Abstract
Unlike cell lines, human hematopoietic stem cells (HSCs) are less efficiently transduced with HIV-1 vectors, potentially limiting this approach. To investigate which step (internalization, reverse transcription, nuclear transport, and integration) limits lentiviral transduction, we evaluated the kinetics of lentiviral transduction in human CD34+ cells. We transduced HeLa and CD34+ cells with self-inactivating HIV-1 vector at low and 10-fold higher MOIs, and evaluated vector amounts at various timepoints based upon the rationale that if a given step was not limiting, 10-fold greater vector amounts would be obtained at the 10-fold higher MOI. We observed slower internalization (>60 minutes), a peak of reverse transcription at 24 hours, and completion of integration at 3 days in CD34+ cells. In HeLa cells, vector amounts at high MOI achieved ~10-fold greater values over all timepoints. When compared to HeLa cells, CD34+ cells had a larger difference of vector amounts between high and low MOIs at 2–6 hours and a smaller difference at 12 hours to 10 days, revealing a limitation in human CD34+ cell transduction around 12 hours, which corresponds to reverse transcription. In serial measurements of reverse transcription at 24 hours, vector amounts didn’t decrease once detected among CD34+ cells. When using an HSC expansion medium, we observed less limitation for starting reverse transcription and more efficient transduction among CD34+ cells in vitro and in xenografted mice. These data suggest that initiation of reverse transcription mainly limits lentiviral transduction for human CD34+ cells. Our findings provide an avenue for optimizing human CD34+ cell transduction.
Keywords: lentiviral vector, human hematopoietic stem cells, transduction efficiency
Introduction
Hematopoietic stem cell (HSC) targeted gene therapy has the potential to correct various types of disorders, and recently several groups reported efficacy in clinical trials, mainly for hereditary immunodeficiency diseases [1–6]. However, improvement of transduction efficiency for human HSCs remains crucial for further development of gene therapy especially for non-immunodeficiency diseases, such as the hemoglobin disorders [7, 8]. Human immunodeficiency virus type 1 (HIV-1) based lentiviral vectors were pursued for efficient transduction for HSCs and long-term gene expression in vivo due to the ability of HIV-1 vectors to transduce non-dividing cells and integrate into host cell chromosomes [9–11]. However, even when using HIV-1 vectors, transduction efficiency for human HSCs is less than various cell lines and mouse HSCs [12–14]. Generally, HIV-1 vectors can transduce almost 100% of cells in various cell lines (including HeLa cells), while around 10–40% of transduction efficiency is achieved among human CD34+ cells, even at high multiplicity of infection (MOI) [14]. Therefore, we sought to investigate the step(s) accounting lower transduction efficiency for human CD34+ cells with an HIV-1 vector.
In contrast to earlier versions of HIV-1 vectors, current versions contain self-inactivating (SIN) long-terminal repeats (LTRs) to inactive the LTRs after integration. They are pseudotyped with other envelopes, such as a vesicular stomatitis virus glycoprotein G (VSVG) envelope [9], to allow transduction of not only T-lymphocytes but also other types of cells, including human CD34+ cells [15].
Before integrated HIV-1 provirus translates viral genes in infected cells, it involves four essential steps: (1) internalization of genomic RNA from the HIV-1 virion into cells, (2) reverse transcription of genomic RNA into DNA, (3) transport of genomic DNA from the cytoplasm to the nucleus, and (4) integration of genomic DNA into the host cell chromosomes [16]. We focused on these 4 steps to investigate which step(s) limit lentiviral transduction for human CD34+ cells with HIV-1 vectors.
Methods
Lentiviral transduction for HeLa cells and human CD34+ cells
An HIV-1 based lentiviral vector encoding enhanced green fluorescent protein (GFP) with a VSVG envelope was prepared, as previously described [14, 15, 17]. Lentiviral titers (transduction units/ml) were calculated by the proportion of GFP-positive cells (%GFP) using a HeLa cell line, as previously described [12, 14]. This viral titer was used to calculate MOI for both HeLa and CD34+ cells. HeLa cells (5×104) were plated in 12 well dish containing Dulbecco's Modified Eagle Medium supplemented with 10% fetal bovine serum in triplicate (n=3). After overnight culture, HeLa cells were transduced with the HIV-1 vector at MOI 0.5 or 5 with 8µg/ml polybrene. After one day exposure (and every 2–3 days), the media were changed to fresh media.
Human CD34+ cells were enriched from peripheral blood stem cells mobilized by granulocyte colony-stimulating factor under a protocol approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Disease [13, 14]. Human CD34+ cells (1×105) were cultured on RetroNectin (Takara, Shiga, Japan)-coated 12 well plates containing X-VIVO10 media (Lonza, Allendale, NJ, USA) with stem cell factor (SCF), fms-related tyrosine kinase 3 ligand (FLT3L), and thrombopoietin (TPO) (all 100ng/ml; R&D Systems, Minneapolis, MN, USA) in triplicate (n=3) [13, 14]. After overnight prestimulation, human CD34+ cells were transduced with a GFP-expressing HIV-1 vector at MOI 5 or 50 with fresh X-VIVO10 media containing the same cytokines. After one day exposure (and every 2–3 days), the media were changed to fresh media with the same cytokines. Additionally, a Stemline II medium (Sigma-Aldrich, St. Louis, MO, USA) was utilized instead of an X-VIVO10 medium to compare transduction efficiency for human CD34+ cells. X-VIVO10 and Stemline II serum-free media contain human serum albumin and no cytokines. Additionally, the X-VIVO10 medium contains human insulin and transferrin. Both media were supplemented with SCF, FLT3L, and TPO.
%GFP was evaluated by flow cytometry (FACSCalibur; BD Biosciences, Franklin Lakes, NJ, USA) for HeLa cells (14 days after viral exposure) and human CD34+ cells (2–3 days after viral exposure).
Reverse transcription (RT) and quantitative polymerase chain reaction (qPCR)
For analysis of vector RNA and DNA amounts in transduced HeLa and human CD34+ cells, transduced cells were collected at different time points from 5 minutes to 10 days. Total RNA and DNA were extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and QIAamp DNA Blood Mini Kit (Qiagen), respectively. Extracted RNA was reverse transcribed into complementary DNA (cDNA) using reagents and random hexamers from SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies, Grand Island, NY, USA). Using cDNA and extracted DNA, HIV-1 specific sequences (around HIV-1 packaging signal) were amplified using LV2 probe and primers, as previously described [12, 18]. The vector DNA amounts in human CD34+ cells were evaluated by SIN-LTR probe and primers (detecting integrated SIN HIV-1 LTR), which were designed to detect integrated vector DNA but not vector plasmids to prevent overestimation from vector plasmid contamination [19]. TaqMan Ribosomal RNA control reagents (Applied Biosystems, Foster City, CA, USA) were used for standardization. Vector genome amounts were determined using the ΔCt method, compared to RNA in HeLa cells at 5 minutes after viral exposure at MOI 0.5 or DNA in a control cell line including one vector copy per cell [19].
