Summary
Proper adaptation to environmental perturbations is essential for tissue homeostasis. In the intestine, diverse environmental cues can be sensed by immune cells, which must balance resistance to microorganisms with tolerance, avoiding excess tissue damage. By applying imaging and transcriptional profiling tools, we interrogated how distinct microenvironments in the gut regulate resident macrophages. We discovered that macrophages exhibit a high degree of gene-expression specialization dependent on their proximity to the gut lumen. Lamina propria macrophages (LpMs) preferentially expressed a pro-inflammatory phenotype when compared to muscularis macrophages (MMs), which displayed a tissue-protective phenotype. Upon luminal bacterial infection, MMs further enhanced tissue-protective programs, and this was attributed to swift activation of extrinsic sympathetic neurons innervating the gut muscularis and norepinephrine signaling to β2 adrenergic receptors on MMs. Our results reveal unique intra-tissue macrophage specialization and identify neuro-immune communication between enteric neurons and macrophages that induces rapid tissue-protective responses to distal perturbations.
Introduction
Intestinal tissue is continuously exposed to numerous microbe- and food-derived antigens. In order to deal with either harmless or potentially pathogenic stimulation, efficient protective responses (resistance) need to be coupled with tissue tolerance, i.e. the ability to limit disease severity induced by a given pathogen burden or inflammatory response (Raberg et al., 2007). Accordingly, while failure in innate or adaptive immunity leads to recurrent infections, deficient tolerance or tissue repair mechanisms result in immunopathology (Medzhitov et al., 2012; Soares et al., 2014). At the mucosal surfaces, microbial sensing mechanisms regulate tissue repair at steady state, but in the context of infection, resistance mechanisms may lead to excessive inflammation and permanent tissue damage. Although the role of environmental cues in the adaptation of immune cells to these conditions has been increasingly appreciated, the nature of these signals and the mechanisms by which they influence immune cells are still unclear (Ayres et al., 2012; Rakoff-Nahoum et al., 2004).
Tissue-resident macrophages represent a highly heterogeneous cell population able to sense and quickly adapt to environmental cues (Hashimoto et al., 2013; Lavin et al., 2014; Nguyen et al., 2011; Okabe and Medzhitov, 2014; Wang et al., 2015; Wang et al., 2012). A vast network of macrophages populates intestinal tissue, playing either protective or tolerogenic roles, depending on the context (Bogunovic et al., 2009; Denning et al., 2007; Parkhurst et al., 2013; Zigmond et al., 2014). Mucosal, or lamina propria macrophages (LpMs), are located underneath the epithelial layer and are in close proximity to the gut lumen (Farache et al., 2013; Mazzini et al., 2014; Zigmond et al., 2014). Muscularis macrophages (MMs), on the other hand, are located underneath the submucosal region between circular and longitudinal muscle layers, comparatively distant from luminal stimulation (Bogunovic et al., 2009). Early studies suggested that LpMs play an important role by sampling luminal bacteria and initiating adaptive immune responses to clear pathogenic bacteria (Niess et al., 2005). Additionally, LpMs are thought to initiate a cascade of events involved in tolerance to dietary antigens (Hadis et al., 2011; Mazzini et al., 2014). In contrast, a recent study indicated that MMs regulate the activity of enteric neurons and peristalsis, although this macrophage population remains largely uncharacterized (Muller et al., 2014). It also remains to be defined how distinct programs in a specific cell lineage can arise within different compartments of the same tissue.
Using live multi-photon microscopy and tissue-clearing imaging techniques, we observed distinct cell dynamics and morphological features between LpMs and MMs. Unique intra-tissue specialization of these two macrophage populations was confirmed by transcriptional profiling tools, which showed that LpMs preferentially expressed a pro-inflammatory phenotype while MMs displayed a tissue-protective gene-expression profile at steady state. Following luminal infection, gut macrophages exhibited distinct responses according to their location, further reinforcing their steady state tissue signature. This divergent transcriptional profile was in part dependent on norepinephrine signaling via β2 adrenergic receptors (β2ARs), which are highly expressed on MMs. Correspondingly, using a gene reporter and transcriptional profiling we observed that luminal infection activates tyrosine hydroxylase-expressing neurons in the sympathetic ganglia innervating the intestine. This work identifies a mechanism by which interaction between intestinal neurons and macrophages can mediate intra-tissue adaptation in response to distal environmental perturbations, forming a cellular network possibly involved in maintaining the balance between resistance and tolerance.
