Abstract
There is emerging interest in stem cell energy metabolism and its effect on differentiation. Bioenergetic changes in differentiating bone marrow mesenchymal stem cells (MSCs) are poorly understood and were the focus of our study. Using bioenergetic profiling and transcriptomics, we have established that MSCs activate the mitochondrial process of oxidative phosphorylation (OxPhos) during osteogenic differentiation, but they maintain levels of glycolysis similar to undifferentiated cells. Consistent with their glycolytic phenotype, undifferentiated MSCs have high levels of hypoxia-inducible factor 1 (HIF-1). Osteogenically induced MSCs downregulate HIF-1 and this downregulation is required for activation of OxPhos. In summary, our work provides important insights on MSC bioenergetics and proposes a HIF-based mechanism of regulation of mitochondrial OxPhos in MSCs.
Introduction
Bone marrow mesenchymal stem (a.k.a. stromal) cells, hereafter referred to as MSCs, are somatic multipotent progenitors that can differentiate into osteoblasts, adipocytes, and chondrocytes [1]. As the precursors to osteoblasts, MSCs are an important component of bone homeostasis. Bone undergoes constant balanced remodeling by bone-resorbing osteoclasts and bone-forming osteoblasts. During osteoporosis, diabetes, and other metabolic diseases, this balance is disrupted, leading to a decline in bone quality, and increase in fracture potential [2,3]. Decreased osteogenesis due to dysfunction in MSC differentiation is a potential contributor to this problem [4].
The role of cellular metabolism during differentiation is not well studied in stem cell biology. Many types of stem cells rely on glycolysis for energy when undifferentiated, and then later activate the mitochondrial process of oxidative phosphorylation (OxPhos) during differentiation [5]. The data regarding metabolism in MSCs specifically are controversial. Chen et al. reported activation of OxPhos in MSCs during osteogenic differentiation [6]. However, this study did not account for the potential contribution of mitochondrial uncoupling and/or nonmitochondrial oxygen consumption. Without accounting for these factors, the measured increase in oxygen consumption may not be entirely indicative of increased mitochondrial OxPhos. In a recent study, Guntur et al. performed bioenergetic profiling during osteogenic differentiation of calvarial osteoblasts, and also observed activation of OxPhos [7]. However, calvarial osteoblasts have a different embryonic origin than MSCs, and they have different osteogenic potential than cells of mesenchymal origin, which may also affect metabolic activity [8].
Conversely, Pattappa et al. found no change in OxPhos in MSCs during osteogenesis [9]. However, this study used media containing pyruvate that can feed directly into the mitochondrial citric acid cycle. This direct path would bypass any constraints that specifically affect OxPhos. Presence of pyruvate could artificially increase oxygen consumption in undifferentiated cells, thereby nullifying any potential differences observed between undifferentiated and osteogenically induced MSCs (ost-MSCs). Similarly, Esen et al. observed no change in OxPhos upon stimulation of osteogenic differentiation in MSC-like ST2 cells with Wnt, once again in the presence of pyruvate [10].
Other considerations have been overlooked when investigating MSC bioenergetics. Supraphysiological levels of glucose and glutamine, often used in culture media to stimulate MSC expansion, can potentially influence cell metabolism. Bioenergetic profiling often lacks a proper inhibitory analysis. Inhibitory analysis assesses the amount of energy being produced by mitochondrial versus nonmitochondrial respiration, respiration directed toward ATP synthesis, and uncoupled respiration by addition of a series of metabolic inhibitors. Furthermore, mitochondrial morphology has not been well studied in differentiating MSCs. Mitochondrial morphology is a dynamic feature that responds to the metabolic needs of the cell. Mitochondrial elongation and network formation indicate increased OxPhos [11].
In light of conflicting data and lack of mitochondria-specific techniques, we felt there remains a gap in the knowledge of MSC bioenergetics. Here, we comprehensively investigate MSC bioenergetics during differentiation, and potential regulatory mechanisms. We found that OxPhos is upregulated in osteogenically induced MSCs, in a hypoxia-inducible factor 1 (HIF-1)-dependent manner.
Materials and Methods
Materials
Reagents were from Sigma unless otherwise noted. Human MSCs were from Lonza.
MSC culture and differentiation
Human MSCs were purchased from Lonza and expanded in MSC growth medium MSCGM BulletKit (Lonza). For our experiments, we used cells at passage 3 to 7. We determined that under our culture conditions, these MSCs do not change phenotype until passage 10 when downregulation of pluripotency markers Oct4 and Nanog were observed (data not shown). MSC surface markers were detected with fluorescently labeled anti-human antibodies against Sca1, cd29, cd105, cd31, and cd45 (BD Bioscience) using flow cytometry and 12-color FACS Canto II machine (BD Bioscience). At confluency, media was switched to DMEM/low glucose media (Gibco), hereafter referred to as DMEM. Osteogenesis was induced by adding 50 μg/mL ascorbate and 10 mM β-glycerophosphate. Adipogenesis was induced by adding 1 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine, 10 μg/mL insulin, and 1 μM rosiglitazone (Cayman Chemicals), and withdrawing 3-isobutyl-1-methylxanthine and dexamethasone after 2 days. Chondrogenesis was induced with pellet culture in Chondrocyte Differentiation Medium (Lonza) supplemented with 10 ng/mL TGF-β3 for 21 days. All media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin mixture (Gibco). To confirm differentiation, cells were stained with osteoblast-specific alkaline phosphatase (ALP) and Alizarin Red (ARed) stains, adipocyte-specific Oil Red O stain, or chondrocyte-specific alcian blue/hematoxylin (ABH) stain.