In addition, we evaluated serial steps of reverse transcription in transduced HeLa and CD34+ cells at 24 hours after viral exposure [20], in which we detected an early phase of reverse transcription with early LTR probe and primers (HIV-R forward primer: 5’-AGA TCT GAG CCT GGG AGC-3’, SIN-LTR probe: 5’-ACA CTA CTT GAA GCA CTC AAG GCA AGC-3’, and SIN-LTR reverse primer: 5’-GTC TGA GGG ATC TCT AGT TAC C-3’) [19], middle phases of reverse transcription with GFP probe and primers [14, 21] and LV2 probe and primers [12, 18], and a late phase of reverse transcription with late LTR probe and primers (HIV-R forward primer, SIN-LTR probe, and HIV-PS reverse primer: 5’-CCT CTG GTT TCC CTT TCG C-3’) [19].
Nuclear isolation from transduced HeLa cells and human CD34+ cells
We transduced HeLa cells at MOI 5 and human CD34+ cells at MOI 50 with the HIV-1 vector in the same manner. Nuclear fractions in transduced HeLa and human CD34+ cells were isolated using a sucrose gradient, as previously described [22]. DNA was extracted from nuclear fractions, and vector signals were amplified by qPCR.
Humanized xenograft mouse model
We used male NOD/SCID/IL2Rγnull mice (NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ; Jackson Laboratory, Bar Harbor, ME), following the guidelines set out by the Public Health Services Policy on Humane Care and Use of Laboratory Animals under a protocol approved by the Animal Care and Use Committee of the National Institute of Diabetes and Digestive and Kidney Diseases, as previously described [13]. Mice were injected with 35mg/kg busulfan (Busulfex; PDL BioPharma, Redwood City, CA) two days before transplantation (n=3–4) [23]. Following overnight prestimulation, human CD34+ cells (2×106 cells per mouse) were transduced with the GFP-expressing HIV-1 vector at MOI 50 in X-VIVO10 media or Stemline II media with SCF, FLT3L, and TPO, and the next day, these cells were injected into the tail vein. The transduction conditions for CD34+ cells were optimized using HSC transplantation models for both xenograft mice and rhesus macaques [13, 14]. Following transplantation, peripheral blood samples were obtained to evaluate %GFP among human CD45+ cells (human CD45-PE antibody, clone HI30; BD Biosciences) over a period of 24 weeks. Three to four mice were evaluated for each group at all experimental time points.
Statistical Analysis
Statistical analyses were performed using the JMP 11 software (SAS Institute Inc., Cary, NC, USA). Two averages were evaluated by the student’s t-test. The ratios of vector amounts at high MOI to low MOI were evaluated by two-way analysis of variance. A p value of <0.01 or 0.05 was deemed significant. Standard errors of the mean are shown as error bars in all figures. We transduced HeLa cells and a single donor of CD34+ cells in triplicate (n=3) and performed qPCR for each DNA extracted from the transduced cells in triplicate.
Results
Evaluation of RNA internalization with an HIV-1 vector in HeLa and human CD34+ cells
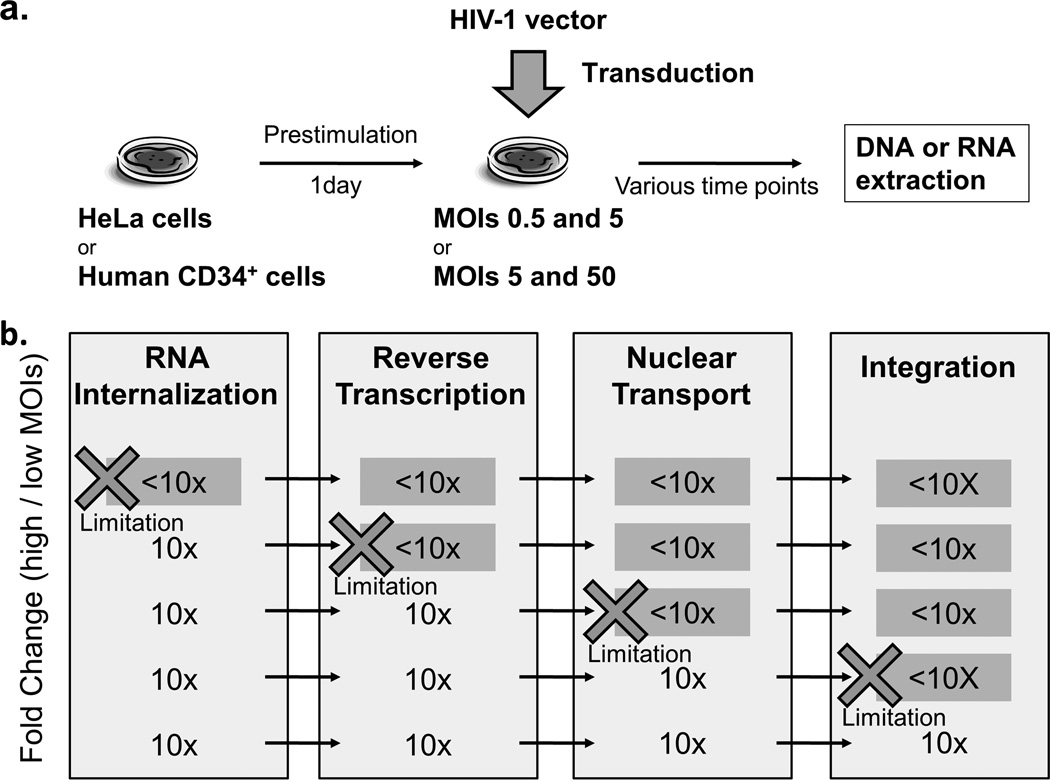
To determine whether a limitation step exists in lentiviral transduction for human CD34+ cells, we evaluated kinetics of vector amounts in both HeLa cells and human CD34+ cells, which were transduced with a GFP-expressing HIV-1 based lentiviral vector at low and 10-fold higher MOIs (Figure 1a). We transduced HeLa cells at low MOI 0.5 and high MOI 5, while human CD34+ cells were transduced at low MOI 5 and high MOI 50 (based on our prior work which demonstrated lentiviral transduction in HeLa cells was much more efficient than in human CD34+ cells [13]). Vector genome amounts were determined by qPCR using RNA and DNA samples extracted from the transduced cells over time. We assayed 4 transduction steps (internalization, reverse transcription, nuclear transport, and integration) based upon the rationale that if a given step was not limiting, 10-fold greater vector amounts would be obtained from samples transduced at the 10-fold higher MOI (Figure 1b). The ratios of vector genome at high MOI to low MOI were compared between HeLa and human CD34+ cells.
Figure 1. Approach to determine a limitation step in human CD34+ cell transduction.
(a) We evaluated vector amounts in both HeLa and human CD34+ at various time points, which were transduced with a self-inactivating HIV-1 vector at low and 10-fold higher multiplicity of infection (MOI, 0.5 and 5 for HeLa cells, and 5 and 50 in CD34+ cells). Amounts of vector genomes were determined by quantitative PCR using RNA and DNA samples. (b) We assayed each of four transduction steps (internalization, reverse transcription, nuclear transport, and integration) based upon the rationale that if a given step was not rate limiting, a 10-fold greater value would be obtained at the 10-fold higher MOI. The ratios of vector genome amounts at high to low MOIs were compared between HeLa cells and CD34+ cells to determine whether a limiting step exists for CD34+ cell transduction. When there are potential limiting steps in transduction for human CD34+ cells, the ratio of vector genome amounts at high to low MOIs would be <10-fold.
