Abstract
Mammalian tissue size is maintained by slow replacement of de-differentiating and dying cells. For adipocytes, key regulators of glucose and lipid metabolism, the renewal rate is only 10% per year. We used computational modeling, quantitative mass spectrometry, and single-cell microscopy to show that cell-to-cell variability, or noise, in protein abundance acts within a network of more than six positive feedbacks to permit pre-adipocytes to differentiate at very low rates. This reconciles two fundamental opposing requirements: High cell-to-cell signal variability is needed to generate very low differentiation rates, whereas low signal variability is needed to prevent differentiated cells from de-differentiating. Higher eukaryotes can thus control low rates of near irreversible cell fate decisions through a balancing act between noise and ultrahigh feedback connectivity.
Understanding how large populations of cells direct small subsets to a different state is crucial for understanding the differentiation of precursor cells and stem cells, as well as cancer progression (1). Adipocyte differentiation is of fundamental importance for disease-associated conditions such as insulin resistance, obesity, and cancer and is also an accessible experimental system for investigating mammalian cell fate decisions. Adipocytes have a large pool of precursor cells that reside in the fat tissue–about one for every five differentiated cells (2)–which means that only about 1 cell out of 60 embarks on a differentiation path during the up to 12 days it takes to differentiate (3) (supplementary text). These pre-adipocytes differentiate through a bistable switch mechanism with a single threshold for activation in each cell that involves positive feedback between two key transcription factors, CCAAT/enhancer binding protein α (CEBPA) and peroxisome proliferator-activated receptor γ (PPARG) (Fig. 1A) (4). In a bistable system, if the precursor population were truly uniform, there would be a single critical threshold stimulus below which 100% of the cells remain precursor cells and above which 100% of the cells differentiate. Thus, it is a conundrum how a graded response is observed experimentally (Fig. 1B) (5), with weak stimuli inducing very low differentiation rates that increase as the stimuli intensity increases rather than causing all cells to convert at one critical stimulus threshold.
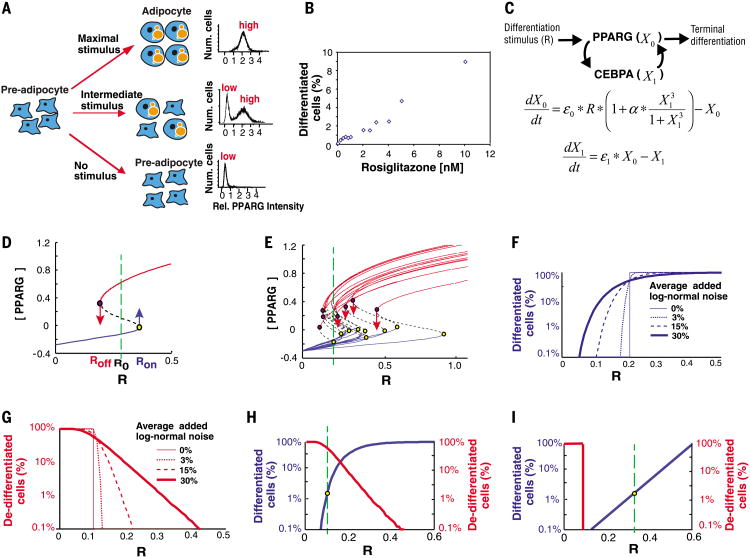
Fig. 1. Description of the control problem.
