Abstract
STUDY HYPOTHESIS
Region-specific transcriptional profiling of tissues and cultured epithelial cells from the human epididymis will predict functional specialization along the duct.
STUDY FINDING
We identified the molecular signature driving functions of the caput, corpus and cauda epithelium, and determined how these differ to establish the regional differentiation of the organ.
WHAT IS KNOWN ALREADY
The epithelium lining the human male genital ducts has a critical role in fertility. In particular, it controls the luminal environment in the epididymis, which is required for normal sperm maturation and reproductive competence. Studies in many animal species have largely informed our understanding of the molecular basis of epididymis function. However, there are substantial differences between species.
STUDY DESIGN, SAMPLES/MATERIALS, METHODS
Using RNA sequencing on biological replicates, we described gene expression profiles for tissue from each region of the epididymis and cultured epithelial cells derived from these regions. Bioinformatic tools were then utilized to identify differentially expressed genes (DEGs) between tissues and cells from the caput, corpus and cauda.
MAIN RESULTS AND THE ROLE OF CHANCE
The data showed that the caput is functionally divergent from the corpus and cauda, which have very similar transcriptomes. Interrogation of DEGs using gene ontology process enrichment analyses showed that processes of ion transport, response to hormone stimulus and urogenital tract development are more evident in the caput, while defense response processes are more important in the corpus/cauda. Consistent with these regional differences in epididymis function, we observed differential expression of transcription factors in the caput and corpus/cauda.
LIMITATIONS, REASONS FOR CAUTION
Cultured caput, corpus and cauda cells may not faithfully represent the same cells in the intact organ, due to loss of hormonal signals from the testis and communication from other cell types.
WIDER IMPLICATIONS OF THE FINDINGS
Our data provide a molecular characterization that will facilitate advances in understanding human epididymis epithelium biology in health and disease. They may also reveal the mechanisms coordinating epididymis luminal environment and sperm maturation.
LARGE SCALE DATA
Data deposited at http://www.ncbi.nlm.nih.gov/geo/GSE72986.
STUDY FUNDING AND COMPETING INTEREST(S)
This work was supported by the National Institutes of Health: R01HD068901 (PI: A.H.). The authors declare no conflict of interest.
Keywords: epididymis epithelium, caput, corpus, cauda, RNA-seq, differential gene expression, luminal environment, sperm maturation, transcriptional network
Introduction
The epithelial layer lining the epididymis has region-specific properties, with substantial functional diversity in the caput (head), corpus (body) and cauda (tail). Each of these segments has a specific role in ensuring that spermatozoa acquire full motility and fertility. Processes within the caput and corpus coordinate early and late sperm maturation, respectively, while the cauda provides a storage location for the mature male gametes. These functions are well studied in rodents (Turner et al., 2003) or large animal models (Guyonnet et al., 2009) but are less well characterized in humans. Epithelial cells within each region of the epididymis have a unique transcriptome and distinct functions (Turner et al., 2003; Cornwall, 2009). The different gene expression patterns along the epididymis are established and maintained by specific transcription factor (TF) networks that coordinate region-specific functions. The sequential changes in the epididymis luminal environment, which are critical for sperm maturation, depend on the expression of a broad spectrum of genes including cell structural proteins, ion channels and transporters and secreted proteins. Region-specific gene expression in the mouse, rat and pig epididymis was shown by microarray analysis of RNA extracted from intact tissue (Jervis and Robaire, 2001; Johnston et al., 2005; Guyonnet et al., 2009). Similarly, the transcriptome of human epididymis tissue segments was investigated by microarrays (Zhang et al., 2006; Dube et al., 2007; Thimon et al., 2007) and showed many differences from the rodent profiles. We recently reported a protocol for the culture of epithelial cells derived from the human epididymis and demonstrated their differentiated function by expression of specific proteins and robust trans-epithelial resistance (Leir et al., 2015). In order to reveal the gene expression repertoire of caput, corpus and cauda epithelial cells, we performed RNA sequencing (RNA-seq) analysis of cultured cells. Moreover, gene expression was compared between intact tissue and cultured cells derived from caput and cauda. These data enhance our understanding of the molecular basis for regional differences in epididymis function and provide a framework for novel experimental approaches to characterize the role of the epididymis in human sperm maturation.
Materials and Methods
Preparation of primary cultures
Human epididymis tissue was obtained with Institutional Review Board approval from four patients (UC05, UC06, UC08, UC09, range: 22–36 years) undergoing inguinal radical orchiectomy for a clinical diagnosis of testicular cancer. Each patient gave informed consent for use of their tissue samples. No patients were taking hormones or drugs with androgenic effects. None of the epididymides had extension of the testicular cancer and all were freshly placed into normal saline until further processing within 2–5 h after surgery. Caput, corpus and cauda tissue were dissected as described previously (Dacheux et al., 2006). Adult human epididymis epithelial (HEE) cultures were established using methods described previously (Leir et al., 2015). The culture media did not contain any additional androgens.
RNA sequencing
Epididymal tissue was placed in Hanks' balances salt solution and fat and connective tissue were removed. The three anatomical regions: caput, corpus and cauda, were separated and segments of each snap frozen in liquid nitrogen. RNA was extracted using Trizol (Life Technologies, LT) as per the manufacturers protocol. RNA quality was confirmed by Nanodrop measurement of OD 260/280 and 260/230 ratios and the RNA was stored at −80°C under ethanol. RNA-seq libraries were prepared using the TruSeq RNA Sample Preparation Kit v2 as per the manufacturer's Low-Throughput protocol (Illumina). The libraries were sequenced on Illumina HiSeq2500 machines. Data were analyzed using TopHat and Cufflinks (Trapnell et al., 2012). All data are deposited at GEO: (http://www.ncbi.nlm.nih.gov/geo/GSE72986). Gene ontology process enrichment analysis (Huang da et al., 2009) was performed to identify statistically significant biological processes associated with the DEGs (as shown by both P-value and false discovery rate, FDR).
Quantitative reverse transcription–PCR
Reverse transcription and quantitative PCR (RT–qPCR) was performed by standard protocols. Briefly, cDNA was synthesized using a TaqMan Reverse Transcription kit (Applied Biosystems) with random hexamers, and qPCR experiments were carried out with SYBR Green master mixes. The sequences of the primer pairs specific for each target gene are listed in Supplementary data, Table SXII and expression was normalized to β2-microglobulin (β2M).
Immunocytochemistry
Primary adult HEEs were grown to confluence on 12 mm circular glass coverslips and fixed with 3% paraformaldehyde for 15 min. The cells were then permeabilized with 0.1% Triton X-100 for 15 min, followed by blocking in 1% bovine serum albumin for 30 min prior to immunocytochemistry. Antibodies against aquaporin 1 (AQP1) (OWL ID 33651), Kir5.1 (OWL ID 33663), NBCn1 (OWL ID 33579) and MMP7 (OWL ID 2960) were purchased from One World Lab, San Diego and N-cadherin antibody (sc-59987) was from Santa Cruz Biotechnology. Alexa Fluor 488-conjugated anti-rabbit IgG or Alexa Fluor 549 conjugated anti-mouse IgG (Jackson Immunoresearch) were used as the secondary antibody. Cells were counterstained with 6-diamidino-2-phenylindole (DAPI) (LT) and mounted using FluorSave (Calbiochem). Samples were then analyzed using a Leica DMR microscope.
Results
RNA-seq was performed on libraries generated from caput, corpus and cauda-derived cultured cells at passage 2 or 3, from four donors and caput, corpus and cauda tissue from two of the same donors. These libraries generated ∼1.9–3.9 × 107 reads per library from the cells (95–99% mapping to the genome) and ∼1.4–3.9 × 107 from the tissue samples (84–99% mapped) (Supplementary data, Table SI). Raw reads were aligned to the genome with TopHat and gene expression values were processed using Cufflinks as Fragments Per Kilobase per Million mapped fragments (FPKM) and complied into an expression value matrix. FPKM values were then subject to principle component analysis (Supplementary data, Fig. SI), which revealed that while caput, corpus and cauda cell samples, respectively, from UC05, UC06 and UC09 clustered together, UC08 was an outlier and thus was not included in further analyses. RNA-seq data from the three biological replicas (UC05, UC06 and UC09) of caput, corpus and cauda were pooled for further analysis (Supplementary data, Table SII).
Differential gene expression profiles for cultured cells from caput, corpus and cauda regions
Next, we used Cufflinks to determine differentially expressed genes (DEGs) between caput, corpus and cauda cells, combined from the three donors. A number of interesting features are evident. First, the gene expression profiles of corpus and cauda are remarkably similar and both differ from the caput to a similar degree. Of the ∼40 genes that are differentially expressed between corpus and cauda (Supplementary data, Table SIII), 8 are small RNAs (microRNAs or small nucleolar RNAs). Among others DEGs of interest, that are highly expressed only in corpus are BPI fold containing family B, member 1 (BPIFB1) a component of the BPI/LBP/PLUNC superfamily cluster that is involved in the innate immune response (Bingle and Craven, 2002), and microseminoprotein, beta- (MSMB), a member of the immunoglobulin binding factor family with inhibin-like activity, that is known to be secreted into seminal plasma by the prostate epithelium (Lilja and Abrahamsson, 1988).
Among the more than 1600 DEGs between caput and corpus/cauda are many that are relevant to the specific functions of the caput. These include FXYD domain containing ion transport regulator 2 (FXYD2), which encodes the sodium/potassium transporting ATPase subunit gamma, a critical regulator of epithelial ion transport and solute carrier family 34 (Type II sodium/phosphate cotransporter) member 2 (SLC34A2), which is a pH-sensitive sodium-dependent phosphate cotransporter.