We evaluated transduction efficiency in both HeLa and human CD34+ cells using %GFP determined by flow cyotometry. Theoretically, following lentiviral transduction at MOI 0.5, around half of exposed HeLa cells would obtain one (or sometimes two) copy(s) of vector with GFP expression. We observed 50.8±2.7% at MOI 0.5 and 90.8±2.1% at MOI 5 in HeLa cells, while in human CD34+ cells, %GFP was 8.8±0.5% at MOI 5 and 38.9±0.6% at MOI 50. When we compared %GFP between both cells at the same MOI 5, 10.3-fold higher %GFP was observed in HeLa cells as compared to human CD34+ cells (p<0.01), confirming less efficient transduction in human CD34+ cells with an HIV-1 vector. HeLa cells can be suitable for an efficient transduction control for comparison to human CD34+ cells.
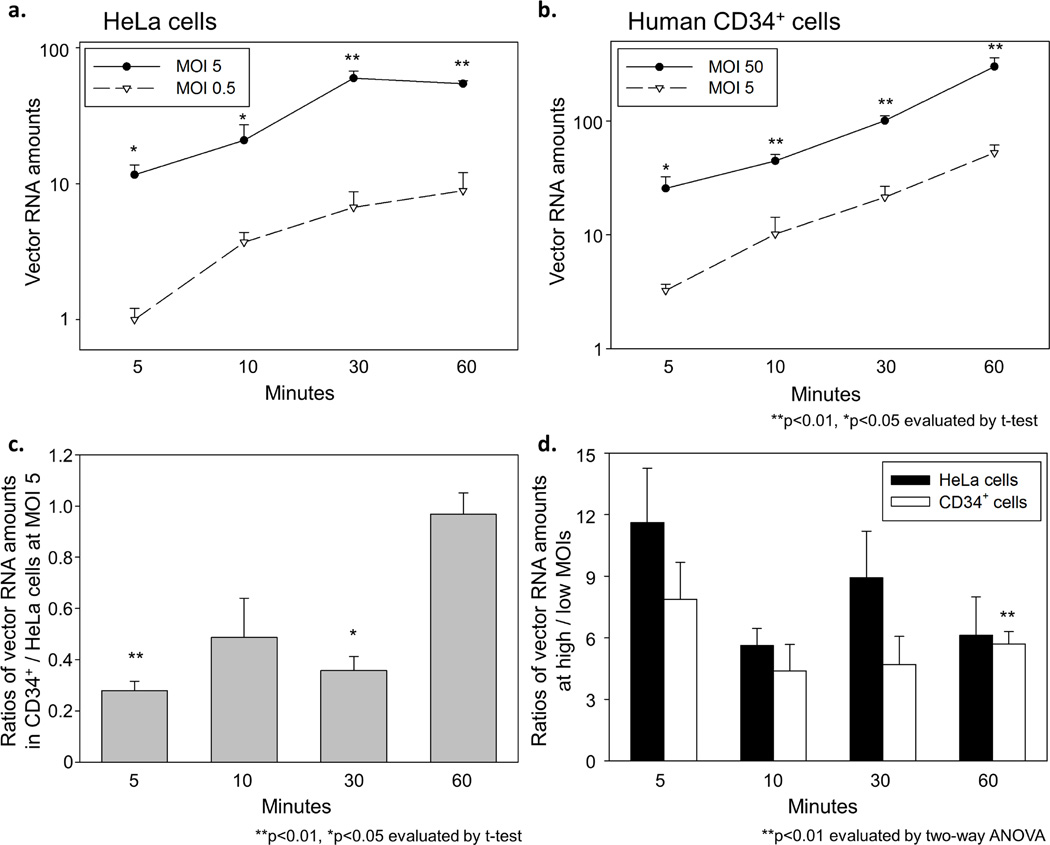
To compare internalization of the vector genome between HeLa and human CD34+ cells, we determined vector RNA amounts at 5, 10, 30, and 60 minutes after viral exposure by using RT-qPCR. The vector RNA amounts in HeLa cells rapidly increased for 30 minutes after viral exposure and then plateaued at 30 minutes (Figure 2a). In human CD34+ cells, a gradual increase of vector RNA amounts was observed at both MOIs over 60 minutes (Figure 2b). We then compared vector RNA amounts of CD34+ cells to HeLa cells at the same MOI 5, which resulted in lower vector RNA amounts (2.1–3.6 fold) at 5–30 minutes (p<0.05 at 5 and 30 minutes) and equivalent vector RNA amounts (1.0-fold) at 60 minutes (Figure 2c), suggesting slower internalization in human CD34+ cells as compared to HeLa cells. When we compared vector RNA amounts at high to low MOIs for both cells, vector RNA amounts at high MOI achieved around 10-fold (or slightly less than 10-fold) greater values than at low MOI over 60 minutes (5.6–11.6 fold in HeLa cells and 4.4–7.9 fold in CD34+ cells) (Figure 2d). Equivalent or slightly smaller differences between high and low MOIs were observed in human CD34+ cells at all time points, compared to HeLa cells (p<0.01 at 60 minutes). These data suggest that despite slower internalization of vector genomic RNA, similar amounts of vector RNA can enter human CD34+ cells, as compared to HeLa cells. High MOI transduction can partially overcome the restriction in vector internalization for both cells. Internalization may not be a major limiting step in human CD34+ cell transduction.
Figure 2. Evaluation of vector RNA internalization in transduced HeLa and human CD34+ cells.
(a) We determined vector RNA amounts in the cells for 5–60 minutes after viral exposure. In HeLa cells, vector RNA amounts rapidly increased and then plateaued at 30 minutes. (b) In human CD34+ cells, vector RNA amounts gradually increased over 60 minutes. (c) We calculated vector RNA ratios of CD34+ cells to HeLa cells at same MOI 5, which resulted in low vector RNA ratios (0.28–0.49) at 5–30 minutes and around 1 of vector RNA ratio at 60 minutes. (d) When we calculated the ratios of vector RNA amounts at high to low MOIs, the vector RNA ratios were slightly less than 10-fold in both cells over 60 minutes. Equivalent or slightly lower ratios were observed in human CD34+ cells, compared to HeLa cells. These data suggest that similar amounts of vector genome can enter human CD34+ cells even with slower internalization, as compared to HeLa cells.
