Summary
Recent studies have suggested that antibody-mediated protection against the Ebolaviruses may be achievable, but little is known about whether or not antibodies can confer cross-reactive protection against viruses belonging to diverse Ebolavirus species, such as Ebola virus (EBOV), Sudan virus (SUDV) and Bundibugyo virus (BDBV). We isolated a large panel of human monoclonal antibodies (mAbs) against BDBV glycoprotein (GP) using peripheral blood B cells from survivors of the 2007 BDBV outbreak in Uganda. We determined that a large proportion of mAbs with potent neutralizing activity against BDBV bind to the glycan cap and recognize diverse epitopes within this major antigenic site. We identified several glycan cap-specific mAbs that neutralized multiple ebolaviruses including SUDV, and a cross-reactive mAb that completely protected guinea pigs from the lethal challenge with heterologous EBOV. Our results provide a roadmap to develop a single antibody-based treatment effective against multiple Ebolavirus infections.
Introduction
The genus Ebolavirus, family Filoviridae, contains three viral species that are known to cause large deadly disease outbreaks in Africa: Zaire ebolavirus represented by Ebola virus (EBOV), Sudan ebolavirus, represented by Sudan virus (SUDV) and Bundibugyo ebolavirus, represented by Bundibugyo virus (BDBV). The most recent EBOV outbreak has caused more than 28,000 cases and more than 11,000 deaths (according to the October 14th, 2015 WHO Ebola Situation Report). While there is no FDA-approved treatment for filovirus infections, several experimental therapeutics against EBOV are being investigated, including small interfering RNAs (Geisbert et al., 2010; Thi et al., 2015), antisense oligonucleotides (Warren et al., 2010; Warren et al., 2015), a nucleoside analog (Warren et al., 2014), therapeutic vaccines (Feldmann et al., 2007; Geisbert et al., 2008) and monoclonal antibody (mAb) cocktails (Olinger et al., 2012; Qiu et al., 2012; Qiu et al., 2014). Of these, preliminary treatment studies suggest that the effect of the ZMapp mAb cocktail exceeded the efficacy and treatment window of other experimental therapeutics described so far (Qiu et al., 2014).
The ZMapp cocktail is composed of three EBOV glycoprotein (GP)-specific mAbs (designated c13C6, c2G4 and c4G7) that were isolated initially from mice (Qiu et al., 2011; Wilson et al., 2000), chimerized with human antibody constant regions, and then produced in Nicotiana benthamiana (Qiu et al., 2014). Single-particle EM reconstructions of these mAbs in complex with EBOV surface protein have revealed key sites of vulnerability on the EBOV GP (Murin et al., 2014). One such site lies within GP base at the GP1/GP2 interface; two out of three mAbs from the ZMapp cocktail (c2G4 and c4G7) bind to overlapping epitopes located in this region. The third mAb from the ZMapp cocktail, c13C6, binds a second antigenic site, which is located in the glycan cap region. GP base region-specific mAbs 2G4 and c4G7 displayed high neutralization activity in vitro (IC50 < 0.1 μg/mL), whereas the glycan cap specific mAb c13C6 weakly neutralized EBOV only in the presence of complement (IC50 > 1.0 μg/mL) (Qiu et al., 2014). The lower in vitro neutralization activity of glycan cap-specific antibodies may be due to the removal of the glycan cap by host proteases (Chandran et al., 2005; Cote et al., 2011; Misasi et al., 2012) inside the endosome before GP engagement with the Niemann-Pick C1 receptor (Carette et al., 2011; Cote et al., 2011).
The ability of mAbs to bind to conserved neutralizing epitopes present on the surface of highly variable viral proteins has been documented extensively for HIV (Burton et al., 2012), influenza viruses (Pappas et al., 2014), dengue virus (Rouvinski et al., 2015), paramyxoviruses (Corti et al., 2013), and alphaviruses (Fox et al., 2015). Despite similar requirements for virus entry into the cell (Misasi et al., 2012), GPs from BDBV, EBOV and SUDV strains differ by over 30% at the amino acid level (Towner et al., 2008). This overall genetic divergence between species of genus Ebolavirus has hampered the development of ebolavirus cross-neutralizing Abs. The key components of multiple antibody cocktails developed over the last decade neutralize only viruses of species Zaire ebolavirus. A weakly neutralizing mAb c13C6 was shown to cross-react with SUDV GPs (Wilson et al., 2000), but it is unknown whether this mAb can neutralize SUDV. Recently, several studies have shown that cross-reactive antibodies in serum can be elicited during natural infection in humans or vaccination of animals. The serum of individuals who survived BDBV, EBOV or SUDV infections contained ebolavirus cross-reactive IgG, but not IgM (Macneil et al., 2011). Other studies demonstrated that mice immunized with a vaccine bearing the GP of EBOV, generated cross-reactive polyclonal mAbs to other ebolaviruses such as BDBV and SUDV (Meyer et al., 2015; Ou et al., 2012). Four broadly reactive non-neutralizing mAbs were isolated in mice after vaccinating animals with recombinant vesicular stomatitis virus (rVSV) expressing EBOV GP and then boosting initial immune response with the heterologous virus containing SUDV GP (Hernandez et al., 2015). The epitopes recognized by such cross-reactive mAbs are unknown.
In this study, we isolated a large panel of BDBV-specific and ebolavirus cross-reactive mAbs from B cells of survivors of BDBV infection. The results show that a large proportion of mAbs with potent neutralizing activity against BDBV bind to the glycan cap and recognize diverse epitopes within this major antigenic site. We identified several glycan cap-specific mAbs that neutralized multiple Ebolavirus species and a cross-reactive mAb that completely protected guinea pigs from the lethal challenge with heterologous EBOV when used as monotherapy. Several of these naturally occurring antibodies exhibit the most potent protective capacity reported, and they possessed unprecedented cross-reactivity for multiple Ebolavirus species including SUDV for which neutralizing human mAbs have not been reported.