Next, the DEGs for each comparison (caput and corpus/cauda) were analyzed using a gene ontology process enrichment analysis DAVID (Huang da et al., 2009) and the top 20 most enriched processes are shown in Table I (caput enriched) and Table II (corpus/cauda enriched). Statistical significance is shown by both P-value and false discovery rate value (FDR). The full analyses are shown in Supplementary data, Tables SIV and SV. Among highly significant caput over-represented processes are response to hormone stimulus (GO:0048545, GO:0009625), which includes many critical cellular hormone receptors. Other enriched processes with very significant P-values are plasma membrane (GO:0005886, GO:0004459), which encompasses many proteins involved in ion transport and ion exchange across the epididymis cells.
Table I.
Top 20 statistically over-represented processes from David analysis of DEGs that are more abundant in caput than corpus/cauda cells.
| Gene ontology term | P-value | FDR |
|---|---|---|
| GO:0044421∼extracellular region part | 2.01E−12 | 2.75E−09 |
| GO:0005578∼proteinaceous ECM | 5.62E−10 | 7.67E−07 |
| GO:0031012∼ECM | 1.16E−09 | 1.58E−06 |
| GO:0048545∼response to steroid hormone stimulus | 4.51E−08 | 7.84E−05 |
| GO:0005886∼plasma membrane | 9.57E−08 | 1.31E−04 |
| GO:0044459∼plasma membrane part | 2.03E−07 | 2.78E−04 |
| GO:0009719∼response to endogenous stimulus | 7.21E−07 | 1.26E−03 |
| GO:0009725∼response to hormone stimulus | 7.34E−07 | 1.28E−03 |
| GO:0005615∼extracellular space | 7.99E−07 | 1.09E−03 |
| GO:0007155∼cell adhesion | 9.62E−07 | 1.67E−03 |
| GO:0022610∼biological adhesion | 9.79E−07 | 1.70E−03 |
| GO:0043627∼response to estrogen stimulus | 2.76E−05 | 4.80E−02 |
| GO:0005576∼extracellular region | 4.15E−05 | 5.66E−02 |
| GO:0010033∼response to organic substance | 5.16E−05 | 8.99E−02 |
| GO:0042995∼cell projection | 1.42E−04 | 1.94E−01 |
| GO:0001655∼urogenital system development | 1.92E−04 | 3.34E−01 |
| GO:0001822∼kidney development | 2.29E−04 | 3.97E−01 |
| GO:0031226∼intrinsic to plasma membrane | 2.51E−04 | 3.43E−01 |
| GO:0005887∼integral to plasma membrane | 4.09E−04 | 5.58E−01 |
| GO:0016323∼basolateral plasma membrane | 4.55E−04 | 6.19E−01 |
Table II.
Top 20 statistically over-represented processes from David analysis of DEGs that are more abundant in corpus/cauda than caput cells.
| Gene ontology term | P-value | FDR |
|---|---|---|
| GO:0007398∼ectoderm development | 2.06E−15 | 3.79E−12 |
| GO:0008544∼epidermis development | 2.40E−14 | 4.30E−11 |
| GO:0009611∼response to wounding | 3.00E−10 | 5.38E−07 |
| GO:0044421∼extracellular region part | 4.27E−10 | 5.82E−07 |
| GO:0006952∼defense response | 5.27E−09 | 9.46E−06 |
| GO:0005576∼extracellular region | 8.81E−09 | 1.20E−05 |
| GO:0043588∼skin development | 3.13E−08 | 5.62E−05 |
| GO:0031012∼ECM | 3.20E−08 | 4.36E−05 |
| GO:0006955∼immune response | 7.58E−08 | 1.36E−04 |
| GO:0009615∼response to virus | 1.15E−07 | 2.07E−04 |
| GO:0042127∼regulation of cell proliferation | 1.95E−07 | 3.49E−04 |
| GO:0030855∼epithelial cell differentiation | 3.15E−07 | 5.65E−04 |
| GO:0042981∼regulation of apoptosis | 4.41E−07 | 7.91E−04 |
| GO:0043067∼regulation of programmed cell death | 6.27E−07 | 1.12E−03 |
| GO:0010941∼regulation of cell death | 7.25E−07 | 1.30E−03 |
| GO:0005578∼proteinaceous ECM | 8.64E−07 | 1.18E−03 |
| GO:0010033∼response to organic substance | 1.66E−06 | 2.97E−03 |
| GO:0001817∼regulation of cytokine production | 3.28E−06 | 5.88E−03 |
| GO:0044459∼plasma membrane part | 4.28E−06 | 5.83E−03 |
| GO:0030198∼ECM organization | 5.37E−06 | 9.63E−03 |
Also of interest are processes of urogenital system development (GO:0001655 and G0:0001822). In contrast, over-represented processes in the corpus/cauda include response to wounding (GO:0009611), defense response (GO:0006952 and GO:0006955, both innate and acquired immunity) and the response to viruses (GO:0009615). Processes of apoptosis are also significantly over-represented in the corpus/cauda (GO:0042981, GO:0043067, GO:0010941). Several processes are over-represented in both Tables I and II, such as extracellular region (GO:0044421) and extracellular matrix (ECM) (GO:0031012), which include many collagens and adhesion molecules, which show diverse expression profiles in the caput and corpus/cauda.
Next, we examined in more detail the significant processes identified by the DAVID analysis to extract data of specific relevance to epididymis epithelial function (Tables III–VIII, and Supplementary data, Tables SIV–SVIII, which show FPKM values for individual genes). Since the DAVID bioinformatics resource is not current, we also used g:Profiler (Reimand et al., 2007) to validate the DAVID gene ontology analysis results. Genes that were more highly expressed in caput cells than in corpus and cauda were submitted to the gProfiler analysis tool, and out of the 20 most significant terms identified by DAVID, 17 were also identified with g:Profiler (not shown). Also the region-specific distribution of individual genes was confirmed by RT–qPCR (Fig. 1).
Table III.
Differential distribution of ion channels and transporters in cells from caput, corpus and cauda (gene expression values as FPKM).
| Function | Gene ID | Gene name | Caput | Corpus | Cauda |
|---|---|---|---|---|---|
| Water channel | AQP1 | Aquaporin 1 | 7.3 | 0.4 | 0.6 |
| Calcium channel | CACNA1D | Calcium channel, voltage-dependent, L type, alpha 1D subunit | 2.1 | 0.2 | 0.2 |
| CACNA1H | Calcium channel, voltage-dependent, T type, alpha 1H subunit | 3.1 | 0.4 | 0.3 | |
| Chloride channel | CFTR | Cystic fibrosis transmembrane conductance regulator | 23.5 | 1.1 | 0.6 |
| CLIC6 | Chloride intracellular channel 6 | 15.5 | 0.5 | 0.2 | |
| Sodium/potassium-transporter | FXYD2,FXYD6, FXYD6-FXYD2 | FXYD domain containing ion transport regulator 2 | 930.4 | 2.4 | 0.7 |
| Potassium channel | KCNJ16 | Potassium inwardly rectifying channel, subfamily J, member 16 | 66.3 | 8.6 | 8.8 |
| KCNJ15 | Potassium inwardly rectifying channel, subfamily J, member 15 | 44.7 | 9.4 | 8.4 | |
| KCNIP3 | Kv channel interacting protein 3, calsenilin | 36.8 | 12.2 | 12.6 | |
| KCTD3 | Potassium channel tetramerization domain containing 3 | 27.6 | 13.4 | 12.9 | |
| KCNC3 | Potassium voltage-gated channel, Shaw-related subfamily, member 3 | 14.0 | 3.6 | 2.9 | |
| KCNK3 | Potassium channel, subfamily K, member 3 | 11.1 | 1.3 | 1.8 | |
| KCNK5 | Potassium channel, subfamily K, member 5 | 7.5 | 1.5 | 1.5 | |
| KCNS3 | Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 | 5.2 | 1.3 | 1.3 | |
| KCND3 | Potassium voltage-gated channel, Shal-related subfamily, member 3 | 3.8 | 0.3 | 0.3 | |
| Exchangers | SLC13A5 | Solute carrier family 13 (sodium-dependent citrate transporter), member 5 | 5.7 | 0.5 | 0.5 |
| SLC17A1 | Solute carrier family 17 (sodium phosphate), member 1 | 2.4 | 0.0 | 0.0 | |
| SLC34A2 | Solute carrier family 34 (sodium phosphate), member 2 | 259.5 | 3.2 | 1.2 | |
| SLC40A1 | Solute carrier family 40 (iron-regulated transporter), member 1 | 9.8 | 2.5 | 2.2 | |
| SLC4A4 | Solute carrier family 4, sodium bicarbonate cotransporter, member 4 | 3.2 | 0.8 | 1.0 | |
| SLC4A7 | Solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 17.0 | 5.4 | 5.5 | |
| SLCO2B1 | Solute carrier organic anion transporter family, member 2B1 | 13.0 | 3.4 | 4.7 |
Table VIII.