Evaluation of reverse transcription, nuclear transport, and integration with an HIV-1 vector in HeLa and human CD34+ cells
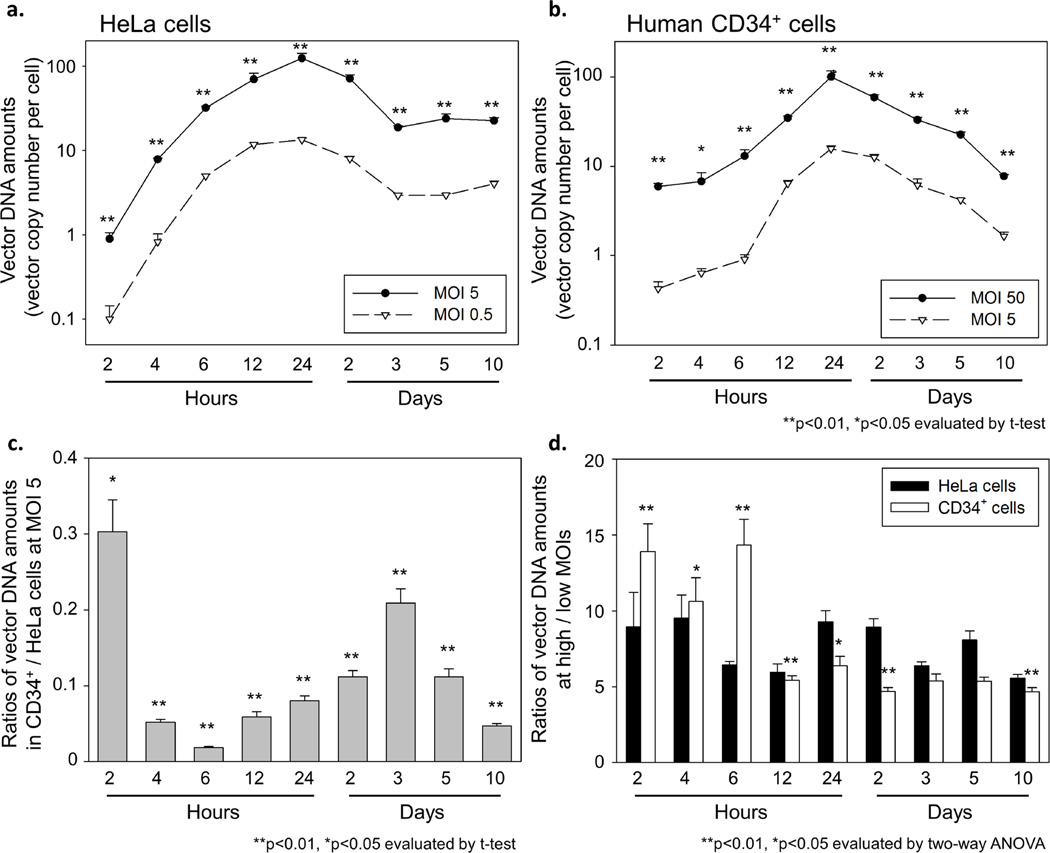
To compare reverse transcription, nuclear transport, and integration of vector genome between HeLa and human CD34+ cells, we determined vector DNA amounts from 2 hours to 10 days after viral exposure by using qPCR. In HeLa cells, the vector DNA amounts peaked at 24 hours at both high and low MOIs (MOI 0.5 and 5), corresponding to reverse transcription, and reached a plateau level at 3 days at both MOIs, corresponding to nuclear transport and integration (Figure 3a). In human CD34+ cells, we observed a gradual increase of vector DNA amount with a peak at 24 hours at both MOIs (MOI 5 and 50), and the vector DNA amounts decreased until 10 days at both MOIs (Figure 3b). When we compared vector DNA amounts in CD34+ cells versus HeLa cells at the same MOI 5, much lower vector DNA amounts (3.3–55.3 fold) were observed in human CD34+ cells (Figure 3c), which represented a larger difference than vector RNA amounts between HeLa and human CD34+ cells (1.0–3.6 fold), suggesting that a step in reverse transcription limits lentiviral transduction for human CD34+ cells. The difference of vector DNA amounts between HeLa and CD34+ cells increased at 6 hours and decreased until 3 days, maybe due to slower internalization and/or slower reverse transcription. When we compared vector DNA amounts at high to low MOIs, vector DNA amounts at high MOI in HeLa cells remained around 10-fold (or slightly less than 10-fold) greater than at low MOI over 10 days (5.6–9.5 fold) (Figure 3d). In human CD34+ cells, the difference of vector DNA amounts between high and low MOIs was larger at 2–6 hours (p<0.05) and smaller at 12 hours to 10 days (p<0.05 except 3 and 5 days), compared to HeLa cells. These data suggest that human CD34+ cell transduction is limited by a step in reverse transcription around 12 hours after viral exposure.
Figure 3. Evaluation of vector DNA amounts in transduced HeLa and human CD34+ cells.
(a) To evaluate reverse transcription, nuclear transport, and integration, we determined vector DNA amounts from 2 hours to 10 days after viral exposure. In HeLa cells, vector DNA amounts peaked at 24 hours (corresponding to reverse transcription), and reached a plateau level at 3 days (nuclear transport and integration). (b) In human CD34+ cells, we observed a slower increase of vector DNA amounts with a peak at 24 hours, and the vector DNA amounts decreased until 10 days. (c) When we calculated vector DNA ratio of CD34+ cells to HeLa cells at same MOI 5, the vector DNA ratios were much lower than vector RNA ratio of CD34+ cells to HeLa cells at same MOI. The vector DNA ratios decreased for 6 hours and increased until 3 days, maybe due to slower internalization and/or reverse transcription. (d) When we calculated the ratios of vector DNA amounts at high to low MOIs, the vector DNA ratios in HeLa cells remained slightly less than 10-fold over 10 days. In human CD34+ cells, higher vector DNA ratios at 2–6 hours and lower ratios at 12 hours to 10 days were observed, as compared to HeLa cells. These data suggest that a step in reverse transcription around 12 hours is a major limit in lentiviral transduction for human CD34+ cells.
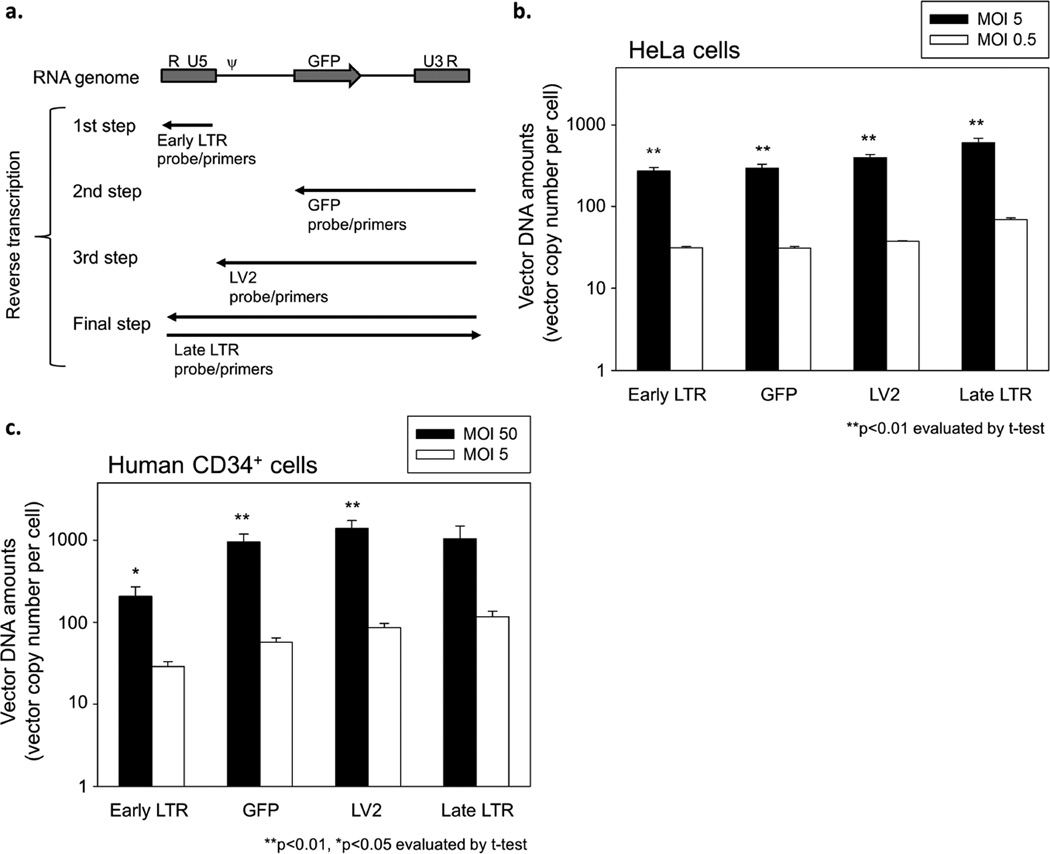
To evaluate serial steps of reverse transcription in lentiviral transduction, we designed specific probe and primers, which can detect an initial phase of reverse transcription with early LTR probe and primers, the middle phases with GFP or LV2 probe and primers, and the final phase with late LTR probe and primers (Figure 4a). Using these probes and primers, we evaluated vector DNA amounts in both HeLa cells (MOI 0.5 and 5) and human CD34+ cells (MOI 5 and 50) at 24 hours after viral exposure. Similar amounts of vector DNA were observed among all probe and primer sets in both HeLa and CD34+ cells at the same MOI (Figures 4b and c). The vector amounts at high MOI achieved around 10-fold greater values than at low MOI among all probe and primers in both HeLa and CD34+ cells. These data suggest that no limiting step exists once reverse transcription starts, and initiation of reverse transcription may limit lentiviral transduction in human CD34+ cells.