(A) Schematic of the bistable switch from pre-adipocyte to adipocyte. Right, bimodality in single cell abundance of PPARG. (B) Control of very low rates of adipocyte differentiation by increasing the stimulus, R, applied to mouse OP9 preadipocyte cells. Here, R is rosiglitazone, a PPARG agonist, which directly induces adipogenesis in these cells (7). (C) Quantitative model of the protein network controlling the terminal differentiation decision in adipocytes. ε0 and ε1 represent noise in the abundance of X0 and X1, respectively, α is a feedback amplification term experimentally measured to be ∼15 (7). (D) Steady-state plot of a one-feedback loop system with cooperativity, n = 3. As the receptor stimulus R is increased, Ron is the level of R at which the modeled cell triggers into the differentiated state (yellow dot). As R is decreased, Roff is the level of R at which the cell loses the differentiated state (purple dot). R0 is the level of basal receptor activity. (E) Sample steady-state plots of the system in (D) but with 30% log-normal noise randomly added to each simulation. At the stimulus intensity marked by the green line, only one cell differentiated but several differentiated cells de-differentiated (red arrows). (Fand G) The relationship between Rand number of cells differentiating or de-differentiating becomes more graded as more noise is added. Each curve in these plots summarizes the Ron (F) or Roff (G) values obtained from 20,000 simulations. Noise was added randomly to each simulation to result in an average of no noise (thin solid line) up to 30% log-normal noise (thick solid line). (H) Plot showing the probabilities of triggering differentiation (blue) versus losing the differentiated state (red) as a function of the stimulus intensity R [20,000 simulations of the system described in (E)]. Such a system would be unable to maintain tissue size because, at the rate of preadipocyte differentiation observed in vivo (1.65%, as marked by dashed green line), ∼80% of adipocytes would de-differentiate. (I) Plot showing ideal condition which allows for graded control of low rates of differentiation but with no de-differentiation.
By extension of results from bacterial differentiation (6), it is plausible that the observed regulated mammalian differentiation rates are generated by noise, or cell-to-cell variability, in the abundance of key regulatory proteins (7–10). Noise in the cell population makes it possible for only a small fraction of cells to differentiate at low receptor stimuli, R, but can also interfere with maintenance of the differentiated state by temporarily lowering the abundance of a key regulatory protein below a critical threshold. This dual effect of noise may explain why de-differentiation occurs in adipose (11), pancreatic beta (12), neuronal (13), epithelial (14), and cardiac cells (15). Thus, organisms have a fundamental optimization problem. They need noise to be able to regulate fractional rates of differentiation, but this same noise can also cause unwanted and stochastic de-differentiation.
We explored these competing requirements computationally. We used a working model that includes the established feedback between PPARG (X0) and CEBPA (X1) as a driver for adipogenesis (Fig. 1C) (4). Figure 1D shows a steady-state representation of this model in which the positive feedback loop generates bistability. If an activating receptor input R exceeds a critical threshold Ron, the cell switches to the differentiated state. If R were later to decrease below Roff, the differentiated cell would drop back into the undifferentiated state. Irreversible differentiation requires that there is a basal activity R0 that remains higher than Roff when the stimulus is turned off.
The typically observed noise for mammalian protein abundance is 30% and stems from intrinsic noise in the synthesis and degradation of mRNA and proteins (16). Our simulations demonstrated that noise added to each of the two feedback components enabled receptor stimuli to control low fractional rates of differentiation: For a specific receptor stimulus (marked with a green line in Fig. 1E), only 1 out of 15 cells switched to the differentiated state. Plots in which different amounts of noise were stochastically added to each simulation showed that the ability to regulate the rate of differentiation by increasing receptor stimuli markedly improved as the amount of noise increased (Fig. 1F). Nevertheless, this same noise also caused increasing fractions of cells in the population to lose the differentiated state over time, even for high-stimulus amplitudes (Fig. 1, E and G). The constraints that allow regulation of low rates of differentiation and also locking of cells in the differentiated state can be appreciated in a combined graph (Fig. 1H). To lose less than 0.33% of the differentiated cells and thereby maintain the 10% yearly rate of mature adipocyte turnover (5, 17), the minimal basal receptor activity R0 value must be higher than 0.4. At the same time, a much smaller R0 of 0.1 is required to regulate the observed in vivo rate of pre-adipocyte differentiation of less than 2% every 12 days (Fig. 1H, left axis, and supplementary text). Thus, this system cannot be optimized because R0 cannot be both more than 0.4 and less than 0.1, simultaneously. An ideal system must instead have differentiation and de-differentiation rates closer to the ones shown in the scheme in Fig. 1I. Although cell death may also contribute to maintaining constant cell numbers in tissues, recent studies indicate that de-differentiation might be an equally or more relevant process (12–15).
