SUMMARY
Stress granules are mRNA-protein granules that form when translation initiation is limited and are related to pathological granules in various neurodegenerative diseases. Super-resolution microscopy reveals stable substructures referred to as cores within stress granules that can be purified. Proteomic analysis of stress granule cores reveals a dense network of protein-protein interactions, links between stress granules and human diseases, and identifies ATP-dependent helicases and protein remodelers as conserved stress granule components. ATP is required for stress granule assembly and dynamics. Moreover, multiple ATP-driven machines affect stress granules differently; with the CCT complex inhibiting stress granule assembly, while the MCM and RVB complexes promote stress granule persistence. Our observations suggest that stress granules contain a stable core structure surrounded by a dynamic shell with assembly, disassembly and transitions between the core and shell modulated by numerous protein and RNA remodeling complexes.
INTRODUCTION
Eukaryotic cells contain non-membrane bound organelles consisting of mRNA and protein, referred to as mRNP granules. mRNP granules can affect mRNA function and localization (Buchan et al., 2014). Stress granules are mRNP granules that form when translation initiation is limiting (Buchan and Parker, 2009). Stress granules, or related assemblies, have been proposed to play a role in neurodegenerative diseases since mutations that cause disease can increase stress granule formation or persistence (Li et al., 2013; Ramaswami et al., 2013). However, the composition and physical properties of stress granules are largely unknown.
Several observations suggest that intrinsically disordered regions (IDRs) on RNA binding proteins play a role in stress granule assembly. First, IDRs of TIA and FUS have been shown to affect targeting of TIA and FUS to stress granules (Gilks et al., 2004; Kato et al., 2012). Second, the assembly of yeast P bodies is promoted by IDRs on several proteins (Decker et al., 2007; Reijns et al., 2008). Since IDRs can also be predicted to have a high probability of forming β-amyloid fibers and such domains on RNA binding proteins self-assemble into fibers in vitro (Eisenberg and Jucker, 2012; Guo et al., 2011; Kato et al., 2012), this has led to a model whereby stress granule assembly is driven by homo- and/or heterotypic interactions between IDRs and/or prion-related domains on RNA binding proteins (Buchan, 2014).
One hypothesis is that RNP granules are liquid-liquid phase separations, where high concentrations of assembly components reach a critical threshold and then spontaneously assemble into RNP granules through weak multivalent interactions (Brangwynne et al., 2011; Weber and Brangwynne, 2012). This is suggested by RNP granules being dynamic by FRAP (reviewed in Buchan and Parker, 2009), which is explained in the phase separation model by intrinsic weak interactions and surface exchange. In addition, RNP granules are observed to display liquid-like behavior of fission and fusion, and they flow like liquids under mechanical pressure (Brangwynne et al., 2009; Weber and Brangwynne, 2012). Moreover, at high concentrations, IDRs on RNA binding proteins can promote phase separations in vitro (Elbaum-Garfinkle et al., 2015; Nott et al., 2015; Lin et al., 2015; Molliex et al., 2015).
We provide evidence that stress granules contain a dynamic shell-like structure surrounding stable cores. Mass spectroscopy of purified yeast and mammalian stress granule cores identified new components of stress granules, including numerous ATP-dependent helicases and protein remodelers. ATP is required for stress granule assembly and dynamics with the Chaperonin-Containing T complex (CCT complex), a well known protein chaperone complex (Leitner et al., 2012) inhibiting stress granule assembly, while the MCM and RVB helicase complexes, which are known to remodel DNA and/or RNA-protein complexes (Labib et al., 2000; Shen et al., 2000; Shin and Kelman, 2006), promote stress granule persistence. Our observations suggest that stress granules contain a stable core structure and a more dynamic shell with assembly, disassembly and transitions between the core and shell modulated by numerous protein and RNA remodeling complexes. Perturbation of specific steps in these stress granule dynamics could lead to aberrant granules and thereby contribute to degenerative disease.
RESULTS
Stress granules are stable ex vivo
One model for mRNP granule assembly is tjhat they represent liquid-liquid phase separation events where multiple weak interactions between components lead to a spontaneous liquid-liquid phase separation event when the components are at a high enough concentration (Brangwynne et al., 2009; Elbaum-Garfinkle et al., 2015; Weber and Brangwynne, 2012). If stress granules were such phase separations, they would be expected to disassemble when present in dilute lysates. Thus, we prepared lysates from stressed and unstressed yeast cells carrying Pab1-GFP (a stress granule marker (Buchan et al., 2008)) and examined the lysates microscopically for stress granules.
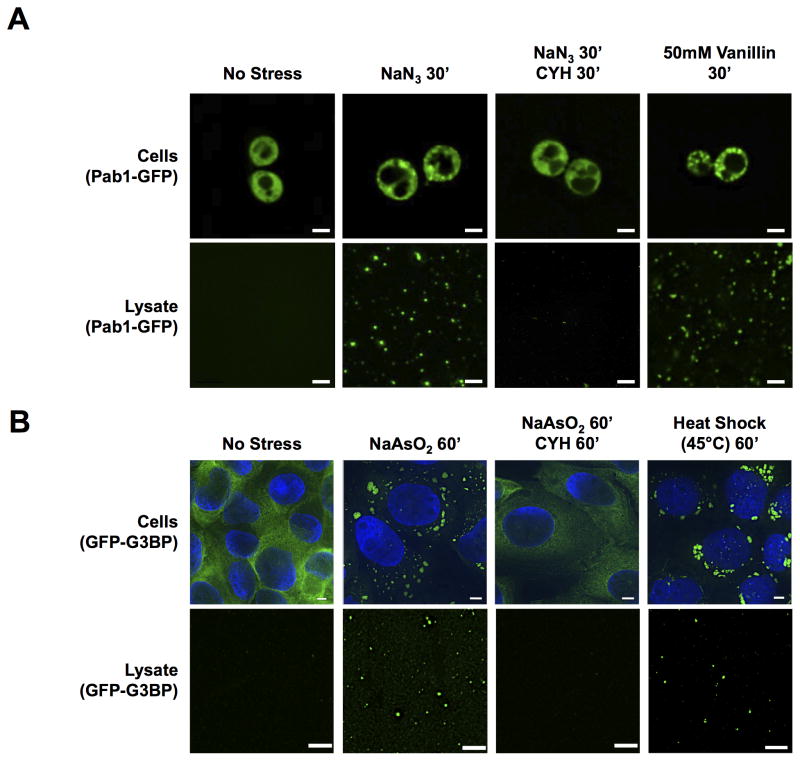
We observed that when yeast cells were treated with Sodium Azide (NaN3, which induces stress granules (Buchan et al., 2011)) prior to lysis, GFP-positive granules were observed in the lysate. GFP granules were absent without stress (Figure 1A). Induction of stress granules with Vanillin (Iwaki et al., 2013) also produced GFP granules in lysates.
Figure 1. Stress Granules are Stable in Cell Lysates.
(A) Pab1-GFP carrying yeast cells and cell lysates from cells ± treatment with NaN3 and ± treatment with Cycloheximide (CYH) or vanillin. (B) GFP-G3BP U-2 OS cells and cell lysates ± treatment with NaAsO2, or heat shock for 60min, ± treatment with CYH. All scale bars are 2μm. See also Figures S1, S3.