Results
Isolation of Human MAbs
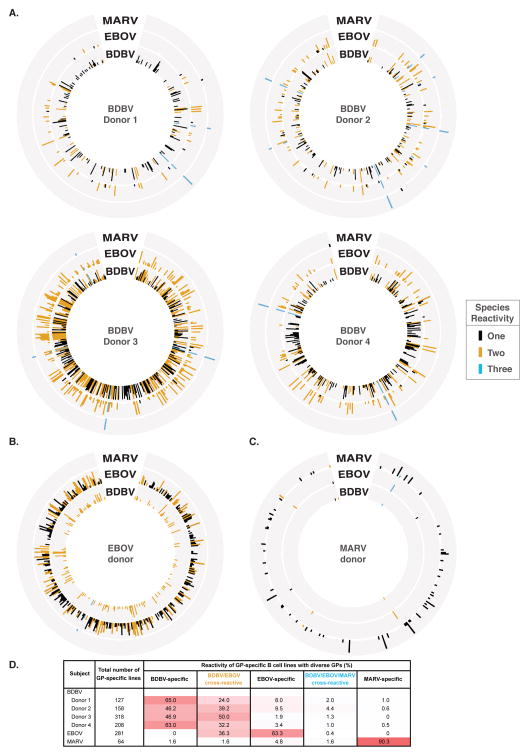
To generate human cell lines secreting human mAbs to BDBV, we transformed peripheral blood B cells from seven survivors of the 2007 Uganda BDBV outbreak with Epstein-Barr virus, as described in the Experimental Procedures section. To determine the breadth of antibody response in survivors of ebolavirus infection, we screened supernatants from EBV-transformed B cell lines for binding to GPs from diverse representatives of filovirus species: BDBV, EBOV or MARV (Figure 1A and S1). We also used the same GP panel to screen supernatants from transformed B cell lines derived from a survivor of the 2014 EBOV outbreak (Figure 1B) or from a donor who survived MARV infection (Figure 1C). We color coded GP-reactive supernatants based on the cross-reactivity pattern: species-specific cell lines are highlighted in black; cross-reactive lines to 2 or 3 species are shown in yellow or blue, respectively (Figure 1A–C and S1). While approximately half of GP-specific B cell lines obtained from BDBV survivors produced antibodies specific to BDBV GP, 24–50% of GP-reactive B cell culture supernatants also cross-reacted with EBOV GP (Figure 1A, D). Similarly, 36% of GP-specific B cell lines obtained from the EBOV survivor cross-reacted with the heterologous BDBV GP (Figure 1B, D). Despite the apparent presence of B cells encoding cross-reactive antibodies in survivors of BDBV or EBOV infections to GPs from heterologous Ebolavirus species, we detected a very limited cross-reactivity with GPs from MARV, which belongs to a different genus in the family Filoviridae (Figure 1A, D). In line with this finding, 90% of GP-reactive B cell lines obtained from the MARV survivor reacted with autologous GP, and only 2% of antigen-specific B cell lines produced Ebolavirus cross-reactive Abs (Figure 1C, D). The limited cross-reactivity of mAbs to GPs from Ebolavirus and Marburgvirus species likely is due in part to low sequence conservation between GPs from two genera (only 27% amino acid identity between BDBV and MARV GP) as well as differences in epitope availability caused by different positions of the mucin-like domains on the GP surface of Ebolavirus and Marburgvirus (Flyak et al., 2015; Fusco et al., 2015; Hashiguchi et al., 2015).
Figure 1. Cross-reactive B cell responses in filovirus immune donors.
Supernatants from EBV-transformed PBMC samples isolated from survivors were screened in ELISA binding assays using BDBV, EBOV or MARV GPs (A–C). Results for four BDBV survivors (A), one EBOV survivor (B) or one MARV survivor (C) are shown. Height of the bars indicates OD405 nm values in ELISA binding to full-length extracellular domain of GP of the indicated virus species. Reactive supernates are color-coded based on the cross-reactivity pattern: species-specific cell lines are highlighted in black; cross-reactive lines to 2 or 3 species are shown in yellow or blue, respectively. Previous work has shown that the amino acid sequence of GP differs between BDBV and EBOV by over 34%, and between BDBV and MARV by over 72%. (D) Percentages of lines secreting antibodies specific to BDBV, EBOV or MARV GPs, or cross-reactive antibodies to BDBV and EBOV (designated BDBV/EBOV) or BDBV, EBOV and MARV (designated BDBV/EBOV/MARV) are shown. Increasing intensity of the pink cell fill color corresponds to increasing reactivity for indicated virus. See also Figure S1.
Binding and Neutralizing Activity of Human MAbs
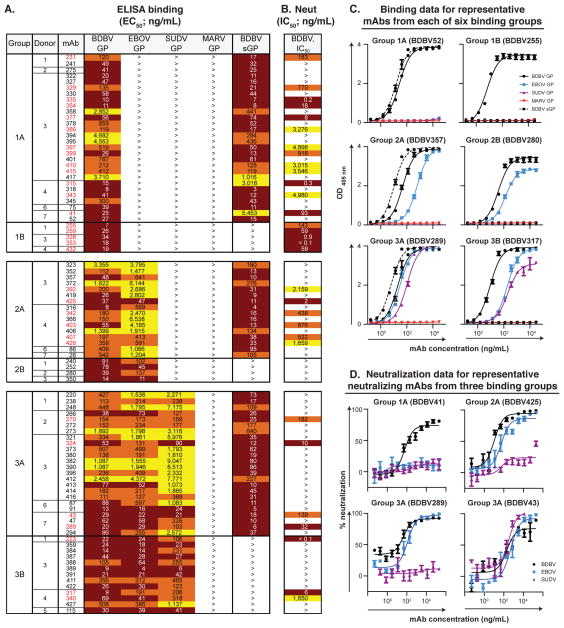
We fused transformed cells from B cell lines producing BDBV GP-reactive Abs with myeloma cells and generated 90 cloned hybridomas secreting BDBV GP-reactive human mAbs. In order to determine the breadth of mAb binding, we screened the mAbs in ELISA binding assays using recombinant GPs from multiple filoviruses: BDBV, EBOV, SUDV or MARV GPs. While 33 Abs recognized only the autologous BDBV GP (designated Groups 1A, 1B), 20 Abs recognized both BDBV and EBOV GPs (Groups 2A, B), and 37 Abs recognized all three GPs from BDBV, EBOV and SUDV (Groups 3A, B) (Figure 2A, C and Data S1). The relative proportions of antibodies that recognize glycoproteins from 1, 2 or 3 Ebolavirus species did not correlate fully with the B cell line frequencies in the initial screen, which can be explained by our prioritization on recovery of a high number of cross-reactive mAbs. We were not able to isolate Abs that bind to the heterologous MARV GP (Figure 2A, C and Data S1). We further characterized the binding of species-specific or cross-reactive mAbs to recombinant GPs by performing a binding assay with the recombinant form of GP that is secreted from the cell to the extracellular space during natural infection (sGP, secreted GP) (Sanchez et al., 1996; Volchkov et al., 1995). While the Ebolavirus GP is a trimer, sGP forms dimers in which each protomer shares only the amino-terminal 295 amino acids with GP. The majority of mAbs recognized epitopes shared between BDBV GP and BDBV sGP (designated Groups 1A, 2A or 3A) (Figure 2A, C). We also identified antibodies that bound to BDBV GP but failed to bind BDBV sGP in ELISA (designated Groups 1B, 2B or 3B) (Figure 2A, C). Antibodies from Groups 1B, 2B or 3B also bound the recombinant GP form that lacks highly glycosylated mucin-like domains (BDBV GPΔmuc), suggesting that mAbs from these three groups target epitopes outside of mucin-like domains (Figure S2).
Figure 2. Cross-neutralizing antibodies from survivors of natural BDBV infection.
(A) Heat map showing the binding of BDBV mAbs to a panel of filovirus GPs. The EC50 value for each GP-mAb combination is shown, with dark red, orange, yellow, or white shading indicating high, intermediate, low, or no detectable binding, respectively. EC50 values greater than 10,000 ng/mL are indicated by the > symbol. NAb names are highlighted in red. (B) Heat map showing the neutralization potency of BDBV GP-specific mAbs against BDBV. The IC50 value for each virus-mAb combination is shown. IC50 values greater than 10,000 ng/mL are indicated by the > symbol. Neutralization assays were performed in triplicate. (C) Binding of representative mAbs from six distinct binding groups to the filovirus GP. (D) Neutralization activity of representative neutralizing mAbs from three binding groups against BDBV, EBOV or SUDV. Error bars represent the SE of the experiment, performed in triplicate. See also Table S1, Data S1 and Figures S2 to S3.