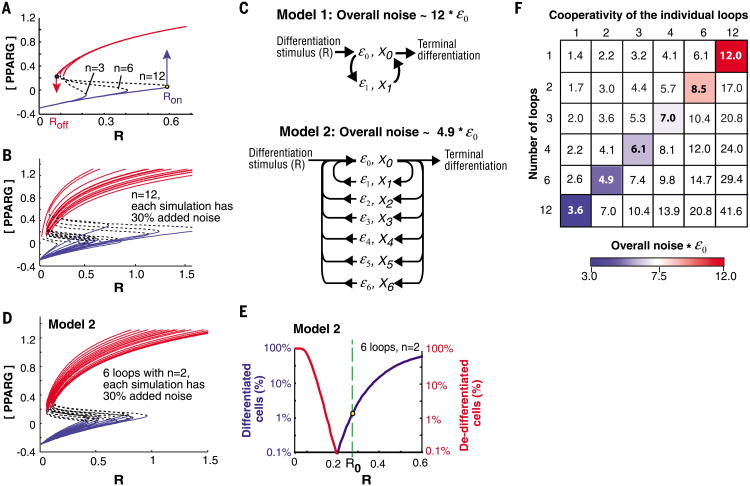
Regulatory systems with bistability typically have cooperativities of 2 to 4, although in a few cases higher cooperativities have been found (18). As shown in Fig. 2A, increasing the cooperativity from 3 to 6, and to 12, broadened the window between Roff and Ron. However, the higher cooperativity combined with the co-dependence between the feedback loop mediators, X0 and X1, resulted in a large increase in the total parameter noise of the system (Fig. 2B). Thus, increasing the cooperativity in a single loop system improves the system's performance but still does not allow optimization (Fig. 2B and fig. S1).
Fig. 2. A protein architecture that can regulate a 10% annual renewal rate.
(A) Steady-state plots when the cooperativity, n, is increased. (B) Sample steady-state plots for a system with a single high cooperative feedback loop (n = 12) and 30% log-normal noise added to each simulation. (C) Schematic of two system architectures that have the same overall cooperativity but different overall noise. Model 1 has one feedback loop with a cooperativity of 12. Model 2 has six positive feedback loops, each with a cooperativity of 2. (D and E) Simulation results for model 2. An average of 30% log-normal noise was randomly added to each simulation. (D) Sample steady-state curves. Plotting the curves with R on a log-scale shows that the variability in Ron and Roff is similar (fig. S1C). (E) Results of 20,000 simulations showing that such a system can maintain tissue size. At the low differentiation rate needed to renew adipose tissue (1.65%, yellow dot), less than 0.1% of differentiated cells would lose the differentiated state. (F) The matrix shows the overall system noise as a function of number of feedback loops versus cooperativity of the individual loops. The colored boxes mark systems with the same total cooperativity (n = 12), but with decreasing system noise as the number of feedback loops increases.
We explored whether increasing the number of feedback loops would help. Dual positive feedback, with one of the feedbacks being much slower, allows high-frequency signaling noise to be rejected (19). We noticed in our simulations that a system with two loops connected in an AND-gate configuration reduced the rate of de-differentiation by a mechanism that does not require different time constants of the loops. The improvement in system performance was still insufficient to allow optimization, so we tested whether a large number of feedbacks might be even better (model 2 in Fig. 2C). A system architecture with ultrahigh feedback connectivity–that is, more than six positive feedbacks, each having a cooperativity of only two–resulted in an optimized system that successfully balanced very low rates of cell differentiation with stability of the differentiated state (Fig. 2, D and E). There are two main reasons for this marked improvement in performance of this ultrahigh feedback loop system: First, by connecting different loops required for adipogenesis in an AND-gate configuration, the cooperativities of the feedback loops can be added together to give a high overall system cooperativity. Second, the noise in protein abundance in each feedback loop is independent and random from each other, leading to a reduction in overall system noise compared with a system with fewer feedback loops and the same overall cooperativity (Fig. 2, C and F, and supplementary text).
We wanted to test our prediction that adipocyte differentiation requires ultrahigh feedback connectivity. To identify feedback loops and evaluate their importance, one needs a method to perturb a protein in the network and then measure the effect of that perturbation over time on many other proteins, that is, the candidate feedback loop partners. We used mouse OP9 cells, a bone marrow-derived adipocyte model, as a cell model that shares key regulatory processes with adipogenesis in vivo (7, 20–23). We selected 60 nuclear proteins–including PPARG, CEBPB, CEBPA, and other proteins implicated as important in adipogenesis from the literature or from our small interfering RNA (siRNA) knockdown screening data (table S1)–as our candidate network to identify the prevalence of feedback loops coupled to PPARG and used selected reaction monitoring mass spectrometry (SRM-MS) to quantitatively measure the abundance of these proteins (figs. S2 to S8) (24, 25).