Four observations argue that the granules observed in yeast cell lysates are related to stress granules. First, in lysates, Pab1-GFP foci co-localize with other mCherry tagged stress granule markers (Ded1 and Pbp1, Figure S1A). Second, these foci stain with an Alexa647-labeled oligo(dT) probe indicating they contain poly(A+) RNA (Figure S1A). The presence of RNA in these granules is also supported by the observation of poly(A+) RNA in ‘stress granule core enriched fractions’ (Figure S4A) as detected by northern blot analysis using an oligo(dT) probe (Figures S1B), and by staining Pab1-GFP foci with RNA specific dyes in lysates (data not shown). Third, inhibiting stress granule assembly in vivo by the addition of cycloheximide during stress leads to a large decrease in the number of GFP granules observed in lysates (Figure 1A). Fourth, the GFP-tagged Ded1Δ141-150 protein, which causes constitutive stress granules (Hilliker et al., 2011), was in granules in lysates even in the absence of stress, while wild type Ded1-GFP was not (Figure S1C).
We also observed stable assemblies in lysates from U-2 OS mammalian cells expressing a GFP-G3BP fusion (Ohn et al., 2008) (Figure 1B). Evidence that these GFP-G3BP foci are related to stress granules is: 1) they co-localize with known stress granule proteins by antibody staining (PABP1, Figure S1D); 2) they are only present after stress (Figure 1B); 3) their induction is inhibited by addition of cycloheximide (Figure 1B) and they contain poly(A+) RNA as detected by hybridization with Alexa647-labeled oligo(dT) (Figure S1D). Foci of GFP-G3BP are also observed in lysates after heat shock (Figure 1B), which induces mammalian stress granules (Buchan et al., 2013). Thus, at least a core structure of stress granules from yeast and mammalian cells is stable in lysates.
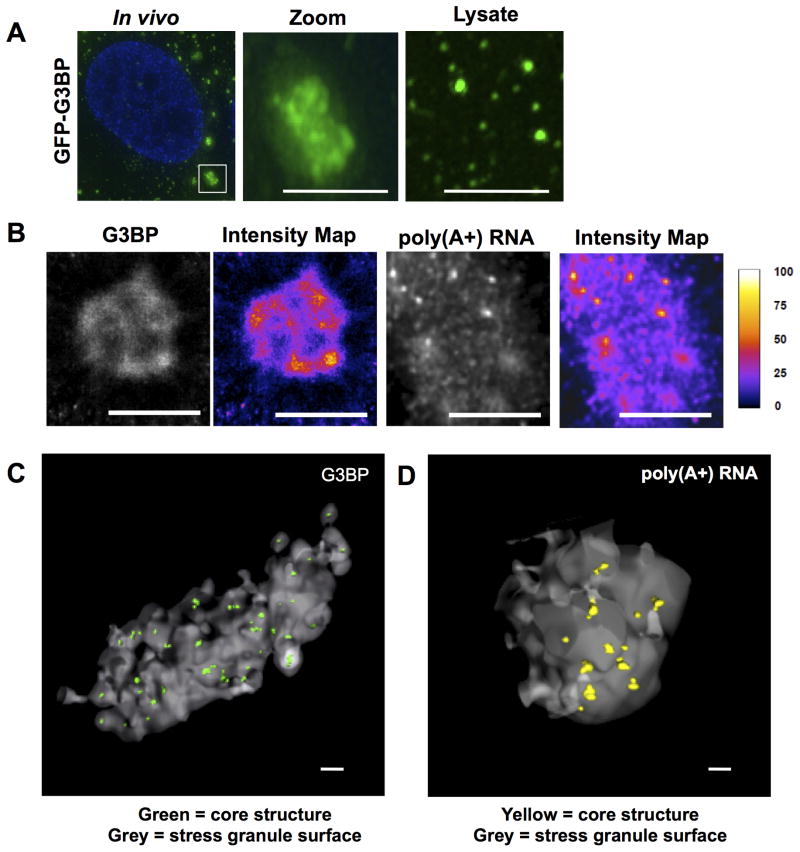
The GFP-G3BP foci in mammalian lysates were smaller than stress granules in cells (Figure 2A) suggesting the stable GFP-G3BP foci observed in lysates might represent a stable ‘core structure’ containing G3BP. Examination of stress granules at higher magnification (Figure 2A), or using super-resolution microscopy (STORM – Stochastic Optical Reconstruction Microscopy or SIM – Structured Illumination Microscopy), reveals G3BP is non-uniform in stress granules with clusters of higher concentration (Figures 2B, S2B & S2C). Similar clusters of higher concentration are also observed in STORM analysis of poly(A+) RNA, as detected by oligo(dT) hybridization (Figure 2B). Non-uniform distribution of PABP1 was also observed in stress granules by SIM (Figure S2B). We refer to these sites of high concentration of RNA or proteins in stress granules as “cores”, and the surrounding less concentrated material as the “shell”.
Figure 2. Super Resolution Microscopy of Mammalian Stress Granules.
(A) Comparison of size between granules in vivo and GFP-G3BP foci in lysates. Middle panel shows zoomed inset. Third panel shows foci in lysate. Scale bars are 2μm. (B) STORM images of stress granules in vivo, showing IF for GFP-G3BP (Alexa647-αGFP) and FISH for poly(A+) RNA (Alexa647-oligo(dT). Intensity map represents relative grey scale intensity. Scale bars represent 2μm. (C) 3D STORM image of a stress granule in vivo, showing IF for GFP-G3BP (Alexa647-αGFP). Grey surface represents surface of a stress granule. Cores are shown in green. Scale bar represents 500nm. (D) 3D STORM image of a stress granule in vivo for poly(A+) RNA (Alexa647-oligo(dT). Grey surface represents surface of a stress granule. Cores are shown in yellow. Scale bar represents 500nm. See also Figure S2 and Movie S1.
We refined the analysis of stress granule substructure by 3D STORM analysis of G3BP and poly(A+) RNA using the Double-Helix Point Spread Function (DHPSF) technique, which enables high precision 3D localization over an extended depth range (Pavani and Piestun, 2008; Pavani et al., 2009) (Figures 2C & 2D; Movie S1). 3D STORM analysis of G3BP and poly(A+) RNA in vivo (at a resolution of ~30nm) revealed cores that had an average diameter of 190.8 ± 28.3 nm for G3BP and 175.3 ± 24.0 nm for poly(A+) RNA. Since this size is much larger than individual RNPs (~30 nm diameter for ribosome), it suggests that each core could contain several mRNPs. The number of cores is linearly correlated with stress granule volume (average of 4.7 cores/μm3 granule volume, r2 = 0.90) demonstrating the consistent nature of these substructures. Based on the density of signal, G3BP and poly(A+) RNA are ~13X more concentrated in the shell than the surrounding cytosol and then concentrated ~30–40X from the shell to the core substructure (Figure S2A).