Figure 4. Evaluation of serial steps of reverse transcription in transduced HeLa and human CD34+ cells.
(a) We designed specific probe/primers to detect serial steps of reverse transcription in lentiviral transduction, in which we can evaluate an initial phase of reverse transcription by early LTR probe/primers, the middle phases by GFP probe/primers and LV2 probe/primers, and the final phase by late LTR probe/primers. Using these probe/primers, we evaluated vector DNA amounts in both HeLa cells (MOI 0.5 and 5) and human CD34+ cells (MOI 5 and 50) at 24 hours after viral exposure. (b and c) We detected similar amounts of vector DNA among all probe/primer sets in both HeLa cells (b) and CD34+ cells (c) at the same MOI. The ratios of vector amounts at high MOI to low MOI were around 10 among all probe/primers in both HeLa and CD34+ cells. These data suggest that no limiting step exists once reverse transcription starts, and initiation of reverse transcription (conversion from RNA to DNA) may limit lentiviral transduction for human CD34+ cells. ψ: packaging signal.
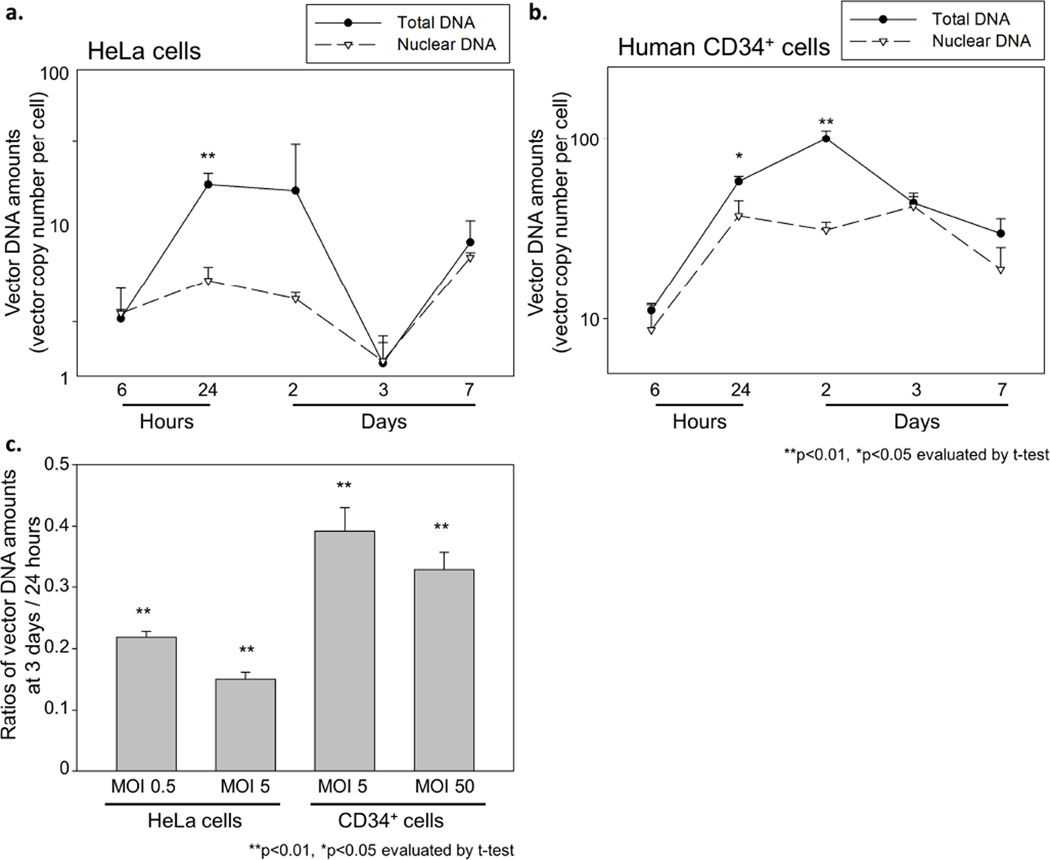
To determine the timing of nuclear transport and integration, we compared vector DNA amounts between total DNA and nuclear DNA. Nuclear DNA was extracted from nuclear fractions of both HeLa cells (MOI 5) and CD34+ cells (MOI 50) after viral exposure, isolated by a sucrose gradient. In both HeLa and CD34+ cells, we observed lower vector DNA amounts in nuclear DNA at 24 hours to 2 days as compared to total DNA (p<0.05 except HeLa cells at 2 days), while the vector amounts were equivalent between nuclear DNA and total DNA at 3–7 days (Figures 5a and b), suggesting that nuclear transport and integration are completed by around 3 days. To evaluate efficiency of nuclear transport and integration, we compared vector DNA amounts at 3 days (after integration) to 24 hours (a peak of reverse transcription), which was in 15–22% in HeLa cells and 33–39% in human CD34+ cells (Figure 5c). These data suggest similar (or slightly higher) efficiency of nuclear transport and integration in human CD34+ cells, as compared to HeLa cells. Nuclear transport and integration are not a major limiting step in lentiviral transduction for human CD34+ cells.
Figure 5. Evaluation of nuclear transport of vector DNA.