For clonogenic capacity assay, MSCs were plated at clonal density (1.6 cells/cm2), grown for 21 days, and then stained with crystal violet.
All cells were cultured under normoxic conditions.
Real-time reverse transcriptase polymerase chain reaction
Total RNA was isolated using the RNeasy kit (Qiagen) and reverse transcribed into cDNA using iScript cDNA synthesis kit (Bio-Rad). cDNA was subjected to real time reverse transcriptase polymerase chain reaction (RT-PCR) using primer pairs: ALP (5′-TGC AGT ACG AGC TGA ACA GGA ACA-3′ and 5′-TCC ACC AAA TGT GAA GAC GTG GGA-3′), runt-related transcription factor 2 (RUNX2) (5′-TCC GGA ATG CCT CTG CTG TTA TGA-3′ and 5′-ACT GAG GCG GTC AGA GAA CAA ACT-3′), peroxisome proliferator-activated receptor gamma (PPARG) (5′-GGA AGA CCA CTC CCA CTC CT-3′ and 5′-GGA GAT GCA GGC TCC ACT TT-3′), transcription factor SOX-9 (SOX9) (5′-AGT ACC CGC ACT TGC ACA AC-3′ and 5′-CCG TTC TTC ACC GAC TTC CT-3′), collagen type II A (COL2A) (5′-ATT GTA GGA CCC AAA GGA CCT C-3′ and 5′-CAA AGT TTC CAC CAA GAC CAG G-3′), NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 (NDUSF5) (5′-GCA GCG GGA TAA GCT GAT AA-3′ and 5′-GTG GAG AAC AGA GAA CAG GAA A-3′), NADH dehydrogenase [ubiquinone] iron-sulfur protein 6 (NDUSF6) (5′-AGA CAG CAC CAC CAC TAG A-3′ and 5′-AAT ACC CTT CGC ACA GAA CC-3′), NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 (NDUSB10) (5′-GTG ACC CTC GTG AGA GAA TTT-3′ and 5′-CAC TGC ATT TCG GCT TCA TAC-3′), NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11 (NDUSB11) (5′-AGC TGA GAG GCT TGT GAA ATA C-3′ and 5′-CTT AGC AAC TGG TCA CTC ATC C-3′), and beta actin (ACTB) (5′-AGC CAT GTA CGT TGC TAT CC-3′ and 5′-CGT AGC ACA GCT TCT CCT TAA T-3′). Real-time RT-PCR was performed in the RotorGene system (Qiagen) using SYBR Green (Quanta). The expression of genes of interest was normalized to the expression of ACTB.
Bioenergetic profiling
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using Seahorse XF24 (Seahorse Bioscience). Cells were plated on Seahorse 24-well plates 48 h before experiment at a density of 4 × 104 cells/well. Immediately before the experiment, media was replaced with DMEM-XF24 containing 5 mM glucose, 1 mM glutamine, 1% serum, and no pyruvate. A baseline measurement of OCR and ECAR was taken, and then an inhibitory analysis was performed using injections of oligomycin (Olig) at 1 μM, FCCP at 0.5 μM, and antimycin A (AntA) at 1 μM [12]. After analysis, cells were trypsinized and counted. The following OxPhos and glycolytic indexes were calculated: basal respiration (OCRpre-Olig − OCRpost-AntA), ATP-linked respiration (OCRpre-Olig − OCRpost-Olig), maximal respiration (OCRpost-FCCP − OCRpost-AntA), respiratory capacity (OCRpost-FCCP − OCRpre-Olig), proton leak (OCRpost-Olig − OCRpost-AntA), basic glycolysis (ECARpre-Olig), glycolytic capacity (ECARpost-Olig), and glycolytic reserve (ECARpost-Olig − ECARpre-Olig). ATP was measured using the CellTiter-Glo kit (Roche).
Mitochondrial morphology assessment
To visualize mitochondria, cells were stained with MitoTracker Green (MTG; Invitrogen) and imaged as described in our previous work [13]. MTG signal tracing was done using ImageJ software. Electron microscopy (EM) was performed as described in our previous work [13]. Mitochondrial form factor (FF) was calculated according to the formula FF = p2/4πA, where “p” is the perimeter and “A” is the area of mitochondria.
Mitochondrial biogenesis assays
Total DNA was isolated and mitochondrial DNA (mtDNA) was assessed with quantitative polymerase chain reaction: copy number of the mtDNA-encoded gene for the ND1 subunit of NADH dehydrogenase (ND1) was normalized to gDNA-encoded ACTB as described in our previous work [14].
RNA sequencing (RNAseq) analysis was used to assay expression of indicated mitochondrial structural and regulatory factors and factors involved in mitochondrial biogenesis.
RNAseq analysis
Total RNA was isolated with the RNeasy kit (Qiagen) and processed in the University of Rochester Genomic Core. The TruSeq RNA Sample Preparation Kit V2 (Illumina) was used for library construction. The amplified libraries were hybridized to the Illumina single end flow cell and amplified using the cBot (Illumina). Reads were generated and aligned to the organism-specific reference genome. A separate analysis was performed to include mtRNA reads according to the manufacturer's instructions. Data were analyzed using Ingenuity Pathway Analysis software under the University of Rochester group license.