We determined how the sizes of the GFP-G3BP foci in lysates compare to the core structures seen in STORM images. Examination of the size of the GFP-G3BP foci in U-2 OS cell extracts on a Nanosight Nanoparticle Tracking Analysis system (Figure S2E) revealed a median size distribution of 233.1 ± 18.6 nm (N = 5 runs). This is similar to the size of the core structures observed in STORM images (190.8 ± 28.3) (Figure S2F). Moreover, if the GFP tagged protein only occupies part of the core structure, the apparent volume occupied by GFP by microscopy will be smaller than the hydrodynamic volume measured by particle tracking. This suggests that the core structures from mammalian cells are stable in extracts, while the more diffuse shell disassembles under dilute conditions. Yeast stress granules were measured to be 208.9 ± 56.9 nm in size (median size, N = 8 runs) as assessed by the Nanosight (Figure S2E) and 179.5 +/− 28.9 nm by SIM analysis in cells (Figures S2F & S2G). This suggests that yeast stress granules may be dominated by the core structure, while the larger mammalian stress granules contain multiple cores encased in a larger more unstable shell (see discussion).
Both yeast and mammalian stress granule cores are stable in vitro (yeast granules at 30°C and mammalian granules at 37°C) for at least an hour (Figure S3A). In lysates, they are also resistant to 1M NaCl, 50mM EDTA, or 2M Urea (Figures S3B & S3C; data not shown), arguing they are not assembled solely by electrostatic interactions. Disrupting weak hydrophobic interactions by the addition of 1,6 hexanediol is also not sufficient to disassemble cores in vitro (Figure S3D) (Patel et al., 2007; Ribbeck and Görlich, 2002). However, the addition of both 1M NaCl and 1,6 hexanediol did reduce the number of mammalian cores observed in vitro (Figure S3D) arguing their stability is due to a combination of both electrostatic and hydrophobic interactions. Both mammalian and yeast stress granule cores are largely resistant to RNase treatment in lysates (Figure S3E), suggesting that once formed, protein-protein interactions play a role in maintaining their integrity (see below). Both yeast and mammalian cores are unstable in 2% SDS (Figure S3F), suggesting highly stable β-zipper interactions between prion domains is not the basis of core stability.
Mass spectrometric analysis of yeast stress granule cores
To identify proteins within stress granule cores, we first enriched preparations for cores by successive differential centrifugation followed by affinity purification (Figures S4A & S4B).
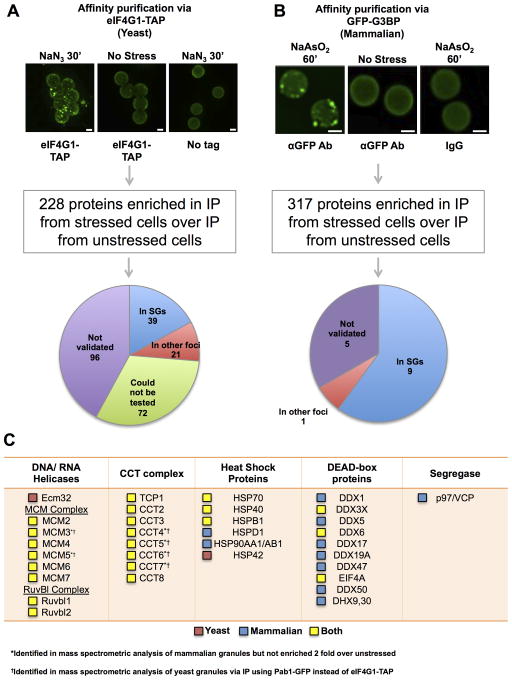
For purification of yeast stress granule cores, rabbit IgG conjugated epoxy Dynabeads were used to pull down cores via a C terminal TAP tag on eIF4G1, a component of yeast stress granules (Brengues and Parker, 2007), from cells also expressing Pab1-GFP. Mass spectrometry identified 228 possible core proteins, as assessed by ≥ 2 fold more peptides as compared to a pull down from an unstressed control, and absence from an untagged control (Figures 3A & S4A, Table S1). This list of proteins was enriched for known stress granule components (e.g. Pab1, Ded1 and 40S Ribosomal proteins (p-value = 1.34 × 10−13).
Figure 3. Mass Spectrometric Analysis of Stress Granule Cores.
(A) Dynabeads bound to immunopurified yeast stress granule cores are shown. Pie chart shows GFP-tagged proteins localized to stress granules by overlap with Ded1-mCherry (see Supplemental Experimental Procedures). (B) Stress granule core enriched fraction from GFP-G3BP U-2 OS cells incubated with αGFP antibody, immunopurified using Protein A Dynabeads. Picture shows isolated granule cores bound to Dynabeads. Pie chart shows result of testing the co-localization of 15 putative stress granule proteins with GFP-G3BP by IF. (A) and (B) Scale bars are 2μm. (C) Table showing various conserved ATPases in stress granules. Red box indicates protein present only in the yeast granule proteome, blue indicates protein present only in the mammalian granule proteome and yellow indicates protein is conserved in both proteomes. See also Figure S4 and Tables S1, S2, S3.
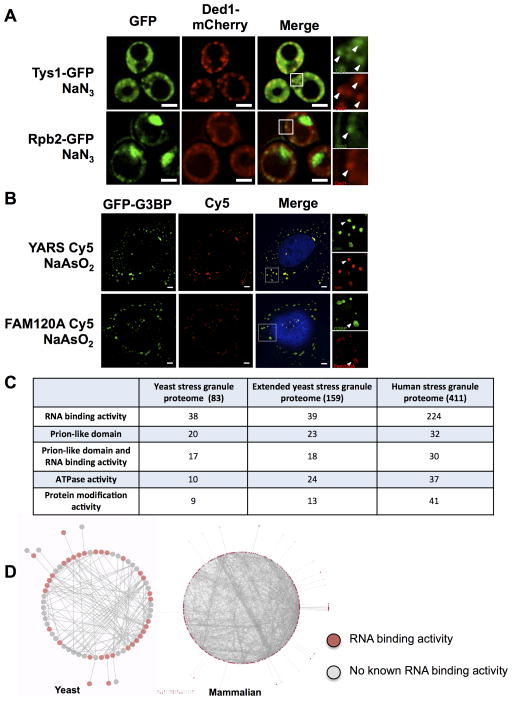
Validation using GFP-tagged versions of 156 of enriched proteins revealed that some showed biased localization in stress granules during stress. Fifty-nine proteins formed foci overlapping to some extent with Ded1-mCherry. Statistical analysis allowed us to rigorously conclude that at least 39 of these 59 overlap with Ded1 and are thus stress granule proteins (Figure 3A; Supplemental Experimental Procedures). These include several novel stress granule components, such as Rpb2, Tys1 and Gus1 (Figures 4A & S5A; Table S1). Seventy-six proteins did not show enrichment in foci during stress. Some of these proteins could localize to stress granules, but not be sufficiently enriched over the cytoplasm to form foci that are significantly brighter than the signal due to proteins distributed in the bulk cytosol. To test this possibility, we examined 2 such proteins (Mcm2 and Mcm4), to see if they concentrated in granules in lysates, where the concentration outside of granules is diluted, allowing greater sensitivity. Even though Mcm2-GFP and Mcm4-GFP were not enriched above the cytoplasmic distribution at Ded1-mCherry foci in cells (data not shown), we observed that they were clearly present in Ded1-mCherry foci in lysates (Figure S4D), demonstrating that there are components of stress granules that are not enriched above the bulk cytosol but still present in these assemblies. Moreover, mixing of two cell extracts with different tagged components of stress granules did not show re-assortment in lysates (Figure S4E), arguing that the composition of these assemblies is largely maintained in cell extracts.
