Abstract
Neurospheres are widely used to propagate and investigate neural stem cells (NSCs) and neural progenitors (NPs). However, the exact cell types present within neurospheres are still unknown. To identify cell types, we used single-cell mRNA profiling of 48 genes in 187 neurosphere cells. Using a clustering algorithm, we identified 3 discrete cell populations within neurospheres. One cell population [cluster unsorted (US) 1] expresses high Bmi1 and Hes5 and low Myc and Klf12. Cluster US2 shows intermediate expression of most of the genes analyzed. Cluster US3 expresses low Bmi1 and Hes5 and high Myc and Klf12. The mRNA profiles of these 3 cell populations correlate with a developmental timeline of early, intermediate, and late NPs, as seen in vivo from the mouse brain. We enriched the cell population for neurosphere-forming cells (NFCs) using morphological criteria of forward scatter (FSC) and side scatter (SSC). FSC/SSChigh cells generated 2.29-fold more neurospheres than FSC/SSClow cells at clonal density. FSC/SSChigh cells were enriched for NSCs and Lewis-X+ve cells, possessed higher phosphacan levels, and were of a larger cell size. Clustering of both FSC/SSChigh and FSC/SSClow cells identified an NFC cluster. Significantly, the mRNA profile of the NFC cluster drew close resemblance to that of early NPs. Taken together, data suggest that the neurosphere culture system can be used to model central nervous system development, and that early NPs are the cell population that gives rise to neurospheres. In future work, it may be possible to further dissect the NFCs and reveal the molecular signature for NSCs.
Introduction
Neural stem cells (NSCs) are undifferentiated cells of the central nervous system (CNS), which can self-renew and are multipotent (for recent review see Ref. [1]). They are present throughout CNS development and are maintained in the adult brain in specific locations. NSCs hold great potential to be harnessed for treatment of neurological diseases [2] and understanding neurodevelopment. During development of the forebrain, neurogenesis occurs first followed by gliogenesis. In rodents, neurogenesis begins at E12 and peaks around E15. Before the onset of neurogenesis, neuroepithelial cells, the primary NSCs, divide symmetrically to expand the NSC pool at the ventricular zone (VZ). At the onset of neurogenesis, the neuroepithelial cells divide asymmetrically to produce radial glial cells (RGCs). Neuroepithelial cells and RGCs are collectively known as the apical progenitors (for recent review see Ref. [3]). RGCs then migrate into the subventricular zone (SVZ) to produce basal progenitors, which in turn differentiate into neurons. Astrocytes and oligodendrocytes are then formed at around E16 and around birth, respectively [4,5]. In vivo, neuroepithelial cells and RGCs are widely regarded as NSCs that give rise to the 3 major neural lineages—astrocytes, oligodendrocytes, and neurons.
During the early 1990s, Reynolds and Weiss demonstrated that NSCs can be cultured in vitro in the presence of epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) to form free-floating spherical structures called neurospheres [6–8]. Apart from NSCs, neural progenitors (NPs) can also give rise to neurospheres [9]. Since the pioneering work of Reynolds and Weiss, neurospheres have been widely used as the main culture system for propagating and studying NSCs. Thus, it is crucial that we have a better understanding of the neurosphere culture system.
A variety of selection criteria have been used to enrich for NSCs from neurospheres. These criteria include (i) Morphological. It has been shown that during fluorescence-activated cell sorting (FACS), NSCs tend to have high values for the parameters forward scatter (FSC), denoting cell size, and side scatter (SSC), denoting cell granularity [10,11]. (ii) Dye exclusion. It has been demonstrated that the fluorescent DNA-binding dye Hoechst 33342 selects for a cell population termed as the side population that enriches for NSCs [12]. (iii) Surface markers. CD133/prominin and Lewis-X have been widely used to enrich for NSCs [13–16]. Other candidate NSC markers include syndecan-1, Notch-1, and integrin-beta1 [17].
The NSC frequency can be defined in vitro as the product of neurosphere-forming units (NFUs, neurospheres/100 cells plated) and the percentage of multipotent neurospheres (as measured under clonal conditions and single nsph differentiation; see Refs. [18,19] for details). When determining multipotency of a cell giving rise to a neurosphere, it is crucial to ensure that the neurosphere is clonal (derived from that one cell and not from an aggregate of cells/neurospheres). In bulk neurosphere culture, aggregation occurs [20–22], which makes it difficult for the neurospheres to remain clonal. Hence, determining multipotency under bulk conditions is not accurate. Louis et al. have recently introduced the big neurosphere assay, where only NSCs are suggested to give rise to neurospheres >2 mm in diameter when cultured in a collagen-containing semisolid matrix for 3 weeks [23]. Under clonal conditions, NSC frequencies in neurosphere culture have been reported to be between 0.07% and 9% depending on the exact growth conditions [18,19,23].
Even though neurospheres have been widely used to investigate NSCs, the exact identity of the cell types present within neurospheres is unknown. Further, the mRNA profiles that define an NSC or a neurosphere-forming cell (NFC) are also unknown. In this study, we used single-cell mRNA profiling of 48 genes associated with NSCs/NPs to identify cell types in neurospheres. We could delineate 3 cell populations that follow a developmental timeline. One population [cluster unsorted (US) 1] expresses high Bmi1 and Hes5 and low Myc and Klf12. Cluster US2 shows intermediate expression of most genes analyzed. Cluster US3 expresses low Bmi1 and Hes5 and high Myc and Klf12. We then go on to identify an NFC cluster by performing single-cell mRNA profiling after enriching for NFCs using morphological criteria of FSC and SSC. We observe that the NFCs express high levels of mRNAs encoding mitogen receptors, such as fibroblast growth factor receptor 1 (FGFR1), FGFR2, and EGF receptor (EGFR), and transcription factors, such as Hes5 and Gli2, proteins which are implicated in self-renewal and proliferation of NSCs. Finally, we demonstrate that early NPs are the cell population that gives rise to neurospheres, and this population is likely to contain NSCs.
Materials and Methods
Isolation and culturing of NSCs and NPs
All animal experiments were approved by the Biomedical Research Council Singapore, Institutional Animal Care and Use Committee (IACUC), in accordance with national guidelines. NSCs and NPs were isolated and cultured as previously described [18]. Cerebral cortices of E14.5 C57BL/6 mouse embryos were excised and triturated into a single-cell suspension. Dissociated cells were seeded at 2×104 cells/mL in 10-cm culture dishes (Nunc) in an NSC growth medium [Dulbecco's modified Eagle's medium (DMEM)/nutrient mixture F-12 (1:1) medium (Invitrogen), B27 supplement (Invitrogen), 20 ng/mL EGF (Peprotech), 10 ng/mL bFGF (Peprotech), and 1% penicillin/streptomycin (Invitrogen)] to grow as neurospheres at 37°C in a 5% CO2 atmosphere in a humidified incubator. Neurospheres were passaged every 5–7 days.
Single-cell mRNA profiling and data processing
A TaqMan assay pool was prepared by adding each of the 48 TaqMan assays (20×; Applied Biosystems) to a final concentration of 0.2× for each assay. Neurospheres were dissociated, and single cells were sorted by FACS directly into 10 μL of RT-PreAmp Master Mix [5 μL CellsDirect 2× Reaction Mix (Invitrogen), 2.5 μL 0.2× Assay pool, 0.5 μL SuperScript® III RT/Platinum® Taq mix (Invitrogen), and 2 μL TE buffer]. Cells were frozen at −80°C and thawed to induce lysis. cDNAs of the 48 genes were generated by sequence-specific reverse transcription (50°C for 20 min) and reverse transcriptase inactivation (95°C for 2 min). Following which, sequence-specific preamplification (18 cycles at 95°C for 15 s and 60°C for 4 min) was performed. The preamplified cDNA was diluted 5-fold and used for single-cell mRNA profiling in 48.48 dynamic arrays on a BioMark system (Fluidigm). Single-cell mRNA profiling was run using the BioMark Data Collection software (Fluidigm), and Ct values were calculated using the BioMark Real-time polymerase chain reaction analysis software (Fluidigm). Cells with a Ct value for the endogenous control β-actin between 15 and 25 were considered for analysis. Ct values for a specific cell were normalized to the endogenous control by subtracting the Ct value of β-actin for the same cell. The assumed baseline Ct value is 31.