Western blotting
Cell lysates were lyzed and subjected to 8% (for HIF-1 detection) or 4%–12% (for all others) sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by transfer to polyvinylidene difluoride and blocking in 5% dry milk. For HIF-1 detection, blots were probed with rabbit anti-human HIF-1α antibody (NB100-449; Novus Biologicals) at 1 μg/mL and horseradish peroxidase (HRP) conjugated goat anti-rabbit antibody (Santa Cruz) at 0.5 μg/mL and developed with Clarity ECL Substrate (Bio-Rad). To verify equal loading blots were either stained with Ponceau S (Bio-Rad) when 40 kDa band migrated out of a gel or reprobed with anti-β-actin antibody (Sigma) at 0.5 μg/mL. For detection of mitochondrial proteins, blots were probed with Total OXPHOS Human Antibody Cocktail (ab110411; Abcam) diluted 1:200 in 5% BSA/PBST and HRP-conjugated rabbit anti-mouse antibody (Bio-Rad) at 0.2 μg/mL and developed with West Femto Substrate (Thermo Scientific). Blots were then reprobed for β-actin. Bands were measured with densitometry using ImageJ software. Signal was normalized to total protein Ponceau S signal or to β-actin.
HIF-1α electrophoretic mobility shift assay
Nuclear extracts were prepared using the NE-PER kit (Thermo Scientific) according to the manufacturer's protocol. Nuclear extract protein concentration was determined using the Bradford Assay. For HIF-1α electrophoretic mobility shift assay (EMSA), preannealed IRDye 700 oligonucleotides from LI-COR Biosciences were used. EMSA was preformed based on the LI-COR Biosciences instructions. Two micrograms of nuclear extract was incubated with 50 fmoles of the probe in binding buffer. All samples were incubated in the dark for 30 min and then 10× Orange loading dye (LI-COR Biosciences) was added to each sample. Samples were resolved on a nondenaturing 4% acrylamide gel at 70 V in TAE buffer. Gels were imaged and analyzed using an Odyssey imager (LI-COR Biosciences) with high quality resolution.
Statistical analysis
Three to five independent experiments were done to derive each panel of the paper. Data were analyzed using Prism 5.01 (GraphPad Software). Mean values and standard errors were calculated, and the statistical significance (P ≤ 0.05) established using either Student's t-test when two variables were compared or one-way analysis of variance when more than two variables were compared.
Results
Characterization of human MSCs
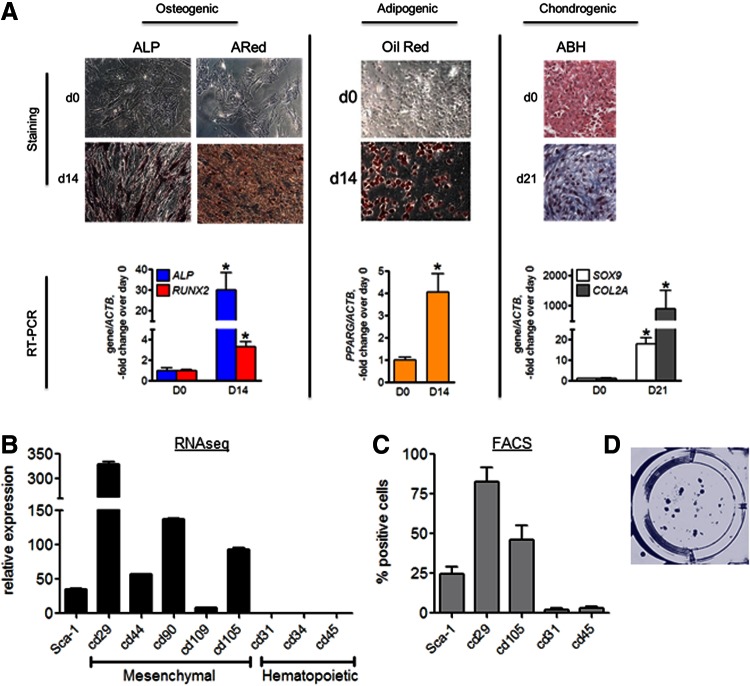
We first confirmed that human MSCs from Lonza have the known characteristics attributed to MSCs [15]. Cells were incubated in osteogenic, adipogenic, or chondrogenic media and analyzed with lineage-specific staining and real-time RT-PCR. After 14 days in osteogenic media, more than 90% of cells stained positive for ALP and ARed, indicating high ALP activity and mineralization, respectively (Fig. 1A). We also observed significant upregulation of osteoblastic genes, ALP and RUNX2. After 14 days in adipogenic media, ∼70% of cells deposited lipid droplets shown by Oil Red O staining, and expressed high levels of the adipogenic marker gene, PPARG (Fig. 1A). After 21 days in chondrogenic media, pellet culture stained with ABH appeared blue/purple indicating presence of glycosaminoglycans/proteoglycans, and we also observed significant increase in chondrogenic markers SOX9 and COL2A (Fig. 1A).
FIG. 1.