Figure 4. Key Findings from the Stress Granule Proteome.
(A) Fluorescence microscopy of cells with Ded1-mCherry and Tys1-GFP or Rpb2-GFP after NaN3 treatment. (B) IF in NaAsO2 stressed GFP-G3BP U-2 OS cells using antibodies against YARS or FAM120A detected by a Cy5 labeled secondary antibody. Arrowheads within magnified areas indicate examples of overlap. Scale bars are 2μm. (C) Properties of the yeast and mammalian stress granule proteomes. (D) Yeast and mammalian stress granule ‘connectomes’ showing physical interactions amongst stress granule proteins. Red nodes represent known RNA binding proteins. See also Figure S5 and Tables S1, S2.
To the list of proteins identified and validated in stress granules, we added proteins known to be in stress granules but not enriched in our mass spectrometric analysis to create an initial list of yeast stress granule proteins (Table S1). Most known stress granule proteins not identified in our mass spectrometric analysis are translation factors (e.g. eIF4E and eIF3), which would not be enriched since they co-IP with eIF4G1 even in the absence of stress granule formation. We confirmed that some of these proteins are observed in stress granule cores in lysates by microscopy (Figure S4F). We also created an additional extended stress granule proteome by including “provisional stress granule components” (proteins we could not validate due to the absence of detectable GFP signal in cells, and proteins observed to localize in granules in vitro) (Table S1).
Mammalian stress granule cores
We purified mammalian stress granule cores from Sodium Arsenite (NaAsO2) stressed U-2 OS cells using a series of differential centrifugations and then affinity purification of GFP-G3BP (Figures 3B, S4B & S4C). As granules observed in lysates might only be stable cores of granules and may no longer contain some granule components post lysis (Figures 2A & S2), mass spectrometry was performed on granules prepared from both formaldehyde cross-linked cells and non cross-linked cells. The former approach is likely to capture weak protein-protein interactions that might be lost during purification without crosslinking. Mass spectrometry analysis identified 317 proteins from cross-linked and 139 proteins from non cross-linked cells as having ≥ 2 fold spectral counts over IP from unstressed cells (Figure 3B). A similar set of proteins was identified from both cross-linked and non cross-linked cells (p-value of overlap between the two lists <10−99), with the vast majority of proteins seen in the cross-linked sample also detected in the uncross-linked experiment, but with insufficient peptides to make our statistical cut-offs. As cross-linked stress granule preps are likely to more fully represent the stress granule proteome, this list was used for all subsequent analyses. This list of proteins is significantly enriched for previously identified stress granule proteins (p-value = 7.30 × 10−61).
To validate the localization of proteins identified by mass spectroscopy in mammalian stress granules, we used immunofluorescence (IF) in NaAsO2 stressed cells to observe the in vivo localization of 15 proteins not previously known to localize to granules. We observed that 9 out of these 15 proteins overlapped with G3BP granules under stress, and are new stress granule components (Figures 3B, 4B & S5B, Table S2). One protein (RALY) was observed to be in P bodies. P bodies are known to physically associate with stress granules in vivo (Kedersha et al., 2005), which could explain the identification of this protein in pull downs from cross-linked cells. Similar to some yeast granule components, a subset of the remaining 4 proteins might localize to stress granules, but not be sufficiently enriched over the cytoplasm to show foci by IF in vivo. Consistent with this possibility, MCM2, which did not show enrichment in stress granules in cells by IF, was observed to localize to cores in lysates (Figure S4G). Based on this validation, we suggest a large fraction of proteins identified by mass spectrometry are stress granule components. By adding known stress granule components not identified by mass spectrometry to this list, we created the first stress granule proteome for a mammalian cell (Table S2).
Properties of the Stress Granule Proteome
Analysis of the yeast and mammalian stress granule proteomes reveals a significant overlap between their compositions (Figure 2C; Table S3) (p-value = 2.10 × 10−99). Both yeast and mammalian stress granules are enriched in translation factors (yeast corrected p-value = 7.37 × 10−24; mammalian corrected p-value = 3.88 × 10−76) and RNA binding proteins (yeast: corrected p-value = 6.95 × 10−8; mammals: corrected p-value = 8.84 × 10−31) (Figure 4C). This list is also enriched for proteins with predicted Prion Like Domains (PrLDs) (yeast: p-value = 1.81 × 10−18; mammalian: p-value = 9.61 × 10−13). Proteins with mRNA binding activity and proteins with predicted PrLDs are highly overlapping (17/20 stress granule proteins with a predicted PrLD have known mRNA binding activity in yeast; p-value = 5.53×10−5, 30/32 of mammalian stress granule proteins with predicted PrLD bind mRNA; p-value = 1.77 × 10−6). Surprisingly, almost half of the stress granule components neither bind mRNA nor contain PrLDs (42/83 in yeast and 185/411 in mammalian stress granules).
The stress granule proteome forms a dense network of protein-protein interactions
Both yeast and mammalian stress granule proteomes form a dense network of protein-protein interactions (Figure 4D). On average, each mammalian and yeast stress granule protein physically interacts with 8.32 and 4.25 other stress granule proteins respectively, higher than expected by chance (p-value = 5.77 × 10−54 and p-value < 10−100). These observations suggest multiple and specific protein-protein interactions function in stress granule assembly.
A similar bioinformatics analysis of previously known yeast P body proteins revealed that in contrast to stress granules, a majority of P body proteins have mRNA binding activity (~73%), which suggests that mRNA-protein interactions may contribute more to P body structure than stress granule structure (Table S1). However, like stress granule proteins, P body proteins also displayed a high degree of interconnectivity (on average, each P body protein interacts with 7.65 other P body proteins, data not shown). Interestingly, in a force-directed layout of a network consisting of both yeast P body and stress granule proteins, P body and stress granule proteins were segregated (Figure S5C). Also, proteins that localize to both granules (such as Dhh1 and Scd6) appeared at the interface of these two populations (Buchan and Parker, 2009). This predicted docking/overlap is consistent with the frequent overlap and/or docking of stress granules with P bodies seen in yeast (Buchan et al., 2008) (Figure S5C).
Connections of mammalian stress granules to human disease
The mammalian stress granule proteome reveals new connections between stress granules and disease. Numerous known stress granule proteins associated with Amyotrophic Lateral Sclerosis (ALS) or Frontal Temporal Lobar Dementia (FTLD) (Li et al., 2013; Ramaswami et al., 2013) are detected in our preparations (Figure S5D; Table S2). In addition, we identify new stress granule components DPYSL3, DCTN1, and TUBA4A, which have been suggested to modify ALS susceptibility (Blasco et al., 2013; Münch et al., 2004; Smith et al., 2014).
We identify 75 stress granule components linked to 153 Mendelian diseases (Table S2) with enrichments in three broad disease categories: neurological (e.g. motor neuron disease, p-value = 10−8), muscular (e.g. myopathy, p-value = 10−5), and cardiovascular (e.g. cardiomyopathy, p-value = 10−6) (Figure S5D). Several newly identified stress granule proteins, when mutated cause Charcot-Marie-Tooth neuropathy (CMT), the most common hereditary neuromuscular disease. For example, dominant mutations in the tyrosyl-tRNA synthetase (YARS) and the chaperone HSPB1 both cause CMT (Evgrafov et al., 2004; Jordanova et al., 2006) and are localized to stress granules in yeast and mammalian cells (Figures 4A & 4B; Table S3). Intriguingly, mutations in YARS and HSPB1 both cause adult-onset CMT with a predominant motor neuropathy and motor neuron involvement (Rossor et al., 2012), suggesting that aberrant stress granule formation may be a broader theme in central and peripheral nervous system diseases.