Clustering of cells based on their mRNA profile
Cells were clustered using nonmetric multidimensional scaling (nMDS) and Model-Based Clustering (Mclust). After dimension reduction by nMDS, Mclust was performed partitioning cells into clusters. The R-packages neatmap and mclust were used for performing nMDS and Mclust, respectively (for information on nMDS, see Refs. [24,25]).
Cell sorting
Dissociated cells were centrifuged, resuspended in phosphate-buffered saline (PBS), and sorting was carried out on an UV FACSAria flow cytometer (BD Biosciences). Viable cells were sorted into an NSC growth medium or RT-PreAmp Master Mix.
Neurosphere formation assay
Sorted cells were plated at low density (1×103 cells/mL in a 24-well plate) and clonal density (1 cell/well in a 96-well plate) and cultured for 7 days to form neurospheres. For low-density cultures, the neurosphere number and size were scored automatically using a high-content screening microscope (Zeiss Axiovert) and Metamorph software. For clonal density cultures, the neurospheres were scored manually. For secondary sphere formation, neurospheres derived from control and FACS-sorted cells were harvested, dissociated, replated at low density (1×103 cells/mL), and grown for 7 days.
Neurosphere differentiation and immunocytochemistry
Single neurospheres were transferred to each well of a 50-well coverglass (Sigma) coated with poly-l-lysine (0.01%; Sigma) and laminin (10μg/mL; Invitrogen). Neurospheres were differentiated for 4 days in a differentiation medium [DMEM/F-12 (1:1) medium (Invitrogen), B27 supplement (Invitrogen), 0.5% fetal bovine serum (Invitrogen), and 1% penicillin/streptomycin (Invitrogen)]. Cells were stained with mouse anti-O4 IgM (1:300; Chemicon), mouse anti-βIII-tubulin (Tuj1) IgG2a (1:500; Covance), and rabbit anti-glial fibrillary acidic protein (GFAP) IgG (1:1,000; Dako). The secondary antibodies used were Alexa-Fluor-488 goat anti-mouse IgM (1:500; Invitrogen), Alexa-Fluor-594 goat anti-mouse IgG2a (1:500; Invitrogen), and Alexa-Fluor-647 donkey anti-rabbit IgG (1:500; Invitrogen). Images were taken using the Olympus point-scanning FV-1000 confocal microscope, and the number of unipotent, bipotent, and tripotent neurospheres was scored.
Timelapse for cell size to neurosphere formation correlation
Cells were dissociated from neurospheres and plated at low density (1×103 cells/mL) in a 96-well plate. Differential interference contrast imaging was used to image cells in each experiment. Images were taken every 2 h at 20× magnification using a CoolSnap HQ CCD camera over 5 days and analyzed.
Phosphacan immunocytochemistry
FSC/SSChigh and FSC/SSClow cells were plated on coverslips coated with poly-l-lysine and left for at least 1 h to adhere to the coverslips. The primary antibody used was mouse anti-phosphacan (1:10; Millipore), and the secondary antibody used was Alexa-Fluor-488 goat anti-mouse (1:500; Invitrogen). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen). The images were captured using a Zeiss AxioVision microscope, and the fluorescence intensity was analyzed using Metamorph software.
Lewis-X staining
Dissociated cells were blocked with 3% bovine serum albumin for 15 min and incubated with anti-Lewis-X antibody (BD Biosciences) tagged with fluorescein isothiocyanate (FITC) for 15 min and washed with PBS. Fluorescence intensity was analyzed using the 3-laser analyzer (BD LSR II).
Statistical analysis
Results are presented as mean±standard deviation (SD). Two-tailed Student's t- test was used as the test for significance where means of 2 groups are compared. One-way ANOVA with post hoc comparisons using Bonferroni test was used for significance where means of more than 2 groups are compared. P values stated in the figures are *P≤0.05, **P≤0.01, and ***P≤0.001.
Results
Single-cell mRNA profiling identifies broadly 3 cell populations in neurospheres: early, intermediate, and late NPs
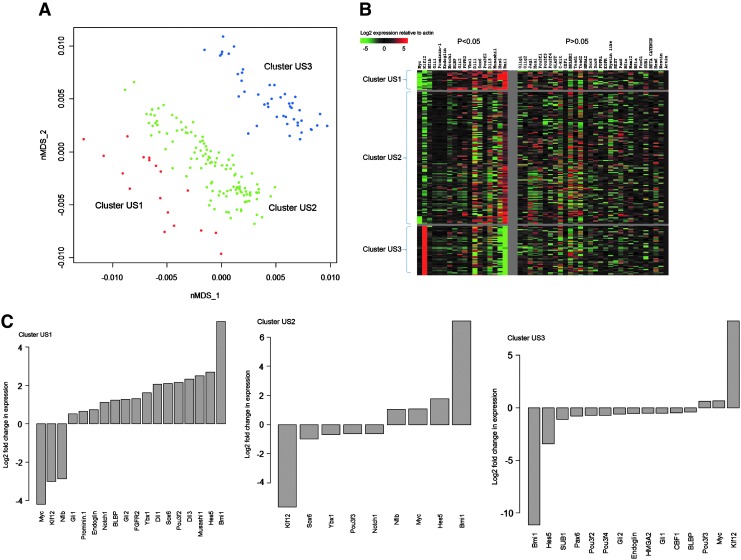
To investigate the different progenitor subclasses in the neurospheres, we profiled, in parallel, at the single-cell level (Fig. 1), 48 genes, which include genes from major signaling pathways (Notch, Wnt, and Shh signaling), classes of genes such as POU (Pit1, Oct1, Unc86) factors, and basic helix-loop-helix (bHLH) factors that have been closely associated with NSCs (Supplementary Table S1 Supplementary Data are available online at www.liebertpub.com/scd). We profiled a total of 187 cells from passage 2 neurospheres and clustered the cells using nMDS and Mclust. Three cell clusters were derived—US1, US2, and US3 (Fig. 2A). The percentage of total cells in cluster US1, US2, and US3 was 9.7%, 65.6%, and 24.7%, respectively.