To evaluate the inhibitory activity of isolated mAbs, we tested mAbs in a BDBV neutralization assay. Thirty-one of the 90 BDBV GP-reactive mAbs had half-maximal inhibitory concentration (IC50) values < 10 μg/mL, and we defined these as neutralizing antibodies (nAbs) (Figure 2B where nAb names are highlighted in red, and S3). Several nAbs displayed an extremely high neutralizing potency, with IC50 values below 1 ng/mL (Figure 2B). Eighteen out of 31 nAbs bound only to BDBV GP in ELISA, 6 nAbs recognized BDBV and EBOV GPs and the remaining 7 nAbs bound to GPs from representatives of three Ebolavirus species: BDBV, EBOV and SUDV. These results suggested that cross-reactive mAbs in our panel might possess neutralizing activity to multiple ebolaviruses. To test this hypothesis, we screened BDBV425 (a Group 2A nAb) in an EBOV neutralization assay as the nAb with the lowest EC50 value to the heterologous EBOV GP, and we determined that BDBV425 neutralized the heterologous EBOV. Encouraged by this result, we tested nAbs from Groups 3A and 3B in EBOV or SUDV neutralization assays to determine whether cross-reactive nAbs can neutralize three Ebolavirus species. We found two cross-reactive nAbs from Group 3A (BDBV43 and BDBV324) that neutralized all three ebolaviruses: BDBV, EBOV and SUDV (Figure 2D, BDBV43). The remaining 5 nAbs from Group 3A and 3B neutralized BDBV and EBOV but failed to neutralize SUDV (Figure 2D, BDBV289). Analysis of the Ab heavy-chain variable domain sequences for 26 nAbs revealed that all BDBV-specific and cross-reactive nAbs were encoded by unique Ab genes (Table S1).
Major Antigenic Sites Recognized by Human MAbs
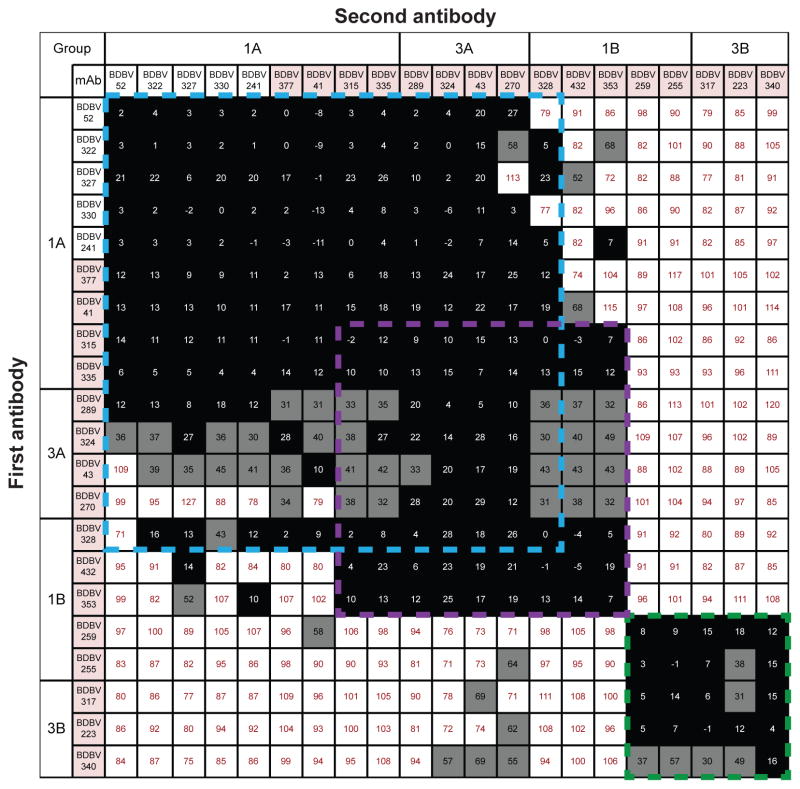
To determine whether Abs from distinct binding groups targeted different antigenic regions on the BDBV GP surface, we performed a quantitative competition-binding assay using a real-time biosensor. We tested 4 BDBV nAbs from binding Group 1A, 5 nAbs from binding Group 1B, 4 nAb from Group 3A and 3 nAbs from Group 3B in a tandem blocking assay in which BDBV GP was attached to the biosensor. We also tested 5 non-neutralizing antibodies from Group 1A to determine whether non-neutralizing antibodies target a unique epitope on GP surface. Non-neutralizing and neutralizing mAbs from Group 1A and nAbs from Group 3A blocked binding of each other to the GP antigen and segregated into a single competition-binding group (Figure 3). These results suggest that mAbs from Group 1A and Group 3A target a single antigenic region that contains epitopes shared between GP and sGP (Figure 2A). NAbs from Group 3B that did not recognize sGP in ELISA (Figure 2A) segregated into a separate competition-binding group. Group 1B antibodies were interesting in that two nAbs in this group competed for binding with Group 3B nAbs, while three nAbs from the group competed for binding with antibodies from Group 3A (Figure 3). These findings suggested that there are at least two major antigenic regions recognized by human BDBV nAbs, based on competition-binding studies. The first major antigenic region contains epitopes which both sGP and GP share (recognized by mAbs from Groups 1A, 3A) as well as epitopes that are present only on the GP surface (recognized by three mAbs from Group 1B). The second major antigenic region contains only epitopes that are present on the GP surface, but not sGP (recognized by two mAbs from Group 1B and three mAbs from Group 3B).
Figure 3. BDBV-neutralizing antibodies target at least two distinct antigenic regions of the GP surface.
Data from competition-binding assays using non-neutralizing mAbs from binding Group 1A (white background) and neutralizing mAbs from binding Groups 1A, 1B, 3A or 3B (pink background). Numbers indicate the percent binding of second mAb in the presence of the first mAb, compared to binding of un-competed second mAb. MAbs were judged to compete for the same site if maximum binding of second mAb was reduced to <30% of its un-competed binding (black boxes with white numbers). MAbs were considered non-competing if maximum binding of second mAb was >70% of its un-competed binding (white boxes with red numbers). Grey boxes with black numbers indicate an intermediate phenotype (competition resulted in between 30 and 70% of un-competed binding). Blue, purple, and green dashed lines indicate what appear to be major competition groups; the blue and purple groups overlap substantially but not completely.
Diverse Patterns of Molecular Recognition Defined by Negative Stain Electron Microscopy
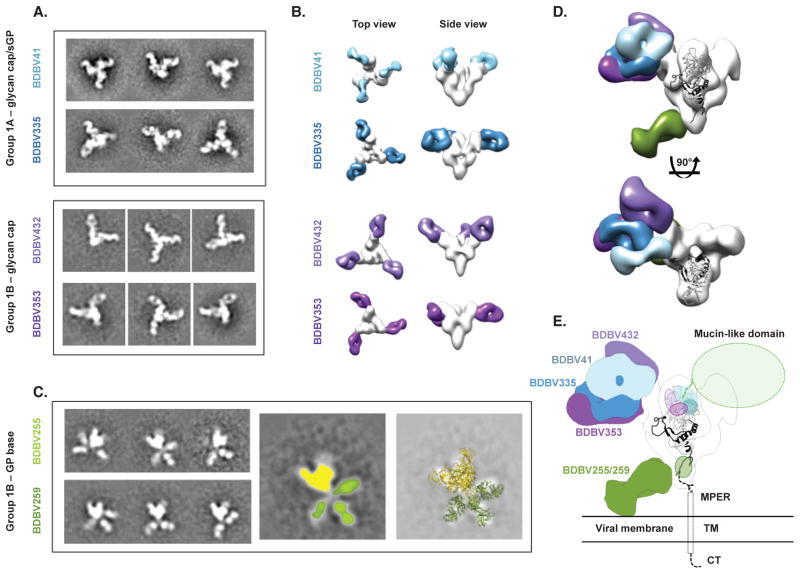
To determine the location of the two major antigenic regions targeted by human BDBV nAbs, we performed negative-stain single-particle electron microscopy (EM) studies using antibodies from Groups 1A and 1B. The EM class averages and reconstructions showed clearly that the two major antigenic regions, defined in competition-binding experiments, corresponded to two distinct sites on GP surface: the glycan cap and the GP base.