Differential distribution of cell junction-related genes from caput, corpus and cauda (gene expression values as FPKM).
| Family | Gene ID | Gene name | Caput | Corpus | Cauda | Family | Gene ID | Gene name | Caput | Corpus | Cauda |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gap junction | GJA1 | Gap junction protein, alpha 1 | 134.1 | 30.6 | 30.6 | Gap junction | GJA5 | Gap junction protein, alpha 5, 4 | 0.1 | 9.4 | 6.9 |
| GJC1 | Gap junction protein, gamma 1 | 3.6 | 1.0 | 1.1 | GJB3 | Gap junction protein, beta 3 | 17.2 | 55.8 | 54.1 | ||
| Nectins | PVRL3 | Poliovirus receptor-related 3 | 15.5 | 4.2 | 4.1 | GJB4 | Gap junction protein, beta 4 | 1.1 | 6.1 | 6.0 | |
| Other | AIF1L | Allograft inflammatory factor 1-like | 58.3 | 7.2 | 6.1 | GJB5 | Gap junction protein, beta 5 | 3.2 | 35.2 | 33.4 | |
| ARHGAP24 | Rho GTPase activating protein 24 | 3.8 | 1.0 | 0.9 | GJD3 | Gap junction protein, delta 3 | 3.9 | 9.0 | 9.5 | ||
| ARHGAP26 | Rho GTPase activating protein 26 | 7.5 | 2.2 | 2.2 | Nectins | PVRL1 | Poliovirus receptor-related 1 (herpesvirus entry mediator C) | 7.3 | 30.9 | 31.5 | |
| ARHGAP4 | Rho GTPase activating protein 4 | 8.2 | 0.9 | 0.9 | PVRL4 | Poliovirus receptor-related 4 | 3.76 | 24.25 | 24.62 | ||
| KCNJ15 | Potassium inwardly rectifying channel, subfamily J, member 15 | 44.7 | 9.4 | 8.4 | Other | AMOT | Angiomotin | 5.5 | 18.5 | 15.1 | |
| LAMC1 | Laminin, gamma 1 (formerly LAMB2) | 298.6 | 48.9 | 52.6 | CALB2 | Calbindin 2 | 3.7 | 35.9 | 35.0 | ||
| NOX4 | NADPH oxidase 4 | 1.0 | 0.3 | 0.2 | DLG1 | Discs, large homolog 1 (Drosophila) | 14.5 | 46.1 | 38.7 | ||
| RHOU | Ras homolog gene family, member U | 4.5 | 0.8 | 0.8 | DSC3 | Desmocollin 3 | 0.6 | 4.0 | 4.8 | ||
| TNS1 | Tensin 1 | 11.0 | 3.6 | 4.9 | LMO7 | LIM domain 7 | 12.1 | 36.1 | 32.1 | ||
| PANX2 | Pannexin 2 | 1.0 | 4.8 | 4.9 | |||||||
| PCDH1 | Protocadherin 1 | 18.0 | 96.7 | 88.9 |
The left panel shows genes that are more highly expressed in caput and the right those in corpus and cauda.
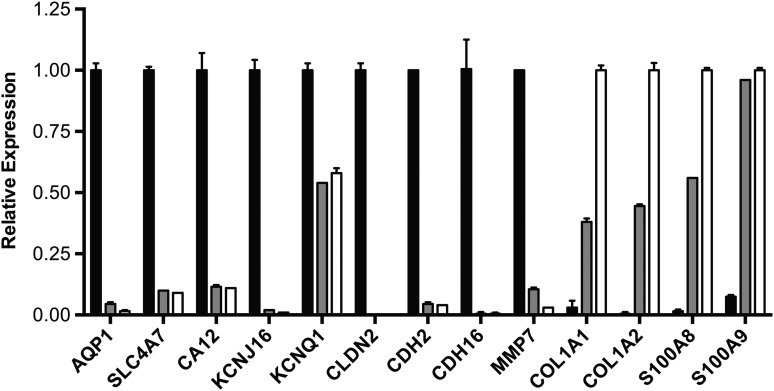
Figure 1.
Region-specific gene expression measured by RT–qPCR. Expression in caput (black bar), corpus (gray bar) and cauda (white bar) cells, is shown relative to β2M. Primers are listed in Supplementary data, Table SXII. Aquaporin 1 (AQP1), Solute Carrier Family 4, Sodium Bicarbonate Cotransporter, Member 7 (SLC4A7), Carbonic Anhydrase XII (CA12), Potassium Channel, Inwardly Rectifying Subfamily J, Member 16 (KCNJ16), Claudin 2 (CLDN2), Cadherin 2, Type 1, N-Cadherin (Neuronal) (CDH2), Cadherin 16, KSP-Cadherin (CDH16), Matrix Metallopeptidase 7 (MMP7), Collagen, Type I, Alpha-1 and -2 (COL1A1 and COL1A2), S100 Calcium Binding Protein-A8 and -A9 (S100A8 and S100A9).
Gene transcripts and processes over-represented in caput cells in comparison to corpus/cauda cells
Ion and solute transport
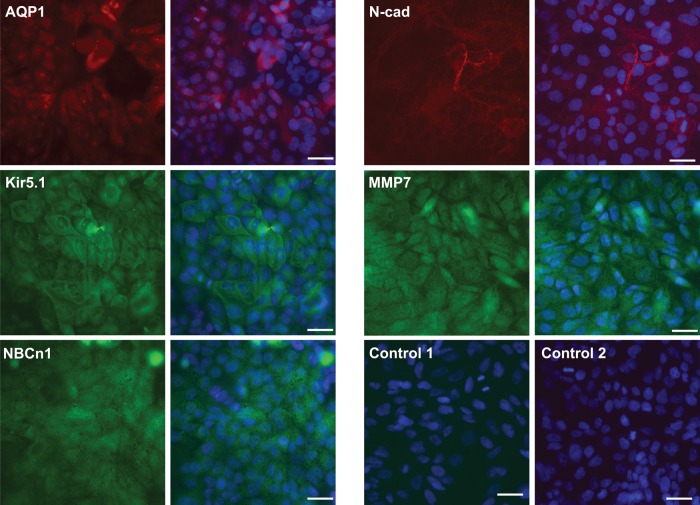
One of the critical functions of the epididymis epithelium, particularly in the caput, is the maintenance of an appropriate luminal environment for normal sperm maturation. The luminal fluid is controlled by a complex network of apical anion channels, (including CFTR), associated chloride:bicarbonate exchangers, cation channels (including potassium channels and basolateral sodium:potassium:chloride exchangers) and AQPs. Many of these proteins were more abundant in the caput than the corpus/cauda (Table III). These observations were verified in cells from a different tissue donor, by RT–qPCR (Fig. 1). AQP1, solute carrier family 4, sodium bicarbonate cotransporter, member 7 (SLC4A7) and potassium inwardly rectifying channel, subfamily J, member 16 (KCNJ16) were all shown to be more abundantly expressed in caput cells (Fig. 1) as was carbonic anhydrase XII (CA12) a driver of cellular bicarbonate secretion. Potassium voltage-gated channel, KQT-like subfamily, member 1 (KCNQ1) provided a control for a gene that is uniformly expressed along the epididymis (Fig. 1). The localization within caput cells of AQP1, NBCn1 and Kir5.1 proteins (encoded by the AQP1, SLC4A7 and KCNJ16 genes, respectively) was also shown by immunofluorescence (Fig. 2).
Figure 2.
Immunostaining of functionally important epididymal proteins in caput cells. Immunofluorescence using antibodies to aquaporin 1 (AQP1), potassium channel, inwardly rectifying subfamily j, member 16 (Kir5.1), solute carrier family 4, sodium bicarbonate cotransporter, member 7 (NBCn1), N-cadherin (N-cad) and matrix metallopeptidase 7 (MMP7) in passage 3 caput cells. Alexa Fluor 488-conjugated anti-rabbit IgG (green) or Alexa Fluor 549 conjugated anti-mouse IgG (red) secondary antibodies. Nuclei were counterstained with 6-diamino-2-phenylindole (blue). Control 1: normal rabbit serum primary and Alexa Fluor 488-conjugated anti-rabbit IgG secondary. Control 2: normal mouse IgG1 primary and Alexa Fluor 549-conjugated anti-mouse IgG secondary, respectively. Size bar = 30 μm.
Response to hormone stimulus
Several processes that are highly over-represented in caput cells involve response to hormone stimulus, including both steroid hormones (FDR = 7.84 × 10−5) and all hormones (FDR = 0.0013). Inspection of the genes contributing to these processes (Table IV) revealed a number of relevant hormone receptor genes including the androgen receptor (AR), the estrogen receptor (ESR) and the glucagon receptor (GCGR). In addition, one of the most abundant transcripts in caput epithelial cells was that encoded by the secreted phosphoprotein 1 (SPP1) gene also known as osteopontin, which is known to be androgen regulated (Luedtke et al., 2002). SPP1 probably has a role in removing minerals from the epididymis fluid, consistent with its role in the kidney (Shiraga et al., 1992). Serpin peptidase inhibitor clade A (alpha-1 antiproteinase, antitrypsin), Member 1 (SERPINA1), also named alpha-1-antitrypsin, was also particularly highly expressed in caput cells, as were the solute carrier family 34 (Type II sodium/phosphate co-transporter), member 2 (SLC34A2), which encodes a pH-sensitive sodium-dependent phosphate transporter (Xu et al., 1999) and chemokine (C-C) motif ligand 2 (CCL2).
Table IV.
