Rohlin et al., 2011) has recently reported the first evidence of promoter 1B involvement in FAP which was associated with a partial deletion of this region. Validation studies using TaqMan CN assays confirmed the CN loss in the affected patient. As the patient’s two affected sons were also verified to carry the same CN loss confirming that the variant was transmitted from one generation to the next, this CN is likely to be the cause of disease in all the affected family members. Our study further supports the role of APC promoter 1B inactivation in FAP development, which is reinforced by the finding that the CNV is transmitted across generations and is suggested to segregate with the expected phenotype. It should also be noted that deep intronic mutations (smaller than the level of detection) in the APC gene and low level somatic mosaicism are reported to account for a proportion of polyposis patients(Spier et al., 2012, Aretz et al., 2007) and may remain a possible unexplored cause of disease in a fraction of the other patients in this cohort.
In this study we also compared CNV data from the patient cohort to the DGV, a much larger control database than that available from the current study. A list of 49 rare genes was revealed that were likely to be associated with disease. In silico analysis was undertaken in search for biologically meaningful relationships among these 49 genes to provide insight into their potential contribution to disease. Several cytogenetic bands that were enriched in the analysis have previously been associated with CRC (20q13, 20q, 10q25, 6q, 11q23, 1q32 and 5q31)(Houlston et al., 2010, Jia et al., 2013, Jiao et al., 2012, Peters et al., 2012, Peters et al., 2013, Tenesa et al., 2008, Cui et al., 2011). Of particular interest was the enrichment of 11q23 which has recently been reported to harbour risk variants for genetically unexplained colorectal adenomatous polyposis(Hes et al., 2014). The findings in the current study provide further support for the possible involvement of this region disease.
Annotation of the 42 miRs proposed to target the enriched 3’UTR miR target region of the 49 rare genes further identified the miR-15 family to be overrepresented which is particularly interesting given this family is has been reported to be associated with tumour suppression(Calin et al., 2002, Roccaro et al., 2009). In CRC more specifically, targeting the miRs miR-15 and miR-16 has been suggested as an effective mechanism to inhibit the growth of CRCs(Dai et al., 2012).
In conclusion, this study has revealed a number of CNVs which may contribute to the identification of genes and genomic regions associated with polyposis development and/or progression. Microarray analysis has identified several previously reported CRC susceptibility genes affected by CNVs in several patients, including MLH1, CTNNB1 and APC. We have also identified chromosome 18 to be a region of interest since loss of 18p11.32 in multiple unrelated patients is associated with a lncRNA that may be involved in disease development. Overall the results of this study provide further evidence for the involvement of CNVs in the aetiology of polyposis.
Author contributions
ALM conducted the experiments, performed data analysis/interpretation and wrote the first draft of the manuscript.
BAT-P and T-JE provided expertise in data analysis and interpretation as well as revising the manuscript.
PM provided statistical expertise.
ADS enabled patient recruitment into this study.
GNH provided critical review of the manuscript and helped design the experiments.
RJS conceived the study, designed the experimental approach and reviewed and approved the final version of the manuscript prior to submission.
Additional Information
This work has been supported by the following funding bodies and Institutions: Australian Rotary Health/Rotary District 9650, the Commonwealth Scientific and Industrial Research Organization (CSIRO), the University of Newcastle and the Hunter Medical Research Institute.
Competing Financial Interests
The author(s) declare no competing financial interests.