Comparison of the structures of glycan cap-directed mAbs from Group 1A with those in Group 1B revealed that the antibodies have partially overlapping epitopes, but approach the glycan cap at distinct angles (Figure 4A, B, Figure S4). We fitted a previously determined atomic resolution structure of Sudan virus (SUDV) GPΔmuc (Dias et al., 2011), which reveals more residues of the glycan cap region than the equivalent EBOV structure, into the envelope of GP from the EM reconstructions and determined the regions targeted by each mAb (Figure 4D, E). BDBV335, which binds GP and sGP equally well, mainly targets a region between residues 274–282. This region appears well defined in the BDBV335 EM map, indicated by the large lobe on the outside of the glycan cap that closely resembles that region in the GP crystal structure. When viewed along the three-fold axis of GP, BDBV41 binds to the right of BDBV335, further up on the glycan cap, close to a loop that extends from residue 266 to 277. Consistent with this position, we passaged a chimeric VSV in which the G protein was replaced with BDBV GP as a sole surface protein (VSV/BDBV-GP) in the presence of mAb BDBV41 to generate a neutralization escape mutant virus that was completely resistant to the antibody and that possessed two amino acid substitutions: G271R and T272S (Figure S5). The mutation at the 272 position likely explains why BDBV41 is a Group 1 antibody, i.e., only recognizes BDBV (with T272) but not EBOV or SUDV (which have the alternate residue K272). BDBV41 also may make contacts with a loop that extends toward the mucin-like domains, from residue 309 to 312 or further in regions that were unresolved in the GP crystal structure. BDBV432 binds to the left of BDBV335, at the top of a helix-loop at residues 259–266, and likely makes extensive contacts with a loop from residues around 302–312. Despite a lack of binding to sGP, BDBV432, as well as BDBV353, bind in the glycan cap region, suggesting that these mAbs make contacts with residues that are exclusive to GP.
Figure 4. BDBV-neutralizing antibodies bind to the glycan cap or base region of GP.
(A) Shown are negative-stain electron microscopy reference-free 2D class averages of Group 1A antibodies that bind both the glycan cap of GP and sGP, and Group 1B antibodies that bind the glycan cap of GP but not sGP. BDBV GP or GPΔmuc was used to generate complexes. (B) 3D reconstructions of glycan cap binders from Groups 1A and 1B reveal that these antibodies bind the glycan cap at overlapping but distinct epitopes. Top (left) and side (right) views of the complexes are shown. (C) Reference free 2D class averages of Group 1B antibodies (left) reveals that these antibodies bind an epitope below the base of GP that is flexible. In the middle image, GP is colored yellow and each Fab colored green. The right-hand panel illustrates a superimposition of crystal structures of SUDV GPΔmuc (PDB 3VEO) and Fabs (PDB 3CSY) to demonstrate how Fabs may bind to GP. (D) The composite model delineates the epitopes of the glycan cap mAbs in Group 1A or 1B. Side (above) and top (below) views are shown. (E) Docking a crystal structure of SUDV GPΔmuc (PDB 3VEO) (Bale et al., 2012), which contains a more complete model of the glycan cap region targeted by Group 1A/B mAbs, reveals how Group 1A/B mAbs target a broad region in the GP1 centered on the glycan cap, near the beginning of the mucin-like domains. Group 1B mAbs that target the base likely bind to a loop near the membrane proximal external region (MPER) that is flexible and has not yet been resolved at high resolution. TM = transmembrane region; CT = cytoplasmic tail. See also Figure S4.
The other antibodies in Group 1B bind to an epitope at the base of GP. These antibodies, including BDBV255 and BDBV259, bind further down on GP than has been observed previously with murine mAbs, possibly contacting residues within GP2 that are part of the membrane proximal external region (MPER) (Figure 4C, D, E). These antibodies were refractory to a reconstruction by EM due to predominant side views of the particles and also apparent flexibility. The class averages, however, clearly show that these antibodies bind an epitope that extends down below the base of GP. Three Fabs can be seen in some of the class averages, indicating that despite the apparent small size of this region, three antibodies can be accommodated on one GP trimer. Although the Fabs adopt various positions in each class average, there is not a continuous range of flexibility since the Fabs themselves are well resolved. These antibodies may require the full MPER and transmembrane (TM) regions, as well as a membrane, in order to bind stably. These features are all lacking in the current recombinant protein used here, a soluble form of the extracellular domain of GP. While the GP2 region is well conserved across the filoviruses, these BDBV-specific mAbs likely bind non-conserved regions in GP2 proximal to the TM region.
Epitope Mapping of Group 3A MAbs Using Saturation Mutagenesis and Negative Stain Electron Microscopy
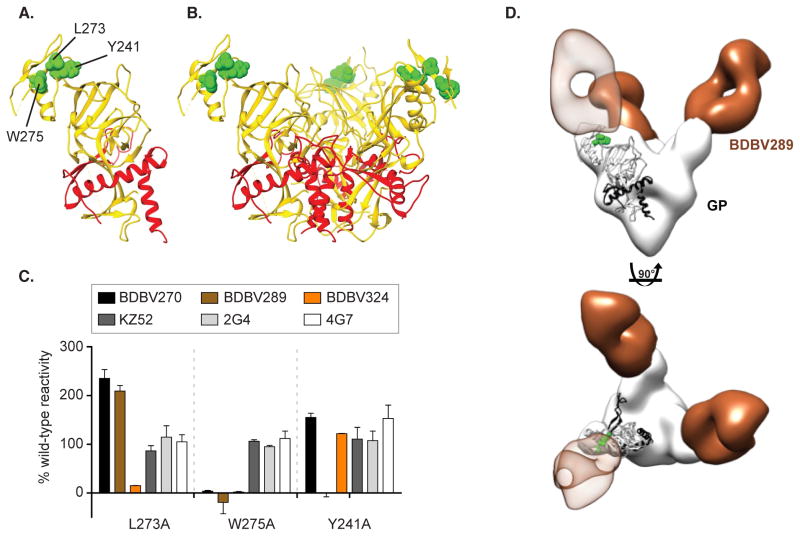
As the Group 3A (cross-reactive) nAbs competed for binding with Group 1A (BDBV-specific) nAbs (Figure 3), we hypothesized that some structural features of the glycan cap are conserved between GPs from multiple Ebolavirus species. We sought to identify critical amino acids that defined epitopes for three Group 3 nAbs (BDBV270, BDBV289, and BDBV324) using a comprehensive EBOV GP alanine-scanning mutation library. Epitope mapping identified critical residues for binding by each nAb, W275 for BDBV270, W275 and Y241 for BDBV289, W275 and L273 for BDBV324. Residues for which mutation reduced binding of three nAbs from Group 3A were visualized on the surface of the high-resolution structure of EBOV GP (PDB ID 3CSY). This finding suggests that each of these antibodies recognize overlapping epitopes in the GP glycan cap (Figure 5A, B). The previously described murine nAbs 2G4 and 4G7 and the human nAb KZ52 were shown previously to bind the base region of the GP (Lee et al., 2008; Murin et al., 2014) and mutations of the W275 or L273 residues did not reduce the binding of these nAbs (Figure 5C). We passaged VSV/BDBV-GP in the presence of BDBV223 or BDBV289 in an attempt to generate escape mutant viruses, but could not detect neutralization resistant viruses. An isolate passaged in the presence of BDBV223 with a R574H polymorphism in heptad repeat 1 (HR1) region was identified, and for BDBV289 an isolate with an I584M polymorphism in the HR1 region alone or in combination with an E149K substitution in the receptor-binding domain. However, none of these mutations was associated with the ability of those viruses to resist neutralization by the corresponding mAb.