Differential distribution of genes related to response to hormone stimulus in cells from caput, corpus and cauda (gene expression values as FPKM).
| Gene ID | Gene name | Caput | Corpus | Cauda |
|---|---|---|---|---|
| CDH2 | Cadherin 2, type 1, N-cadherin (neuronal) | 26.8 | 6.7 | 8.9 |
| VGF | VGF nerve growth factor inducible | 39.3 | 0.6 | 0.3 |
| PDGFB | Platelet-derived growth factor beta polypeptide | 23.0 | 6.4 | 6.7 |
| SLC34A2 | Solute carrier family 34 (sodium phosphate), member 2 | 259.5 | 3.2 | 1.2 |
| STEAP2 | Six transmembrane epithelial antigen of the prostate 2 | 10.3 | 3.4 | 3.4 |
| PARP1 | Poly (ADP-ribose) polymerase 1 | 23.4 | 10.7 | 11.5 |
| CFTR | Cystic fibrosis transmembrane conductance regulator | 23.5 | 1.1 | 0.6 |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | 21.3 | 8.2 | 8.2 |
| SRD5A2 | Steroid-5-alpha-reductase, alpha polypeptide | 2.4 | 0.3 | 0.2 |
| SOCS3 | Suppressor of cytokine signaling 3 | 22.2 | 10.3 | 10.2 |
| SHH | Sonic hedgehog homolog (Drosophila) | 7.2 | 0.6 | 0.5 |
| PTGS1 | Prostaglandin-endoperoxide synthase 1 | 4.2 | 1.2 | 1.5 |
| CA2 | Carbonic anhydrase II | 40.7 | 9.8 | 7.2 |
| ACADS | Acyl-co-enzyme A dehydrogenase, C-2 to C-3 short chain | 21.9 | 5.7 | 6.2 |
| AR | Androgen receptor | 2.8 | 0.4 | 0.2 |
| TNFRSF11B | Tumor necrosis factor receptor superfamily, member 11b | 1.4 | 3.7 | 1.9 |
| ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) | 97.4 | 24.5 | 27.5 |
| ENO2 | Enolase 2 (gamma, neuronal) | 24.9 | 3.3 | 4.5 |
| CA9 | Carbonic anhydrase IX | 70.1 | 16.3 | 13.9 |
| AQP1 | Aquaporin 1 | 7.3 | 0.4 | 0.6 |
| SH2B2 | SH2B adaptor protein 2 | 4.4 | 0.8 | 1.0 |
| PRKCA | Protein kinase C, alpha | 11.0 | 3.5 | 3.8 |
| SERPINA1 | Serpin peptidase inhibitor, clade A, member 1 | 638.4 | 12.7 | 15.3 |
| CCL2 | Chemokine (C-C motif) ligand 2 | 159.6 | 7.0 | 3.1 |
| ESR1 | Estrogen receptor 1 | 1.4 | 0.1 | 0.1 |
| SPP1 | Secreted phosphoprotein 1 | 4700.8 | 41.9 | 33.6 |
| PLA2G4A | Phospholipase A2, Group IVA | 4.7 | 1.5 | 1.4 |
| ADCY5 | Adenylate cyclase 5 | 4.2 | 0.2 | 0.2 |
| GCGR | Glucagon receptor | 6.7 | 0.2 | 0.1 |
| GSTM3 | Glutathione S-transferase mu 3 (brain) | 16.1 | 6.4 | 6.3 |
| NEFL | Neurofilament, light polypeptide | 4.1 | 0.6 | 0.5 |
| ARSB | Arylsulfatase B | 8.7 | 3.5 | 3.7 |
| FADS1 | Fatty acid desaturase 1 | 32.0 | 9.7 | 10.9 |
Urogenital tract development
To examine further the identification of urogenital track development processes among the caput-enriched processes, we sorted the DEG list according to previous data on their role in kidney development (Chai et al., 2013; Marcotte et al., 2014) (Table V). Of particular note were the paired-box TFs paired box 2(PAX2), consistent with our previous observations of the over-representation of PAX2-binding sites in open chromatin from epididymis cells (Bischof et al., 2013; Browne et al., 2014) and PAX8. Also showing marked caput enrichment are transcripts of genes encoding many growth factors and their receptors including fibroblast growth factors (FGF18 and FGFR4). Other caput enriched TFs are considered further below in the context of the region-specific epididymis transcriptional network.
Table V.
Differential distribution of genes related to urogenital tract development in cells from caput, corpus and cauda.
| Function | Gene ID | Gene name | Caput | Corpus | Cauda |
|---|---|---|---|---|---|
| AR related | AR | Androgen receptor | 2.85 | 0.40 | 0.23 |
| SRD5A2 | Steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) | 2.37 | 0.28 | 0.23 | |
| Outgrowth and branching of ureteric bud | GLI2 | GLI family zinc finger 2 | 0.86 | 0.15 | 0.21 |
| GREM1 | Gremlin 1, cysteine knot superfamily, homolog | 9.97 | 2.75 | 3.10 | |
| PAX2 | Paired box 2 | 19.78 | 4.95 | 4.36 | |
| PAX8 | Paired box 8 | 140.81 | 54.31 | 52.95 | |
| SALL1 | Sal-like 1 | 24.19 | 0.58 | 0.45 | |
| SHH | Sonic hedgehog homolog | 7.17 | 0.64 | 0.48 | |
| Others | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 60.88 | 15.29 | 14.13 |
| CA2 | Carbonic anhydrase II | 40.72 | 9.79 | 7.21 | |
| CDC25B | Cell division cycle 25 homolog B | 35.46 | 12.72 | 14.07 | |
| CDH6 | Cadherin 6, type 2, K-cadherin | 72.83 | 2.40 | 1.34 | |
| CFTR | Cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | 23.50 | 1.08 | 0.59 | |
| DZIP1 | DAZ interacting protein 1 | 17.94 | 3.83 | 4.78 | |
| FGF18 | Fibroblast growth factor 18 | 10.08 | 0.62 | 0.11 | |
| FGF9 | Fibroblast growth factor 9 (glia-activating factor) | 1.87 | 0.10 | 0.13 | |
| FGFR4 | Fibroblast growth factor receptor 4 | 9.38 | 0.44 | 0.45 | |
| FOXP2 | Forkhead box P2 | 0.92 | 0.07 | 0.03 | |
| HHIP | Hedgehog interacting protein | 6.90 | 0.43 | 0.35 | |
| HNF1A | HNF1 homeobox A | 3.59 | 0.23 | 0.16 | |
| HNF1B | HNF1 homeobox B | 49.17 | 11.95 | 12.04 | |
| HOOK1 | Hook homolog 1 | 17.34 | 3.14 | 2.63 | |
| MAPK8IP3 | Mitogen-activated protein kinase 8 interacting protein 3 | 40.01 | 17.71 | 17.43 | |
| PCSK5 | Proprotein convertase subtilisin/kexin type 5 | 8.16 | 0.83 | 0.73 | |
| PDE3A | Phosphodiesterase 3A, cGMP-inhibited | 1.60 | 0.28 | 0.22 | |
| PLA2G4A | Phospholipase A2, Group IVA (cytosolic, calcium-dependent) | 4.69 | 1.49 | 1.40 | |
| POU3F3 | POU class 3 homeobox 3 | 32.39 | 1.73 | 0.98 | |
| SPP1 | Secreted phosphoprotein 1 | 4700.7 | 41.88 | 33.60 | |
| SPRY2 | Sprouty homolog 2 | 33.62 | 9.12 | 9.03 | |
| STRA8 | Stimulated by retinoic acid gene 8 homolog | 2.82 | 0.03 | 0.04 | |
| VANGL2 | Vang-like 2 | 6.19 | 2.09 | 2.57 | |
| VGF | VGF nerve growth factor inducible | 39.28 | 0.63 | 0.32 | |
| WNT7A | Wingless-type MMTV integration site family, member 7A | 87.12 | 24.98 | 19.36 | |
| WWTR1 | WW domain containing transcription regulator 1 | 21.94 | 10.42 | 10.34 | |
| YBX2 | Y box binding protein 2 | 4.72 | 0.93 | 0.74 |
Genes classified according to their role in kidney development (Chai et al., 2013; Marcotte et al., 2014) (gene expression values as FPKM).
Gene transcripts and processes over-represented in corpus/cauda cells in comparison to caput cells
Defense response
Over-represented within the corpus/cauda-specific genes are multiple components of the innate and acquired immune responses to a wide spectrum of agents, including bacteria and viruses. A subset of genes that are involved in innate immunity are shown in Table VI to illustrate the marked distribution of these transcripts within different regions of the epididymis. Of particular note were the toll-like receptors (TLRs), cell surface receptor TLR2 and the endosomal receptor TLR3, which recognize bacterial and viral motifs, respectively (West et al., 2006). Also over-represented were interleukin and antibacterial peptide genes such as S100 calcium binding protein (S100A7, A8 and A9), that encode peptides inhibiting bacterial adhesion cell division at the mucosal surface (Moncrief et al., 1990), and indoleamine 2,3-dioxygenase 1 (IDO1). IDO1 has a role in epididymis immune tolerance toward sperm and also causes immunosuppression to prevent inflammation of the epididymis (Jrad-Lamine et al., 2013). The DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (DDX58), which plays a major role in sensing viral nucleic acids during infection, was also differentially expressed in corpus/cauda. Higher expression of the S100A8 and A9 genes in the corpus/cauda, was confirmed by RT–qPCR (Fig. 1).
Table VI.