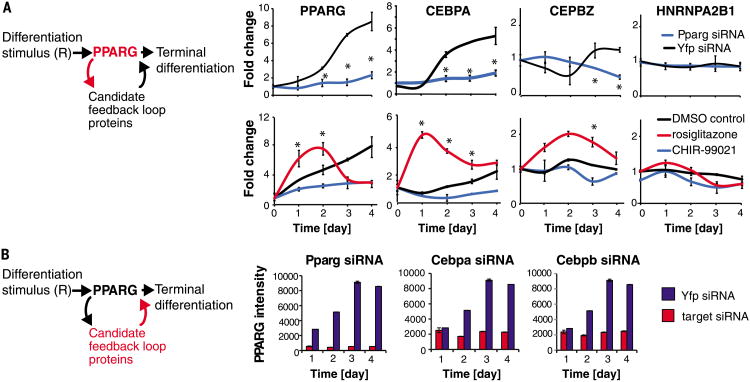
For a protein to be classified as being in a feedback loop with PPARG, it had to meet four criteria. First, because the abundance of PPARG changes dramatically during adipogenesis, we assumed that the abundance of a protein in a feedback loop with PPARG also had to change over the time course of adipogenesis (figs. S9 and S10). Second, to confirm a link from PPARG to the candidate feedback loop protein, changing PPARG abundance with siRNA or small molecules needed to result in a corresponding change in the abundance of the candidate feedback loop protein (Fig. 3A and figs. S11 and S12). Third, to confirm a link from the candidate protein back to PPARG, changing the abundance of the candidate protein by siRNA-mediated depletion needed to result in a corresponding change in the abundance of PPARG (Fig. 3B and fig. S13). As a fourth criterion, titration of the PPARG activator rosiglitazone onto undifferentiated cells at basal conditions had to result in a direct change of expression of the candidate protein (Fig. 4A and fig. S14). Out of 60 candidate proteins (table S1), we identified seven positive feedback mediators of PPARG that satisfied these four criteria: three previously unknown positive feedback mediators (the CCAAT-box DNA binding protein CEBPZ, the fatty-acid binding protein FABP4, and the phosphatidic acid phosphohydrolase enzyme LPIN1), two new mediators of double negative feedback loops that function analogously to positive feedback loops (the actin-binding protein FLNA and transcription factor RUNX2), and validated two known positive feedback mediators (the transcription factors CEBPB and CEBPA) (7) (Fig. 4B).
Fig. 3. A method to systematically uncover feedback loops in a protein network.
(A) SRM-MS was used to measure changes in nuclear protein concentrations over the timecourse of adipogenesis in response to siRNA or chemical perturbations to PPARG (see also figs. S10 to S12). OP9 preadipocytes were induced to differentiate into mature adipocytes in 4 days by addition of the adipogenic cocktail (dexamethasone, isobutylmethylxanthine, and insulin). (Top) Cells were transfected with siRNA targeting PPARG (blue) or yellow fluorescent protein (YFP) as a control (black) 24 hours before addition of the adipogenic factors. (Bottom) A PPARG agonist (rosiglitazone) or PPARG inhibitor (CHIR-99021) was added together with the adipogenic mix. All values were normalized to the control value at day 0. Each data point is the average of three biological replicates (error bars show standard deviation). A protein was classified as regulated by PPARG activity if its abundance in the perturbed versus control (YFP) samples varied significantly at one or more time points, P < 0.05 (*) calculated using the student's t test, two-tailed, two-sample equal variance. (B) siRNA-mediated depletion of the candidate feedback loop partners was used to determine which ones regulate PPARG. The knockdown effect in OP9 cells was measured by immunocytochemistry staining for PPARG. Each bar in the plots represents ∼10,000 cells. Error bars show standard error. See also fig. S13.