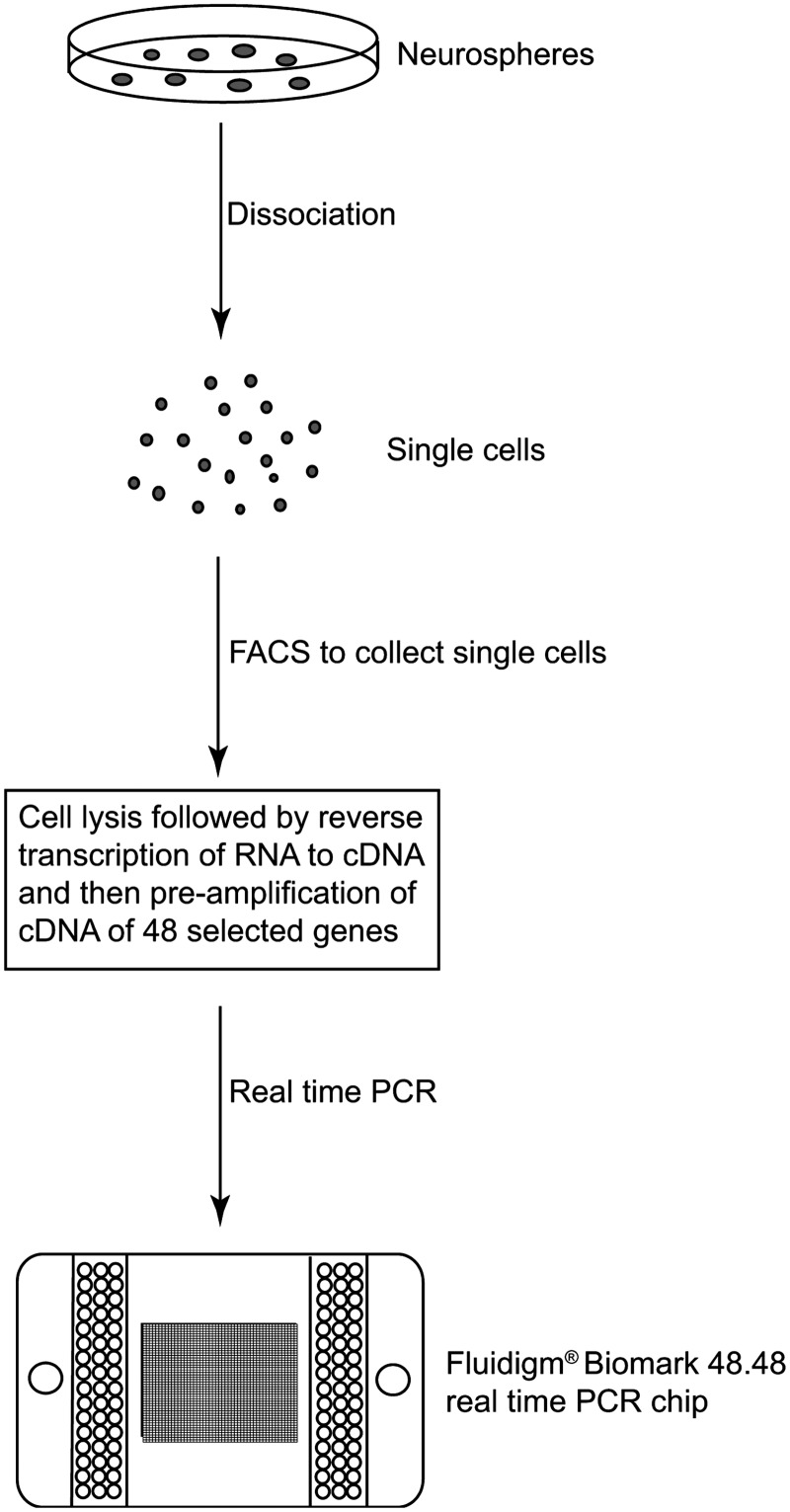
FIG. 1.
Workflow for single-cell mRNA profiling. Neurospheres are dissociated into single cells and sorted by FACS. Each sorted cell is then lysed before reverse transcription and real-time PCR are performed on the single cells to analyze the expression of 48 selected genes. FACS, fluorescence-activated cell sorting; PCR, polymerase chain reaction.
FIG. 2.
Single-cell mRNA profiling analysis of unsorted neurosphere cells. (A) Forty-eight-dimensional mRNA profiling data from passage 2 unsorted neurosphere cells were compressed to 2 dimensions by nMDS. Axes for the 2 dimensions are labeled as nMDS_1 and nMDS_2. Mclust was then used to cluster cells based on the mRNA profile of 48 genes. Three clusters were derived—US1, US2, and US3. (B) Heatmap showing the mRNA profile of the different clusters. Each row represents one cell, and each column represents one gene. Genes under the P<0.05 section are genes that show a statistically significant change in expression in cluster US1 compared to clusters US2 and US3 combined. (C) Bar plots showing a log2-fold change in gene expression for genes that show significant change in expression (P<0.05) in a cluster in comparison with the remaining clusters combined. nMDS, nonmetric multidimensional scaling; US1, unsorted 1.
We sought the identity of the 3 clusters based on their mRNA profiles. To better visualize the mRNA profile of each of the clusters, a heatmap was ordered from the expression data (Fig. 2B). Bar plots showing the genes that were significantly expressed at a higher or lower level in a specific cluster compared to the other 2 clusters combined were derived from the expression data (Fig. 2C).
Previously, Kawaguchi et al. used single-cell mRNA profiling to characterize the different progenitor subclasses that exist in vivo in the embryonic mouse brain [26]. They identified 3 groups of progenitors—apical progenitors, young basal progenitors, and basal progenitors. The apical and young basal progenitors reside in the VZ and are at an early stage of development. From their data, the apical progenitors on average had high expression of NSC/RGC markers such as Musashi1 and BLBP, Notch signaling-related genes such as Notch1 and Hes5, FGF signaling-related genes such as FGFR2, and Hedgehog signaling-related genes. The young basal progenitors had a strikingly high expression of the Delta signal Dll1. NSC/RGC markers, Notch, FGF, and Hedgehog signaling-related factors are critical in maintaining the apical and young basal progenitors in their early undifferentiated state, and Delta signals maintain neighboring progenitors at their early developmental stage. From our data, we observed that similar to the apical and young basal progenitors, cluster US1 had high expression of Musashi1, BLBP, Notch1, Hes5, and FGFR2; Hedgehog signaling-related genes such as Gli1 and Gli2; and Delta signals Dll1 and Dll3 (Fig. 2B, C). This suggests that cluster US1 comprises of cells similar to the apical and young basal progenitors and is likely to be at an early stage of development. Cluster US1 also had high expression of NSC-related genes such as Bmi1, POU3f2, and Prominin1. Knockout and knockdown studies have shown that Bmi1 is critical for NSC self-renewal and maintenance of the NSC pool in vitro and in vivo [27–31]. POU3f2 has been implicated in repression of NSC differentiation [32,33], and Prominin1 has been shown to enrich for NSCs [16]. Since NSCs are the earliest cells during development, it further supports that cells in cluster US1 are at an early stage of development.
Several genes such as Bmi1, Hes5, POU3f2, Gli1, Gli2, and BLBP, which were expressed at a higher level in cluster US1, were expressed at a lower level in cluster US3, and genes such as Myc and Klf12, which were expressed at a lower level in cluster US1, were expressed at a higher level in cluster US3 (Fig. 2B, C). Thus, it seems that cluster US3 has a near opposite profile to that of cluster US1. In addition, cluster US3 had low expression of Pax6, whose expression has been shown to decrease over development when RGCs divide to form intermediate progenitor cells [34]. Taken together, our data suggest that cells in cluster US3 are at a later stage of development.
Cluster US2 seems to adopt characteristics of cluster US1 in its high expression of Bmi1 and Hes5 and low expression of Klf12 and characteristics of cluster US3 in its high expression of Myc (Fig. 2B, C). Moreover, cluster US2 showed significant changes in only 9 genes compared to cluster US1 and US3 combined. This indicates that many of the other genes are probably expressed at an intermediate level compared to clusters US1 and US3. Therefore, cells in cluster US2 are likely to be cells transiting between clusters US1 and US3 and could be at an intermediate stage of development. In essence, single-cell mRNA profiling of cells from neurospheres indicates the presence of 3 progenitor subclasses, which follow a developmental timeline—early, intermediate, and late.
To test whether the differences in mRNA levels are due to actual differences between clusters or due to stochastic noise, we used violin plots to analyze the expression data (Supplementary Fig. S1). A multimodal distribution in these plots indicates gene expression differences between clusters, whereas stochastic noise exhibits a unimodal distribution. The plot for β-actin is unimodal, indicating little variation between the cells, whereas the plots for genes such as Bmi1 and Hes5 are multimodal, indicating clusters with distinct expression of these genes.