New conserved components of stress granules
Comparison of the yeast and mammalian stress granule proteomes identified new conserved members of stress granules (Table S3). For example, tRNA synthetases are found in stress granules in both yeast and mammals (Figures 4A & 4B; Table S3), consistent with tRNA synthetases binding and regulating mRNAs (Castello et al., 2012; Frugier and Giegé, 2003; Mitchell et al., 2013). Ribosome biogenesis factors are conserved components of stress granules (NOP58, FTSJ3, NOP2 and FBL in mammalian cells; Kap123, Rio2, Fap7 and Cam1 in yeast), which might reflect a role for stress granule formation in the down-regulation of ribosome biogenesis during stress (Tsang and Zheng, 2007). Finally, a number of ATP dependent protein/nucleic acid remodeling complexes including protein chaperones (e.g. Hsp70/Hsp40 and CCT complex), and multiple RNA and DNA helicases (e.g. DEAD-box proteins, MCM and RVB helicases) are found in yeast and mammalian stress granules (Figures 3C, S4D, S4G, S5A, S7G & S7H; Tables S1 & S2).
Granule assembly and dynamicity require ATP
The presence of numerous ATPases in stress granule cores suggested that stress granules might be modulated by the activity of these ATPases. Thus, we examined the contribution of ATP to stress granule assembly and dynamics by adding 2-deoxyglucose (2DG) and Carbonyl Cyanide m-Chlorophenyl Hydrazone (CCCP) to block the glycolytic pathway and oxidative phosphorylation respectively, thereby depleting ATP. These treatments led to a reduction in cellular ATP levels within 5 minutes as assessed by a GFP-based ATP sensor (Figure S6A) (Berg et al., 2009).
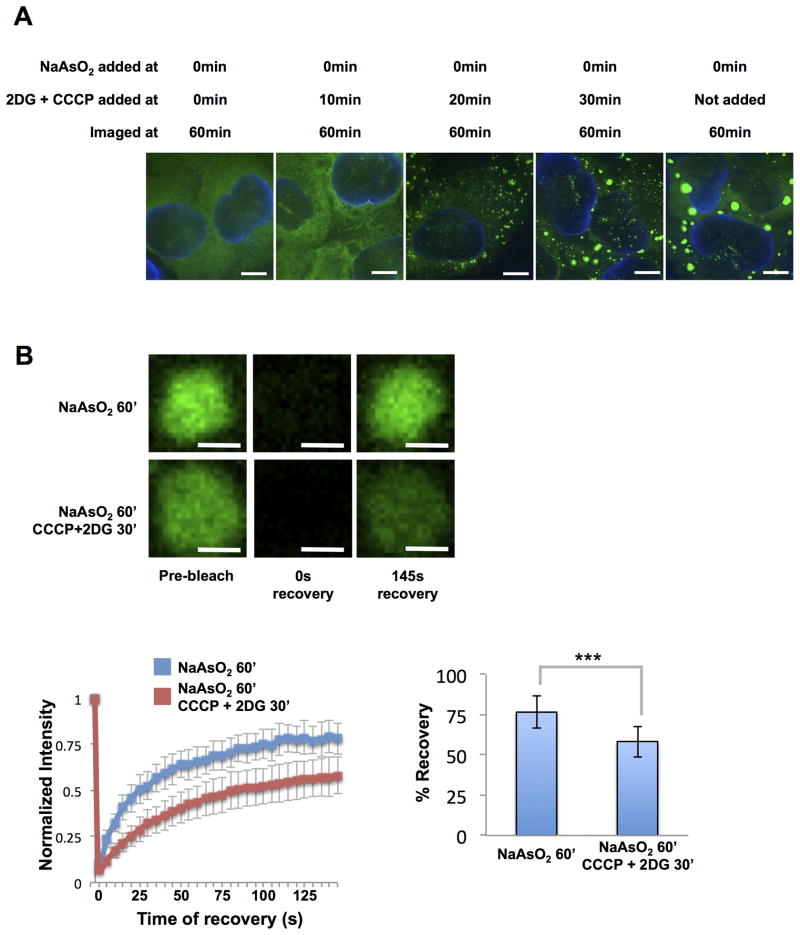
We observed that ATP is required for stress granule formation since the addition of 2DG and CCCP at the same time, or 10min after NaAsO2, completely blocked stress granule assembly (Figure 5A). Small granules were observed when ATP production was blocked 20min after the addition of NaAsO2, even though stress granules had not begun to form in cells where ATP was not depleted (Figure S6B). Beyond 30min of treatment with NaAsO2, the addition of CCCP and 2DG made little difference to granule formation (Figures 5A and S6B). This indicates that ATP dependent events are required for stress granule formation, which could be movement of mRNPs to sites of stress granule formation by motors (Loschi et al., 2009), or remodeling of mRNPs to load critical components, promoting stress granule assembly on untranslating mRNAs.
Figure 5. ATP is Required for Stress Granule Assembly and Dynamicity in vivo.
(A) Different times at which 2DG and CCCP were added to these cells are indicated. Scale bars are 5μm. (B) Granules shown prior to, at 0s after and at 145s after photobleaching. Cells were either treated for 60min with NaAsO2 or with NaAsO2 with 2DG and CCCP added 30min after the addition of NaAsO2. Graph shows recovery curves as an average of 15 granules ± standard deviation. Scale bars are 2μm. Bar graph shows average total percentage recovery from 15 granules ± standard deviation. *** p-value <0.0001. See also Figure S6 and Movies S2, S3.
Two observations indicate ATP affects the dynamics of stress granules post-assembly. First, granules in cells treated with NaAsO2 for 60 minutes were observed to move and fuse in the cytoplasm (Movie S2), while granules in ATP depleted cells were static and showed no fusion events (Movie S3). Second, FRAP analysis revealed that stress granules in cells treated with 2DG and CCCP showed reduced total recovery of GFP-G3BP signal and increased t1/2 of recovery (t1/2 = 37.5s) as compared to cells with normal levels of ATP (t1/2 = 22.5s; p-value < 0.0001) (Figure 5B). Thus, ATP is required to fully maintain the exchangeable pool of G3BP in stress granules and to allow for liquid-like behavior of these assemblies.
CCT, MCM and RVB ATPase complexes modulate stress granule assembly/disassembly
The modulation of stress granule assembly and dynamics by ATP argues that cellular ATPases affect stress granules. We determined the roles of some of the new conserved stress granule ATPases by examining stress granule formation (following 30′ of NaN3 treatment) and disassembly (after washing out the NaN3) in yeast strains with temperature sensitive or hypomorphic alleles in the CCT, MCM and RVB complexes.