Figure 5. Epitope mapping of Group 3A mAbs using saturation mutagenesis and negative stain electron microscopy.
Epitope residues for three nAbs from Group 3A (BDBV270, BDBV289 and BDBV324) were identified as those for which mutation to alanine specifically reduced binding of these antibodies (A–B). GP residue W275 was common to all three nAbs, while L273 was specific for BDBV324, and Y241 was specific for BDBV289. The mutated residues are shown in space filling forms on a ribbon diagram of the EBOV GP structure, based on PDB 3CSY. (C) Binding values for nAbs and previously isolated mAbs KZ52, 2G4 and 4G7 to library clones with mutations at residues L273, W275 and Y241. The Ab reactivities against each mutant EBOV GP clone were calculated relative to reactivity with wild-type EBOV GP. (D) BDBV289 (brown) binds at the top of the viral GP near the glycan cap region. Complexes are of BDBV antibody Fab fragments bound to BDBV GPΔTM with side view (top panel) or top view (bottom panel). A representative Fab crystal structure is fit in the Fab density for each reconstruction (from PDBID 3CSY). A monomer of the GP trimer crystal structure (PDBID 3CSY) is also fit in the GP density, with white corresponding to GP1 and black to GP2. Two critical residues for binding by BDBV289 (W275 and Y241, determined using saturation mutagenesis) are highlighted in green. See also Figure S5.
We further characterized the antibody BDBV289 by single particle EM studies of antibody in complex with GP. BDBV289 binds the glycan cap region of GP, centered on the residues W275 and Y241 (Figure 5D, S4). The angle of approach resembles that of the mAb 1H3 from the antibody cocktail ZMab, although 1H3 is specific to EBOV and is weakly neutralizing (Murin et al., 2014; Qiu et al., 2011). Further, BDBV289 also binds sGP, which shares the first 295 amino acids of GP1 with GP, including the glycan cap region (Sanchez et al., 1996; Volchkov et al., 1995). Therefore, despite previous hypotheses that propose that sGP is an immune decoy and that cleavage of the glycan cap prevents neutralizing antibodies from binding this region (Mohan et al., 2012; Murin et al., 2014), we have now identified several antibodies that challenge these ideas. Interestingly, BDBV289 targets an overlapping epitope with antibodies that we identified to be specific to BDBV and that do not bind sGP (Figure 4). Therefore, the glycan cap region is a major antigenic site that contains epitopes with subtle features that influence sGP and GP binding, neutralization and species cross-reactivity of targeting mAbs.
Therapeutic Efficacy of Human MAbs in Small Animal Models of EBOV Infection
To determine the therapeutic activity of cross-neutralizing Abs, we tested several antibodies in mice. We focused on cross-reactive antibodies, and we studied the heterologous effect of BDBV survivor mAbs against EBOV challenge. We selected two nAbs from Groups 3A (BDBV289) and 3B (BDBV223) that bound non-overlapping antigenic regions in the competition-binding experiments (Figure 3). Seven week-old BALB/c mice received 100 μg of antibody by the IP route 1 or 3 days after inoculation with 1,000 PFU of mouse-adapted EBOV, strain Mayinga (Bray et al., 1998). BDBV223 and BDBV289 reduced disease and protected mice from death when delivered one day after challenge with EBOV (Figure 6). We did not observe a therapeutic effect in the mice receiving the antibodies three days after the challenge.
Figure 6. Survival and clinical signs of EBOV-inoculated mice treated with BDBV mAbs.
Groups of 5 mice in each group were injected with individual mAbs by the intraperitoneal route 1 day after EBOV challenge, using 100 μg of mAb per treatment. Animals treated with dengue virus-specific human mAb 2D22 served as controls. (A) Kaplan-Meier survival curves. (B) Body weight. (C) Illness score.
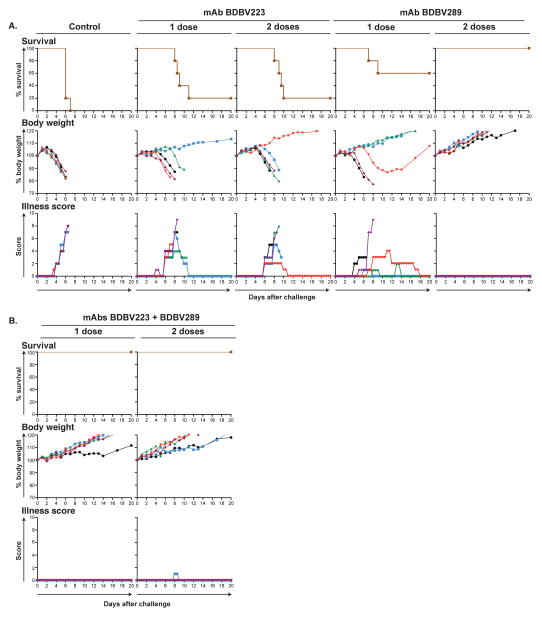
Finally, we set out to test in vivo efficacy of the cross-reactive nAbs BDBV289 and BDBV223 using a guinea pig model of EBOV infection. Five to six week-old guinea pigs, strain Hartley, were injected with 5 mg of antibody by the IP route once (day 1) or twice (days 1 and 3) after inoculation with 1,000 PFU of guinea pig-adapted EBOV, strain Mayinga. BDBV223 provided marginal protection, as only 1 out of 5 animals survived the lethal challenge (Figure 7). Surprisingly, a glycan cap-specific nAb, BDBV289, fully protected guinea pigs when delivered twice after the virus challenge. The protective efficiency of BDBV289 with a single treatment against a heterologous EBOV (Figure 7, 3 animals out of 5 survived) was higher than a protective efficiency of the equivalent glycan-cap-specific mAb c13C6, a component of the ZMapp cocktail (1 animal out of 6 survived) (Qiu et al., 2014). To determine whether a combination of two mAbs that target two neutralizing epitopes on EBOV GP surface confer better protection than treatment with a single mAb alone, we tested the combination of BDBV223 and BDBV289 in guinea pigs. The combination of two antibodies provided complete protection against a heterologous EBOV with only a single treatment (Figure 7). We isolated viral RNA from blood of representative animals that were treated with mAbs BDBV223 or BDBV289 but died, and then sequenced the genes encoding the GP (Table S2). Several polymorphisms were detected, but none appeared to be directly related to the epitope specificity of the mAb used for treatment.
Figure 7. Survival and clinical signs of EBOV inoculated guinea pigs treated with BDBV mAbs.