Derivation of suitable populations to identify an NFC cluster
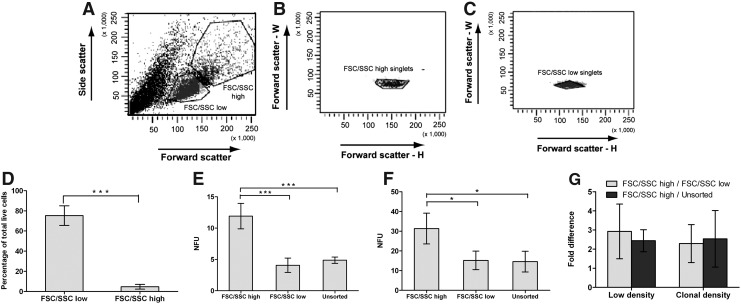
We sought to identify which of the 3 developmental subclasses, early, intermediate, or late, gives rise to neurospheres (ie, NFC cluster). The percentage of NFCs in the unsorted population is 4.88% at low density and 14.6% at clonal density (Fig. 3E, F). The reason why a lower percentage of NFCs is obtained at low density is unclear, although cell aggregation at low density may be a contributory factor. At both densities, NFCs are not well represented in the unsorted population, and thus using this population to detect an NFC cluster would be difficult. Instead, a population with increased neurosphere-forming potential should help identify an NFC cluster. Earlier studies have shown enrichment of NSCs based on cell size and granularity [10,11]. Therefore, we used morphological criteria of FSC (cell size) and SSC (granularity) to derive a population with increased neurosphere-forming potential. Gates were set up to classify passage 2 cells as FSC/SSChigh or FSC/SSClow cells, and these gates were used consistently for all subsequent experiments (Fig. 3A). Using the FSC/SSC profile, only singlets were gated and used for all experiments (Fig. 3B, C). We observed that majority of neurosphere cells were FSC/SSClow cells (∼75.2%), and a small percentage was FSC/SSChigh cells (∼4.6%) (Fig. 3D).
FIG. 3.
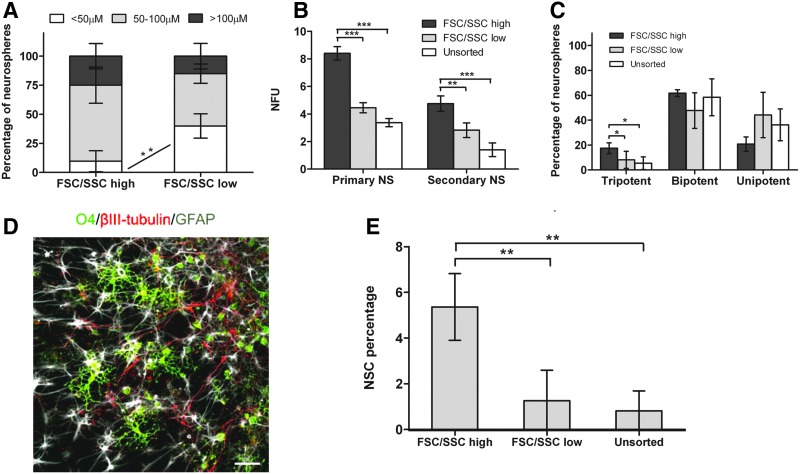
Sorting of neurosphere cells based on morphological characteristics and neurosphere-forming potential of the sorted populations. (A) FSC/SSC profile of passage 2 E14.5 neurosphere cells. FSC/SSChigh and FSC/SSClow populations were defined based on FSC/SSC signal intensity. (B) FSC/SSChigh singlets and (C) FSC/SSClow singlets were gated and sorted for the experiments to avoid doublets. (D) Percentage of FSC/SSChigh and FSC/SSClow population based on total live-cell population (mean±SD; n=5; ***P≤0.001). (E) Neurosphere-forming potential expressed as NFU, which refers to the number of neurospheres formed per 100 cells plated. NFA was done at low density (1000 cells/mL) (mean±SD; n=5; ***P≤0.001) and (F) at clonal density (1 cell/well) after sorting cells into FSC/SSChigh and FSC/SSClow populations (mean±SD; n=4; *P≤0.05). (G) Fold difference in neurosphere formation for FSC/SSChigh compared with FSC/SSClow cells or unsorted cells at low and clonal density (mean±SD; n≥4). FSC, forward scatter; SSC, side scatter; E, embryonic day; NFU, neurosphere-forming unit; NFA, neurosphere formation assay; SD, standard deviation.
FSC/SSChigh population is enriched for NFCs
From the neurosphere formation assay (NFA), the percentage of NFCs was greater in the FSC/SSChigh population (11.91%±2.03%) than that in the FSC/SSClow population (4.07%±1.15%) and the unsorted population (4.88%±0.50%) at low density (Fig. 3E). At low density, cell aggregation may occur, which would complicate neurosphere counting. To avoid the problem of aggregation, the NFA was also done at clonal density (1 cell/well). Similar to at low density, the percentage of NFCs was greater in the FSC/SSChigh population (31.3%±7.79%) than that in the FSC/SSClow population (15.2%±4.71%) and the unsorted population (14.6%±5.25%) at clonal density (Fig. 3F). There was thus a 2.29-fold and a 2.54-fold higher neurosphere formation in the FSC/SSChigh population compared to the FSC/SSClow population and unsorted population, respectively, at clonal density (Fig. 3G).
Characterization of FSC/SSC-sorted populations
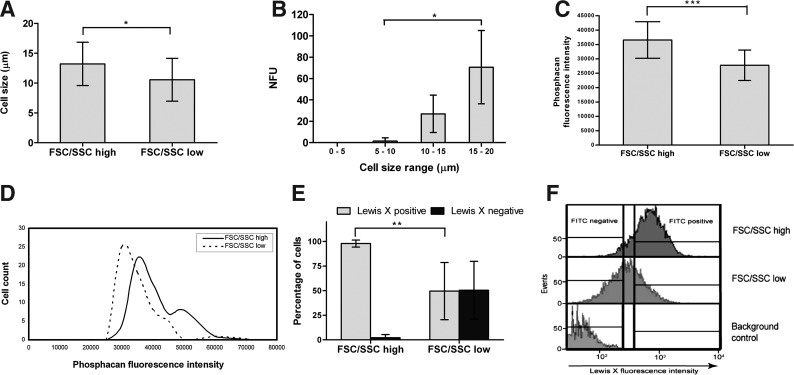
Specific features of the FSC/SSC-sorted populations were investigated, and the following characteristics were observed: (i) the FSC/SSChigh cells have on average a larger cell size than the FSC/SSClow cells (Fig. 4A). We also observed that larger cells tend to lead to higher NFU than smaller cells (Fig. 4B). (ii) We analyzed expression of 2 surface molecules closely associated with NSCs—phosphacan [19,35,36] and Lewis-X [13,15]. The average expression of phosphacan (Fig. 4C, D) and Lewis-X (Fig. 4E, F) was higher in the FSC/SSChigh population than that in the FSC/SSClow population. Almost all of the FSC/SSChigh cells (97.86%±3.53%) were Lewis-X+ve (Fig. 4E). (iii) NSC characteristics: (a) the FSC/SSChigh population gave rise to a lower percentage of small neurospheres (<50 μM) and seemingly a higher percentage of large spheres (>50 μM) than the FSC/SSClow population (Fig. 5A), indicating greater proliferation ability. (b) Secondary neurosphere formation was significantly higher in the FSC/SSChigh population than that in the FSC/SSClow and unsorted population, indicating greater self-renewal potential in the FSC/SSChigh population (Fig. 5B). (c) A higher percentage of neurospheres from the FSC/SSChigh cells were multipotent compared to the FSC/SSClow cells (Fig. 5C, D). (d) The NSC frequency, calculated as a product of the NFU at clonal density and percentage of multipotent neurospheres [18,19], was higher in the FSC/SSChigh population (5.36%±1.46%) than that in the FSC/SSClow population (1.26%±1.34%) and the unsorted population (0.82%±0.87%) (Fig. 5E). Taken together, the data show that the FSC/SSChigh population is enriched significantly for NSCs.
FIG. 4.
Characterization of FSC/SSC-sorted populations. (A) Average cell size of FSC/SSChigh and FSC/SSClow cells (mean±SD; n=22 and n=15 for FSC/SSChigh and FSC/SSClow cells respectively; *P≤0.05). (B) Correlation of neurosphere formation to cell size range (mean±SD; n=4; *P≤0.05). (C) Fluorescence intensity of 57 FSC/SSChigh and 1564 FSC/SSClow cells was recorded after phosphacan staining (mean±SD; ***P≤0.001). (D) Fluorescence intensity distribution of phosphacan staining in FSC/SSChigh and FSC/SSClow cells. (E) Percentage of Lewis-X- positive and negative cells in the FSC/SSChigh and FSC/SSClow populations (mean±SD; n=4; **P≤0.01). (F) Fluorescence intensity distribution of Lewis-X in the FSC/SSChigh and FSC/SSClow populations. Cells were stained with FITC-conjugated anti-Lewis-X antibody, and the fluorescence intensity of 1000 cells per population was recorded. Passage 2 cells were used for the experiments. FITC, fluorescein isothiocyanate.