Results
Distinct morphological features and cell dynamics inherent to lamina propria and muscularis macrophages
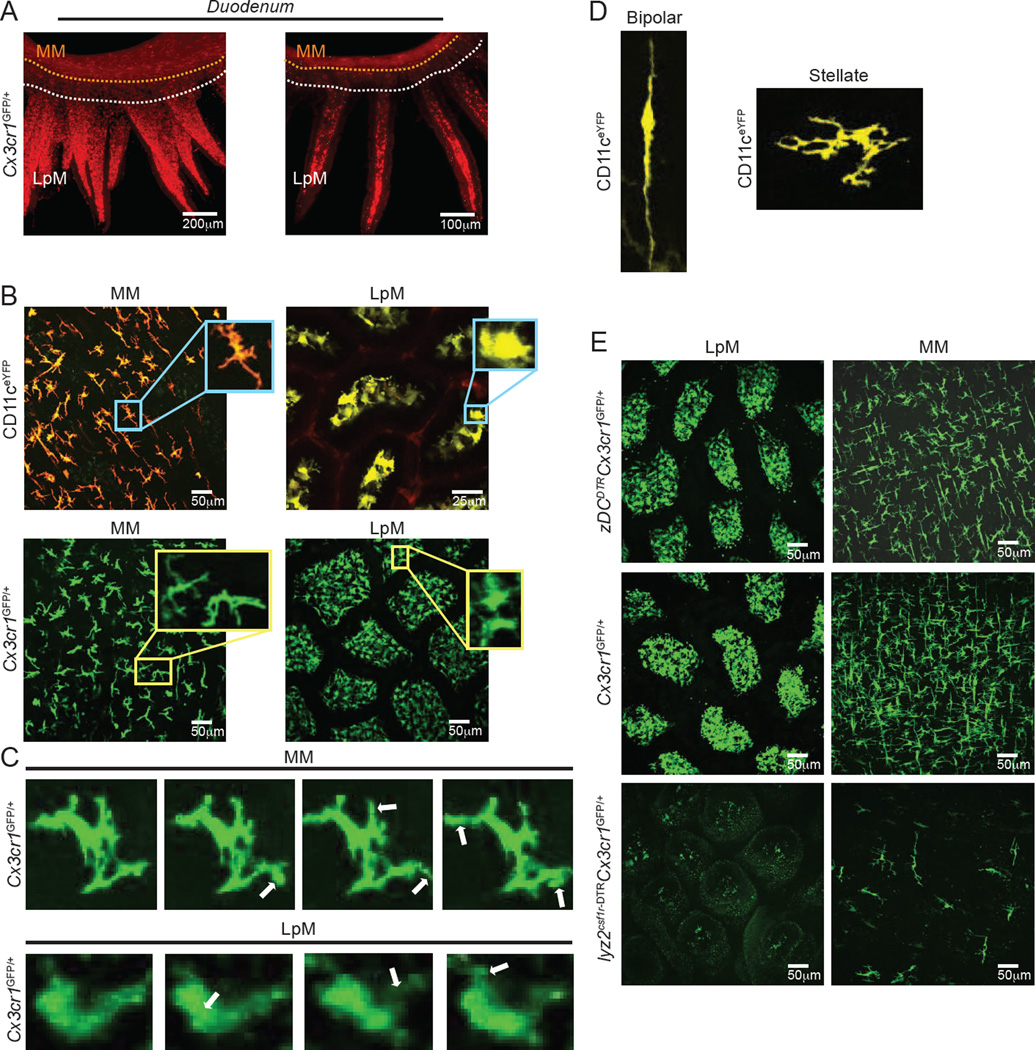
To obtain a deep-tissue, 3D view of gut-resident macrophage distribution within the intact intestinal tissue, we performed whole-mount immunolabeling, utilizing a tool referred to as immunolabeling-enabled three-dimensional imaging of solvent-cleared organs (iDISCO) (Renier et al., 2014). Small intestine sections from Cx3cr1GFP/+ macrophage reporter mice (Niess et al., 2005) were stained with anti-GFP antibodies and visualized using light-sheet microscopy. Resulting images revealed dense macrophage networks throughout the tissue layers and particularly concentrated in the lamina propria (LpMs) and muscularis (MMs) regions, suggesting compartmentalization of gut macrophage populations (Movie S1A and Figure 1A). To gain insight into cell dynamics in these two distinct layers of the intestinal wall of live animals, we utilized intravital multi-photon microscopy (IVM) (Farache et al., 2013). We analyzed ItgaxeYFP (CD11ceYFP) and Cx3cr1GFP/+ reporter mice side-by-side, which allow the visualization of intestinal antigen-presenting cells (APCs) and macrophage populations, respectively (Farache et al., 2013; Niess et al., 2005). Comparison between macrophages (CD11c+ and/or CX3CR1+) residing in the lamina propria and muscularis compartments revealed distinct morphologies inherent to these populations, including varied displacement and dendrite extension patterns. LpMs exhibited slow displacement, while MMs were primarily static, and MMs possessed greater dendrite ramifications than LpMs but reduced dendritic extension movements (Movies S1B and S1C and Figures 1B and 1C). Among the MM population, we observed at least two morphologically distinct sub-populations: bipolar and stellate cells (Phillips and Powley, 2012) (Movies S1D and S1E and Figure 1D). In general, bipolar cells produced small pseudopodia along the length of the cell body, approximately 0.2–0.8µm in size. Stellate cells also exhibited these pseudopodia, but they also displayed constant extensions and retractions of their dendritiform processes. These observations suggest that macrophages within distinct compartments of the gut display different morphologies and cellular dynamics.
Figure 1. MMs Differ from LpMs in Morphology and Cell Dynamics.