We compared the vector DNA amounts between total DNA and nuclear DNA in transduced HeLa cells (a) and human CD34+ cells (b). Nuclear DNA was extracted from the nuclear fractions of transduced cells, which were isolated by a sucrose gradient. In both cells, the vector amounts in nuclear DNA were lower than total DNA at 24 hours to 2 days, and similar at 3–7 days, suggesting that nuclear transport and integration are completed until 3 days. (c) We calculated vector DNA ratios of 3 days (after integration) to 24 hours (a peak of reverse transcription), and observed similar DNA ratios between HeLa and CD34+ cells. These data suggest that efficiency of nuclear transport and integration is similar between HeLa and CD34+ cells.
Hematopoietic stem cell expansion increases reverse transcription in human CD34+ cells
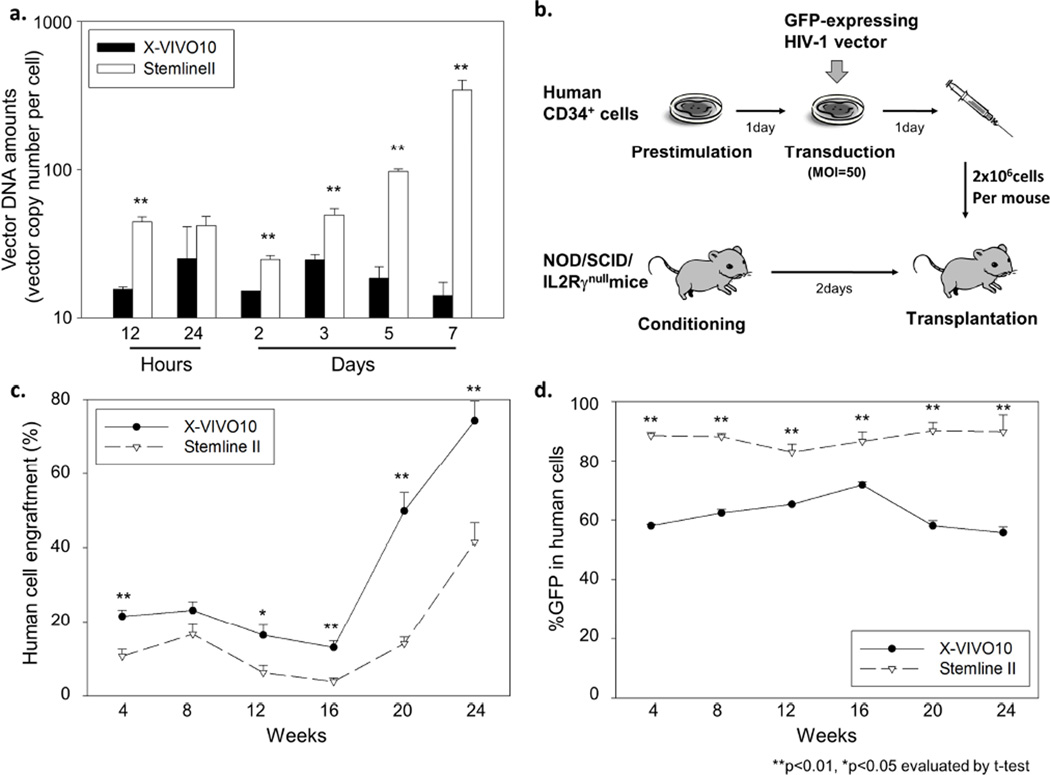
To compare lentiviral transduction between different culture conditions among the same donor’s human CD34+ cells, we utilized an X-VIVO10 medium (as our standard) or a Stemline II medium (with greater expansion), and evaluated vector DNA amounts in transduced CD34+ cells from 12 hours to 7 days after viral exposure. In CD34+ cells transduced in Stemline II media, 2-fold higher cell expansion was observed 3 days after viral exposure, compared to X-VIVO10 media (4.08±0.36 fold vs. 1.89±0.36 fold, p<0.01). Lentiviral transduction in Stemline II media resulted in higher vector DNA amounts in CD34+ cells in vitro at all time points, even at 12 hours, as compared to X-VIVO10 media (p<0.01 at all time points except 24 hours, Figure 6a). These data suggest that more efficient reverse transcription results from human CD34+ cell expansion during transduction.
Figure 6. A hematopoietic stem cell expansion medium increases reverse transcription in human CD34+ cells.
(a) We compared vector DNA amounts in human CD34+ cells transduced in an X-VIVO10 medium (our standard) or a Stemline II medium (greater expansion). Transduction in Stemline II media resulted in higher vector DNA amounts even at 12 hours after viral exposure, suggesting stronger reverse transcription in Stemline II media. (b) Human CD34+ cells were transduced with a GFP-expressing HIV-1 vector at MOI 50, and the transduced cells were transplanted into immunodeficient mice (NOD/SCID/IL2Rγnull mice) following sublethally conditioning. (c and d) We evaluated the proportions of human CD45-positive cells (human cell engraftment) and GFP-positive cells (%GFP) in peripheral blood cells of xenografted mice. Ex vivo transduction in Stemline II media resulted in higher %GFP in human cells and lower human cell engraftment.
In addition, to evaluate transduction efficiency for human hematopoietic repopulating cells, we transduced human CD34+ cells ex vivo with a GFP-expressing HIV-1 vector at MOI 50 in either X-VIVO10 or Stemline II medium, and the transduced cells were transplanted into immunodeficient mice (NOD/SCID/IL2Rγnull mice) (Figure 6b). The proportion of human CD45-positive cells (human cell engraftment) and %GFP in human cells were evaluated in peripheral blood cells every 4 weeks until 24 weeks after transplantation. Ex vivo transduction in Stemline II media resulted in higher %GFP in human cells at all time points, compared to X-VIVO10 (p<0.01), while human cell engraftment was reduced by Stemline II media (p<0.05 at all time points except at 8 weeks, Figures 6c and d). These data suggest that a Stemline II HSC expansion medium can enhance reverse transcription, which results in more efficient transduction in human CD34+ cells in vitro and in xenografted mice; however, this occurred at the cost of reduced engraftment ability.
Discussion
In this study, we demonstrate the kinetics of HIV-1 based lentiviral vector transduction in human CD34+ cells at various time points between low and high MOIs, and show that initiation of reverse transcription is a major limiting step in human CD34+ cell transduction, as compared to HeLa cells. The timing of nuclear transport and integration was evaluated by vector amounts in nuclear DNA in human CD34+ cells. Human CD34+ cell culture in Stemline II HSC expansion media enhanced reverse transcription as compared to our standard X-VIVO10 media, which resulted in more efficient transduction for human CD34+ cells in vitro and in xenografted mice.
Internalization in HIV-1 infection was reported to occur during 1 hour after in vitro exposure [24], while internalization in vesicular stomatitis virus is a fast process within 2–3 minutes [25–27]. Before starting the current work, we expected that internalization of the HIV-1 vector should occur in a few minutes, since our HIV-1 vector was pseudotyped with a VSVG envelope. To our surprise, internalization of the HIV-1 vector was slower in both HeLa cells (~30 minutes, Figure 2a) and human CD34+ cells (>60 minutes, Figure 2b). When we calculated the vector RNA ratios at high to low MOIs (Figure 2d), the vector RNA ratios were slightly less than 10-fold in both cells (5.6–11.6 fold in HeLa cells and 4.4–7.9 fold in CD34+ cells), and similar or slightly lower in human CD34+ cells than HeLa cells. These data suggest that the internalization step slightly limits lentiviral transduction; however, it is not a major limiting step for human CD34+ cell transduction.