FIG. 5.
NSC characteristics of FSC/SSChigh and FSC/SSClow cells. (A) Size categorization of neurospheres derived from the FSC/SSChigh and FSC/SSClow cells. Neurospheres below 50 μm, 50–100 μm, and above 100 μm in diameter were counted (mean±SD; n=4; **P≤0.01). (B) Secondary NFA done at low density (1000 cells/mL) for the FSC/SSChigh, FSC/SSClow, and unsorted population (mean±SD; n=4; **P≤0.01, ***P≤0.001). (C) Scoring of the number of unipotent, bipotent, and tripotent neurospheres after single neurosphere differentiation (mean±SD; n=4; *P≤0.05). (D) Immunostaining of a single multipotent neurosphere: O4 (green), βIII-tubulin (red), GFAP (gray). Scale bar=50 μm. Image was taken with a 20× objective. (E) NSC percentage calculated by multiplying NFU at clonal density and percentage of tripotent neurospheres (mean±SD; n=4; **P≤0.01). Passage 2 cells were used for the experiments. NSC, neural stem cell; NS, neurospheres; GFAP, glial fibrillary acidic protein.
Single-cell mRNA profiling of FSC/SSC-sorted cells identifies an NFC cluster
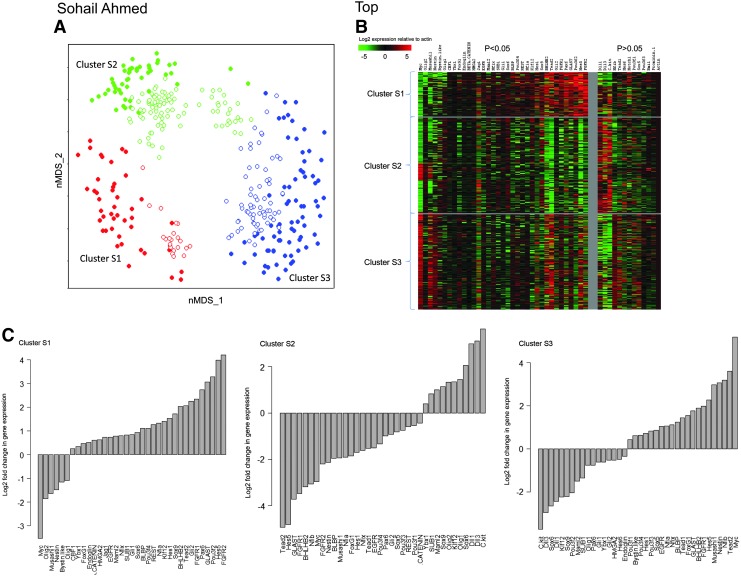
We performed single-cell mRNA profiling on near equal numbers of passage 2 FSC/SSChigh cells (170 cells) and passage 2 FSC/SSClow cells (181 cells), combined the expression data, and clustered the cells. nMDS clustering revealed 3 clusters—S (sorted) 1, S2, and S3 (Fig. 6A). Heatmap and expression bar plots derived from the expression data showed that 35 genes were significantly expressed at a higher or lower level in cluster S1 compared to clusters S2 and S3 combined (Fig. 6B, C). Violin plots show that the differences in mRNA levels between clusters are not due to stochastic noise (Supplementary Fig. S2). Cluster S1 had high expression of mitogen-related genes such as FGFR2, FGFR1, and EGFR, Notch signaling-related genes such as Hes5, Hes1, and Jag1, Hedgehog signaling-related genes such as Gli1 and Gli2. Cluster S1 also had low expression of Myc, Olig1, and Olig2. Interestingly, cluster S1 had high expression of some NSC-related genes such as GLAST and BLBP and low expression of other NSC-related genes such as Musashi1 and Nestin. Cluster S2 had high expression of certain early developmental genes such as Dll1 and Dll3 and had low expression of other early developmental genes such as Hes5 and FGFR2. Cluster S3 had low expression of certain early developmental genes such as Dll1, Dll3, and Pax6 and had high expression of Myc.
FIG. 6.
Single-cell mRNA profiling analysis of FSC/SSChigh and FSC/SSClow cells. (A) Forty-eight-dimensional mRNA profiling data from passage 2 FSC/SSChigh and FSC/SSClow cells were compressed to 2 dimensions by nMDS. Axes for the 2 dimensions are labeled as nMDS_1 and nMDS_2. Mclust derived 3 clusters from FSC/SSChigh (filled circles) and FSC/SSClow (open circles) cells—cluster S1 (red), S2 (green), and S3 (blue). (B) Heatmap showing the mRNA profile of the different clusters. Each row represents 1 cell, and each column represents 1 gene. Genes under the P<0.05 section are genes that show a statistically significant change in expression in cluster S1 compared to clusters S2 and S3 combined. (C) Bar plots showing log2-fold change in gene expression for genes that show significant change in expression (P<0.05) in a cluster in comparison with the remaining clusters combined. S, sorted.
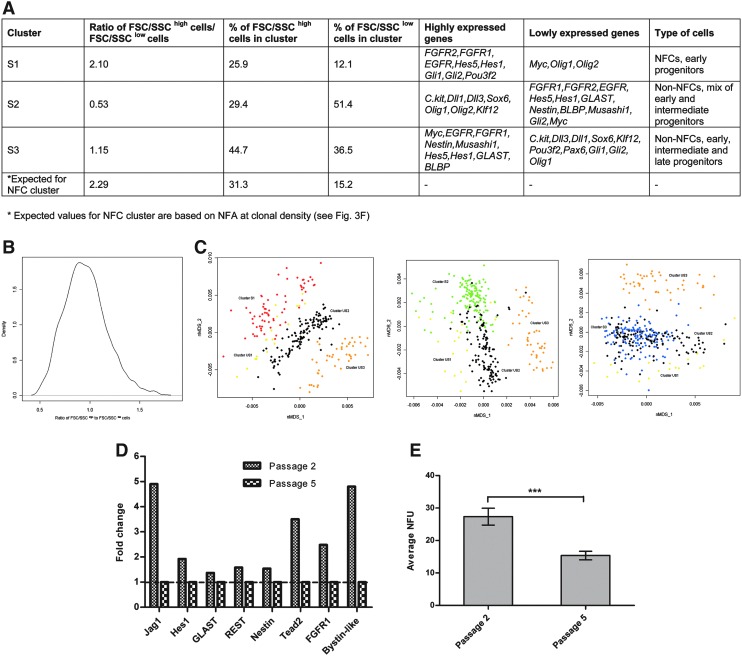
The FSC/SSChigh population had a 2.29-fold higher NFC percentage than the FSC/SSClow population at clonal density (Fig. 3G). If all the NFCs from the FSC/SSChigh and FSC/SSClow population were to fall in 1 cluster (ie, NFC cluster), the ratio of FSC/SSChigh to FSC/SSClow cells in this cluster would be about 2.29. We therefore checked the ratio of FSC/SSChigh to FSC/SSClow cells in the 3 clusters and found that cluster S1 had a ratio of 2.1 compared to clusters S2 and S3, which had a ratio of 0.53 and 1.15, respectively (Fig. 7A). This suggests that cluster S1 is most likely the NFC cluster. Further analysis revealed that 25.9% of all FSC/SSChigh cells and 12.1% of all FSC/SSClow cells analyzed fell in cluster S1 (Fig. 7A). These percentages fit well with the percentage of NFCs in the FSC/SSChigh cells (31.3%) and FSC/SSClow cells (15.2%) derived from the NFA (Fig. 3F). On the other hand, the percentages obtained from clusters S2 and S3 did not fit well with the data from the NFA. We also analyzed the likelihood that cluster S1 is formed by chance. Random clustering of FSC/SSChigh and FSC/SSClow cells was performed several times, and the resultant distribution curve showed that the likelihood of obtaining a cluster with the FSC/SSChigh to FSC/SSClow ratio 2.1 by chance is not statistically significant (Fig. 7B). Our data strongly suggest that cluster S1 is the NFC cluster.