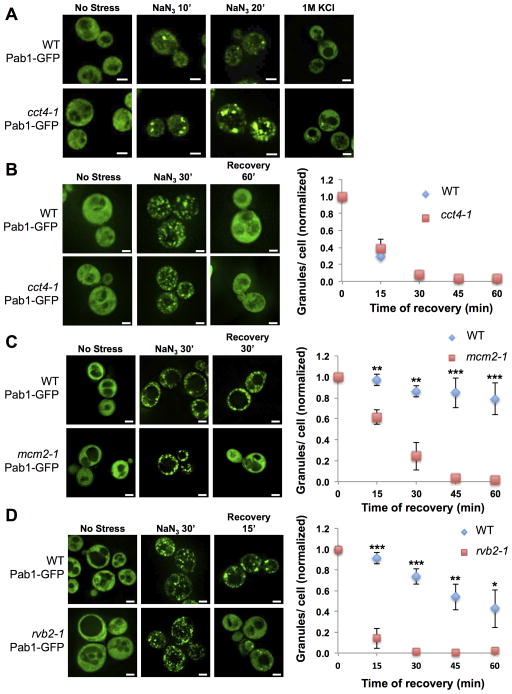
Strains with the temperature sensitive cct4-1 allele formed larger and more stress granules when stressed as compared to isogenic wild-type controls at the non-permissive temperature (Figure 6A). This phenotype is more evident after shorter treatments with NaN3 (10min, 20min) or under conditions of milder stress such as 1M KCl. Hypomorphic alleles in the ATPase active sites of the CCT subunits (D to E mutations in CCT2, CCT3, CCT5, CCT6, CCT7 and CCT8) also led to the formation of increased numbers of stress granules (Figures S7A, S7B). However, granule disassembly after washing out NaN3 remained unaffected in cct4-1 cells (Figure 6B) and in cells with the hypomorphic alleles (data not shown) as compared to wild type cells. These observations argue that the CCT complex inhibits the assembly of stress granules.
Figure 6. Inhibition of Cct4, Mcm2 or Rvb2 ATPase Activity Affects Stress Granule Assembly/Disassembly.
(A) WT or cct4-1 cells carrying Pab1-GFP imaged after shifting to 37°C (60min) either before NaN3 stress, after NaN3 treatment for 10min, after NaN3 treatment for 20min or with 1M KCl for 30min. (B) WT or cct4-1 cells carrying Pab1-GFP imaged prior to stress, after 30min of NaN3, after 60min of recovery at 37°C. Graph shows mean from 3 experiments ± standard deviation. (C) WT and mcm2-1 yeast cells with Pab1 GFP prior to stress, after stress (30min), and upon recovery from stress. Graph shows mean from 3 experiments ± standard deviation. (D) Same as (C) but for rvb2-1 and the corresponding WT yeast cells. * p-value < 0.05. ** p-value < 0.01. *** p-value < 0.0001. White scale bars indicate 2μm. See also Figure S7.
In contrast, yeast strains with temperature sensitive alleles in the MCM (mcm2-1) or RVB (rvb2-1) complexes formed a similar number of granules as wild type cells at the non-permissive temperature during stress (Figures 6C, 6D & S7C). However, granules in the mcm2-1 and rvb2-1 cells disassembled faster than in wild-type controls upon recovery from stress (Figures 6C & 6D). siRNA knockdown of MCM and RVB components in U-2 OS cells also led to a faster loss of stress granules during recovery (Figures S7D, S7E, & S7F). These effects on granule dynamics are likely to be direct since we confirmed their localization to both yeast and mammalian stress granules (Figures S4D, S4G, S7G & S7H). The MCM and RVB complexes are generally thought to act on DNA to remodel chromatin for both DNA synthesis and transcription (Labib et al., 2000; Shen et al., 2000). However, a role for these complexes in modulating RNP granules is rational since the MCM complex can bind RNA in vitro (Shin and Kelman, 2006), RUVBL2 (one of the two components of the human RVB complex) cross links to mRNA in vivo (Castello et al., 2012), and the RVB complex has been suggested to modulate snoRNP assembly (Kakihara and Houry, 2012). Given that the MCM and RVB complexes are suggested to serve as remodelers of protein-nucleic acid complexes, they may limit stress granule disassembly by either promoting a late step in granule assembly leading to more stable granules, or by inhibiting granule disassembly.
DISCUSSION
Stress granules contain core structures
Several observations suggest that stress granules contain stable cores and a dynamic shell. First, super-resolution microscopy of mammalian stress granules reveals regions of higher concentrations of G3BP, PABP1 or poly(A+) RNA, of approximately 200 nm diameter within stress granules. These cores are surrounded by the bulk of the stress granule wherein components are at lower concentrations (Figure S2A). Second, stress granules typically contain both mobile and immobile fractions as assessed by FRAP (Figure 5B) revealing that proteins within these granules can be in two distinct states. Third, a substructure of stress granules is stable in lysates (Figures 2 & S2), and this substructure is of similar size (as assessed by Nanoparticle Tracking Analysis) as the cores observed in super-resolution microscopy. An attractive hypothesis is that the cores within stress granules are relatively stable while the shell of mammalian stress granules is highly dynamic. Since yeast stress granules are of similar size in cells and in lysates, yeast stress granules may have proportionally smaller dynamic shells than mammalian stress granules. The presence of stable cores in stress granules argues that these assemblies are not solely liquid-liquid phase separations, which is consistent with their limited circularity when examined at higher resolution (Figures S2C & S2D).
Three observations suggest that the liquid-liquid phase separation aspect of a stress granule is limited to the “shell” surrounding the stable core structures. First, several IDRs on RNA binding proteins can promote liquid-liquid phase separations in vitro (Elbaum-Garfinkle et al., 2015; Nott et al., 2015; Lin et al., 2015; Molliex et al., 2015) indicating a propensity of these domains to promote this separation. Second, even when cells are depleted of ATP we still observe that a fraction of GFP-G3BP is rapidly exchanging, which is consistent with surface exchange from a phase separation based on dynamic weak interactions, although we cannot formally rule out that some exchange is due to residual ATP. Third, a phase-separated shell would be expected to disassemble in lysates, thereby providing a biochemical explanation for the smaller size of stress-dependent structures in lysates. Finally, the fact that yeast stress granules, which we suggest are primarily stable cores, are less sensitive than mammalian stress granules to 1,6 hexanediol in cells (Kroschwald et al., 2015), is consistent with hexanediol disrupting weak interactions that allow liquid-liquid phase separations, but not disrupting stable core structures (Figure S3D). Stable cores within granules surrounded by a dynamic shell may be a common feature of RNP granules since the nucleolus is suggested to consist of both a liquid-liquid phase separated shell and a stable fibrillarin core (Brangwynne et al., 2011).
The formation of stable cores within stress granules could be important for assembling mRNPs into a more stable structure allowing tighter control of mRNP localization. In addition, cores might be comprised of related mRNPs and thereby allow their coordinate regulation, analogous to the clustering of co-regulated genes in the nucleus (Rinn and Guttman, 2014). This type of organization may explain the clustering of specific mRNAs seen in Drosophila maternal mRNP granules (Trcek et al., 2015).