(A) Whole-mount imaging (iDISCO) of gut macrophages in the duodenum section isolated from Cx3cr1GFP/+ reporter mouse and stained with anti-GFP. Left, still image from a 3-D reconstruction (see Movie S1A). Right, Imaris-generated orthogonal slice. (B) Images from IVM of the ileum muscularis (left panels) and lamina propria (right panels) of live CD11ceYFP (upper panels) and Cx3cr1GFP/+ (lower panels) mice. Insets depict different morphological features of APCs in each region (see also Movies S1B and S1C). (C) Images from IVM of the ileum muscularis (upper panels) and lamina propria (lower panels) of live Cx3cr1GFP/+ mice. White arrows indicate morphologic changes over 15–20 min. (D) Confocal images from the ileum muscularis isolated from CD11ceYFP mice depicting different morphological features of MMs: bipolar cell (left panel) and stellate cell (right panel) (see also IVM Movies S1D and S1E). (E) Images from IVM of the ileum lamina propria (left column) and muscularis (right column) of Cx3cr1GFP/+ (middle row), zDCDTRCx3cr1GFP/+ (upper row) and lyz2csf1r-DTRCx3cr1GFP/+ (lower row) mice 12h post final DT administration. (A), Image is representative of 2 similar experiments with sections obtained from duodenum, jejunum, and ileum; (B-D), images are representative of at least 5 similar experiments; (E), images are representative of 3 similar experiments. See also Movie S1 and Figure S1.
Since, in addition to macrophages, the gut wall also contains several DC subpopulations (Bogunovic et al., 2009; Schreiber et al., 2013), we complemented this cell surface marker–based lineage classification with complementary ontogeny strategies. We targeted the macrophage/monocyte lineage using Lyz2Cre × Csf1rlsl-DTR (Lyz2Csf1r-DTR) and the pre-DC derived lineage using Zbtb46DTR (zDCDTR) mice (Schreiber et al., 2013). Administration of diphtheria toxin (DT) to Lyz2Csf1r-DTR mice leads to broad, rapid depletion of monocytes, monocyte– and yolk sac–derived tissue macrophages and inflammatory DCs that lasts for at least 36h. Complementarily, DT-injected zDCDTR mice show rapid and virtually complete loss of classical DCs (cDCs) (Schreiber et al., 2013). We generated bone marrow chimeras (BMC) from Cx3cr1GFP/+ reporter mice crossed to lyz2csf1r-DTR, zDCDTR or WT controls to allow CX3CR1+ APCs not targeted by DT to be visualized by the expression of GFP. At 12h post final DT injection, we observed a drastic reduction in GFP+ cells in the muscularis and lamina propria of lyz2csf1r-DTR when compared to WT controls or zDCDTR mice, indicating that the great majority of CX3CR1+ cells in these layers belongs to the monocyte/macrophage lineage (Figure 1E). In contrast, DT administration to CD11ceYFPlyz2csf1r-DTR BMC resulted in a drastic reduction in YFP+ cells in the muscularis, but only mild reduction in YFP+ in the lamina propria (Figure S1). These data indicate that CX3CR1-based strategies faithfully label macrophages throughout the gut tissue, while CD11c also labels non-monocyte/macrophages in the lamina propria and can therefore only be used to visualize MMs.
By combining methods to visualize deep-tissue structures with intersectional genetics and live imaging, these experiments establish tissue segregation, distinct morphology and cell dynamics of macrophages residing within distinct microenvironments in the intestinal tissue.
Intra-tissue specialization of lamina propria and muscularis macrophages
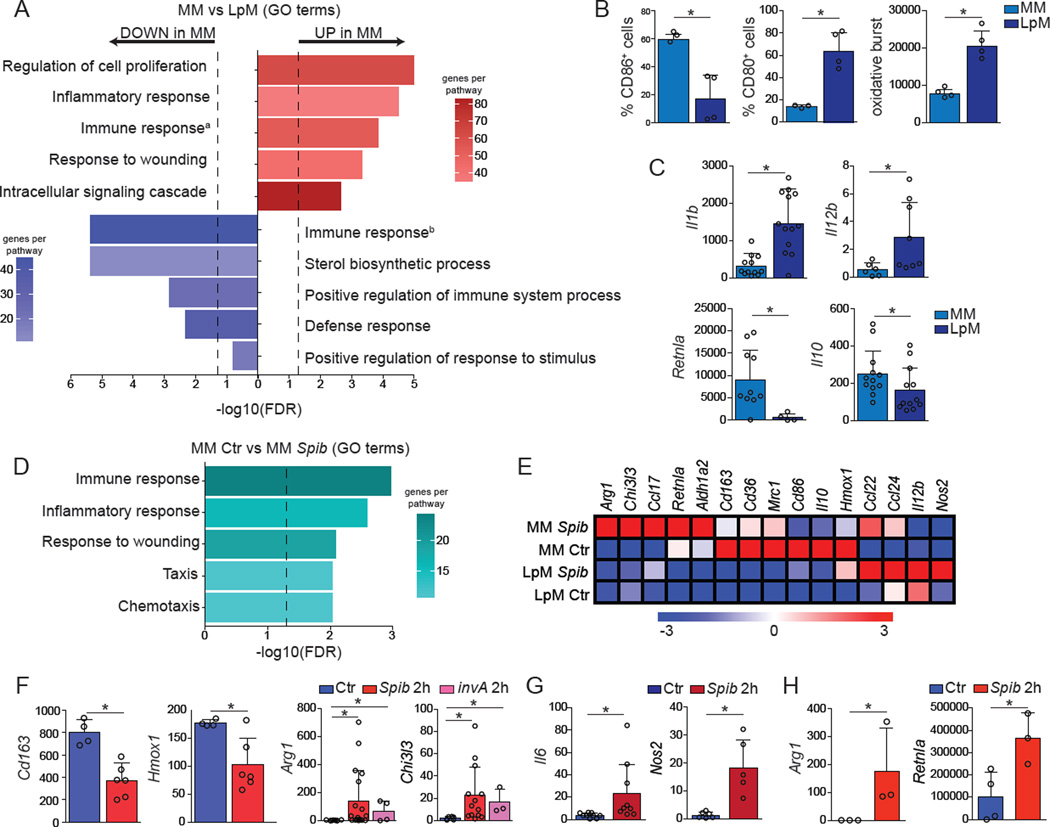