Groups of 5 guinea pigs per group were injected with individual mAbs by the intraperitoneal route 1 day or 1 and 3 days after EBOV challenge, using 5 mg of individual mAb (A) or 5 mg of the combination of two mAbs per treatment (B), as indicated. Animals treated with dengue virus-specific human mAb 2D22 served as controls. The survival curves are based on morning and evening observations. Mortality in the morning is shown in whole day numbers, in the evening in 1/2 day values. The body weight and illness scores are shown with one value per day. See also Table S2.
Discussion
This study reveals that natural BDBV infection in humans triggers the development of ebolavirus cross-reactive antibodies that target epitopes on the GP surface that are conserved in diverse species of genus Ebolavirus. During these studies we isolated 90 human mAbs from humans following BDBV infection and found 57 cross-reactive mAbs that recognized heterologous EBOV GPs. Remarkably, some of the isolated cross-reactive mAbs not only bound but also neutralized multiple Ebolavirus species. The majority of cross-reactive mAbs neutralized BDBV and EBOV, but we also isolated two antibodies that displayed potent neutralizing activity against representatives of three Ebolavirus species – BDBV, EBOV and SUDV. We tested two cross- neutralizing mAbs in mice and guinea pigs and showed that they protected animals from lethal challenge with a heterologous species of EBOV. These data suggest that cross-neutralizing mAbs can be used to develop a universal treatment against multiple ebolaviruses, and implies that highly immunogenic vaccines with proper presentation of GP from one species could induce some measure of cross-protection against viruses of the other species. The ability of these mAbs to bind and neutralize a broad range of Ebolavirus species also suggest that such antibodies might offer protection against emerging filoviruses in the future.
Our study also highlights the neutralization and protective potencies of human glycan cap- specific antibodies. It has been suggested previously that glycan cap-binding mAbs may not neutralize well because cathepsins remove this region during viral entry (Murin et al., 2014). However, several of the BDBV glycan cap-specific mAbs isolated here exhibit very potent neutralizing activity, and they recognize diverse epitopes within this major antigenic site. Furthermore, a single glycan cap-specific nAb, BDBV289, provided complete protection in EBOV-challenged guinea pigs. The mechanism used by glycan cap-binding mAbs to neutralize the virus in vitro is unclear. The glycan cap-specific antibodies described here bind to sites distant from the putative cathepsin cleavage site (located at residue 200), so they are unlikely to interfere with GP cleavage. While the amino acid sequence of the GP1 region is generally less well conserved than that of GP2 in viruses of diverse filovirus species, the five neutralizing glycan cap mAbs studied with EM imaging here target conserved residues, indicating that these regions may be important to the viral lifecycle. Therefore, these mAbs may block some yet undefined function of the glycan cap.
Several antibody-based treatments provided a complete species-specific protection from EBOV in non-human primate model of infection (Qiu et al., 2014). However, antibody-based therapeutics against other members of the Ebolavirus genus, such and BDBV and SUDV, are not yet available. While one strategy would be to develop separate antibody treatments for each filovirus infection, an alternative strategy would be to have a universal treatment effective against diverse Ebolavirus species. The development of universal antibody treatments for ebolaviruses seems inevitable, given recent progress in the identification of broad and potent neutralizing antibodies against viruses that exhibit more antigenic diversity than the filoviruses such as HIV (Burton et al., 2012), influenza viruses (Pappas et al., 2014), dengue virus (Rouvinski et al., 2015), alphaviruses (Fox et al., 2015), and paramyxoviruses (Corti et al. 2013). Our results provide a roadmap to develop a single antibody-based treatment effective against multiple Ebolavirus infections. We propose that the principal components of such treatment should include cross-neutralizing mAbs that target conserved elements of the non-overlapping major neutralizing antigenic sites on the GP surface.
EXPERIMENTAL PROCEDURES
Donors
De-identified peripheral blood mononuclear cells (PBMCs) from 7 survivors of the 2007 BDBV outbreak in Uganda (Towner et al., 2008) were obtained from a repository at Makerere University (Kampala, Uganda) managed in collaboration with the U.S. Military HIV Research Program MHRP, which is part of the Walter Reed Army Institute of Research. PBMCs were obtained after informed consent from a U.S. survivor of Ebola virus Zaire (EBOV) infection who was infected while delivering health care in Liberia during the 2014 Ebola virus outbreak with Makona virus. Cells from the EBOV survivor were obtained about 11 weeks after infection and about 7 weeks after discharge from hospital, following several negative PCR tests for presence of virus. PBMCs were obtained from a U.S. survivor of Marburg virus (MARV) infection who developed the disease in early 2008 following exposure to fruit bats in the Python Cave in Queen Elizabeth National Park, Uganda. This donor’s clinical course was documented previously (Centers for Disease and Prevention, 2009), and we have previously reported isolation of human antibodies from this donor (Flyak et al., 2015). Peripheral blood from the donor was obtained in 2012, four years after the illness, following informed consent. The studies were approved by the Vanderbilt University Institutional Review Board.
Viruses
BDBV strain 200706291 Uganda was isolated originally from the serum of a patient during the first recorded outbreak caused by this virus (Towner et al., 2008) and passaged three times in Vero E6 cells. The virus was provided originally by the Special Pathogens Branch of the U.S. Centers for Disease Control and Prevention (CDC) and deposited at the World Reference Center of Emerging Viruses and Arboviruses (WRCEVA), housed at the University of Texas Medical Branch (UTMB), Galveston, TX. The chimeric EBOV/BDBV-GP, EBOV/MARV-GP and EBOV/SUDV-GP constructs expressing eGFP were obtained by replacing the gene encoding EBOV GP with that of BDBV, MARV or SUDV, respectively (Ilinykh P., unpublished data), and passaged two times in Vero E6 cells. Additional details are reported in Supplemental Experimental Procedures.
Generation of human hybridomas secreting monoclonal antibodies (mAbs)
Human hybridomas were generated as described previously (Flyak et al., 2015). In brief, previously cryopreserved samples were transformed with Epstein-Barr virus, CpG and additional supplements. After 7 days, cells from each well of the 384-well culture plates were expanded into four 96-well culture plates using cell culture medium containing irradiated heterologous human PBMCs (recovered from blood unit leukofiltration filters, Nashville Red Cross) and incubated for an additional four days. Plates were screened for BDBV GP antigen-specific antibody-secreting cell lines using enzyme-linked immunosorbent assays (ELISAs). Cells from wells with supernates reacting with antigen in an ELISA were fused with HMMA2.5 myeloma cells using an established electrofusion technique (Yu et al., 2008).
Human mAb and Fab production and purification
After fusion, hybridoma cell lines were cloned by single-cell fluorescence-activated cell sorting and expanded in post-fusion medium as previously described (Flyak et al., 2015). HiTrap Protein G or HiTrap MabSelectSure columns were used to purify antibodies from filtered supernates. Fab fragments were generated by papain digestion, as described previously (Flyak et al., 2015).