Characterization of MSCs. (A) Left: Undifferentiated (D0) and osteogenically induced cells (D14) were stained with ALP-specific stain and Alizarin Red (ARed). Real-time RT-PCR analysis of ALP and RUNX2 expression normalized to ACTB. Middle: Undifferentiated (D0) and adipogenically induced cells (D14) were stained with Oil Red O. Real-time RT-PCR analysis of PPARG expression normalized to ACTB. Right: Undifferentiated (D0) and chondrogenically induced cells (D21) were stained with ABH. Real-time RT-PCR analysis of SOX9 and COL2A expression normalized to ACTB. Cell staining panels are representatives of four. Data in graphs are mean ± SEM (n = 4). *P < 0.05 as determined with t-test. (B) RNAseq analysis of surface markers expression in undifferentiated MSCs. Data are mean ± SEM (n = 3). (C) Flow cytometry analysis of surface markers in undifferentiated MSCs. Data are mean ± SEM (n = 5). (D) Clonogenic capacity assay. Cells were plated at clonogenic density of 1.6/cm2 and stained with Crystal Violet at D21 (n = 6). ABH, alcian blue/hematoxylin; ALP, alkaline phosphatase; D0, day 0; D14, day 14; D21, day 21; MSC, mesenchymal stem (stromal) cells; RNAseq, RNA sequencing; RT-PCR, real time reverse transcriptase polymerase chain reaction. Color images available online at www.liebertpub.com/scd
Undifferentiated MSCs expressed standard mesenchymal cell surface markers, and lacked hematopoietic markers, as shown by RNAseq (Fig. 1B) and with flow cytometry analysis (Fig. 1C). When plated as clonal density, these cells were able to form colonies stained by crystal violet (Fig. 1D).
These data confirm the ability of Lonza human MSCs to undergo multilineage differentiation, in addition to showing mesenchymal surface markers, and clonogenic potential in our culture system.
Bioenergetic profiling of MSCs during osteogenic differentiation
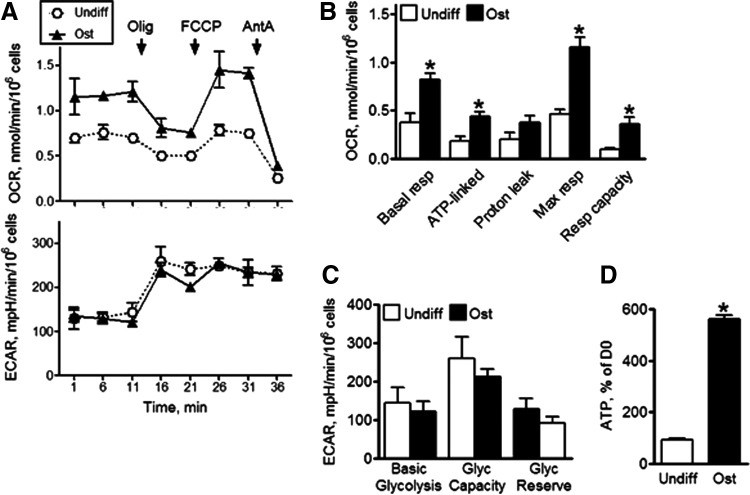
Next, using the Seahorse XF24 analyzer we performed bioenergetic profiling of undifferentiated MSCs in comparison to ost-MSCs. Seahorse XF24 allows simultaneous measurement of mitochondrial function via OCR and glycolysis via ECAR due to lactate production. To avoid any potential artifacts caused by media composition, we performed the bioenergetic profiling using media with physiological concentrations of glucose and glutamine and no pyruvate. Figure 2A shows OCR traces for undifferentiated and ost-MSCs. We also performed the inhibitory analysis, which allowed calculation of the OxPhos indexes shown in Fig. 2B. Figure 2A and B show significant increases in basal and ATP-linked (mitochondrial) respiration during osteogenesis, while maximal respiration levels were increased relative to undifferentiated MSCs. Additionally, we performed bioenergetic profiling of MSCs after 7 days in osteogenic media and did not observe any significant changes relative to undifferentiated MSCs, and after 21 days producing results similar to the day 14 time point (data not shown).
FIG. 2.
Bioenergetic profiling of MSCs during differentiation. (A) OCR and ECAR were analyzed using Seahorse XF24. OCR was measured at baseline and after addition of Olig, FCCP, and AntA. (B) Mitochondrial respiratory indexes were calculated as described in the Materials and Methods section. (C) Glycolytic indexes were calculated as described in the Materials and Methods section. (D) ATP was measured in cell lysates with bioluminescence assay and expressed as % of values at D0. Data are mean ± SEM (n = 4). *P < 0.05 as determined with t-test. AntA, antimycin A; ECAR, extracellular acidification rate; OCR, oxygen consumption rate; Olig, oligomycin; Ost, osteogenically differentiated MSCs; Undiff, undifferentiated MSCs.
ECAR data indicated that both basal glycolytic rate and glycolytic capacity remained unchanged during osteogenesis (Fig. 2A, C). Consistent with the observed levels of OxPhos activity, which is the more efficient ATP production method, ATP output was significantly induced in ost-MSCs (Fig. 2D).
Based on these findings, we established that MSCs indeed undergo distinct bioenergetic changes during osteogenic differentiation, activating the mitochondrial process of OxPhos.