Innate immune response genes showing differentially high expression in the corpus and cauda (gene expression values as FPKM).
| Function | Gene ID | Gene name | Caput | Corpus | Cauda |
|---|---|---|---|---|---|
| Receptor-like proteins | TLR2 | Toll-like receptor 2 | 5.5 | 24.7 | 18.3 |
| TLR3 | Toll-like receptor 3 | 3.8 | 15.2 | 12.6 | |
| DDX58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | 6.0 | 61.9 | 53.3 | |
| CALCOCO2 | Calcium binding and coiled-coil domain 2 | 15.4 | 35.8 | 34.1 | |
| IFIH1 | Interferon induced with helicase C domain 1 | 5.8 | 56.8 | 48.1 | |
| PGLYRP4 | Peptidoglycan recognition protein 4 | 0.1 | 2.4 | 3.4 | |
| Adapter protein | MYD88 | Myeloid differentiation primary response gene (88) | 17.2 | 53.4 | 47.2 |
| Other IL1 signaling components | IL1B | Interleukin 1, beta | 23.9 | 64.3 | 68.0 |
| IRAK2 | Interleukin-1 receptor-associated kinase 2 | 9.8 | 20.5 | 22.8 | |
| IL1A | Interleukin 1, alpha | 10.0 | 80.9 | 82.1 | |
| IL1RL1 | Interleukin 1 receptor-like 1 | 3.1 | 75.1 | 64.1 | |
| IL1RN | Interleukin 1 receptor antagonist | 18.3 | 219.2 | 241.0 | |
| Interleukin signaling | IL18R1 | Interleukin 18 receptor 1 | 0.9 | 5.1 | 4.8 |
| IL20RB | Interleukin 20 receptor beta | 1.4 | 20.4 | 22.3 | |
| Cytokine | IL17C | Interleukin 17C | 1.6 | 7.1 | 5.9 |
| Chemokines | CXCL10 | Chemokine (C-X-C motif) ligand 10 | 1.4 | 41.4 | 46.6 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | 0.5 | 15.7 | 16.3 | |
| Antibacterial/antifungal/antimicrobial activity | HIST1H2BC | Histone cluster 1, H2bc | 4.5 | 59.9 | 41.8 |
| HIST1H2BK | Histone cluster 1, H2bk | 23.4 | 99.4 | 89.8 | |
| HIST2H2BE | Histone cluster 2, H2be | 2.1 | 15.6 | 13.5 | |
| RNASE7 | Ribonuclease, RNase A family, 7 | 0.7 | 8.8 | 7.8 | |
| IDO1 | Indoleamine 2,3-dioxygenase 1 | 14.6 | 105.1 | 89.7 | |
| Anti-bacterial peptides | S100A7 | S100 calcium binding protein A7 | 15.4 | 76.6 | 107.0 |
| S100A8 | S100 calcium binding protein A8 | 56.6 | 194.8 | 321.9 | |
| S100A9 | S100 calcium binding protein A9 | 414.4 | 2190.1 | 2464.7 | |
| Antiviral activity/response to viruses | IRF7 | Interferon regulatory factor 7 | 18.4 | 72.8 | 63.9 |
| MX1 | MX Dynamin-Like GTPase 1 | 30.5 | 356.5 | 328.4 | |
| MX2 | MX Dynamin-Like GTPase 2 | 0.2 | 14.8 | 12.6 | |
| RSAD2 | Radical S-adenosyl methionine domain containing 2 | 5.0 | 117.1 | 97.2 | |
| SAMHD1 | SAM domain and HD domain 1 | 4.9 | 27.0 | 23.1 | |
| Antigen presentation | TAP2 | Transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | 19.8 | 78.7 | 67.5 |
Several defensins, including DEF4A/4B and DEFB1, which are also critical components of the innate immune response, were abundantly expressed in caput, corpus and cauda cells, and did not show regional localization.
Apoptosis
Multiple processes related to apoptosis were also enriched in corpus and cauda cells. As an example of many similar processes, genes contributing to the GO:0042981 processes (regulation of apoptosis) are shown in Supplementary data, Table SVI. These include both positive and negative regulators of apoptosis of multiple different classes.
Processes showing enrichment in both caput and corpus/cauda through different DEGs
Several gene ontology processes showed highly significant representation in both caput and corpus/cauda cells as a result of a marked regional distribution of gene expression. Examples include genes involved in processes of adhesion, cell junctions and ECM.
Adhesion and cell junctions
Table VII shows the diverse gene expression values in caput, corpus and cauda for key components of cell adhesion, including cadherins, collagens, integrins, laminins and nectins. Several cadherins, claudins and laminins were much more abundant on caput cells. Among the collagens, it is of note that those more abundant in caput cells are primarily non-fibrillar, while the fibrillar collagens, such as those encoded by COL1A1 and COL5A1, were over-expressed in corpus and cauda cells. Regional expression of claudin 2 (CLDN2), cadherin 2, type 1, N-cadherin (CDH2) and cadherin 16, KSP-cadherin (CDH16), collagen, type I, alpha 1 and 2 (COL1A1/2) was confirmed by RT–qPCR (Fig. 1) and N-cadherin protein expression was shown by immunofluorescence in Fig. 2.
Table VII.
Differential distribution of cell adhesion/tight junctions-related genes from caput, corpus and cauda (gene expression values as FPKM).
| Family | Gene ID | Gene name | Caput | Corpus | Cauda | Family | Gene ID | Gene name | Caput | Corpus | Cauda |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cadherins | CDH16 | Cadherin 16, KSP-cadherin | 38.08 | 0.96 | 0.77 | Cadherins | CDH3 | Cadherin 3, type 1, P-cadherin (placental) | 26.44 | 149.95 | 131.65 |
| CDH2 | Cadherin 2, type 1, N-cadherin | 26.75 | 6.66 | 8.86 | DSC3 | Desmocollin 3 | 0.56 | 4.05 | 4.78 | ||
| CDH6 | Cadherin 6, type 2, K-cadherin | 72.83 | 2.40 | 1.34 | Proto-cadherins | PCDH1 | Protocadherin 1 | 18.01 | 96.66 | 88.90 | |
| CDHR1 | Protocadherin 21 | 7.86 | 1.69 | 2.29 | PCDH7 | Protocadherin 7 | 0.83 | 6.24 | 5.30 | ||
| Proto-cadherins | PCDH20 | Protocadherin 20 | 1.30 | 0.05 | 0.03 | Collagens | COL1A1 | Collagen, type I, alpha 1 | 13.76 | 82.28 | 171.03 |
| Collagens | COL13A1 | Collagen, type XIII, alpha 1 | 5.74 | 0.54 | 0.74 | COL3A1 | Collagen, type III, alpha 1 | 2.39 | 13.03 | 24.90 | |
| COL14A1 | Collagen, type XIV, alpha 1 | 1.03 | 0.05 | 0.04 | COL4A6 | Collagen, type IV, alpha 6 | 1.12 | 15.24 | 16.14 | ||
| COL18A1 | Collagen, type XVIII, alpha 1 | 177.58 | 22.15 | 19.59 | COL5A1 | Collagen, type V, alpha 1 | 0.87 | 16.60 | 36.62 | ||
| COL27A1 | Collagen, type XXVII, alpha 1 | 13.13 | 3.05 | 3.55 | COL5A3 | Collagen, type V, alpha 3 | 0.09 | 1.31 | 2.05 | ||
| COL28A1 | Collagen, type XXVIII, alpha 1 | 1.12 | 0.28 | 0.28 | COL6A2 | Collagen, type VI, alpha 2 | 1.73 | 11.60 | 16.61 | ||
| Integrins | ITGA7 | Integrin, alpha 7 | 1.47 | 0.32 | 0.32 | COL6A3 | Collagen, type VI, alpha 3 | 0.93 | 9.30 | 16.52 | |
| ITGB2 | Integrin, beta 2 | 9.39 | 1.77 | 1.93 | COL8A2 | Collagen, type VIII, alpha 2 | 0.40 | 1.63 | 2.03 | ||
| EFCAB13,ITGB3 | Integrin, beta 3 | 70.21 | 11.08 | 14.77 | Integrins | ITGB4 | Integrin, beta 4 | 42.96 | 163.15 | 145.82 | |
| Laminins | LAMB1 | Laminin, beta 1 | 166.30 | 46.68 | 48.46 | ||||||
| LAMC1 | Laminin, gamma 1 | 298.61 | 48.92 | 52.55 | |||||||
| Claudins | CLDN11 | Claudin 11 | 16.36 | 4.75 | 5.62 | ||||||
| CLDN2 | Claudin 2 | 79.03 | 0.36 | 0.26 | |||||||
| CLDN3 | Claudin 3 | 49.14 | 3.66 | 2.90 |
The left panel shows genes that are more highly expressed in caput and the right those in corpus and cauda. (Nectins, which are also involved in cell adhesion and are differentially expressed, are shown in Table VIII.)
Among cell-junction proteins, many of which also have a role in cell adhesion, both gap junction proteins and nectins showed a very different distribution along the epididymis (Table VIII). Gap junction protein alpha 1 (GJA1) was most abundant in caput cells, while gap junction proteins beta 3 and 5 (GJB3/5) were more highly expressed in corpus and cauda cells.
Cell matrix
Many cell matrix associated genes showed regional distribution along the epididymis epithelium (Supplementary data, Tables SVII and SVIII). These included matrix metallopeptidase (MMPs), glypicans, serpins, mucins and an extensive group of more than 80 other ECM proteins that are caput enriched and a further 95 that are enriched in corpus/cauda. The caput-enriched expression of matrix metallopeptidase 7 (MMP7) and the corpus/cauda-enriched expression of COL1A1/A2 were confirmed by RT–qPCR (Fig. 1). Localization of MMP7 in caput cells was shown by immunofluorescence in Fig. 2.
Regional distribution of TFs in caput, corpus and cauda
The extensive differences between gene expression patterns in the caput, corpus and cauda cells led us to examine the network of TFs that might be driving this regional distribution. To identify TFs and chromatin modifying factors among the DEGs, we intersected this list with the factors included in the Dharmacon human siRNA libraries for TFs. Table IX shows the top 20 most differentially expressed TFs together with several additional factors of biological relevance to epididymis epithelial function. The complete list of DEGs encoding TFs is presented in Supplementary data, Table S-IX. As seen for other proteins, gene expression profiles in the corpus and cauda were very similar, with no differentially expressed TFs between the two cell types. TFs differentially expressed between caput and corpus, or caput and cauda overlap (∼80%). We identified 32 TFs that were significantly more abundant in caput cells and 58 that were higher levels in corpus and cauda cells (Supplementary data, Table S-IX). Among TFs that were more abundant in caput cells was the AR, with 7.2- and 12.4-fold higher expression levels than in corpus and cauda, respectively. These data are consistent with a predominant role for the caput epithelium in AR-regulated processes of sperm maturation (O'Hara et al., 2011; Krutskikh et al., 2011). Also higher in caput cells was PAX2. Mutations in PAX2 are known to be associated with kidney and urogenital tract abnormalities (Eccles and Schimmenti, 1999; Bower et al., 2012) and its binding sites are over-represented in open chromatin from primary epididymis epithelial cells (Bischof et al., 2013; Browne et al., 2014). Other TFs of interest that were differentially expressed in caput include SRY (Sex Determining Region Y)-Box 17 and Spalt-like TF 1 (SALL1). SOX17 has an important role in heart and definitive gut endoderm but mutations in this TF cause autosomal dominant vesicoureteral reflux syndrome (OMIM:61374) (Gimelli et al., 2010), which has a phenotype of functional abnormalities in the kidneys, bladder and ureters. SALL1 is the most differentially expressed TF between caput (where it is more abundant) and corpus. SALL1 is a zinc finger transcriptional repressor that may be part of the NuRD histone deacetylase complex. An earlier name for SALL1 was Epididymis Secretory Protein Li 89 and mutations in this TF are associated with two inherited disorders, Townes–Brocks syndrome (TBS) (Kohlhase et al., 1998) and branchio-oto-renal syndrome (BOR) (Engels et al., 2000), both developmental defects which affect multiple organ systems including the anus (TBS) and the kidneys (BOR).