FIG. 7.
Characteristics of clusters S1, S2, and S3. (A) The ratio of FSC/SSChigh cells to FSC/SSClow cells and the percentage of FSC/SSChigh cells and FSC/SSClow cells in each cluster were calculated. The genes that show significant changes in expression in each cluster are shown. (B) Distribution curve reflecting the likelihood of obtaining clusters with a specific FSC/SSChigh-to-FSC/SSClow cell ratio. Single-cell mRNA data from passage 2 FSC/SSChigh and FSC/SSClow cells were pooled, and 66 cells (number of cells in cluster S1) were randomly selected to form a cluster, and the ratio of FSC/SSChigh to FSC/SSClow cells was calculated. This procedure was repeated 1000 times to construct the distribution curve. Values 0.5 and 1.5 on the x-axis are the boundaries at the 99% significance level. Note that the ratio 2.1 does not fall within this region, indicating that cluster S1 is not formed by chance. (C) nMDS mapping of clusters S1 (red), S2 (green), and S3 (blue) onto clusters US1 (yellow), US2 (black), and US3 (orange). The proposed type of cells in clusters S1, S2, and S3 [as interpreted from (C)] is shown in (A). (D) Single-cell mRNA profiling was performed on 190 passage 2 and 191 passage 5 neurosphere cells. Bar plots show the expression level of the 8 genes that were expressed at a higher level by at least 1.3-fold in passage 2 compared to passage 5 cells. The expression fold change in passage 2 cells was calculated relative to passage 5 cells (given a value of 1). (E) Neurosphere-forming potential expressed as NFU, which refers to the number of neurospheres formed per 100 cells plated. NFA was done at clonal density (1 cell/well) after FACS sorting passage 2 and passage 5 cells (mean±SD; n=4; ***P≤0.001).
Cluster S1 cells are early NPs
We tried to determine the developmental profile of cluster S1 cells by mapping cluster S1 cells onto the early, intermediate, and late cells from the unsorted population (Fig. 7C). Most, if not all of cluster S1 cells mapped closely to the early cell cluster (cluster US1), indicating that cluster S1 comprises of early NPs. Cluster S2 cells mapped onto both the early and the intermediate clusters, whereas cluster S3 cells mapped onto the early, intermediate, and late clusters.
Since cluster S1 comprises of early NPs, we deduced that NFCs are early NPs. We validated this deduction through single-cell mRNA profiling of passages 2 and 5 neurosphere cells. Passage 2 cells are assumed to have more early NPs than passage 5 cells, which develop across their longer time in culture. Single-cell mRNA profiling revealed that 8 genes (Jag1, Hes1, GLAST, REST, Nestin, Tead2, FGFR1, and Bystin-like) were expressed at a higher level by at least 1.3-fold in passage 2 cells compared to passage 5 cells (Fig. 7D). Similarly, 6 of these genes (Jag1, Hes1, GLAST, REST, Tead2, and FGFR1) were also expressed at a higher level in cluster S1 (Fig. 6C), highlighting similar gene expression patterns in passage 2 cells (enriched with early NPs) and cluster S1 cells (proposed NFCs). In addition, passage 2 cells have a significantly higher neurosphere formation than passage 5 cells, suggesting that the higher neurosphere formation could be due to the enrichment of early NPs in passage 2 cells (Fig. 7E).
Discussion
Neurospheres derived from the mouse brain have been used to follow NSC behavior. To investigate the progenitor subclasses present in neurospheres, we performed single-cell mRNA profiling of 48 genes in individual passage 2 cells derived from E14.5 mouse neurospheres. We show that there are broadly 3 progenitor subclasses—clusters US1, US2, and US3. In a recent study, single-cell mRNA profiling of 11 genes on adult mouse neurosphere cells did not detect any subpopulations within the neurospheres [37]. However, it has to be noted that only few of the genes profiled were NSC-/NP-associated genes and that could be a reason why no subpopulation was detected. Here we profiled 48 genes associated with NSCs/NPs, and the finding that there are at least 3 progenitor subclasses makes this as the first study that shows the cellular makeup of neurospheres.
Previous studies have performed single-cell mRNA profiling on primary cells directly derived from the brain, hence focusing on in vivo expression dynamics of NPs [26,38–40]. Single-cell mRNA profiling showed that primary cells from the mouse embryonic brain can be classified into apical, young basal, and basal progenitors [26]. The apical and young basal progenitors reside in the VZ and are at an earlier stage of development, whereas the basal progenitors reside in the SVZ and are at a later stage of development. Cluster US1 had a similar mRNA profile as the apical and young basal progenitors in its high expression of Hes5, Musashi1, FGFR2, BLBP, Notch1, Gli2, and Dll1. These genes are critical in maintaining early progenitors, suggesting that cluster US1 is at an early developmental stage. On the other hand, cluster US3 had a similar mRNA profile as the basal progenitors in its low expression of Hes5, Pax6, and BLBP, indicating that this cluster is at a late developmental stage. Cluster US2 seems to be a transition cluster between clusters US1 and US3. Therefore, similar to the in vivo subclasses, the in vitro subclasses each seems to define a developmental timepoint. This would give valuable insights into the dynamics of gene regulation and the functions of genes that are not well studied in the context of CNS development. The resemblance of the neurosphere progenitor subclasses to the in vivo progenitor subclasses demonstrates that the neurosphere makeup reflects CNS development, and therefore can be used to model it.
The proportion of cells in cluster US2 (65.6%) is higher than that in cluster US1 (9.7%) and US3 (24.7%). It could be possible that the rate of transition from early to intermediate developmental cells is faster than the rate of transition from intermediate to late developmental cells, resulting in accumulation and greater proportion of intermediate cells.
A key observation in our work is that the early progenitor cluster had high expression of genes associated with the Notch pathway such as Notch1, Hes5, Musashi1, Dll1, and Dll3. The intermediate progenitor cluster seems to lose Notch1 expression, and the late progenitor cluster had low expression of the Notch effector Hes5 and failed to have high expression of any of the Notch pathway-related genes. This indicates that the Notch-signaling pathway is critical in maintaining the early progenitor subclass, and loss of Notch signaling probably converts the early progenitors to intermediate and then to late progenitors. This corroborates well with the in vivo scenario where loss of Notch signaling converts apical progenitors to basal progenitors [26]. This further suggests that neurospheres can reflect the in vivo signaling dynamics, a criterion for a good CNS model.
Another key observation in our work is that among all the genes analyzed, Bmi1, Hes5, Klf12, and Myc were 4 genes that showed the most drastic difference in expression between the early and late progenitors. Studies have shown that the first 2 genes Bmi1 [27–31] and Hes5 [41] are critical for NSC maintenance. These studies suggest that downregulation of these 2 genes promotes maturation of the early progenitors to late progenitors. This supports our data, as Bmi1 and Hes5 expression decreased from early to late progenitors. The third gene Klf12 is a transcription factor that represses expression of AP-2 alpha protein, which in turn plays a role in neural tube development [42,43]. The early progenitors had low expression of Klf12 and possibly high expression of AP-2, which perhaps maintains the early progenitor pool. The fourth gene Myc is known to regulate neural precursor cell fate, cell cycle, and metabolism [44,45]. Since our data show that Myc expression increased from early to late progenitors, Myc could possibly be involved in maturation of NPs. This is consistent with a recent study that showed that brains from Myc knockout mice lack specific mature neuronal subtypes [44]. Overall, we propose that expression levels of Bmi1, Hes5, Myc, and Klf12 can reflect whether a cell is an early or late NP in vitro. Intriguingly, it could be possible that these 4 genes may regulate each others' expression to drive maturation of an early NP to a late NP.