Expression and purification of filovirus GPs
BDBV GP ectodomain (BDBV GP, residues 1–637) or the secreted glycoprotein dimer (BDBV sGP, residues 1–316) were used to screen supernatants of transformed B cells. Recombinant glycoproteins were engineered with a C-terminal double strep tag and cloned into a modified pMTpuro vector for expression in Drosophila S2 cells. Briefly, plasmids were transfected into S2 cells using Effectene reagent (Qiagen) followed by stable cell selection with 6 μg/mL puromycin. S2 cells first were cultured in Schneider’s medium supplemented with 10% (v/v) FCS (Lonza), and later adapted to Insect Xpress medium for large-scale expression in 2L shaker flasks. Stable cells were induced with 0.5 mM CuSO4 and harvested after 4 to 5 days at 27°C. Tangential flow filtration then was used to buffer exchange the supernatants into 100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 15 μg/mL avidin pH 8.0, and target proteins were purified using Streptactin Superflow affinity (Qiagen). GP ectodomains were purified further with S200 size exclusion chromatography (SEC); sGP was purified with S75 SEC. Recombinant ectodomains for EBOV, SUDV or MARV were designed and expressed similarly.
Screening and half maximal effective concentration (EC50) ELISA binding analysis
Soluble forms of the full-length extracellular domain of BDBV, EBOV, SUDV or MARV GPs or the sGP form of BDBV GP were coated overnight onto 384-well plates at 1 μg/mL. For screening ELISA, 10 μL of supernate from a well of a tissue-culture plate were transferred to each well of a 384-well ELISA plate. For EC50 binding analysis by ELISA, purified antibodies were applied to the plates at a concentration range of 30 μg/mL to 170 ng/mL, using three-fold serial dilutions. The presence of antibodies bound to the GP was determined using goat anti- human IgG alkaline phosphatase conjugate and p-nitrophenol phosphate substrate tablets, with optical density read at 405 nm after 120 minutes. A non-linear regression analysis was performed on the resulting curves using Prism version 5 (GraphPad) to calculate EC50 values. The Circos software package was used for data visualization (Krzywinski et al., 2009).
EBOV and MARV neutralization experiments
Isolated mAbs were screened initially in a high-throughput neutralization assay using EBOV/BDBV-GP with or without 5% guinea pig complement (MP Biomedicals, Santa Ana, CA) (Ilinykh P., unpublished data). The mAbs that exhibited neutralizing activity also were screened for neutralization of eGFP-expressing EBOV (Towner et al., 2005). Several mAbs were tested for neutralization of EBOV/SUDV-GP and EBOV/MARV-GP by the same approach. Additional information is in Supplemental Experimental Procedures.
Biolayer interferometry competition binding assay
Competition binding studies using biolayer interferometry and biotinylated BDBV GP (EZ-link® Micro NHS-PEG4-Biotinylation Kit, Thermo Scientific #21955) (5 μg/mL) were performed on an Octet RED biosensor (ForteBio, Menlo Park, CA), as described previously (Flyak et al., 2015). In brief, the antigen was immobilized onto streptavidin-coated biosensor tips. After a brief washing step, biosensor tips were immersed first into the wells containing first antibody at a concentration of 100 μg/mL and then into the wells containing a second mAb at a concentration of 100 μg/mL. The percent binding of the second mAb in the presence of first mAb was determined by comparing the maximal signal of the second mAb applied after the first mAb complex to the maximal signal of the second mAb alone.
Sequence analysis of antibody variable region genes
Antibody variable gene sequence analysis was performed as previously described (Flyak et al., 2015). Heavy chain antibody variable region sequences were analyzed using the IMGT/V-Quest program (Brochet et al., 2008; Giudicelli et al., 2011).
Electron microscopy and sample preparation
Fabs were added in 10 molar excess to BDBV GPdMuc and subsequently purified and stained as previously described (Murin et al., 2014).
Image processing of protein complexes
Particles were automatically picked using DoG Picker (Voss et al., 2009) and particle stacks were generated using Appion (Lander et al., 2009). Subsequently, reference-free 2D class averages were generated using iterative MRA/MSA (van Heel et al., 1996). Non-GP complexes and those with a clear lack of full saturation by Fab were removed to generate a final stack for reconstructions. In some cases, orientation bias or flexibility of Fabs prevented convergence of an acceptable model, although examination of class averages allowed a general assignment of the epitope. Final stack class averages were used to generate initial models using EMAN2 common lines (Tang et al., 2007). A model matching its reference projections was further refined using the entire raw particle stack with EMAN2, as described previously (Murin et al., 2014). For the BDBV41 reconstruction, the EMAN2 reconstruction lacked important features that were present in the class averages, indicating that perhaps some particles lacked full Fab saturation. In order to circumvent this problem, we utilized the Relion package, which allows 3D-classification to remove particles that may only contain 2 Fabs, significantly improving the quality of the final EM map (Scheres, 2012). Modeling fitting and EM figures were generated using UCSF Chimera (Pettersen et al., 2004).
Epitope mapping using an EBOV GP alanine-scan mutation library
Comprehensive high-throughput alanine scanning (‘shotgun mutagenesis’) was carried out on an expression construct for EBOV GP (Yambuku-Mayinga variant GP; Uniprot accession number Q05320). Additional details are reported in Supplemental Experimental Procedures.
In vivo testing
The animal protocols for testing of mAbs in mice and guinea pigs were approved by the Institutional Animal Care and Use Committee of the University of Texas Medical Branch at Galveston. Seven-week-old BALB/c mice (Charles River Laboratories) were placed in the ABSL-4 facility of the Galveston National Laboratory. Groups of mice at 5 animals per group were injected with 1,000 PFU of the mouse-adapted EBOV by the intraperitoneal route. Twenty-four or seventy four hours later, animals were injected with individual mAbs by the intraperitoneal route using 100 μg per treatment. Animals treated with the antibody specific to dengue virus 2D22 served as controls. Animals were weighed and monitored daily over the two-week period after challenge. Once animals were symptomatic, they were examined no less than twice per day. The disease was scored using the following parameters: dyspnea (possible scores 0–5), recumbency (0–9), unresponsiveness (0–5), and bleeding/hemorrhage (0–5). To test the protective efficacy of mAbs in guinea pigs, five to six week-old animals (strain Hartley) were placed in the ABSL-4 facility of the Galveston National Laboratory. Groups of 5 animals per group were injected with 1,000 PFU of guinea pig-adapted EBOV, by the intraperitoneal route. Twenty-four hours later and 72 hours later, animals were injected with individual mAbs (5 mg per treatment), or a cocktail of two mAbs (2.5 mg of each mAb per treatment). Animals were weighted and monitored daily for 14 days. After animals became symptomatic, they were examined no less than twice per day. The disease was scored using the following parameters: appearance (possible scores 0–3), body condition (0–3), natural behavior (0–3), and provoked behavior (0–3).