To address whether the morphological and cell dynamics characteristics of gut macrophages correspond to divergent tissue-specific adaptation, we analyzed the transcriptomes of LpMs and MMs. RNA-seq analysis of sorted (live, CD45+Lineage−MHCII+CD11b+CD11c+CD103−) cells at steady state revealed significant differences between LpM and MM cells in genes related to immune- and metabolic-processes (Figure S2, Table S1, Figures 2A and 2E). MM preferentially expressed tissue-protective and wound healing genes, such as Retnla (encoding Fizz1), Mrc1, Cd163 and Il10 and increased levels of the co-stimulatory molecule CD86 when compared to macrophages isolated from the lamina propria (Figures 2A–2C and 2E), and overall resemble alternatively activated (M2) macrophages (Van Dyken and Locksley, 2013). Analysis of the ImmGen database (Gautier et al., 2012), as well as flow cytometry and qPCR further confirmed this trend, also indicating that LpMs preferentially expressed pro-inflammatory, or M1, genes with increased oxidative burst and increased CD80 expression when compared to MMs (Figures 2B and 2C).
Figure 2. MMs and LpMs Exhibit Distinct Gene Expression Signatures.
(A–G) MMs and LpMs were isolated from the small intestine muscularis externa and mucosal layers, respectively, of WT mice and sorted as live (Aqua−) CD45+Lin−MHCII+F4/80+CD11B+ CD11C+CD103−. (A) Annotated gene ontology (GO) biological processes were assigned to genes differentially expressed by MMs when compared to LpMs, determined by RNA-seq. (B) Flow cytometry analysis of CD80 and CD86 expression and oxidative burst by MMs and LpMs. (C) Expression of mRNA for Il1b, Il12b, Retnla, and Il10 by MMs and LpMs determined by qPCR, presented relative to housekeeping gene Rpl32 expression. (D) Annotated GO biological processes were assigned to genes differentially expressed by MMs 2 h post intragastric exposure to Spib when compared to MMs isolated from naïve mice, determined by RNA-seq. (E) Heat map of representative M1– and M2–related gene were assigned to genes differentially expressed by MMs and LpMs 2 h post intragastric exposure to Spib, determined by RNA-seq. (F,G) Expression of mRNA for various genes by (F) MMs and (G) LpMs 2 h post intragastric exposure to Spib or invA Salmonella Typhimurium mutant strains determined by qPCR, presented relative to Rpl32 expression. (H) Expression of polysome-associated mRNA from small intestine muscularis of Lyz2 mice 2 h post intragastric exposure to Spib determined by qPCR, presented relative to Gapdh. (A, D and E), n = 2 per condition; (B), data are representative of at least 2 independent experiments, n = 4; (C), n = 6–12, pooled data; (F and G), n = 4–13; pooled data; (H), n= 3 per condition. Data were analyzed by unpaired T-test and are shown as average±SD, *p ≤ 0.05. See also Table S1, worksheets a–c). See also Figure S2.
The above data demonstrate an intra-tissue specialization among intestinal macrophages with potential for modified responses in specific microenvironments. To determine whether the basal differences between intestinal macrophages are maintained during responses to a pathogen, we sorted LpMs and MMs 2 h post intra-gastric exposure to mutant strains of Salmonella typhimurium, Spib and invA, which exhibit impaired proliferation or invasiveness, respectively, due to mutations in the type-III secretion system (Tsolis et al., 1999). We chose to study nonpathogenic or non-invasive bacteria strains since wild-type (WT) Salmonella Typhimurium quickly invades and damages the intestinal epithelium, hindering our objective to compare macrophages located in different regions of intact intestinal tissue. A 2 h time-point was chosen to represent the earliest detection point for immediate early gene (IEG) responses in macrophages (Ghisletti et al., 2010). Following gavage with the Spib-mutant, we observed significant further upregulation in some M2-associated genes in MMs, particularly Arg1 and Chi3I3 (encoding YM1). While Cd163 and Hmox1 were downregulated, they were still expressed at higher or similar levels than in LpMs, respectively (Table S1, Figures 2D–2F). Of note, whereas we observed variability in the expression of some IEG’s, particularly Arg1, among different animals, MMs from non-infected mice consistently did not express these genes (Figure 2F). In contrast, very few changes in gene expression were observed in LpMs upon intragastric exposure to Spib, and these changes were mostly restricted to M1-associated genes, including Nos2 and Il6 (Figures 2E, 2G).