Simultaneous analysis of multiple genes allows us to study NSC/NP behavior, since behavior is likely to be regulated by many genes. To position single-cell mRNA profiling in the context of NSC/NP behavior, we tried to identify an NFC cluster. We used cell size (FSC) and cell granularity (SSC) to derive 2 populations with different neurosphere-forming potential—the FSC/SSChigh population having a higher neurosphere-forming potential than the FSC/SSClow population. Through time-lapse imaging, we observed that more neurospheres are formed from cells of larger sizes. This suggests that the increased cell size of the FSC/SSChigh population partly contributes to its increased neurosphere formation as compared to the FSC/SSClow population.
Compared to the FSC/SSClow cells, the FSC/SSChigh cells had higher expression of phosphacan and Lewis-X, 2 surface molecules associated with NSCs [13,15,19]. Furthermore, the FSC/SSChigh cells showed higher proliferation, self-renewal capacity, and multipotency than the FSC/SSClow cells. We also found that there is about a 4.3-fold enrichment of NSCs in the FSC/SSChigh population compared to the FSC/SSClow population. These data are consistent with previous studies that show that cell size, granularity, and Lewis-X selection can be used to enrich for NSCs [10,11,13,15].
Single-cell mRNA profiling of the FACS-sorted populations gave rise to 3 clusters—cluster S1, S2, and S3. By comparing the percentage of FSC/SSChigh and FSC/SSClow cells in each cluster with the percentage of NFCs in the FSC/SSChigh and FSC/SSClow population, we propose cluster S1 as the NFC cluster. The mRNA profile of cluster S1 shows that this cluster is highly mitogen responsive, as it had high expression of FGFR1, FGFR2, and EGFR. In addition, cluster S1 also had high expression of the Notch effectors Hes5 and Hes1 [41,46,47] and Sonic Hedgehog signaling-related proteins Gli1 and Gli2 [48–52], all of which are known to induce NSC/NP proliferation. High expression of these genes enables the cluster S1 cells to acquire high proliferative potential and allows these cells to initiate formation of a neurosphere.
Cluster S1 most closely matched the mRNA profile of early NPs, suggesting that early NPs give rise to NFCs. Both early NPs and cluster S1 cells had high expression of FGFR2, Hes5, POU3f2, Gli1, Gli2, BLBP, Sox6, Endoglin, and Ybx1 and low expression of Myc. Moreover, Olig1 and Olig2, genes expressed in mature progenitors committed to oligodendrogliogenesis, were expressed at a lower level in cluster S1. Furthermore, we found that passage 2 cells, which are enriched with early NPs, are also enriched with NFCs and had greater similarity to the mRNA profile of NFCs compared to passage 5 cells.
Our work has provided a clear insight into the distinct subclasses of NPs in neurospheres and their association with different stages of development and behavior in culture. In future studies, the following issues could be addressed using the tools developed here: (i) Using neurospheres to model CNS development. The presence of early, intermediate, and late NPs suggests that developmental timeline is being followed in vitro. Studying the mechanisms of how early NPs develop into late NPs in neurospheres and the interaction between the different progenitor subclasses could help us understand the intricacies of CNS development. (ii) Identification of an NSC cluster. From our data, the percentage of NSCs in the FSC/SSChigh population is only 5.36%. We have found novel NSC markers that along with Lewis-X enrich NSCs to ∼80% (M. Yu and S. Ahmed, unpublished data). Single-cell mRNA profiling of this enriched population could help derive the molecular signature for NSCs. Deciphering the molecular signature of NSCs could be pivotal in understanding the key genes that define an NSC and the regulatory mechanisms and signaling pathways that maintain a cell as an NSC. (iii) Function of novel genes important for NSCs. For instance, the proposed NFC cluster (cluster S1) has high expression of genes such as Nfia and Tead2. Although transgenic mice with deficiency of these genes show malformations of the developing brain and neural tube defects, the roles of Nfia and Tead2 in maintenance of NSCs are not known and could be investigated [53,54].
Conclusion
Neurospheres are composed of 3 major subclasses of NPs, each representing a developmental stage: early, intermediate, and late. The close relationship of these subclasses to ones directly derived from the brain indicates that neurospheres could be used to model embryonic CNS development. We also identified an NFC cluster and propose that early NPs are the cell population that gives rise to neurospheres. Lastly, NSCs are likely to be a subpopulation of the NFC cluster, and thus if we can increase their representation within the population, we should be able to identify NSCs definitively in vitro.
Supplementary Material
Acknowledgments
We thank the Agency for Science, Technology and Research, Singapore, for funding this research. We would also like to thank Dr. Mikael Huss for providing the initial R-script for the clustering analysis and Dr. Guo Guoji for guidance when performing single-cell mRNA profiling.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ahmed S. The culture of neural stem cells. J Cell Biochem. 2009;106:1–6. doi: 10.1002/jcb.21972. [DOI] [PubMed] [Google Scholar]
- 2.Kim SU. Vellis Jd. Stem cell-based cell therapy in neurological diseases: a review. J Neurosci Res. 2009;87:2183–2200. doi: 10.1002/jnr.22054. [DOI] [PubMed] [Google Scholar]
- 3.Farkas LM. Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Levison SW. Chuang C. Abramson BJ. Goldman JE. The migrational patterns and developmental fates of glial precursors in the rat subventricular zone are temporally regulated. Development. 1993;119:611–622. doi: 10.1242/dev.119.3.611. [DOI] [PubMed] [Google Scholar]
- 5.Parnavelas JG. Glial cell lineages in the rat cerebral cortex. Exp Neurol. 1999;156:418–429. doi: 10.1006/exnr.1999.7044. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds BA. Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds BA. Tetzlaff W. Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynolds BA. Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds BA. Rietze RL. Neural stem cells and neurospheres—re-evaluating the relationship. Nat Methods. 2005;2:333–336. doi: 10.1038/nmeth758. [DOI] [PubMed] [Google Scholar]
- 10.Murayama A. Matsuzaki Y. Kawaguchi A. Shimazaki T. Okano H. Flow cytometric analysis of neural stem cells in the developing and adult mouse brain. J Neurosci Res. 2002;69:837–847. doi: 10.1002/jnr.10339. [DOI] [PubMed] [Google Scholar]
- 11.Rietze RL. Valcanis H. Brooker GF. Thomas T. Voss AK. Bartlett PF. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature. 2001;412:736–739. doi: 10.1038/35089085. [DOI] [PubMed] [Google Scholar]
- 12.Kim M. Morshead CM. Distinct populations of forebrain neural stem and progenitor cells can be isolated using side-population analysis. J Neurosci. 2003;23:10703–10709. doi: 10.1523/JNEUROSCI.23-33-10703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capela A. Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–875. doi: 10.1016/s0896-6273(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 14.Uchida N. Buck DW. He D. Reitsma MJ. Masek M. Phan TV. Tsukamoto AS. Gage FH. Weissman IL. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capela A. Temple S. LeX is expressed by principle progenitor cells in the embryonic nervous system, is secreted into their environment and binds Wnt-1. Dev Biol. 2006;291:300–313. doi: 10.1016/j.ydbio.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 16.Corti S. Nizzardo M. Nardini M. Donadoni C. Locatelli F. Papadimitriou D. Salani S. Del Bo R. Ghezzi S, et al. Isolation and characterization of murine neural stem/progenitor cells based on Prominin-1 expression. Exp Neurol. 2007;205:547–562. doi: 10.1016/j.expneurol.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Nagato M. Heike T. Kato T. Yamanaka Y. Yoshimoto M. Shimazaki T. Okano H. Nakahata T. Prospective characterization of neural stem cells by flow cytometry analysis using a combination of surface markers. J Neurosci Res. 2005;80:456–466. doi: 10.1002/jnr.20442. [DOI] [PubMed] [Google Scholar]