Table IX.
Differential expression of TF genes in caput, corpus and cauda cells (gene expression values as FPKM).
| Gene | Caput | Corpus | Cauda | Fold change (caput over corpus) | Gene | Caput | Corpus | Cauda | Fold change (corpus over caput) |
|---|---|---|---|---|---|---|---|---|---|
| SALL1 | 24.19 | 0.58 | 0.45 | 41.52 | CREB3L1 | 0.34 | 5.35 | 6.92 | 15.89 |
| SOX17 | 27.12 | 1.17 | 0.33 | 23.19 | OVOL1 | 0.75 | 10.82 | 9.85 | 14.35 |
| POU3F3 | 32.39 | 1.73 | 0.98 | 18.71 | ETV7 | 0.69 | 9.07 | 7.23 | 13.14 |
| TBX18 | 5.09 | 0.29 | 0.08 | 17.82 | GATA3 | 2.24 | 28.88 | 27.41 | 12.91 |
| HLF | 2.29 | 0.13 | 0.11 | 17.05 | PRDM16 | 0.24 | 2.95 | 3.00 | 12.19 |
| ESR1 | 1.43 | 0.09 | 0.06 | 16.59 | TFAP2A | 2.52 | 29.69 | 29.69 | 11.77 |
| HNF1A | 3.59 | 0.23 | 0.16 | 15.55 | VGLL1 | 3.56 | 40.73 | 32.65 | 11.44 |
| TBX15 | 6.37 | 0.42 | 0.11 | 15.05 | TP63 | 1.88 | 21.49 | 19.65 | 11.42 |
| FOXP2 | 0.92 | 0.07 | 0.03 | 14.11 | TRIM29 | 4.92 | 44.94 | 50.29 | 9.14 |
| MLXIPL | 2.22 | 0.21 | 0.27 | 10.68 | ID3 | 7.24 | 64.52 | 89.27 | 8.91 |
| MSX2 | 1.37 | 0.16 | 0.17 | 8.56 | ANKRD1 | 1.57 | 13.92 | 8.71 | 8.86 |
| POU2F2 | 2.27 | 0.28 | 0.19 | 8.17 | PITX1 | 0.28 | 2.35 | 4.79 | 8.28 |
| TEAD2 | 22.87 | 2.89 | 4.71 | 7.91 | PRDM1 | 0.32 | 2.48 | 2.41 | 7.63 |
| IRX5 | 1.68 | 0.23 | 0.16 | 7.33 | HR | 0.24 | 1.61 | 1.67 | 6.64 |
| AR | 2.85 | 0.40 | 0.23 | 7.19 | SP6 | 1.97 | 12.53 | 12.43 | 6.35 |
| FOSB | 5.71 | 0.81 | 0.85 | 7.06 | HOPX | 1.61 | 10.11 | 12.29 | 6.30 |
| ZNF69 | 8.45 | 1.42 | 1.33 | 5.96 | TBX3 | 2.74 | 17.20 | 22.51 | 6.28 |
| GLI2 | 0.86 | 0.15 | 0.21 | 5.69 | TRIM16 | 10.02 | 53.77 | 47.15 | 5.37 |
| CDKN2C | 8.03 | 1.63 | 2.37 | 4.93 | NR3C2 | 0.44 | 2.36 | 1.71 | 5.36 |
| PAX6 | 0.69 | 0.15 | 0.17 | 4.47 | EYA2 | 0.81 | 3.95 | 4.62 | 4.84 |
| GLIS3 | 13.02 | 3.05 | 3.15 | 4.27 | GRHL2 | 1.86 | 8.99 | 7.88 | 4.84 |
| HNF1B | 49.17 | 11.95 | 12.04 | 4.11 | ATF3 | 10.11 | 48.29 | 47.50 | 4.78 |
The left panel shows genes that are more highly expressed in caput and the right those in corpus and cauda.
Among TFs that were more abundant in corpus and cauda, several are relevant to epididymis function. These include, Ovo-like zinc finger (OVOL1), paired-like homeodomain 2, bicoid class of homeodomain transcription factors (PITX2), activating transcription factor 3 (ATF3), T-Box-3 (TBX3), GATA3 and the nuclear receptors retinoid acid receptor γ (RARG) and retinoid X receptor α (RXRA).
OVOL1 is critical for normal spermatogenesis: Ovol1−/− male mice have greatly decreased sperm production and male fertility (Dai et al., 1998) and this factor has a role in the pachytene progression of male germ cells (Li et al., 2005). Our data suggest an important role in epididymis function. PITX2 regulates procollagen lysyl hydroxylase gene expression and is involved in development of eye, teeth and abdominal organs. Mutations in PITX2 are associated with reproductive system abnormalities in mice (http://www.informatics.jax.org) and Axenfeld–Rieger syndrome (Priston et al., 2001). Mutations in ATF3 are associated with hypospadias (displacement of the urethral opening) (Beleza-Meireles et al., 2008), those in TBX3 are found in several phenotypes of abnormal genital development (Packham and Brook, 2003) and GATA3 mutations may cause renal failure (Van Esch et al., 2000). The nuclear receptors RARG and RXRA dimerize in response to retinoid acid (RA) binding, and occupy RA response elements to regulate RA responsive genes. RARA mutant mice are either infertile or have reduced fertility due to genital duct obstruction (Costa et al., 1997) and our data suggest that this process may also have a role in regional function of the human epididymis epithelium.
Comparison of regional gene expression profiles of epididymis tissues and epithelial cells derived from them
The epithelium lining the epididymis has a pivotal role in sperm maturation, however, in the intact organ it receives many signals from the adjacent tissues. To investigate the additional cell types and functions in the caput, corpus and cauda, we performed RNA-seq on whole tissue from each region. Tissues were from two of the same donors that were used to derive epithelial cell cultures. DEGs between primary cells and intact tissues in each region are shown in Supplementary data, Table SX and indicate quite distinct profiles. There are more than ∼6000 DEGs between cells and tissues in both caput and corpus regions, and ∼3000 DEGs in cauda. Inspection of the DEGs using a gene ontology process enrichment analysis by DAVID confirms the different cellular composition in the tissue samples (Table X) (Supplementary data, Table SXI).
Table X.
Top 20 DAVID gene ontology pathways generated from DEGs in caput tissue compared with caput primary epithelial cells.
| Term | P-value | FDR |
|---|---|---|
| GO:0005576∼extracellular region | 5.92E−16 | 8.10E−13 |
| GO:0005578∼proteinaceous ECM | 4.34E−09 | 6.37E−06 |
| GO:0031012∼ECM | 5.77E−09 | 8.49E−06 |
| GO:0005581∼collagen | 2.44E−08 | 3.58E−05 |
| GO:0044421∼extracellular region part | 4.44E−07 | 6.53E−04 |
| GO:0005583∼fibrillar collagen | 1.04E−06 | 1.54E−03 |
| GO:0005509∼calcium ion binding | 1.87E−06 | 3.07E−03 |
| GO:0019838∼growth factor binding | 1.93E−06 | 3.16E−03 |
| GO:0005201∼ECM structural constituent | 2.39E−06 | 3.91E−03 |
| GO:0042742∼defense response to bacterium | 2.73E−06 | 5.06E−03 |
| GO:0044420∼ECM part | 3.28E−06 | 4.83E−03 |
| GO:0048407∼platelet-derived growth factor binding | 6.03E−06 | 9.87E−03 |
| GO:0007283∼spermatogenesis | 6.63E−06 | 1.23E−02 |
| GO:0048232∼male gamete generation | 6.63E−06 | 1.23E−02 |
| GO:0019953∼sexual reproduction | 9.87E−06 | 1.83E−02 |
| GO:0007276∼gamete generation | 2.09E−05 | 3.87E−02 |
| GO:0030141∼secretory granule | 3.11E−05 | 4.56E−02 |
| GO:0001871∼pattern binding | 3.96E−05 | 6.48E−02 |
| GO:0030247∼polysaccharide binding | 3.96E−05 | 6.48E−02 |
Among genes transcripts enriched in tissue samples from all three regions of the epididymis, were those associated with ECM proteins, which are abundant in connective tissue fibroblasts. These include collagens, matrix metallopeptidase (MMPs) and members of ADAMTS (A Disintegrin And Metalloproteinase with Thrombospondin Motifs) family, all of which are important for the formation and maintenance of the ECM. Gene transcripts that were highly expressed in caput and corpus tissues, though not in the epithelial cells from those regions, include many sperm-specific transcripts, such as several sperm-associated antigen (SPAG) genes and spermatogenesis-associated (SPATA) genes. These data likely reflect the spermatozoa captured in the epididymis at the time of tissue collection. Perhaps unexpectedly, enrichment of sperm-associated gene transcripts was not evident in the cauda tissue, despite the fact that mature sperm may be stored at this site. Also notable among tissue-enriched genes in the caput only, were many involved in the defense response to bacteria. The antimicrobial peptides defensins are a significant contributor to this process, however their cellular origin in the human caput is currently uncharacterized. Although epithelia are one of the main sources of defensins in other organ sites, this may not be the case in the epididymis, where sperm-derived defensins may play a prominent role (Shimizu et al., 2008).