To corroborate observed changes in MM gene expression profile using an independent approach, we conducted cell type-specific ribosomal profiling. This approach eliminates the need for cell sorting to purify sub-populations, thus reducing processing time and variability. We chose to specifically focus on actively translated mRNAs by interbreeding mice expressing Cre under the Lyz2 promoter with mice carrying a targeted knockin into the ribosomal protein L22 gene (RiboTag). Cre-mediated activation in this system creates an HA-tagged ribosomal protein, enabling affinity-tagged purification and identification of actively translated ribosomebound mRNAs in macrophages from intact tissues (Sanz et al., 2009). Analysis of intestinal muscularis from Lyz2RiboTag mice confirmed high levels of active translation of M2-related genes in steady state in MMs, particularly Retnla. Importantly, intragastric inoculation of mice with Spib rapidly led to upregulation of Arg1 as well as RetnIa (Figure 2H). These results suggest that unique microenvironmental cues influence the specialization of gut-resident macrophages and that this specialization is reinforced upon infection.
β2AR–positive muscularis macrophages reside in close proximity to firing neurons
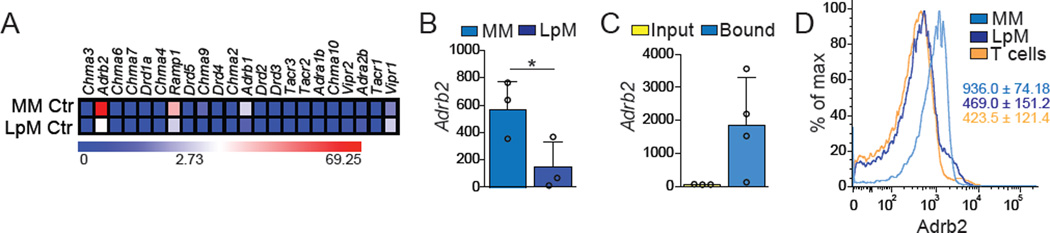
We next asked whether specific environmental signals, particularly soluble factors, were responsible for the distinct gene expression profiles and responses to luminal insults by LpM and MM populations. Gene expression profiling of LpM and MM cells indicated that pro- or anti-inflammatory cytokine-, TLR- or FcR-mediated signaling did not segregate these populations, especially considering the canonical M1 (IFN-γR, IFN-αR, IL-1R, TLR4) or M2 (FcγRIIb, IL-4Rα, IL-10Rα, ST2 (encoded by Il1rl1)) polarizing receptors (Figure S2B) (Gautier et al., 2012). Given their distinct locations near the lumen (LpM) or near neuronal networks (MM) (Kinoshita et al., 2007; Muller et al., 2014; Phillips and Powley, 2012), we next asked whether these macrophage populations differentially expressed neurotransmitter or neuropeptide receptors. MMs expressed high levels of Adrb2 (encoding β2 adrenergic receptors, AR), and this was also among the most significantly differentially-expressed genes between MM and LpM populations in the ImmGen database (Figure 3A) (Gautier et al., 2012). We confirmed these results by performing qPCR on sorted cells isolated from WT mice and on HA-bound polysomes of intestinal muscularis tissue isolated from Lyz2RiboTag mice (Figures 3B and 3C). Furthermore, flow-cytometric analysis of LpMs and MMs isolated from the small intestine of WT mice revealed high levels of membrane β2AR expression in MMs and lower, albeit detectable, expression of β2ARs by LpMs (Figure 3D).
Figure 3. MMs Preferentially Express β2-ARs.