- 18.Gan HT. Tham M. Hariharan S. Ramasamy S. Yu YH. Ahmed S. Identification of ApoE as an autocrine/paracrine factor that stimulates neural stem cell survival via MAPK/ERK signaling pathway. J Neurochem. 2011;117:565–578. doi: 10.1111/j.1471-4159.2011.07227.x. [DOI] [PubMed] [Google Scholar]
- 19.Tham M. Ramasamy S. Gan HT. Ramachandran A. Poonepalli A. Yu YH. Ahmed S. CSPG is a secreted factor that stimulates neural stem cell Survival possibly by enhanced EGFR signaling. PLoS ONE. 2010;5:e15341. doi: 10.1371/journal.pone.0015341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singec I. Knoth R. Meyer RP. Maciaczyk J. Volk B. Nikkhah G. Frotscher M. Snyder EY. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat Methods. 2006;3:801–806. doi: 10.1038/nmeth926. [DOI] [PubMed] [Google Scholar]
- 21.Mori H. Fujitani T. Kanemura Y. Kino-Oka M. Taya M. Observational examination of aggregation and migration during early phase of neurosphere culture of mouse neural stem cells. J Biosci Bioeng. 2007;104:231–234. doi: 10.1263/jbb.104.231. [DOI] [PubMed] [Google Scholar]
- 22.Coles-Takabe BL. Brain I. Purpura KA. Karpowicz P. Zandstra PW. Morshead CM. van der Kooy D. Don't look: growing clonal versus nonclonal neural stem cell colonies. Stem Cells. 2008;26:2938–2944. doi: 10.1634/stemcells.2008-0558. [DOI] [PubMed] [Google Scholar]
- 23.Louis SA. Rietze RL. Deleyrolle L. Wagey RE. Thomas TE. Eaves AC. Reynolds BA. Enumeration of neural stem and progenitor cells in the neural colony-forming cell assay. Stem Cells. 2008;26:988–996. doi: 10.1634/stemcells.2007-0867. [DOI] [PubMed] [Google Scholar]
- 24.Taguchi Y-h. Oono Y. Relational patterns of gene expression via non-metric multidimensional scaling analysis. Bioinformatics. 2005;21:730–740. doi: 10.1093/bioinformatics/bti067. [DOI] [PubMed] [Google Scholar]
- 25.Rajaram S. Oono Y. NeatMap—non-clustering heat map alternatives in R. BMC Bioinformatics. 2010;11:45. doi: 10.1186/1471-2105-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawaguchi A. Ikawa T. Kasukawa T. Ueda HR. Kurimoto K. Saitou M. Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- 27.Molofsky AV. He S. Bydon M. Morrison SJ. Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molofsky AV. Pardal R. Iwashita T. Park IK. Clarke MF. Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruggeman SW. Valk-Lingbeek ME. van der Stoop PP. Jacobs JJ. Kieboom K. Tanger E. Hulsman D. Leung C. Arsenijevic Y. Marino S. van Lohuizen M. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zencak D. Lingbeek M. Kostic C. Tekaya M. Tanger E. Hornfeld D. Jaquet M. Munier FL. Schorderet DF. van Lohuizen M. Arsenijevic Y. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J Neurosci. 2005;25:5774–5783. doi: 10.1523/JNEUROSCI.3452-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fasano CA. Dimos JT. Ivanova NB. Lowry N. Lemischka IR. Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Catena R. Tiveron C. Ronchi A. Porta S. Ferri A. Tatangelo L. Cavallaro M. Favaro R. Ottolenghi S, et al. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279:41846–41857. doi: 10.1074/jbc.M405514200. [DOI] [PubMed] [Google Scholar]
- 33.Josephson R. Muller T. Pickel J. Okabe S. Reynolds K. Turner P. Zimmer A. McKay R. POU transcription factors control expression of CNS stem cell-specific genes. Development. 1998;125:3087–3100. doi: 10.1242/dev.125.16.3087. [DOI] [PubMed] [Google Scholar]
- 34.Englund C. Fink A. Lau C. Pham D. Daza RA. Bulfone A. Kowalczyk T. Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu WL. Fu SL. Wang YX. Li Y. Lu HZ. Xu XM. Lu PH. Chondroitin sulfate proteoglycans regulate the growth, differentiation and migration of multipotent neural precursor cells through the integrin signaling pathway. BMC Neurosci. 2009;10:128. doi: 10.1186/1471-2202-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ida M. Shuo T. Hirano K. Tokita Y. Nakanishi K. Matsui F. Aono S. Fujita H. Fujiwara Y. Kaji T. Oohira A. Identification and functions of chondroitin sulfate in the milieu of neural stem cells. J Biol Chem. 2006;281:5982–5991. doi: 10.1074/jbc.M507130200. [DOI] [PubMed] [Google Scholar]
- 37.Ståhlberg A. Andersson D. Aurelius J. Faiz M. Pekna M. Kubista M. Pekny M. Defining cell populations with single-cell gene expression profiling: correlations and identification of astrocyte subpopulations. Nucleic Acids Res. 2011;39:e24. doi: 10.1093/nar/gkq1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ginsberg SD. Che S. Expression profile analysis within the human hippocampus: comparison of CA1 and CA3 pyramidal neurons. The Journal of Comp Neurol. 2005;487:107–118. doi: 10.1002/cne.20535. [DOI] [PubMed] [Google Scholar]
- 39.Kamme F. Salunga R. Yu J. Tran D-T. Zhu J. Luo L. Bittner A. Guo H-Q. Miller N. Wan J. Erlander M. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tietjen I. Rihel JM. Cao Y. Koentges G. Zakhary L. Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 41.Ohtsuka T. Sakamoto M. Guillemot Fo. Kageyama R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J Biol Chem. 2001;276:30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- 42.Roth C. Schuierer M. Günther K. Buettner R. Genomic structure and DNA binding properties of the human zinc finger transcriptional repressor AP-2rep (KLF12) Genomics. 2000;63:384–390. doi: 10.1006/geno.1999.6084. [DOI] [PubMed] [Google Scholar]
- 43.de Crozé N. Maczkowiak F. Monsoro-Burq AH. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proceedings of the National Academy of Sciences. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wey A. Cerdeno V. Pleasure D. Knoepfler P. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. The Cerebellum. 2010;9:537–547. doi: 10.1007/s12311-010-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wey A. Knoepfler P. c-myc and N-myc promote active stem cell metabolism and cycling as architects of the developing brain. Oncotarget. 2010;1:120–130. doi: 10.18632/oncotarget.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabos P. Kabosova A. Neuman T. Blocking HES1 expression initiates GABAergic differentiation and induces the expression of p21CIP1/WAF1 in human neural stem cells. J Biol Chem. 2002;277:8763–8766. doi: 10.1074/jbc.C100758200. [DOI] [PubMed] [Google Scholar]
- 47.Solecki DJ. Liu X. Tomoda T. Fang Y. Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 48.Galvin KE. Ye H. Erstad DJ. Feddersen R. Wetmore C. Gli1 induces G2/M arrest and apoptosis in hippocampal but not tumor-derived neural stem cells. Stem Cells. 2008;26:1027–1036. doi: 10.1634/stemcells.2007-0879. [DOI] [PubMed] [Google Scholar]
- 49.Matise M. Epstein D. Park H. Platt K. Joyner A. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 50.Stecca B. Ruiz I Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J. 2009;28:663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palma V. Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- 52.Takanaga H. Tsuchida-Straeten N. Nishide K. Watanabe A. Aburatani H. Kondo T. Gli2 Is a novel regulator of Sox2 expression in telencephalic neuroepithelial cells. Stem Cells. 2008;27:165–174. doi: 10.1634/stemcells.2008-0580. [DOI] [PubMed] [Google Scholar]
- 53.Mason S. Piper M. Gronostajski RM. Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- 54.Kaneko KJ. Kohn MJ. Liu C. DePamphilis ML. Transcription factor TEAD2 is involved in neural tube closure. Genesis. 2007;45:577–587. doi: 10.1002/dvg.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.