The reverse comparison looking for DEGs that were more abundant in primary epithelial cell cultures than in the tissues they were derived from predictably identified genes encoding many cell junction proteins and genes involved in cell cycle regulation (data not shown).
Discussion
Understanding the molecular basis of epididymis function is critical to the generation of novel approaches to alleviate phenotypes of male infertility. Investigating individual cells types in the intact organ would provide the most accurate functional characterization of each part of the epididymis. However, it is not possible with current methodologies to obtain sufficient numbers of defined cell populations to perform many genome-wide analyses. To circumvent this limitation, we established differentiated epithelial cell cultures from the human epididymis. Here we describe an in depth analysis of the gene expression repertoire of primary cultures of epithelial cells and intact tissues from each region of the adult human epididymis. We identify the molecular signature driving functions of the caput, corpus and cauda epithelium and determine how these differ to establish the regional differentiation of the organ. The deep sequencing methodologies used here provide data at an unprecedented level of detail and accuracy. Moreover, by including samples from multiple epididymis donors, we are able to account for the considerable individual variation of transcriptomes.
The transcriptional profile of human epididymis tissues, although not cultured epithelial cells, was previously characterized by microarray analysis (Thimon et al., 2007). Those data showed that 1839 genes were differentially expressed between caput and corpus tissue, 1265 between caput and cauda and only 201 between corpus and cauda. Consistent with these microarray results, our RNA-seq data identify few genes that are differentially expressed between the corpus and cauda, while there are many DEGs between these regions and the caput. A comparison of the DEG list between the RNA-seq and microarray analysis of tissue samples shows some similarity. Out of the 30 genes that are up-regulated in caput with a ratio caput/corpus > 10 in the microarray analysis, 21 were also identified by RNA-seq. Moreover, gene ontology pathway analysis of the microarray data coincided with some aspects of our RNA-seq analysis in identifying processes of cell adhesion, which were differentially expressed in the regions of the epididymis. However, probably due to the greatly increased molecular resolution provided by the deep sequencing technology, the RNA-seq dataset revealed many additional DEGs and GO processes that diverged significantly in the caput and corpus/cauda.
The transcriptomes of cultured epithelial cells from the different regions of the epididymis reveal important functions of the organ. Aspects of epididymis biology that are directly relevant to the normal properties of spermatozoa are largely focused in processes enriched in the caput segment. Sperm maturation is intimately dependent on the luminal environment, which requires the coordinated expression of many genes encoding proteins with a role in epithelial transport.
Our data show several cation, anion and solute transport-related processes that are predominant in caput epithelial cells. Potassium transport. High luminal potassium levels in the epididymis were shown to inhibit sperm motility to maintain quiescence during storage and maturation (Wong and Lee, 1983). Potassium channels and transporters within both the epididymis and sperm (such as Na,K-ATPase α4 (Jimenez et al., 2011)) likely contribute to the luminal potassium concentration. Multiple genes encoding potassium channels are differently expressed in caput cells including the inwardly rectifying KCNJ15/16 (encoding Kir4.2 and Kir5.1, respectively). In renal tubular epithelium, Kir5.1 forms heteromeric potassium channels with Kir4.1 (encoded by the KCNJ10 gene), which are sensitive to intracellular pH (pHi) and inhibited by acidification (Tanemoto et al., 2000; Tucker et al., 2000). Another critical potassium channel gene, which is expressed in all regions of the epididymis epithelium, is the voltage-gated KCNQ1 (encoding Kv7.1). In intestinal crypt cells, Kv7.1 together with KCNE3 form a potassium channel that is required for cAMP-stimulated intestinal chloride secretion (Schroeder et al., 2000). Chloride and bicarbonate transport. CFTR has a pivotal role in mediating both chloride and bicarbonate transport across the epididymal epithelium (Harris and Coleman, 1989; Leung et al., 1996; Chan et al., 1996) and it is differentially expressed in caput cells. A similar expression pattern is evident for the SLC4A4 and SLC4A7 genes, which encode an electrogenic (NBCe1) and an electroneutral (NBCn1) sodium-dependent bicarbonate transporter, respectively. Together with CFTR, these transporters may facilitate a low-bicarbonate and acidic environment in the caput lumen, which is critical for sperm maturation and quiescence prior to ejaculation (reviewed by Liu et al., 2012). Again parallels may be drawn with the function of these transporters in the kidney, where NBCn1 has an important role in pHi regulation in renal intercalated cells (Yip et al., 2002). Water channels. Although the epithelium lining the efferent ducts reabsorbs much of the testicular fluid, water continues to be removed from the epididymal lumen (Levine and Marsh, 1971). It is probable that epithelial AQPs play a central role in this process (Hermo and Smith, 2011) hence it was of interest to find AQP9 uniformly expressed along the epididymis while AQP1 expression was limited to the caput epithelium. AQP1 and AQP9 were also identified in previous microarray analyses of whole epididymis human tissue (Zhang et al., 2006; Thimon et al., 2007; Dube et al., 2007).
Also predominant in caput epithelial cells are genes involved in urogenital tract development. Our observation that the genes encoding PAX2 and PAX8 are among the caput DEGs, which was confirmed by western blot (data not shown), are of interest since these two TFs are also central regulators of kidney development (Torres et al., 1995; Bouchard et al., 2002). HNF1β, which is abundant in the caput DEGs, is also important for kidney development, as highlighted by urogenital tract abnormalities in both kidney-specific HNF1β knockout mice (Igarashi et al., 2005) humans with HNF1β mutations (Bellanne-Chantelot et al., 2004). Also important for kidney development, is GREM1, which inhibits BMP4 expression to facilitate initiation of ureteric bud outgrowth and invasion of the mesenchyme (Michos et al., 2007). The detection of DAZ (Deleted in Azospermia)—interacting zinc finger protein 1 (DAZIP1) as a caput-specific DEG is of interest as DAZ genes are important in normal human spermatogenesis (Reijo et al., 1995).
Another notable feature revealed by comparative analysis of the transcriptomes of epithelial cells from the caput, corpus and cauda, is the regional distribution of component of innate immunity. Consistent with the ascending nature of epididymitis, multiple components of the innate immune response are more highly expressed in the corpus/cauda. However, other factors such as specific defensins are abundant throughout the epididymis epithelium, suggesting they may have a constitutive role in monitoring epididymis health.
In conclusion, we provide a comprehensive molecular atlas of the epididymis and its epithelium, which will be valuable to decipher processes of normal epididymis function and aspects of epididymis disease that cause male infertility and other phenotypes.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors' roles
J.A.B., R.Y., S.-H.L., S.E.E. and A.H. acquired, analyzed and interpreted data. J.A.B., R.Y. and A.H. drafted the article. All authors revised and approved the article.
Funding
This work was supported by NIH grant R01HD068901 (PI: A.H.).
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We are grateful to the anonymous men with testicular cancer who consented to provide epididymis tissue for these studies. We thank Timothy Skimina for computer and server support.