(A) Heat maps of genes for neurotransmitter receptors expressed by sorted small intestine MMs and LpMs isolated from naïve WT mice under steady state conditions, determined by RNA-seq. (B) Expression of mRNA for Adrb2 by sorted small intestine MMs and LpMs isolated from naïve WT mice, determined by qPCR, presented relative to Rpl32 expression. (C) Expression of polysome-associated mRNA from small intestine muscularis of Lyz2RiboTag mice, determined by qPCR, presented relative to Gapdh expression. (D) Representative flow cytometry histogram for β2-AR expression by small intestine MMs, LpMs, and peripheral T cells isolated from naïve WT mice. (A), n = 2 per condition; (B), data are representative of 2 independent experiments, n = 3; (C), Input n = 3, Bound n = 4; (D), histograms are representative of 2 independent experiments, n =3. Data were analyzed by unpaired T-test and are shown as average±SD, *p ≤ 0.05. See also Figure S2
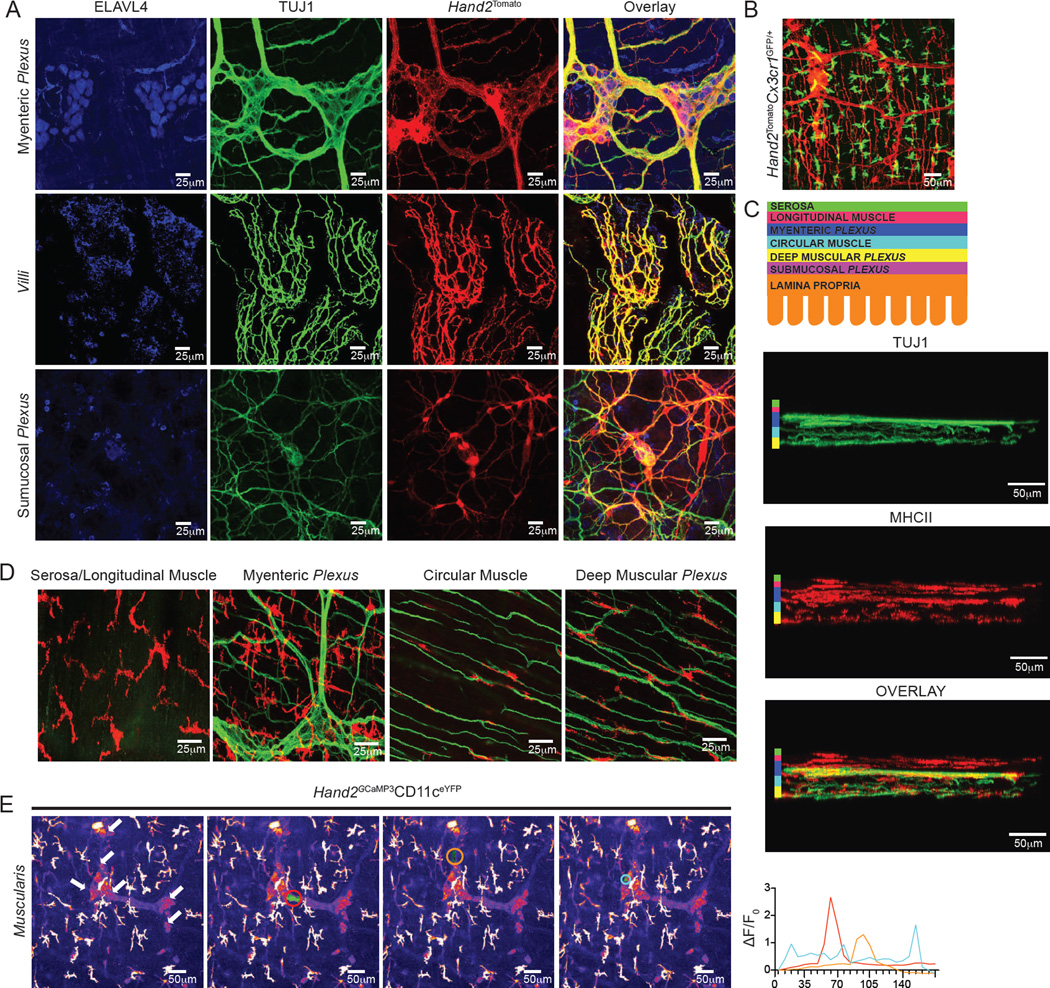
The near immediate gene-expression response of MMs to distal luminal stimuli, along with their selective expression of neurotransmitter receptors, suggested that their specific location in close proximity to neuronal networks could be influencing their activity. We therefore chose to characterize this neuron-rich microenvironment and how it interacts with gut macrophages. To that end, we developed multiple genetic labeling tools, allowing for concomitant and dynamic visualization of these cell types. To gain insight into neuron-macrophage interactions in live animals, we first generated a more specific enteric neuronal-associated reporter by interbreeding Rosa26lsl-tdTomato with Hand2Cre (Hand2Tomato) mice (Hendershot et al., 2007). These mice express Cre recombinase under the promoter of Hand2, a helix-loop-helix transcription factor with an established role in development of the enteric-associated neurons (EAN) (Barron et al., 2011). Cre-mediated removal of the stop cassette generated bright red fluorescence that allowed the visualization and tracking of Hand2-expressing cells, mostly restricted to EAN (Figure 4A). These mice were crossed with Cx3cr1GFP/+ reporter mice for tracking intestinal macrophages, and the resulting offspring were used for concomitant EAN-macrophage visualization. We observed that most of the CX3CR1+ cells in the muscular region were in close proximity to neuronal cell bodies or processes of the myenteric ganglia (Movie S2A and Figure 4B). We distinguished four layers of muscularis macrophages: serosal/longitudinal, myenteric plexus, circular muscle, and deep muscular plexus (Figures 4C and 4D). Serosal macrophages do not appear to be associated with enteric neurons and tend to be larger in size when compared to the other layers (Figure 4D). Myenteric plexus MMs are in close proximity to neuronal cell bodies and some nerve fibers, while MMs within the circular muscle and deep muscular plexus have their cell bodies running in parallel to the nerve fibers (Figure 4D).