References
- Aretz S. Somatic APC mosaicism: a frequent cause of familial adenomatous polyposis (FAP) Hum. Mutat. 2007;28:985–992. doi: 10.1002/humu.20549. [DOI] [PubMed] [Google Scholar]
- Cacev T., Radosevic S., Spaventi R., Pavelic K., Kapitanovic S. NF1 gene loss of heterozygosity and expression analysis in sporadic colon cancer. Gut. 2005;54:1129–1135. doi: 10.1136/gut.2004.053348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.L. A novel germline 1.8-kb deletion of hMLH1 mimicking alternative splicing: a founder mutation in the Chinese population. Oncogene. 2001;20:2976–2981. doi: 10.1038/sj.onc.1204376. [DOI] [PubMed] [Google Scholar]
- Charames G.S. A large novel deletion in the APC promoter region causes gene silencing and leads to classical familial adenomatous polyposis in a Manitoba Mennonite kindred. Hum. Genet. 2008;124:535–541. doi: 10.1007/s00439-008-0579-4. [DOI] [PubMed] [Google Scholar]
- Clendenning M. Mutation deep within an intron of MSH2 causes Lynch syndrome. Familial Cancer. 2011;10:297–301. doi: 10.1007/s10689-011-9427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costi R. Repeated anastomotic recurrence of colorectal tumors: genetic analysis of two cases. World J. Gastroenterol. 2011;17:3752–3758. doi: 10.3748/wjg.v17.i32.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut. 2011;60:799–805. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L. Vector-based miR-15a/16-1 plasmid inhibits colon cancer growth in vivo. Cell Biol. Int. 2012;36:765–770. doi: 10.1042/CBI20110404. [DOI] [PubMed] [Google Scholar]
- De Moor M.H. Genome-wide association study of exercise behavior in Dutch and American adults. Med. Sci. Sports Exerc. 2009;41:1887–1895. doi: 10.1249/MSS.0b013e3181a2f646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon E.R. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- Ferreira M.A. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema I.F. LOVD v.2.0: the next generation in gene variant databases. Hum. Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- Galiatsatos P., Foulkes W.D. Familial adenomatous polyposis. Am. J. Gastroenterol. 2006;101:385–398. doi: 10.1111/j.1572-0241.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Girirajan S., Eichler E.E. Phenotypic variability and genetic susceptibility to genomic disorders. Hum. Mol. Genet. 2010;19:R176–R187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gismondi V. 310 basepair APC deletion with duplication of breakpoint (439ins15del310) in an Italian polyposis patient. Hum. Mutat. 1998;(Suppl. 1):S220–S222. doi: 10.1002/humu.1380110171. [DOI] [PubMed] [Google Scholar]
- Hes F.J. Colorectal cancer risk variants on 11q23 and 15q13 are associated with unexplained adenomatous polyposis. J. Med. Genet. 2014;51:55–60. doi: 10.1136/jmedgenet-2013-102000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzenecker A.M. Downregulation of the ubiquitin-proteasome system in normal colonic macrophages and reinduction in inflammatory bowel disease. Digestion. 2012;86:34–47. doi: 10.1159/000336353. [DOI] [PubMed] [Google Scholar]
- Hirata H. MicroRNA-1826 targets VEGFC, beta-catenin (CTNNB1) and MEK1 (MAP2K1) in human bladder cancer. Carcinogenesis. 2012;33:41–48. doi: 10.1093/carcin/bgr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach R. Discovery of variants unmasked by hemizygous deletions. Eur. J. Hum. Genet. 2012;20:748–753. doi: 10.1038/ejhg.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston R.S. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton S.E. A profile of differentially expressed genes in primary colorectal cancer using suppression subtractive hybridization. FEBS Lett. 1999;463:77–82. doi: 10.1016/s0014-5793(99)01578-1. [DOI] [PubMed] [Google Scholar]
- Jasperson K.W., Tuohy T.M., Neklason D.W., Burt R.W. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W.H. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat. Genet. 2013;45:191–196. doi: 10.1038/ng.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S. Genome-wide search for gene-gene interactions in colorectal cancer. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0052535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepischi A.C. Germline DNA copy number variation in familial and early-onset breast cancer. Breast Cancer Res. 2012;14:R24. doi: 10.1186/bcr3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo K.T. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. 2009;69:4036–4042. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guellec S. CTNNB1 mutation analysis is a useful tool for the diagnosis of desmoid tumors: a study of 260 desmoid tumors and 191 potential morphologic mimics. Mod. Pathol. 2012;25:1551–1558. doi: 10.1038/modpathol.2012.115. [DOI] [PubMed] [Google Scholar]