Previous reports demonstrated that after in vitro HIV-1 exposure, reverse transcription starts at 5–8 hours [28], and viral DNA integration occurs as early as at 18 hours [28]. Separate reports estimated that during in vivo HIV-1 infection of human T-cells, reverse transcription took up to 33 hours, and viral integration, about 5 hours [29]. In addition, slower reverse transcription was reported in non-dividing cells, such as G0-arrested fibroblast cells, macrophages, and quiescent lymphocytes [11, 30, 31], and reverse transcription was detected at 36–48 hours in macrophages in vitro (nearly non-dividing) [30]. We observed an increase of vector DNA amounts started by reverse transcription at 4 hours after viral exposure, and the vector amounts reached a peak at 24 hours, which is consistent with long period of reverse transcription (Figure 3b). These data also suggest that our cytokine stimulation in an X-VIVO10 medium is sufficient to induce HIV-1 reverse transcription in human CD34+ cells, which should be important for efficient transduction. After 24 hours of viral exposure, vector DNA amounts decreased in both cell types, which can be explained by that reverse transcription has completed, and the transduction process has progressed to nuclear transport and integration. When we evaluated the ratios of vector DNA amounts at high to low MOIs, we found that the vector DNA ratios in human CD34+ cells started at 13.9-fold (2 hours) and gradually decreased to 4.7-fold (10 days), while the vector DNA ratios in HeLa cells remained slightly less than 10-fold (5.6 to 9.5-fold) at all time points (Figure 3d). The lower vector DNA ratios at 12 hours to 10 days in human CD34+ cells suggest that reverse transcription occurring around 12 hours is a major limitation in lentiviral transduction for human CD34+ cells. Additionally, we evaluated serial steps of reverse transcription at 24 hours after viral exposure (early, middle, and late steps), which resulted in similar vector amounts among all steps (Figures 4b and c). These data suggest that there is no limiting step during reverse transcription once it starts, and initiation of reverse transcription (conversion from RNA to DNA) may limit lentiviral transduction in human CD34+ cells.
We then performed nuclear separation and evaluated vector DNA amounts in nuclear fractions. In both cells, the vector amounts in nuclear DNAs were lower at 24 hours to 2 days and similar at 3–7 days, as compared to total DNA (Figures 5a and b). These data suggest that no preintegration complex of vector genome remains in transduced cells at 3 days after viral exposure. This is consistent with the data that the vector DNA amounts were stabilized at 3 days in HeLa cells (Figure 3a). To evaluate efficiency of nuclear transport and/or integration, we compared vector DNA amounts of 3 days (after integration) to 24 hours (a peak of reverse transcription), and demonstrated similar efficiency between HeLa cells (0.15–0.22) and CD34+ cells (0.33–0.39) (Figure 5c). These data suggest that both nuclear transport and integration are not major limiting steps in lentiviral transduction for human CD34+ cells.
It is well-known that HIV-1 vectors can transduce non-dividing cells in contrast to γ-retroviral vectors [11, 32], and the HIV-1 central DNA flap is crucial for nuclear transport and transduction in human CD34+ cells [33, 34]. Our HIV-1 vector construct contains the DNA flap component, and we obtained efficient transduction (38.9±0.6%) in human CD34+ cells at MOI 50, suggesting that our HIV-1 vector including the DNA flap transduces CD34+ cells efficiently due to enhanced nuclear transport.
To evaluate lentiviral transduction among the same human CD34+ cells using on another cell culture condition, we compared a Stemline II HSC expansion medium to our a standard X-VIVO10 culture medium (same conditions except culture media). Human CD34+ cell culture in Stemline II media resulted in greater proliferation of cells at 3 days and higher vector DNA amounts even at 12 hours after viral exposure (Figure 6a), suggesting that a Stemline II medium increases reverse transcription as compared to an X-VIVO10 medium. Our human CD34+ cell in vitro and in xenografted mice data (Figures 6c and d) support that higher cell expansion increased reverse transcription, which resulted in more efficient transduction, at the price of lower long term cell engraftment. This is consistent with our previous data, demonstrating that lower engraftment ability in transduced human CD34+ cells by longer ex vivo culture and higher concentrations of cytokine stimulation [13]. In efforts to improve engraftment of genetically modified cells, we (and others) performed ex vivo expansion of transduced CD34+ cells, which results in a significant reduction of engraftment and more efficient transduction [13]. The balance between the positive effect on transduction efficiency and the negative effect on reconstitution ability is important for ex vivo culture of transduced CD34+ cells.
In addition, HIV-1 infection is restricted by innate immune factors, such as tripartite motif-containing protein 5 a (TRIM5α) [35, 36]. TRIM5α is a significant restriction factor for retroviral species by targeting the capsids in a species-specific manner [37]. Rhesus TRIM5α strongly inhibits HIV-1 vector transduction, while an HIV-1 vector can escape from human TRIM5a restriction to achieve transduction [14, 21, 38, 39]. However, we previously demonstrated that human TRIM5a negatively affects transduction efficiency with an HIV-1 vector in human CD34+ cells [38]. These data suggest that human TRIM5a can reduce HIV-1 genomic RNA (packaged in the capsids) in human CD34+ cells, but should not completely deplete the HIV-1 genome. We observed a slower increase of vector RNA amounts in human CD34+ cells (Figure 2b), which may be caused by TRIM5α restriction. High MOI transduction (MOI 50) increased both vector RNA amounts (Figure 2b) and DNA amounts (Figure 3b) in human CD34+ cells, suggesting that greater amounts of HIV-1 vectors can overcome the innate immune restriction in human CD34+ cells.
We observed a plateau level of vector DNA amounts in HeLa cells at 3 days after viral exposure (Figure 3a); however, in human CD34+ cells, vector amounts gradually decreased in X-VIVO10 media (Figures 3b and 6a) but increased in Stemline II media (Figure 6a). The transduced HeLa cells and non-transduced HeLa cells should have similar levels of proliferation, since these cells are a monoclonal cell line, allowing a stable level of vector DNA amounts during long term culture. On the other hand, CD34+ cells contain various populations of hematopoietic progenitor cells and HSCs, and the proliferation level of each cell population should be affected by culture conditions including X-VIVO10 versus Stemline II media. Also, the proliferation levels of transduced CD34+ cells and non-transduced CD34+ cells should be different and affected by culture conditions (X-VIVO10 versus Stemline II), potentially resulting in increasing or decreasing levels of vector DNA amounts for a long term culture.
In summary, we demonstrate kinetics of HIV-1 vector transduction in human CD34+ cells, resulting in slower internalization (>60 minutes), a peak of reverse transcription at 24 hours, and completion of internalization at 3 days. In our analysis, initiation of reverse transcription is a major limiting step in lentiviral transduction for human CD34+ cells. An HSC expansion medium could increase transduction efficiency for human hematopoietic repopulating cells evaluated by humanized xenograft mice, likely due to stronger reverse transcription. These data are helpful for the design of strategies to improve upon lentiviral transduction for human CD34+ cells.
Highlights.
We documented kinetics of HIV-1 vector transduction in human CD34+ cells.
Initiation of reverse transcription limits lentiviral transduction for CD34+ cells.