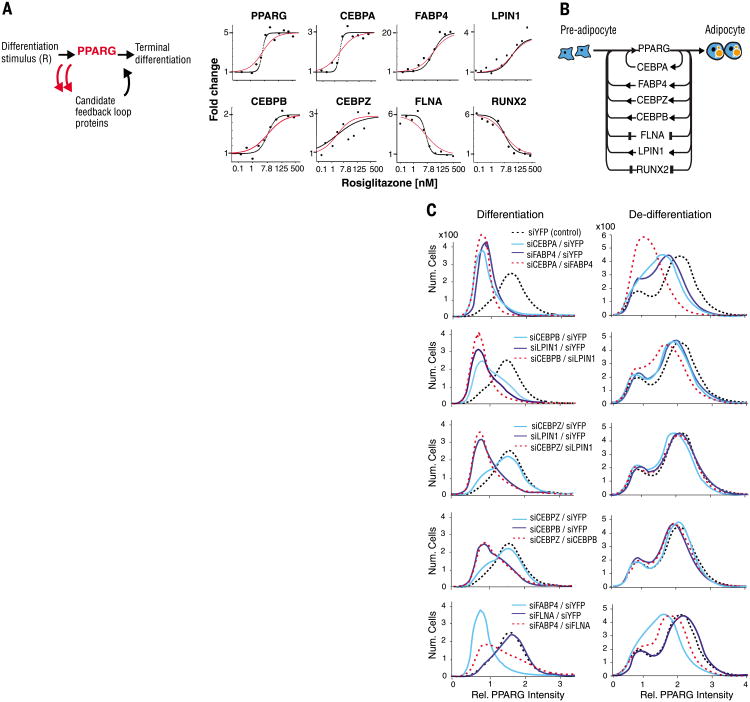
Fig. 4. Differentiation from pre-adipocyte to adipocyte is regulated by a single module consisting of multiple positive-feedback loops connecting back to PPARG.
(A) Test for feedback connectivity by directly activating PPARG. The PPARG agonist rosiglitazone was titrated into the medium of undifferentiated OP9 cells, and the protein abundance of the candidate feedback loop mediators was measured by SRM-MS 48 hours later, a time point at which PPARG is maximally expressed (7). Curves show the best fits to the data using an optimized Hill coefficient (black) versus a Hill coefficient of 1 (red). (B) Schematic of the seven identified feedback loops. (C) Contributions of the identified feedback loops to switching pre-adipocytes (low PPARG peak) to adipocytes (high PPARG peak) and to preventing de-differentiation. To measure contributions to differentiation, we transfected OP9 cells with siRNA 24 hours before the start of the experiment to remove the specified feedback loop components. The cells were then stimulated with 1 μM rosiglitazone for 24 hours and fixed (left column). To measure contributions to de-differentiation, OP9 cells were first differentiated into adipocytes by adding 1 μM rosiglitazone to the culture media for 48 hours, then transfected with the specified siRNA and fixed 24 hours later (right column). PPARG abundance was quantified by immunocytochemistry staining with anti-PPARG and then imaged.
Supporting the assumption that different feedback loops regulate PPARG expression independently, the identified feedback mediators interacted with PPARG in very different ways. For example, the promoter of CEBPZ has several predicted CEBPA binding elements (26), suggesting that the observed up-regulation of CEBPZ expression by PPARG activation is likely through PPARG-mediated increase in accumulation of CEBPA FABP4 likely activates PPARG by transporting PPARG-activating ligands into the nucleus (27). LPIN1 activates PPARG by releasing a corepressor of PPARG (28).
To understand whether the newly identified feedback loops contribute to differentiation, we used siRNA to deplete the feedback loop proteins in undifferentiated OP9 cells; induced differentiation by adding the PPARG activator rosiglitazone for 24 hours (7,29); and then fixed, imaged, and analyzed the cells (Fig. 4C, left column). To understand the feedback loop contributions to preventing de-differentiation, we terminally differentiated OP9 preadipocytes into adipocytes by adding rosiglitazone for 48 hours and then used siRNA to deplete the feedback loop proteins in these cells (Fig. 4C, right column). The effect of the feedback loops on differentiation and de-differentiation was not necessarily symmetrical. For example, depletion of CEBPB or LPIN1 had a greater effect on differentiation than on de-differentiation, suggesting that these loops promote differentiation and are less important for keeping cells in the terminally differentiated state.