- Ligtenberg M.J. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1. Nat. Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- Loewer S. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Shi B., Wang J., Cao Q., Cui Q. TAM: a method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics. 2010;11:419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch H.T., de la Chapelle A. Hereditary colorectal cancer. N. Engl. J. Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- Masson A.L. A catalogue of Copy Number Variants (CNVs) identified in polyposis patients. Data Brief. 2015 (submitted for publication) [Google Scholar]
- McEvoy M. Cohort profile: The Hunter Community Study. Int. J. Epidemiol. 2010;39:1452–1463. doi: 10.1093/ije/dyp343. [DOI] [PubMed] [Google Scholar]
- Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molatore S. MUTYH mutations associated with familial adenomatous polyposis: functional characterization by a mammalian cell-based assay. Hum. Mutat. 2010;31:159–166. doi: 10.1002/humu.21158. [DOI] [PubMed] [Google Scholar]
- Morak M. Biallelic MLH1 SNP cDNA expression or constitutional promoter methylation can hide genomic rearrangements causing Lynch syndrome. J. Med. Genet. 2011;48:513–519. doi: 10.1136/jmedgenet-2011-100050. [DOI] [PubMed] [Google Scholar]
- Nagano T. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Orom U.A. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters U. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum. Genet. 2012;131:217–234. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters U. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807.e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzi A. Relative role of APC and MUTYH mutations in the pathogenesis of familial adenomatous polyposis. Scand. J. Gastroenterol. 2009;44:1092–1100. doi: 10.1080/00365520903100481. [DOI] [PubMed] [Google Scholar]
- Pylkas K. Rare copy number variants observed in hereditary breast cancer cases disrupt genes in estrogen signaling and TP53 tumor suppression network. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccaro A.M. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlin A. Inactivation of promoter 1B of APC causes partial gene silencing: evidence for a significant role of the promoter in regulation and causative of familial adenomatous polyposis. Oncogene. 2011 doi: 10.1038/onc.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustgi A.K. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- Shi Z.Z. Genomic profiling of rectal adenoma and carcinoma by array-based comparative genomic hybridization. BMC Med. Genet. 2012;5:52. doi: 10.1186/1755-8794-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlien A., Malkin D. Copy number variations and cancer susceptibility. Curr. Opin. Oncol. 2010;22:55–63. doi: 10.1097/CCO.0b013e328333dca4. [DOI] [PubMed] [Google Scholar]
- Spier I. Deep intronic APC mutations explain a substantial proportion of patients with familial or early-onset adenomatous polyposis. Hum. Mutat. 2012;33:1045–1050. doi: 10.1002/humu.22082. [DOI] [PubMed] [Google Scholar]
- Stella A. Germline novel MSH2 deletions and a founder MSH2 deletion associated with anticipation effects in HNPCC. Clin. Genet. 2007;71:130–139. doi: 10.1111/j.1399-0004.2007.00745.x. [DOI] [PubMed] [Google Scholar]
- Stenson P.D. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sygut A. Genetic Variations of the CTNNA1 And The CTNNB1 Genes in Sporadic Colorectal Cancer in Polish Population. Pol. Przegl. Chir. 2012;84:560–564. doi: 10.2478/v10035-012-0093-1. [DOI] [PubMed] [Google Scholar]
- Tenesa A. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagalingam S. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat. Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- Trevino L.R. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T. Distinct methylation patterns of two APC gene promoters in normal and cancerous gastric epithelia. Oncogene. 2000;19:3642–3646. doi: 10.1038/sj.onc.1203704. [DOI] [PubMed] [Google Scholar]
- Upadhyay G. Molecular association between beta-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/beta-catenin signaling. Oncogene. 2008;27:5845–5855. doi: 10.1038/onc.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.Y. Anti-proliferation effect of APO866 on C6 glioblastoma cells by inhibiting nicotinamide phosphoribosyltransferase. Eur. J. Pharmacol. 2012;674:163–170. doi: 10.1016/j.ejphar.2011.11.017. [DOI] [PubMed] [Google Scholar]
- Zhang C., Tong J., Huang G. Nicotinamide phosphoribosyl transferase (Nampt) is a target of microRNA-26b in colorectal cancer cells. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0069963. [DOI] [PMC free article] [PubMed] [Google Scholar]