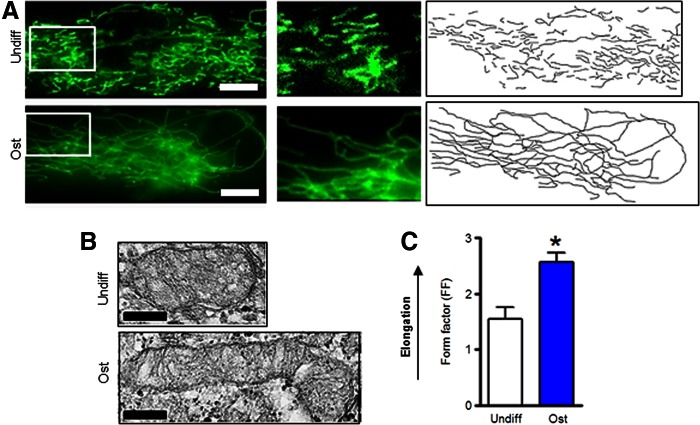
Mitochondrial morphology in MSCs during osteogenic and adipogenic differentiation
Mitochondria change morphology in accordance with the metabolic needs of the cell. We assessed mitochondrial morphology using MTG, fluorescence microscopy, and EM. As evident from Figure 3A, mitochondria in undifferentiated MSCs presented as separate organelles while mitochondria in ost-MSCs formed a network. Formation of this network indicates mitochondrial activation. This is most obvious in the right panels of Fig. 3A showing a tracing of the MTG signal. EM showed that mitochondria in ost-MSCs were elongated (Fig. 3B), which was confirmed with quantitative analysis using a FF measure of mitochondrial elongation (Fig. 3C). Thus, mitochondrial elongation and network formation are consistent with the increased OxPhos observed in ost-MSCs.
FIG. 3.
Mitochondrial morphology in MSCs during differentiation. (A) Cells were stained with MitoTracker Green (MTG) and live-cell fluorescent images were taken. The middle panels are magnified regions of cells shown in the left panels. The right panels show tracing of the MTG signal shown in the left panels. Images are representatives of 30 cells from three different plates. (B) Electron micrographs of MSCs. Images are representatives of 30 cells/group from three different plates. Scale bar is 200 nm. (C) Mitochondrial form factor (FF), a measure of mitochondrial elongation, was calculated using electron microscopy. Data are mean ± SEM (n = 30). *P < 0.05 as determined with t-test. Color images available online at www.liebertpub.com/scd
Mitochondrial biogenesis in MSCs during osteogenic differentiation
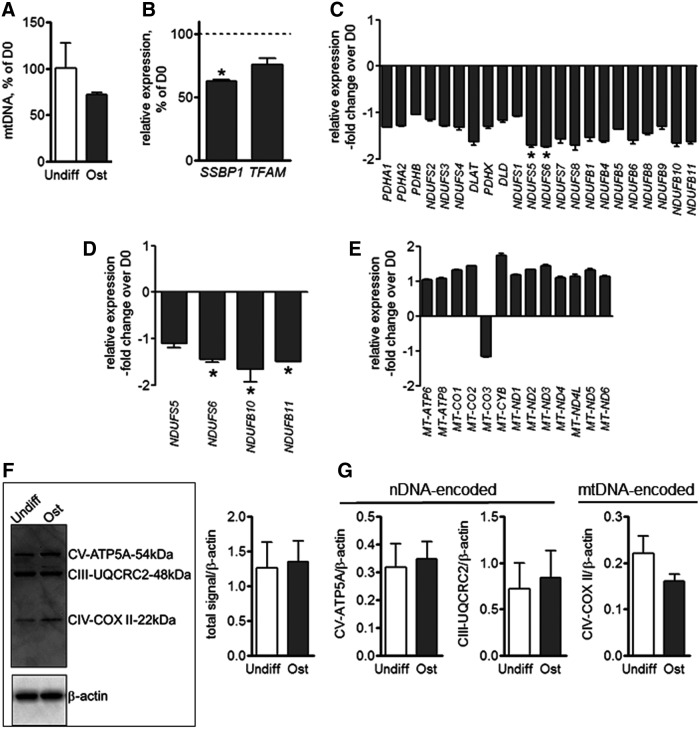
To assess whether the observed increases in mitochondrial activity were due to increases in mitochondrial biogenesis, we measured mtDNA levels and found no significant differences in the amount of mtDNA in ost-MSCs when compared to undifferentiated MSCs (Fig. 4A). We also observed a decrease in expression of a known regulator of mitochondrial biogenesis, SSBP1. (Fig. 4B). Further, RNAseq transriptome analysis showed no significant change in gene expression levels of OxPhos factors in ost-MSCs relative to undifferentiated cells, with the exception of NDUFS5 and 6, which were downregulated (Fig. 4C). These results were further verified using real-time RT-PCR on a selection of these factors, which showed a decrease in expression (Fig 4D). Our RNAseq data were reanalyzed to include mtRNA reads, which is presented in Fig. 4E. Our data show no significant difference in expression of the 13 mitochondrial peptides encoded by mtDNA. To verify expression data by protein level, we performed immunoblotting using a cocktail of antibodies specific for OxPhos (Fig. 4F). The semi-quantitative densitometry analysis of these blots revealed no significant difference in OxPhos protein levels (Fig. 4G). Together these data indicate that mitochondrial biogenesis was not significantly induced during MSC differentiation.
FIG. 4.