At least four of the identified feedback loop proteins–CEBPA, FABP4, CEBPB, and LPIN1– are required for adipogenesis and are thus connected as AND-gates. siRNA-mediated depletion of each of these proteins during adipogenesis resulted in a nearly complete lack of adipogenesis in most cells (enrichment of cells in the low PPARG abundance peak as shown by the solid lines in the top two plots in the left column of Fig. 4C). Nevertheless, the specific regulatory mechanisms by which each of the identified feedback loops acts is likely not the same and may include a mixture of additive (OR-gate) and multiplicative (AND-gate) features. In addition, the differential effects of the feedback loops on adipogenesis indicate that the cooperativity is likely different for the different loops and that our model in which all loops have equal cooperativity is a simplification.
This ultrahigh feedback system with multiple AND-gate feedback loops creates the ultrasensitivity needed to create a robust bistable switch that can be triggered at variable activation thresholds within the cell population and at the same time limits the noise within a range that permits both low rates of differentiation and locking of cells into the differentiated state. Furthermore, a system with a large number of feedback loops with various regulatory proteins can integrate information from a broad range of signaling, metabolic, and stress inputs to regulate and better control the fraction of differentiating cells. The advantages of such a system design suggest that the combination of noise with ultrahigh feedback is a ubiquitous regulatory feature that applies to other cell fate decisions in multicellular organisms. Our integrated approach combining SRM-MS, chemical and genetic perturbations, and single-cell microscopy may be useful to identify and understand such feedback loop circuits in other cell fate decisions.
Supplementary Material
Acknowledgments
We thank T. Meyer, S. Collins, G. Anderson, S. Spencer, together with members of the Teruel lab, for thoughtful comments and discussions. This work is supported by Stanford University New Faculty Startup Funds and NIH P50GM107615 (M.N.T.), Deutsche Forschungsgemeinschaft Fellowship AH 220/1-1 (R.A.), and American Heart Association Fellowship 13POST17090092 (A.O.).
Footnotes
References and Notes
- 1.Zuber J, et al. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tchoukalova YD, Sarr MG, Jensen MD. Am J Physiol. 2004;287:R1132. doi: 10.1152/ajpregu.00337.2004. [DOI] [PubMed] [Google Scholar]
- 3.Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 4.Rosen ED, et al. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spalding KL, et al. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 6.Süel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 7.Park BO, Ahrends R, Teruel MN. Cell Reports. 2012;2:976–990. doi: 10.1016/j.celrep.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanna J, et al. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palani S, Sarkar CA. Cell Reports. 2012;1:215–224. doi: 10.1016/j.celrep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Poloni A, et al. Stem Cells. 2012;30:965–974. doi: 10.1002/stem.1067. [DOI] [PubMed] [Google Scholar]
- 12.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiff T, et al. J Neurosci. 2010;30:905–915. doi: 10.1523/JNEUROSCI.5368-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tata PR, et al. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senyo SE, et al. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niepel M, Spencer SL, Sorger PK. Curr Opin Chem Biol. 2009;13:556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang QA, Tao C, Gupta RK, Scherer PE. Nat Med. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trunnell NB, Poon AC, Kim SY, Ferrell JE., Jr Mol Cell. 2011;41:263–274. doi: 10.1016/j.molcel.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandman O, Ferrell JE, Jr, Li R, Meyer T. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosen ED, MacDougald OA. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 21.Berry R, Jeffery E, Rodeheffer MS. Cell Metab. 2014;19:8–20. doi: 10.1016/j.cmet.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry DC, Stenesen D, Zeve D, Graff JM. Development. 2013;140:3939–3949. doi: 10.1242/dev.080549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolins NE, et al. J Lipid Res. 2006;47:450–460. doi: 10.1194/jlr.D500037-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Abell E, Ahrends R, Bandara S, Park BO, Teruel MN. Proc Natl Acad Sci USA. 2011;108:14485–14490. doi: 10.1073/pnas.1018266108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefterova MI, et al. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayers SD, Nedrow KL, Gillilan RE, Noy N. Biochemistry. 2007;46:6744–6752. doi: 10.1021/bi700047a. [DOI] [PubMed] [Google Scholar]
- 28.Kim HE, et al. Biochem J. 2013;453:49–60. doi: 10.1042/BJ20121598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tontonoz P, Spiegelman BM. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.