Expansion media improve transduction for human hematopoietic repopulating cells.
Acknowledgments
This work was supported by the intramural research program of the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute of Diabetes, Digestive, and Kidney Diseases (NIDDK) at the National Institutes of Health (NIH). We thank Molly E. Evans for editing the manuscript. We thank Kayo Uchida for statistical analysis. We thank Lydia Raines and Anna Shvygin for help in performing experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Aiuti A, Cattaneo F, Galimberti S, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 2.Aiuti A, Slavin S, Aker M, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 3.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 5.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 6.Schwarzwaelder K, Howe SJ, Schmidt M, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Persons DA, Allay ER, Sabatino DE, Kelly P, Bodine DM, Nienhuis AW. Functional requirements for phenotypic correction of murine beta-thalassemia: implications for human gene therapy. Blood. 2001;97:3275–3282. doi: 10.1182/blood.v97.10.3275. [DOI] [PubMed] [Google Scholar]
- 8.Persons DA, Hargrove PW, Allay ER, Hanawa H, Nienhuis AW. The degree of phenotypic correction of murine beta -thalassemia intermedia following lentiviral-mediated transfer of a human gamma-globin gene is influenced by chromosomal position effects and vector copy number. Blood. 2003;101:2175–2183. doi: 10.1182/blood-2002-07-2211. [DOI] [PubMed] [Google Scholar]
- 9.Matrai J, Chuah MK, VandenDriessche T. Recent advances in lentiviral vector development and applications. Mol Ther. 2010;18:477–490. doi: 10.1038/mt.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi H, Smith KA, Mosier DE, Verma IM, Torbett BE. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 11.Naldini L, Blomer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 12.Uchida N, Hanawa H, Dan K, Inokuchi K, Shimada T. Leukemogenesis of b2a2-type p210 BCR/ABL in a bone marrow transplantation mouse model using a lentiviral vector. J Nippon Med Sch. 2009;76:134–147. doi: 10.1272/jnms.76.134. [DOI] [PubMed] [Google Scholar]
- 13.Uchida N, Hsieh MM, Hayakawa J, Madison C, Washington KN, Tisdale JF. Optimal conditions for lentiviral transduction of engrafting human CD34(+) cells. Gene Ther. 2011 doi: 10.1038/gt.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uchida N, Washington KN, Hayakawa J, et al. Development of a human immunodeficiency virus type 1-based lentiviral vector that allows efficient transduction of both human and rhesus blood cells. J Virol. 2009;83:9854–9862. doi: 10.1128/JVI.00357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanawa H, Kelly PF, Nathwani AC, et al. Comparison of various envelope proteins for their ability to pseudotype lentiviral vectors and transduce primitive hematopoietic cells from human blood. Mol Ther. 2002;5:242–251. doi: 10.1006/mthe.2002.0549. [DOI] [PubMed] [Google Scholar]
- 16.Greene WC. The molecular biology of human immunodeficiency virus type 1 infection. N Engl J Med. 1991;324:308–317. doi: 10.1056/NEJM199101313240506. [DOI] [PubMed] [Google Scholar]
- 17.Uchida N, Washington KN, Lap CJ, Hsieh MM, Tisdale JF. Chicken HS4 Insulators Have Minimal Barrier Function Among Progeny of Human Hematopoietic Cells Transduced With an HIV1-based Lentiviral Vector. Mol Ther. 2010 doi: 10.1038/mt.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sastry L, Johnson T, Hobson MJ, Smucker B, Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 2002;9:1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- 19.Uchida N, Evans ME, Hsieh MM, et al. Integration-specific In Vitro Evaluation of Lentivirally Transduced Rhesus CD34(+) Cells Correlates With In Vivo Vector Copy Number. Mol Ther Nucleic Acids. 2013;2:e122. doi: 10.1038/mtna.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 21.Uchida N, Hargrove PW, Lap CJ, et al. High-efficiency transduction of rhesus hematopoietic repopulating cells by a modified HIV1-based lentiviral vector. Mol Ther. 2012;20:1882–1892. doi: 10.1038/mt.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacot S, Coute Y, Belin S, et al. Isolation of nucleoli. Chapter 3:Unit3. Curr Protoc Cell Biol. 2010:36. doi: 10.1002/0471143030.cb0336s47. [DOI] [PubMed] [Google Scholar]
- 23.Hayakawa J, Hsieh MM, Uchida N, Phang O, Tisdale JF. Busulfan produces efficient human cell engraftment in NOD/LtSz-Scid IL2Rgamma(null) mice. Stem Cells. 2009;27:175–182. doi: 10.1634/stemcells.2008-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van 't Wout AB, Lehrman GK, Mikheeva SA, et al. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J Virol. 2003;77:1392–1402. doi: 10.1128/JVI.77.2.1392-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5:e1000394. doi: 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cureton DK, Massol RH, Whelan SP, Kirchhausen T. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 2010;6:e1001127. doi: 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johannsdottir HK, Mancini R, Kartenbeck J, Amato L, Helenius A. Host cell factors and functions involved in vesicular stomatitis virus entry. J Virol. 2009;83:440–453. doi: 10.1128/JVI.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donahue DA, Sloan RD, Kuhl BD, Bar-Magen T, Schader SM, Wainberg MA. Stage-dependent inhibition of HIV-1 replication by antiretroviral drugs in cell culture. Antimicrob Agents Chemother. 2010;54:1047–1054. doi: 10.1128/AAC.01537-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray JM, Kelleher AD, Cooper DA. Timing of the components of the HIV life cycle in productively infected CD4+ T cells in a population of HIV-infected individuals. J Virol. 2011;85:10798–10805. doi: 10.1128/JVI.05095-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Brien WA, Namazi A, Kalhor H, Mao SH, Zack JA, Chen IS. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zack JA, Haislip AM, Krogstad P, Chen IS. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uchida N, Sutton RE, Friera AM, et al. HIV, but not murine leukemia virus, vectors mediate high efficiency gene transfer into freshly isolated G0/G1 human hematopoietic stem cells. Proc Natl Acad Sci U S A. 1998;95:11939–11944. doi: 10.1073/pnas.95.20.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 34.Sirven A, Pflumio F, Zennou V, et al. The human immunodeficiency virus type-1 central DNA flap is a crucial determinant for lentiviral vector nuclear import and gene transduction of human hematopoietic stem cells. Blood. 2000;96:4103–4110. [PubMed] [Google Scholar]
- 35.Nakayama EE, Shioda T. Anti-retroviral activity of TRIM5 alpha. Rev Med Virol. 2010;20:77–92. doi: 10.1002/rmv.637. [DOI] [PubMed] [Google Scholar]
- 36.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 37.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans ME, Kumkhaek C, Hsieh MM, Donahue RE, Tisdale JF, Uchida N. TRIM5alpha variations influence transduction efficiency with lentiviral vectors in both human and rhesus CD34(+) cells in vitro and in vivo. Mol Ther. 2014;22:348–358. doi: 10.1038/mt.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchida N, Hsieh MM, Washington KN, Tisdale JF. Efficient transduction of human hematopoietic repopulating cells with a chimeric HIV1-based vector including SIV capsid. Exp Hematol. 2013;41:779–788. e771. doi: 10.1016/j.exphem.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]