References
- Beleza-Meireles A, Tohonen V, Soderhall C, Schwentner C, Radmayr C, Kockum I, Nordenskjold A. Activating transcription factor 3: a hormone responsive gene in the etiology of hypospadias. Eur J Endocrinol 2008;158:729–739. [DOI] [PubMed] [Google Scholar]
- Bellanne-Chantelot C, Chauveau D, Gautier JF, Dubois-Laforgue D, Clauin S, Beaufils S, Wilhelm JM, Boitard C, Noel LH, Velho G et al. Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Ann Intern Med 2004;140:510–517. [DOI] [PubMed] [Google Scholar]
- Bingle CD, Craven CJ. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum Mol Genet 2002;11:937–943. [DOI] [PubMed] [Google Scholar]
- Bischof JM, Gillen AE, Song L, Gosalia N, London D, Furey TS, Crawford GE, Harris A. A genome-wide analysis of open chromatin in human epididymis epithelial cells reveals candidate regulatory elements for genes coordinating epididymal function. Biol Reprod 2013;89:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by PAX2 and PAX8. Genes Dev 2002;16:2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower M, Salomon R, Allanson J, Antignac C, Benedicenti F, Benetti E, Binenbaum G, Jensen UB, Cochat P, DeCramer S et al. Update of PAX2 mutations in renal coloboma syndrome and establishment of a locus-specific database. Hum Mutat 2012;33:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne JA, Yang R, Song L, Crawford GE, Leir SH, Harris A. Open chromatin mapping identifies transcriptional networks regulating human epididymis epithelial function. Mol Hum Reprod 2014;20:1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai OH, Song CH, Park SK, Kim W, Cho ES. Molecular regulation of kidney development. Anat Cell Biol 2013;46:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HC, Ko WH, Zhao W, Fu WO, Wong PY. Evidence for independent Cl− and HCO3− secretion and involvement of an apical Na(+)-HCO3− cotransporter in cultured rat epididymal epithelia. Exp Physiol 1996;81:515–524. [DOI] [PubMed] [Google Scholar]
- Cornwall GA. New insights into epididymal biology and function. Hum Reprod Update 2009;15:213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa SL, Boekelheide K, Vanderhyden BC, Seth R, McBurney MW. Male infertility caused by epididymal dysfunction in transgenic mice expressing a dominant negative mutation of retinoic acid receptor alpha 1. Biol Reprod 1997;56:985–990. [DOI] [PubMed] [Google Scholar]
- Dacheux JL, Belghazi M, Lanson Y, Dacheux F. Human epididymal secretome and proteome. Mol Cell Endocrinol 2006;250:36–42. [DOI] [PubMed] [Google Scholar]
- Dai X, Schonbaum C, Degenstein L, Bai W, Mahowald A, Fuchs E. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev 1998;12:3452–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube E, Chan PT, Hermo L, Cyr DG. Gene expression profiling and its relevance to the blood-epididymal barrier in the human epididymis. Biol Reprod 2007;76:1034–1044. [DOI] [PubMed] [Google Scholar]
- Eccles MR, Schimmenti LA. Renal-coloboma syndrome: a multi-system developmental disorder caused by PAX2 mutations. Clin Genet 1999;56:1–9. [DOI] [PubMed] [Google Scholar]
- Engels S, Kohlhase J, McGaughran J. A SALL1 mutation causes a branchio-oto-renal syndrome-like phenotype. J Med Genet 2000;37:458–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelli S, Caridi G, Beri S, McCracken K, Bocciardi R, Zordan P, Dagnino M, Fiorio P, Murer L, Benetti E et al. Mutations in SOX17 are associated with congenital anomalies of the kidney and the urinary tract. Hum Mutat 2010;31:1352–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyonnet B, Marot G, Dacheux JL, Mercat MJ, Schwob S, Jaffrezic F, Gatti JL. The adult boar testicular and epididymal transcriptomes. BMC Genomics 2009;10:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Coleman L. Ductal epithelial cells cultured from human foetal epididymis and vas deferens: relevance to sterility in cystic fibrosis. J Cell Sci 1989;92(Pt 4):687–690. [DOI] [PubMed] [Google Scholar]
- Hermo L, Smith CE. Thirsty business: cell, region, and membrane specificity of aquaporins in the testis, efferent ducts, and epididymis and factors regulating their expression. J Androl 2011;32:565–575. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- Igarashi P, Shao X, McNally BT, Hiesberger T. Roles of HNF-1beta in kidney development and congenital cystic diseases. Kidney Int 2005;68:1944–1947. [DOI] [PubMed] [Google Scholar]
- Jervis KM, Robaire B. Dynamic changes in gene expression along the rat epididymis. Biol Reprod 2001;65:696–703. [DOI] [PubMed] [Google Scholar]
- Jimenez T, McDermott JP, Sanchez G, Blanco G. Na,K-ATPase alpha4 isoform is essential for sperm fertility. Proc Natl Acad Sci USA 2011;108:644–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, DiCandeloro P, Wilson E, Kopf GS, Turner TT. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod 2005;73:404–413. [DOI] [PubMed] [Google Scholar]
- Jrad-Lamine A, Henry-Berger J, Damon-Soubeyrand C, Saez F, Kocer A, Janny L, Pons-Rejraji H, Munn DH, Mellor AL, Gharbi N et al. Indoleamine 2,3-dioxygenase 1 (ido1) is involved in the control of mouse caput epididymis immune environment. PloS one 2013;8:e66494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W. Mutations in the SALL1 putative transcription factor gene cause Townes–Brocks syndrome. Nat Genet 1998;18:81–83. [DOI] [PubMed] [Google Scholar]
- Krutskikh A, De Gendt K, Sharp V, Verhoeven G, Poutanen M, Huhtaniemi I. Targeted inactivation of the androgen receptor gene in murine proximal epididymis causes epithelial hypotrophy and obstructive azoospermia. Endocrinology 2011;152:689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leir SH, Browne JA, Eggener SE, Harris A. Characterization of primary cultures of adult human epididymis epithelial cells. Fertil Steril 2015;103:647–54 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AY, Wong PY, Yankaskas JR, Boucher RC. cAMP- but not Ca(2+)-regulated Cl− conductance is lacking in cystic fibrosis mice epididymides and seminal vesicles. Am J Physiol 1996;271:C188–C193. [DOI] [PubMed] [Google Scholar]
- Levine N, Marsh DJ. Micropuncture studies of the electrochemical aspects of fluid and electrolyte transport in individual seminiferous tubules, the epididymis and the vas deferens in rats. J Physiol 1971;213:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Nair M, Mackay DR, Bilanchone V, Hu M, Fallahi M, Song H, Dai Q, Cohen PE, Dai X. Ovol1 regulates meiotic pachytene progression during spermatogenesis by repressing Id2 expression. Development 2005;132:1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja H, Abrahamsson PA. Three predominant proteins secreted by the human prostate gland. Prostate 1988;12:29–38. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang DK, Chen LM. The physiology of bicarbonate transporters in mammalian reproduction. Biol Reprod 2012;86:99. [DOI] [PubMed] [Google Scholar]
- Luedtke CC, McKee MD, Cyr DG, Gregory M, Kaartinen MT, Mui J, Hermo L. Osteopontin expression and regulation in the testis, efferent ducts, and epididymis of rats during postnatal development through to adulthood. Biol Reprod 2002;66:1437–1448. [DOI] [PubMed] [Google Scholar]
- Marcotte M, Sharma R, Bouchard M. Gene regulatory network of renal primordium development. Pediatr Nephrol 2014;29:637–644. [DOI] [PubMed] [Google Scholar]
- Michos O, Goncalves A, Lopez-Rios J, Tiecke E, Naillat F, Beier K, Galli A, Vainio S, Zeller R. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development 2007;134:2397–2405. [DOI] [PubMed] [Google Scholar]
- Moncrief ND, Kretsinger RH, Goodman M. Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J Mol Evol 1990;30:522–562. [DOI] [PubMed] [Google Scholar]
- O'Hara L, Welsh M, Saunders PT, Smith LB. Androgen receptor expression in the caput epididymal epithelium is essential for development of the initial segment and epididymal spermatozoa transit. Endocrinology 2011;152:718–729. [DOI] [PubMed] [Google Scholar]
- Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet 2003;12(Spec No 1):R37–R44. [DOI] [PubMed] [Google Scholar]
- Priston M, Kozlowski K, Gill D, Letwin K, Buys Y, Levin AV, Walter MA, Heon E. Functional analyses of two newly identified PITX2 mutants reveal a novel molecular mechanism for Axenfeld–Rieger syndrome. Hum Mol Genet 2001;10:1631–1638. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet 1995;10:383–393. [DOI] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 2007;35:W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BC, Waldegger S, Fehr S, Bleich M, Warth R, Greger R, Jentsch TJ. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature 2000;403:196–199. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Watanabe Y, Isobe N, Yoshimura Y. Expression of avian beta-defensin 3, an antimicrobial peptide, by sperm in the male reproductive organs and oviduct in chickens: an immunohistochemical study. Poult Sci 2008;87:2653–2659. [DOI] [PubMed] [Google Scholar]
- Shiraga H, Min W, VanDusen WJ, Clayman MD, Miner D, Terrell CH, Sherbotie JR, Foreman JW, Przysiecki C, Neilson EG et al. Inhibition of calcium oxalate crystal growth in vitro by uropontin: another member of the aspartic acid-rich protein superfamily. Proc Natl Acad Sci USA 1992;89:426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanemoto M, Kittaka N, Inanobe A, Kurachi Y. In vivo formation of a proton-sensitive K+ channel by heteromeric subunit assembly of Kir5.1 with Kir4.1. J Physiol 2000;525(Pt 3):587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimon V, Koukoui O, Calvo E, Sullivan R. Region-specific gene expression profiling along the human epididymis. Mol Hum Reprod 2007;13:691–704. [DOI] [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development 1995;121:4057–4065. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SJ, Imbrici P, Salvatore L, D'Adamo MC, Pessia M. pH dependence of the inwardly rectifying potassium channel, Kir5.1, and localization in renal tubular epithelia. J Biol Chem 2000;275:16404–7. [DOI] [PubMed] [Google Scholar]
- Turner TT, Bomgardner D, Jacobs JP, Nguyen QA. Association of segmentation of the epididymal interstitium with segmented tubule function in rats and mice. Reproduction 2003;125:871–878. [DOI] [PubMed] [Google Scholar]
- Van Esch H, Groenen P, Nesbit MA, Schuffenhauer S, Lichtner P, Vanderlinden G, Harding B, Beetz R, Bilous RW, Holdaway I et al. GATA3 haplo-insufficiency causes human HDR syndrome. Nature 2000;406:419–422. [DOI] [PubMed] [Google Scholar]
- West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Ann Rev Cell Dev Biol 2006;22:409–437. [DOI] [PubMed] [Google Scholar]
- Wong PY, Lee WM. Potassium movement during sodium-induced motility initiation in the rat caudal epididymal spermatozoa. Biol Reprod 1983;28:206–212. [DOI] [PubMed] [Google Scholar]
- Xu H, Bai L, Collins JF, Ghishan FK. Molecular cloning, functional characterization, tissue distribution, and chromosomal localization of a human, small intestinal sodium-phosphate (Na+-Pi) transporter (SLC34A2). Genomics 1999;62:281–284. [DOI] [PubMed] [Google Scholar]
- Yip KP, Tsuruoka S, Schwartz GJ, Kurtz I. Apical H(+)/base transporters mediating bicarbonate absorption and pH(i) regulation in the OMCD. Am J Physiol Renal Physiol 2002;283:F1098–F1104. [DOI] [PubMed] [Google Scholar]
- Zhang JS, Liu Q, Li YM, Hall SH, French FS, Zhang YL. Genome-wide profiling of segmental-regulated transcriptomes in human epididymis using oligo microarray. Mol Cell Endocrinol 2006;250:169–177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.