Figure 4. MMs are Closely Associated with Active Neurons.
(A) Confocal images from the myenteric plexus (upper panels), villi (middle panels) and submucosal plexus (lower panels) isolated from naïve Hand2CreRosa26lsl-tdTomato (Hand2Tomato) mice and stained using anti-ELAVL4 (HuC/D) and anti-TUJ1 (βIII tubulin) antibodies. (B) Image from IVM of the ileum muscularis of live Hand2TomatoCx3cr1GFP/+ mice (see Movie S2A). (C, D) Confocal images from the ileum isolated from naïve WT mice stained using anti-TUJ1 (βIII tubulin) and anti-MHCII antibodies. (C, upper panel) Schematic of layers of different small intestine muscularis. (C, lower panel) Four layers of muscularis orthogonal slices are depicted according to the color scheme. (D) Images depict macrophages occupying each of the four layers: serosal/longitudinal muscle, myenteric plexus, circular muscle, and deep muscular plexus. (E) Images from IVM of the ileum muscularis of live Hand2GCaMP3CD11ceYFP mice over 3 min. White arrows indicate neurons labeled with GCaMP3, and colored regions of interest (ROI) highlight neural activity (see also Movie S2B). Corresponding colored traces plot changes in fluorescence (ΔF) of ROI per time frame (in seconds), with an increase in fluorescence indicative of Ca2+ influx and depolarization (right panel). (A), images are representative of 2 similar experiments with sections obtained from duodenum, jejunum, and ileum; (B), image is representative of at least 10 similar experiments; (C–E), images are representative of at least 3 independent experiments. See also Movie S2.
To complement our cell dynamics analysis, we assessed neuronal activation in areas where MMs were co-localized with EAN utilizing a mouse strain containing a genetically-encoded, Cre-dependent calcium indicator (GCaMP3) (Zariwala et al., 2012). Analysis of Hand2Cre × Rosa26lsl-GCaMP3 (Hand2GCaMP3) × CD11ceYFP mice revealed neuronal activity in very close proximity to MMs in the myenteric plexus (Movie S2B and Figure 4E). Taken together, these data indicate that MMs that express β2AR reside in close proximity to active neurons in the intestine.
Rapid activation of catecholaminergic neurons occurs upon luminal infection
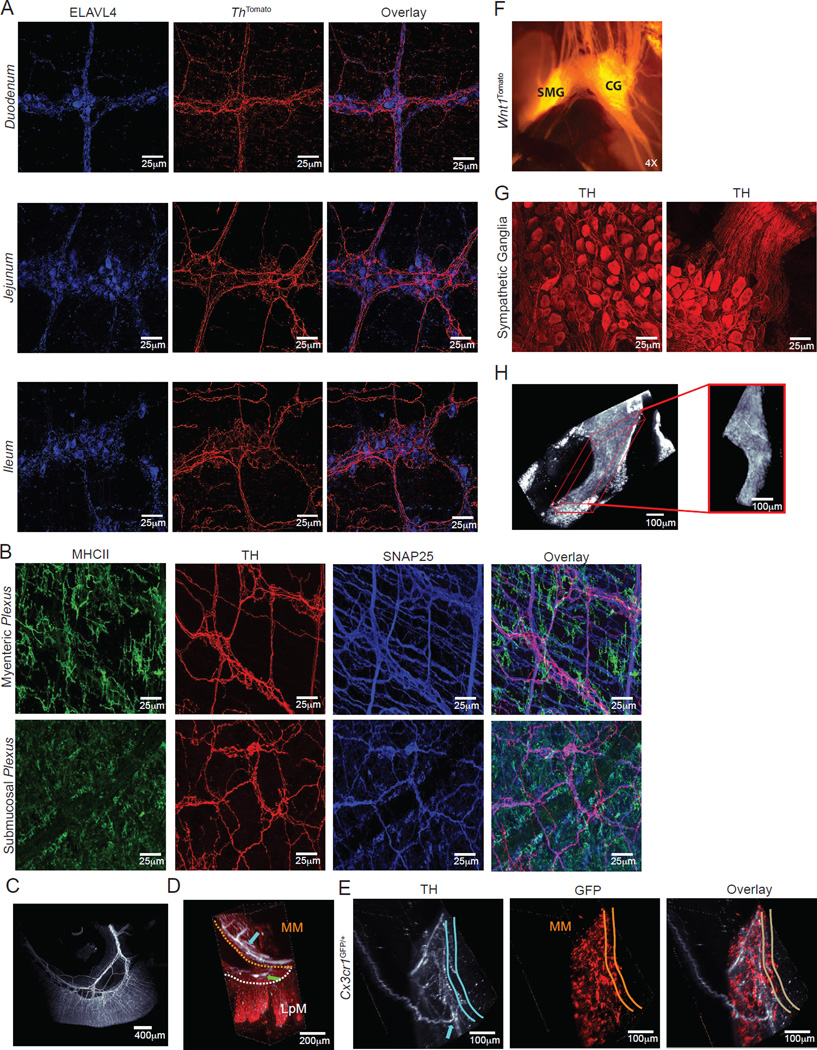
We next sought to characterize the specific neurochemical pathway involved in the activation of β2AR+ MMs by defining the source of norepinephrine (NE) in the gut muscularis. Although neurons are the most studied sources for catecholamines, recent studies have shown that myeloid cells, including macrophages, can also produce these neurotransmitters. Specifically, alternatively-activated macrophages involved in the regulation of thermogenesis were shown to express tyrosine hydroxylase (TH) and produce NE (Nguyen et al., 2011). To better characterize possible neuronal and non-neuronal sources for NE in the gut, we first interbred Rosa26lsl-tdTomato with ThCre mice (ThTomato) as a fate-mapping strategy to visualize cells that express TH at any point during development. Immunofluorescence microscopy analysis of ThTomato mice suggested that neurons, not myeloid cells, were the main source of NE within intestinal tissue, as revealed by extensive Tomato co-localization with the pan-neuronal marker ELAVL4 in the muscularis, and absence of Tomato expression by myeloid cells (Figure 5A), data confirmed by flow cytometric analysis (data not shown). To visualize cells actively expressing TH, we also performed ex vivo staining for TH and additional neuronal or myeloid markers. Again, TH staining was restricted to the neuronal compartment, particularly