Mitochondrial biogenesis in MSCs during differentiation. (A) mtDNA was assayed using qPCR and expressed as % of D0 values. (B) Gene expression of mitochondrial biogenesis factors SSBP1 and TFAM in ost-MSCs relative to undifferentiated MSCs as analyzed with RNAseq. (C) Gene expression of OxPhos structural and regulatory factors in ost-MSCs relative to undiff-MSC as analyzed with RNAseq. (D) Gene expression of OxPhos factors in ost-MSCs relative to undiff-MSC as analyzed with qPCR. (E) Gene expression of mitochondrial RNA as analyzed with RNAseq. (F) Protein analysis of OxPhos factors by western blot. (G) Densitometry analysis of (F). Data are mean ± SEM (n = 3). *P < 0.05 as determined with t-test. mtDNA, mitochondrial DNA; nDNA, nuclear DNA; OxPhos, oxidative phosphorylation; ost-MSCs, osteogenically induced MSCs; qPCR, quantitative polymerase chain reaction.
Involvement of HIF in regulation of mitochondrial function in differentiating MSCs
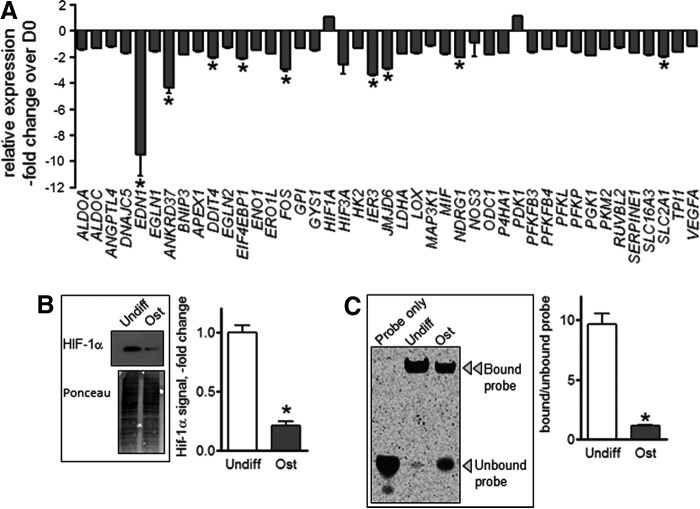
To better understand the mechanism(s) underlying the observed changes in cell bioenergetics during MSC differentiation, we performed pathway analysis using RNAseq-based transcriptomics focusing on known regulators of cellular metabolism such as mTOR, Akt, and HIF-1. Figure 5A shows a dramatic downregulation of HIF-1 signature genes in ost-MSCs relative to undifferentiated MSCs. No such changes were seen in the mTOR or Akt pathways (data not shown). Consistent with the RNAseq data, western blot analysis revealed high HIF-1α protein expression in undifferentiated MSCs, and a marked suppression of HIF-1α in ost-MSCs (Fig. 5B).
FIG. 5.
HIF-1 signaling changes during osteogenic differentiation of MSCs. (A) Expression of HIF signature genes in ost-MSC relative to undiff-MSCs as analyzed with RNAseq (n = 3/each). (B) Western blot of HIF-1α in undifferentiated and ost-MSCs. Ponceau S staining was used to verify equal loading. Densitometry values shown as fold change from undifferentiated MSCs. Blots are representatives of three. (C) Electrophoretic mobility shift assay of HIF-1α. DNA-binding activity using fluorescent probes and ratio of bound to unbound probe signal. Gel is representative of three. Data are mean ± SEM (n = 3). *P < 0.05 as determined with t-test. HIF, hypoxia-inducible factor.
As HIF-1α is a transcription factor, we next looked at DNA binding-activity to determine whether there was increased HIF-1α function in addition to the increased protein expression. EMSA and subsequent analysis of the ratio of bound to unbound probe indicated that HIF-1α-binding activity was high in undifferentiated MSCs, and low in ost-MSCs (Fig. 5C).
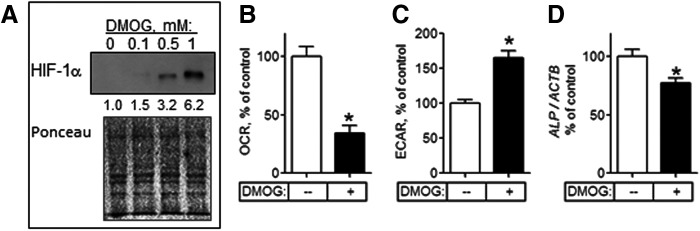
HIF-1 inhibits OxPhos [16–18]. We therefore examined whether HIF-1 would affect metabolism and osteogenesis in MSCs. This was done by stabilizing HIF in ost-MSCs with dimethyloxalylglycine (DMOG). DMOG-mediated inhibition of PHD enzymes prevents HIF-1α hydroxylation thereby disabling VHL binding, ubiquitination, and subsequent proteasomal degradation, thus stabilizing HIF-1α. Dose–response studies revealed that DMOG at 0.5 mM caused significant induction of HIF-1α without cell toxicity (Fig. 6A). Of note, protein loading was lower here than in the experiment shown in Fig. 5B (15 μg/lane in Fig. 6A vs. 25 μg/lane in Fig. 5B), which explains the absence of HIF-1α band in the control sample. We incubated ost-MSCs with DMOG at 0.5 mM for 48 h and performed bioenergetic profiling. Incubation with DMOG led to a significant inhibition of OxPhos in ost-MSCs, illustrated by the decrease in OCR (Fig. 6B). In concert with the observed inhibition of OxPhos, there was activation of glycolysis by DMOG in ost-MSCs as evident from the increased ECAR values (Fig. 6C). Thus, DMOG-mediated induction of HIF-1α led to significant inhibition of OxPhos and a corresponding activation of glycolysis. In addition, the induction of HIF-1α and subsequent inhibition of OxPhos led to downregulation of the osteogenic marker ALP (Fig. 6D) suggesting that low HIF/high OxPhos state is supportive of osteogenesis, and reversal of this state disrupts osteogenesis. These data suggest that OxPhos in ost-MSCs is sensitive to HIF-1 and thus HIF-1α must be downregulated for osteogenesis.