A striking feature of the stress granule proteomes is that the components of granules constitute a dense network of interacting proteins, which has two important corollaries. First, the dense set of protein-protein interactions could form the basis of a redundant set of interactions that drive stress granule or core assembly. This is supported by genetic analyses in yeast, which have failed to identify any single gene that is essential for stress granule formation (Buchan et al., 2008; Yang et al., 2014; Yoon et al., 2010). This interaction network argues that some proteins will be recruited to stress granules independent of binding RNA, which is consistent with the observation that roughly half of the proteins in the yeast or mammalian stress granule proteomes are not known to bind RNA (Castello et al., 2012; Mitchell et al., 2013) (Figure 4C; Tables S1 & S2).
Several observations indicate that stress granules are modulated by a variety of ATP driven remodeling complexes. First, we identified a number of energy driven protein/nucleic acid chaperones or remodeling complexes as stress granule components (Figure 3C). Second, stress granule assembly was dependent on ATP (Figure 5A), and once assembled, stress granules required ATP for movement and fusion (Movies S2 & S3). Third, when ATP is depleted, a greater pool of G3BP in stress granules becomes immobile (Figure 5B). Since many stress granule components have both mobile and immobile pools (reviewed in Buchan and Parker, 2009), we suggest that stress granules consist of both a stable core and a more dynamic pool of molecules which rapidly exchange with the surrounding cytosol. Moreover, a predominant role of ATP would be to maintain a higher percentage of the proteins in the dynamic pool. This role of ATP might counterbalance the propensity of IDRs on granule components to form stable amyloid fibers when present at high concentrations (Kato et al., 2012; Lin et al., 2015; Molliex et al., 2015).
Additional evidence that stress granules are modulated by remodelers comes from genetic experiments. Since strains defective in the CCT complex form more stress granules while granule clearance during recovery is unaffected (Figures 6A & 6B), we suggest the CCT complex plays a role in inhibiting granule assembly, perhaps by limiting interactions between polyQ domains on stress granule components (Tam et al., 2006). In contrast, defects in MCM or RVB complexes lead to normal numbers of stress granules, but faster disassembly during stress recovery (Figures 6C, 6D, S7E & S7F). One possibility is that MCM and RVB complexes normally inhibit granule disassembly by the removal of stress granule disassembly factors. A second possible model is that MCM and RVB complexes could utilize ATP to remodel granule mRNPs to facilitate the binding of proteins required for stable stress granule assembly. Thus the absence of MCM/RVB ATPase activity would lead to the assembly of a granule with fewer interactions, which could disassemble faster than normal granules. In either case, these results highlight that ATP dependent remodeling complexes can both promote and inhibit stress granule persistence.
Other ATPases also affect stress granule dynamics. For example, defects in the AAA ATPase Cdc48/VCP lead to persistence of stress granules during recovery and inhibit autophagy of stress granules (Buchan et al., 2013). Specific mutations in the RNA helicase Ded1 can also trap mRNAs in yeast stress granules and ATP hydrolysis appears to be required for stress granule release of mRNAs into translation (Hilliker et al., 2011). Finally, defects in Hsp70/Hsp40 complexes can lead to persistence of yeast stress granules during recovery (Walters et al., 2015). This diversity of ATPases affecting stress granules suggests that they will work on different stress granule components, and/or may differentially affect shell or core assembly. Interestingly, defects in the yeast Hsp40 protein Ydj1 appear to primarily affect stress granule disassembly to promote translation while defects in the Hsp40 protein Sis1 preferentially affect autophagy of stress granules (Walters et al., 2015). This implies that stress granules will not be uniform assemblies but the action of different remodeling complexes will create a variety of states, with potentially different biological outcomes.
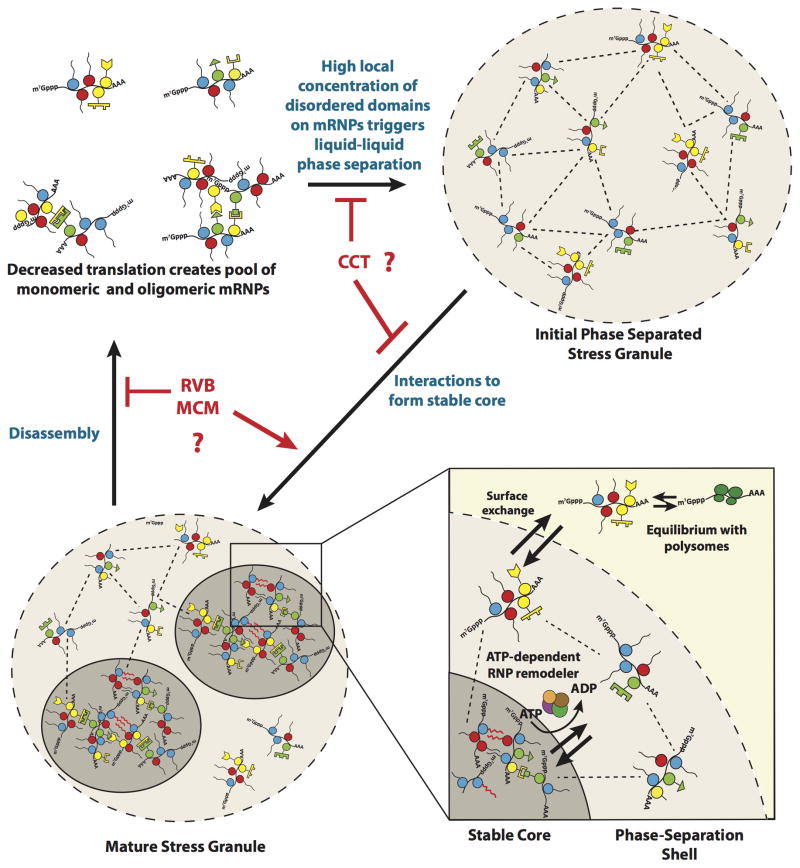
Based on the observations herein, and the literature, a working model for stress granule formation and dynamics can be developed with the following key points (Figure 7). First, following inhibition of translation initiation and ribosome run-off, the pool of untranslating mRNPs bound by a variety of mRNA binding proteins with IDRs increases. The increased concentration of such IDRs on individual, oligomerized, or subcellular concentrations of mRNPs could then lead to a liquid-liquid phase separation (Elbaum-Garfinkle et al., 2015; Nott et al., 2015; Lin et al., 2015; Molliex et al., 2015). Since the higher concentration of proteins within phase separations can promote protein-protein interactions in vitro (Lin et al., 2015; Molliex et al., 2015), one possibility is that the higher concentration of mRNPs within an initial stress granule shell promotes both traditional protein-protein interactions, as well as potential fiber formation by IDRs, thereby leading to the formation of stable cores. However, it remains possible that mRNPs first oligomerize into stable cores followed by the assembly of a shell around these cores and merger into larger microscopically visible stress granules. Protein chaperones and RNA-protein remodeling complexes are hypothesized to maintain a dynamic exchange between the stable core and phase-separated shell of the stress granule. This energy dependent remodeling of the stable core may be critical to limit the formation of extensive amyloid-like fibers by IDRs on RNA binding proteins, which could be toxic (Kim et al., 2013; Li et al., 2013; Ramaswami et al., 2013). During stress recovery, the resumption of translation and continued disassembly of stress granules by various ATPases would lead to components dissipating. An interesting implication of most ATPase machines affecting only assembly or disassembly is that granule assembly and disassembly may be distinct and different processes, which can now be elucidated by understanding the specific roles of different remodeling complexes in stress granule dynamics.