Supplementary Material
Acknowledgments
This project received support from the Defense Threat Reduction Agency (grant HDTRA1-13-1-0034 to JEC and AB), U.S. NIH grant U19 AI109711 (to JEC and AB), U19 AI109762 (to EOS and ABW), NIH contract HHSN272201400058C (to BJD), and R01 AI067927 (to EOS). EOS is an Investigator in the Pathogenesis of Infectious Disease of the Burroughs Wellcome Fund. The project was supported by NCRR Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank the donors, and Cheryl Stewart and Jill Janssen in the Vanderbilt Clinical Trials Center for help in sample acquisition in the U.S. We thank Monica Millard and Hannah Kibuuka of the Makerere University Walter Reed Project (MUWRP) in Kampala Uganda who provided repository samples from the Long-term Bundibugyo Ebola virus survivors study done in collaboration with US Army Medical Research Institute of Infectious Diseases, Ft. Detrick, MD, USA. We thank Frances Smith-House at Vanderbilt and Kathleen Pommert at Scripps for excellent technical support. LB was a summer undergraduate research fellow at the Vanderbilt Vaccine Center, and JAD was a summer undergraduate research fellow at The Scripps Research Institute. We thank Andrew McNeal and Rachel Fong for assistance with epitope mapping. We also thank Dr. Alexander Freiberg (The University of Texas Medical Branch at Galveston) for providing the guinea pig-adapted EBOV strain. Flow cytometry experiments were performed in the VMC Flow Cytometry Shared Resource, supported by NIH grants P30 CA68485 and DK058404. The EM work was conducted at The Scripps Research Institute electron microscopy facility. EM reconstructions have been deposited in the Electron Microscopy Data Bank under the accession codes EMD-6527 through 6532. CDM is supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
Author Contributions. AIF, AB, and JEC conceived the study. AIF, XS, CDM, MLF, HLT, JAD, RL, NK, PAI, HK, LB, GS, CK, CB and TGK performed experiments. AIF, XS, PAI, NK, AB, ED, BJD, JCS, EOS, ABW, AB, and JEC analyzed data. AIF, PAI, AB, and JEC wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bale S, Dias JM, Fusco ML, Hashiguchi T, Wong AC, Liu T, Keuhne AI, Li S, Woods VL, Jr, Chandran K, et al. Structural basis for differential neutralization of ebolaviruses. Viruses. 2012;4:447–470. doi: 10.3390/v4040447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis. 1998;178:651–661. doi: 10.1086/515386. [DOI] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease, C., and Prevention. Imported case of Marburg hemorrhagic fever - Colorado, 2008. MMWR Morb Mortal Wkly Rep. 2009;58:1377–1381. [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013;501:439–443. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, et al. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Jones SM, Daddario-DiCaprio KM, Geisbert JB, Stroher U, Grolla A, Bray M, Fritz EA, Fernando L, Feldmann F, et al. Effective post-exposure treatment of Ebola infection. PLoS Pathog. 2007;3:e2. doi: 10.1371/journal.ppat.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak AI, Ilinykh PA, Murin CD, Garron T, Shen X, Fusco ML, Hashiguchi T, Bornholdt ZA, Slaughter JC, Sapparapu G, et al. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell. 2015;160:893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JM, Long F, Edeling MA, Lin H, van Duijl-Richter MK, Fong RH, Kahle KM, Smit JM, Jin J, Simmons G, et al. Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell. 2015;163:1095–1107. doi: 10.1016/j.cell.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco ML, Hashiguchi T, Cassan R, Biggins JE, Murin CD, Warfield KL, Li S, Holtsberg FW, Shulenin S, Vu H, et al. Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs. PLoS Pathog. 2015;11:e1005016. doi: 10.1371/journal.ppat.1005016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Bausch DG, Feldmann H. Prospects for immunisation against Marburg and Ebola viruses. Rev Med Virol. 2010;20:344–357. doi: 10.1002/rmv.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Daddario-DiCaprio KM, Williams KJ, Geisbert JB, Leung A, Feldmann F, Hensley LE, Feldmann H, Jones SM. Recombinant vesicular stomatitis virus vector mediates postexposure protection against Sudan Ebola hemorrhagic fever in nonhuman primates. J Virol. 2008;82:5664–5668. doi: 10.1128/JVI.00456-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Brochet X, Lefranc MP. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- Hashiguchi T, Fusco ML, Bornholdt ZA, Lee JE, Flyak AI, Matsuoka R, Kohda D, Yanagi Y, Hammel M, Crowe JE, Jr, et al. Structural basis for marburg virus neutralization by a cross-reactive human antibody. Cell. 2015;160:904–912. doi: 10.1016/j.cell.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez H, Marceau C, Halliday H, Callison J, Borisevich V, Escaffre O, Creech J, Feldmann H, Rockx B. Development and Characterization of Broadly Cross-reactive Monoclonal Antibodies Against All Known Ebolavirus Species. J Infect Dis. 2015;212:S410–413. doi: 10.1093/infdis/jiv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macneil A, Reed Z, Rollin PE. Serologic cross-reactivity of human IgM and IgG antibodies to five species of Ebola virus. PLoS Negl Trop Dis. 2011;5:e1175. doi: 10.1371/journal.pntd.0001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Garron T, Lubaki NM, Mire CE, Fenton KA, Klages C, Olinger GG, Geisbert TW, Collins PL, Bukreyev A. Aerosolized Ebola vaccine protects primates and elicits lung-resident T cell responses. J Clin Invest. 2015;125:3241–3255. doi: 10.1172/JCI81532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi J, Chandran K, Yang JY, Considine B, Filone CM, Cote M, Sullivan N, Fabozzi G, Hensley L, Cunningham J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J Virol. 2012;86:3284–3292. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan GS, Li W, Ye L, Compans RW, Yang C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 2012;8:e1003065. doi: 10.1371/journal.ppat.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murin CD, Fusco ML, Bornholdt ZA, Qiu X, Olinger GG, Zeitlin L, Kobinger GP, Ward AB, Saphire EO. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc Natl Acad Sci USA. 2014;111:17182–17187. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinger GG, Jr, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci USA. 2012;109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou W, Delisle J, Jacques J, Shih J, Price G, Kuhn JH, Wang V, Verthelyi D, Kaplan G, Wilson CA. Induction of ebolavirus cross-species immunity using retrovirus-like particles bearing the Ebola virus glycoprotein lacking the mucin-like domain. Virol J. 2012;9:32. doi: 10.1186/1743-422X-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Qiu X, Alimonti JB, Melito PL, Fernando L, Stroher U, Jones SM. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin Immunol. 2011;141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Qiu X, Audet J, Wong G, Pillet S, Bello A, Cabral T, Strong JE, Plummer F, Corbett CR, Alimonti JB, et al. Successful treatment of ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci Transl Med. 2012;4:138ra181. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Thi EP, Mire CE, Lee AC, Geisbert JB, Zhou JZ, Agans KN, Snead NM, Deer DJ, Barnard TR, Fenton KA, et al. Lipid nanoparticle siRNA treatment of Ebola-virus-Makona-infected nonhuman primates. Nature. 2015;521:362–365. doi: 10.1038/nature14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- Towner JS, Sealy TK, Khristova ML, Albarino CG, Conlan S, Reeder SA, Quan PL, Lipkin WI, Downing R, Tappero JW, et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Volchkov VE, Becker S, Volchkova VA, Ternovoj VA, Kotov AN, Netesov SV, Klenk HD. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TK, Warfield KL, Wells J, Swenson DL, Donner KS, Van Tongeren SA, Garza NL, Dong L, Mourich DV, Crumley S, et al. Advanced antisense therapies for postexposure protection against lethal filovirus infections. Nat Med. 2010;16:991–994. doi: 10.1038/nm.2202. [DOI] [PubMed] [Google Scholar]
- Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, Dong L, Retterer CJ, Eaton BP, Pegoraro G, et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren TK, Whitehouse CA, Wells J, Welch L, Heald AE, Charleston JS, Sazani P, Reid SP, Iversen PL, Bavari S. A single phosphorodiamidate morpholino oligomer targeting VP24 protects rhesus monkeys against lethal Ebola virus infection. mBio. 2015;6 doi: 10.1128/mBio.02344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- Yu X, McGraw PA, House FS, Crowe JE., Jr An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods. 2008;336:142–151. doi: 10.1016/j.jim.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.