FIG. 6.
Reactivation of HIF-1 signaling inhibits OxPhos activity in ost-MSCs. (A) Western blot of HIF-1α in ost-MSCs treated with indicated doses of DMOG for 48 h. Ponceau S staining was used to verify equal loading. Numbers under the HIF-1α blot show densitometry values for each band normalized to total protein. Blot is representative of three. OCR (B) and ECAR (C) in ost-MSCs treated with DMOG at 0.5 mM or PBS for 48 h. (D) Real-time RT-PCR analysis of expression of ALP normalized to ACTB in DMOG-treated ost-MSCs. Data in are mean ± SEM (n = 3 to 5). *P < 0.05 as determined with t-test. DMOG, dimethyloxalylglycine.
Discussion
The mechanisms underlying bioenergetic changes in stem cells during differentiation are not well understood. Based on our data, we can conclusively state that OxPhos is activated during osteogenic differentiation of MSCs. Active OxPhos is likely required to meet high ATP demands needed for extensive biosynthesis of extracellular matrix protein during osteogenesis. The observed low basal respiration, and the lack of response to the mitochondrial inhibitors and uncoupler suggest that mitochondria are not yet fully capable of active OxPhos in undifferentiated MSCs.
With regards to glycolysis, we observed no significant changes in ost-MSCs relative to undifferentiated MSCs. Previous studies provide contradictory results: while a decrease in glycolysis was shown by Chen et al. [6], data from Long's group suggest continuous dependence on glycolysis [10]. Our conclusion provides a compromise: while OxPhos is activated during MSC osteogenesis, glycolytic activity does not decrease. This suggests metabolic plasticity and versatility of osteogenic lineage in satisfying cell energy demands. Another consideration is that glycolysis is the preferred method of ATP production in proliferating cells and upon induction of osteogenic differentiation, glycolysis may be required for MSCs to start actively dividing. In agreement with this, our data indicate that OxPhos becomes active only at a stage of osteogenic differentiation when proliferation is less pronounced, that is, after 7 days in osteogenic media. It should be noted that although our study was limited to commercially available hMSCs from Lonza, we are collecting and characterizing primary human MSCs isolated from bone marrow aspirates. In future work, we will evaluate energy metabolism in these primary cells to confirm the current data from Lonza cells. We anticipate similar results in primary cells as previous studies used primary human MSCs [6] and detected similar metabolic changes during osteogenic differentiation.
Because of the clear increase of mitochondrial activity, we also investigated network formation and biogenesis of mitochondria. Presentation of mitochondria as separate organelles is usually associated with low activity while fusion of mitochondria into a network indicates activation [11]. We observed separate organelles in undifferentiated MSCs, but a mitochondrial network in ost-MSCs, which correlates to the measured higher OxPhos activity. One possible explanation for the observed activation of OxPhos in ost-MSCs would be an increase in mitochondrial biogenesis. However, we found no evidence of more mitochondria as measured by mtDNA. The increase in OxPhos during osteogenic differentiation of MSCs is not due to mitochondrial biogenesis. Our current data do not show any differences in mtDNA and mtRNA in MSCs during osteogenic differentiation. Further studies of the role of mtDNA and mtRNA in MSCs are needed, and we plan to fully address this in our future work. Aging, diabetes, and other disorders are known to damage mtDNA, which may lead to mitochondrial impairment and incomplete activation during differentiation, and may, therefore, influence stem cell fate.
The change in bioenergetics during osteogenic differentiation must also have a regulatory mechanism. Our data indicate that HIF-1α might serves as a possible regulator of MSC energy metabolism during osteogenic differentiation. It is most active in undifferentiated MSCs, but is dramatically downregulated in ost-MSC, where OxPhos is activated. HIF-1α is activated via protein stabilization in hypoxia, engaging a transcriptional paradigm that shifts cellular bioenergetics toward anaerobic glycolysis [19]. HIF-1α can also be stabilized in normoxia, a phenomenon called “pseudohypoxia” and frequently observed in cancer cells due to interaction with other signaling pathways, such as Notch and NF-κB [20,21]. Importantly, none of the MSCs in our study were cultured under hypoxic conditions.
HIF-1α-regulated transcription promotes glycolysis and inhibits OxPhos. We have shown that reactivation of HIF-1α in ost-MSCs effectively inhibits OxPhos to the levels found in undifferentiated cells. Earlier works provide opposing views on the role of HIF-1 in osteoblasts; while Regan et al. describe an anabolic effect of HIF-1 [22], Riddle et al. report an inhibitory role of HIF-1 on osteoblast differentiation [23]. Our data provide insight on this controversy: HIF-1α may be required at early stages of osteogenesis to maintain active glycolysis needed for high rates of proliferation. At later stages, when bone matrix protein biosynthesis becomes predominant and proliferation subsides, the demand for active mitochondria increases while the need for HIF-1α diminishes. The question remains of how HIF-1α is downregulated during MSC osteogenesis without changes in oxygenation. Regulation is likely complex and requires further study focused on possible regulators of HIF in normoxia, such as Notch and NF-κB.