Figure 7. Model for Stress Granule Assembly and Dynamicity.
Dashed lines between mRNPs represent weak physical interactions in the Phase-Separated Shell. Red wavy lines represent strong interactions between Prion-Like Domains. Possible sites of activity of the CCT, RVB and MCM complexes are also shown.
EXPERIMENTAL PROCEDURES
Yeast and U-2 OS Growth Conditions and Reagents
Yeast strains, and plasmids are described in Table S4. Standard methods for yeast for growth and manipulation, and microscopy, were used and are described in detail in Supplementary Experimental Procedures. Standard methods were used for the growth and microscopy of U-2 OS cells. Antibodies used for immunofluorescence, and siRNAs used for knock-downs are given in Table S4.
Purification of Stress Granule Cores
Detailed protocols for purification of yeast and mammalian stress granule cores are provided in Supplementary Experimental Procedures. In brief cells were grown, lysed and multiple differential centrifugation spins were used to create a “stress-granule core enriched fraction” from which stress granule cores were affinity purified using antibodies and Dynabeads. Purified stress granule cores were then analyzed by mass spectroscopy to identify protein components (see Supplementary Experimental Procedures for details).
2DG and CCCP treatment and FRAP analysis
U-2 OS cells were grown in 96 well plates. At times indicated, media was replaced with media containing 0.5mM NaAsO2; 200mM 2DG, 100μM CCCP or 0.5mM NaAsO2, 200mM 2DG and 100μM CCCP for the times indicated in Figure 5. Cells were imaged using a Nikon AR1 LSM confocal microscope. For time lapse images, cells were fixed at time points given in Figure 5, as described in Buchan et al (Buchan et al., 2013) (these images were taken using the Deltavision). Fluorescence recovery after photobleaching (FRAP) experiments were done as follows: bleach areas, approximately the size of granule (~2μm), were determined. Images were taken every 5s during recovery. Mean intensity within similar sized areas was determined at each time point using ImageJ (It). Mean intensity within a different part of the cytoplasm was also measured (IBt) to correct for bleaching during image acquisition. Corrected mean intensity at each time point was determined by taking the ratio: It/IBt. Curve for exponential recovery was determined using MATLAB.
Supplementary Material
Acknowledgments
We thank the Drubin, Horovitz. Tye, Kralj and Kohno labs for strains and plasmids. We thank Anne Webb, Carolyn Decker, Sarah Mitchell, David Protter and the Parker Lab for helpful discussions and feedback on the manuscript. We thank the CU Mass Spectrometry Facility; CU Biofrontiers and MCDB microscopy cores. This work was funded by NIH-F30N2093682 (JRW), NSF- IIP- 1353638 (Double Helix LLC), NIH-GM045443 (RP) and the Howard Hughes Medical Institute (RP). SIM in MCDB was made possible by equipment supplements to R01 GM79097 (D. Xue) and P01 GM105537 (M. Winey).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, S.J., J.R.W. and R.P.; Methodology, S.J., J.R.W. and R.P.; Investigation, S.J., J.R.W., R.W.W., A.A. and A.B.; Software, S.J., J.R.W., A.A. and A.B.; Writing – Original Draft, S.J., J.R.W. and R.P.; Writing – Review & Editing, S.J., J.R.W., R.W.W. and R.P.; Funding Acquisition, J.R.W. and R.P.; Supervision, R.P.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nature Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco H, Bernard-Marissal N, Vourc’h P, Guettard YO, Sunyach C, Augereau O, Khederchah J, Mouzat K, Antar C, Gordon PH, et al. A rare motor neuron deleterious missense mutation in the DPYSL3 (CRMP4) gene is associated with ALS. Hum Mutat. 2013;34:953–960. doi: 10.1002/humu.22329. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. mRNP granules: Assembly, function, and connections with disease. RNA Biol. 2014;11 doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Molecular Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon JH, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. Journal of Cell Science. 2011;124:228–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CCH, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL, Schagina O, Verpoorten N, Van Impe K, Fedotov V, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- Frugier M, Giegé R. Yeast aspartyl-tRNA synthetase binds specifically its own mRNA. Journal of Molecular Biology. 2003;331:375–383. doi: 10.1016/s0022-2836(03)00767-8. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Chen Y, Zhou X, Kar A, Ray P, Chen X, Rao EJ, Yang M, Ye H, Zhu L, et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-Box Protein Ded1 Modulates Translation by the Formation and Resolution of an eIF4F-mRNA Complex. Molecular Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki A, Ohnuki S, Suga Y, Izawa S, Ohya Y. Vanillin inhibits translation and induces messenger ribonucleoprotein (mRNP) granule formation in saccharomyces cerevisiae: application and validation of high-content, image-based profiling. PLoS ONE. 2013;8:e61748. doi: 10.1371/journal.pone.0061748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes & Development. 2013;27:2628–2641. doi: 10.1101/gad.227843.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- Kakihara Y, Houry WA. The R2TP complex: discovery and functions. Biochim Biophys Acta. 2012;1823:101–107. doi: 10.1016/j.bbamcr.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4 doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Leitner A, Joachimiak LA, Bracher A, Mönkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;20:814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Molecular Cell. 2015 doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. Journal of Cell Science. 2009;122:3973–3982. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, Prudlo J, Peraus G, Hanemann CO, Stumm G, et al. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Molecular Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Pavani SRP, Piestun R. High-efficiency rotating point spread functions. Opt Express. 2008;16:3484–3489. doi: 10.1364/oe.16.003484. [DOI] [PubMed] [Google Scholar]
- Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N, Twieg RJ, Piestun R, Moerner WE. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proceedings of the National Academy of Sciences. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MAM, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. Journal of Cell Science. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Görlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. The EMBO Journal. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J, Guttman M. RNA Function. RNA and dynamic nuclear organization Science. 2014;345:1240–1241. doi: 10.1126/science.1252966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossor AM, Kalmar B, Greensmith L, Reilly MM. The distal hereditary motor neuropathies. J Neurol Neurosurg Psychiatr. 2012;83:6–14. doi: 10.1136/jnnp-2011-300952. [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Shin JH, Kelman Z. The replicative helicases of bacteria, archaea, and eukarya can unwind RNA-DNA hybrid substrates. J Biol Chem. 2006;281:26914–26921. doi: 10.1074/jbc.M605518200. [DOI] [PubMed] [Google Scholar]
- Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp S, Kenna KP, Scotter EL, Kost J, Keagle P, Miller JW, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84:324–331. doi: 10.1016/j.neuron.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Zheng XFS. TOR-in(g) the nucleus. Cell Cycle. 2007;6:25–29. doi: 10.4161/cc.6.1.3675. [DOI] [PubMed] [Google Scholar]
- Walters RW, Muhlrad D, Garcia J, Parker R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. Rna. 2015 doi: 10.1261/rna.053116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Yang X, Shen Y, Garre E, Hao X, Krumlinde D, Cvijović M, Arens C, Nyström T, Liu B, Sunnerhagen P. Stress granule-defective mutants deregulate stress responsive transcripts. PLoS Genet. 2014;10:e1004763. doi: 10.1371/journal.pgen.1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Choi EJ, Parker R. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J Cell Biol. 2010;189:813–827. doi: 10.1083/jcb.200912019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.