Together, our findings provide important insights on the distinct regulation of bioenergetics in undifferentiated and ost-MSCs. These insights are critical for developing new approaches that will allow manipulation of MSC fate and lineage commitment by changing MSC bioenergetics, presenting new and important therapeutic opportunities.
Acknowledgments
We would like to thank Drs. Regis O'Keefe, Matthew Hilton, Paul Brookes, Sergiy Nadtochiy, and George Porter for fruitful discussions, Dr. Marc Halterman for sharing his resources, and Jerry Madukwe for his assistance. Financial support was from the NIH/NIAMS K01 AR064610, NIH/NIAMS R03 AR061515, and NIH/NIAMS P30 AR061307 grants.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bianco P, Cao X, Frenette PS, Mao JJ, Robey PG, Simmons PJ. and Wang C-Y. (2013). The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat Med 19:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolagas SC. (2000). Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev 21:115–137 [DOI] [PubMed] [Google Scholar]
- 3.Saito M. and Marumo K. (2013). Bone quality in diabetes. Front Endocrinol 4:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker N, Boyette LB. and Tuan RS. (2015). Characterization of bone marrow-derived mesenchymal stem cells in aging. Bone 70:37–47 [DOI] [PubMed] [Google Scholar]
- 5.Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA, Ramalho-Santos J, Van Houten B. and Schatten G. (2011). Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 6:e20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C-T, Shih Y-RV, Kuo TK, Lee OK. and Wei Y-H. (2008). Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 26:960–968 [DOI] [PubMed] [Google Scholar]
- 7.Guntur AR, Le PT, Farber CR. and Rosen CJ. (2014). Bioenergetics during calvarial osteoblast differentiation reflect strain differences in bone mass. Endocrinology 155:1589–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quarto N, Wan DC, Kwan MD, Panetta NJ, Li S. and Longaker MT. (2010). Origin matters: differences in embryonic tissue origin and Wnt signaling determine the osteogenic potential and healing capacity of frontal and parietal calvarial bones. J Bone Miner Res 25:1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattappa G, Heywood HK, de Bruijn JD. and Lee DA. (2011). The metabolism of human mesenchymal stem cells during proliferation and differentiation. J Cell Physiol 226:2562–2570 [DOI] [PubMed] [Google Scholar]
- 10.Esen E, Chen J, Karner CM, Okunade AL, Patterson BW. and Long F. (2013). WNT-LRP5 signaling induces warburg effect through mtorc2 activation during osteoblast differentiation. Cell Metab 17:745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand MD. and Nicholls DG. (2011). Assessing mitochondrial dysfunction in cells. Biochem J 435:297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giang A-H, Raymond T, Brookes P, de Mesy Bentley K, Schwarz E, O'Keefe R. and Eliseev R. (2013). Mitochondrial dysfunction and permeability transition in osteosarcoma cells showing the Warburg effect. J Biol Chem 288:33303–33311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapovalov Y, Hoffman D, Zuch D, de Mesy Bentley KL. and Eliseev RA. (2011). Mitochondrial dysfunction in cancer cells due to aberrant mitochondrial replication. J Biol Chem 286:22331–22338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao L, Gorski JL. and Chen CS. (2011). The Cdc42 guanine nucleotide exchange factor FGD1 regulates osteogenesis in human mesenchymal stem cells. Am J Pathol 178:969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picard M, Shirihai OS, Gentil BJ. and Burelle Y. (2013). Mitochondrial morphology transitions and functions: implications for retrograde signaling? Am J Physiol Regul Integr Comp Physiol 304:R393–R406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goda N. and Kanai M. (2012). Hypoxia-inducible factors and their roles in energy metabolism. Int J Hematol 95:457–463 [DOI] [PubMed] [Google Scholar]
- 17.Papandreou I, Cairns RA, Fontana L, Lim AL. and Denko NC. (2006). HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3:187–197 [DOI] [PubMed] [Google Scholar]
- 18.Semenza GL. (2012). Hypoxia-inducible factors in physiology and medicine. Cell 148:399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semenza GL. (2007). Life with oxygen. Science 318:62–64 [DOI] [PubMed] [Google Scholar]
- 20.Nam SY, Ko YS, Jung J, Yoon J, Kim YH, Choi YJ, Park JW, Chang MS, Kim WH. and Lee BL. (2011). A hypoxia-dependent upregulation of hypoxia-inducible factor-1 by nuclear factor-κB promotes gastric tumour growth and angiogenesis. Br J Cancer 104:166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U. and Bondesson M. (2005). Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell 9:617–628 [DOI] [PubMed] [Google Scholar]
- 22.Regan JN, Lim J, Shi Y, Joeng KS, Arbeit JM, Shohet RV. and Long F. (2014). Up-regulation of glycolytic metabolism is required for HIF1α-driven bone formation. Proc Natl Acad Sci U S A 111:8673–8678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riddle RC, Leslie JM, Gross TS. and Clemens TL. (2011). Hypoxia-inducible factor-1α protein negatively regulates load-induced bone formation. J Biol Chem 286:44449–44456 [DOI] [PMC free article] [PubMed] [Google Scholar]