the fibers near MMs (Figure 5B). To visualize the catecholaminergic innervation throughout the intestinal tissue and how it permeates gut macrophages, we performed iDISCO in small intestinal sections from Cx3cr1GFP/+ reporter mice, co-stained with anti-GFP and anti-TH antibodies. Light-sheet microscopy analysis revealed intense TH+ innervation in the gut muscularis, but sparse labeling in the lamina propria (Movies S3A and S3B and Figures 5C and 5D). We also observed TH+ processes extending from the muscularis region (Movie S3C and Figures 5C and 5E), suggesting an extrinsic nature for this innervation.
Figure 5. Catecholaminergic Innervation of the Intestinal Muscularis.
(A) Confocal images from the duodenum (upper panels), jejunum (middle panels) and ileum (lower panels) isolated from naïve ThCreRosa26lsl-tdTomato (ThTomato) mice and stained using anti-ELAVL4 (HuC/D) antibodies. (B) Confocal images from the myenteric (upper panels) and submucosal plexuses (lower panels) isolated from naïve WT mice and stained using anti-MHCII, anti-SNAP25 and anti-TH antibodies. (C) Whole-mount imaging (iDISCO) from the ileum isolated from naïve WT mouse stained using anti-TH (white) antibodies (see also Movie S3A). (D, E) iDISCO from the ileum isolated from Cx3cr1GFP/+ reporter mouse stained using anti-GFP (red) and anti-TH (white) antibodies. (D) Arrows indicate TH+ processes in the myenteric (light blue) and submucosal (green) plexuses (see also Movie S3B). (E) Light blue arrow indicates TH+ processes innervating the myenteric plexus (see also Movie S3C). (F) Epifluorescent imaging from the celiac (CG) and superior mesenteric ganglia (SMG) of Wnt1CreRosa26lsl-tdTomato (Wnt1Tomato) mice. (G) Confocal images from the CG isolated from naïve WT mice and stained using anti-TH antibodies. Left panel, TH+ cell bodies; right panel, TH+ cell bodies and processes. (H) iDISCO from the CG isolated from WT mouse stained using anti-ELAVL4 antibody, showing the entire ganglia and the surrounding fat tissue (see also Movie S3D). (A–G), images are representative of at least 2 independent experiments with sections obtained from duodenum, jejunum, and ileum; (H), image is representative of at least 14 independent experiments. See also Movie S3 and Figure S3.
After defining catecholaminergic innervation as the probable source of NE in the MM microenvironment, we next characterized the intrinsic (cell bodies that lie within the intestinal tissue) versus extrinsic (cell bodies that lie outside of the intestinal tissue) nature of these neuronal processes (Furness et al., 1999). In order to identify and visualize the sympathetic innervation of the intestine, we developed general fate-mapping strategies by crossing Rosa26lsl-tdTomato with Snap25Cre (pan-neuronal) mice (Harris et al., 2014) or with Wnt1Cre (neural-crest derived) (Danielian et al., 1998) mice, respectively, which revealed the superior mesenteric-celiac ganglia (SMG-CG) (Figure 5F and data not shown). Immunolabeling of the SMG-CG showed a dense network of ELAVL4 and TH+ neurons extending processes towards the gut (Movie S3D and Figures 5G and 5H). Consistent with this, staining for dopamine β-hydroxylase (DBH), which converts dopamine into norepinephrine, revealed punctate labeling restricted to ganglia and nerve fibers in the myenteric and submucosal plexuses, while extrinsic sympathetic neurons displayed signal in their cytoplasm (Figure S3). This observation indicates that at least part of the TH staining observed in the muscularis was of an extrinsic nature, as previously suggested (Li et al., 2010).
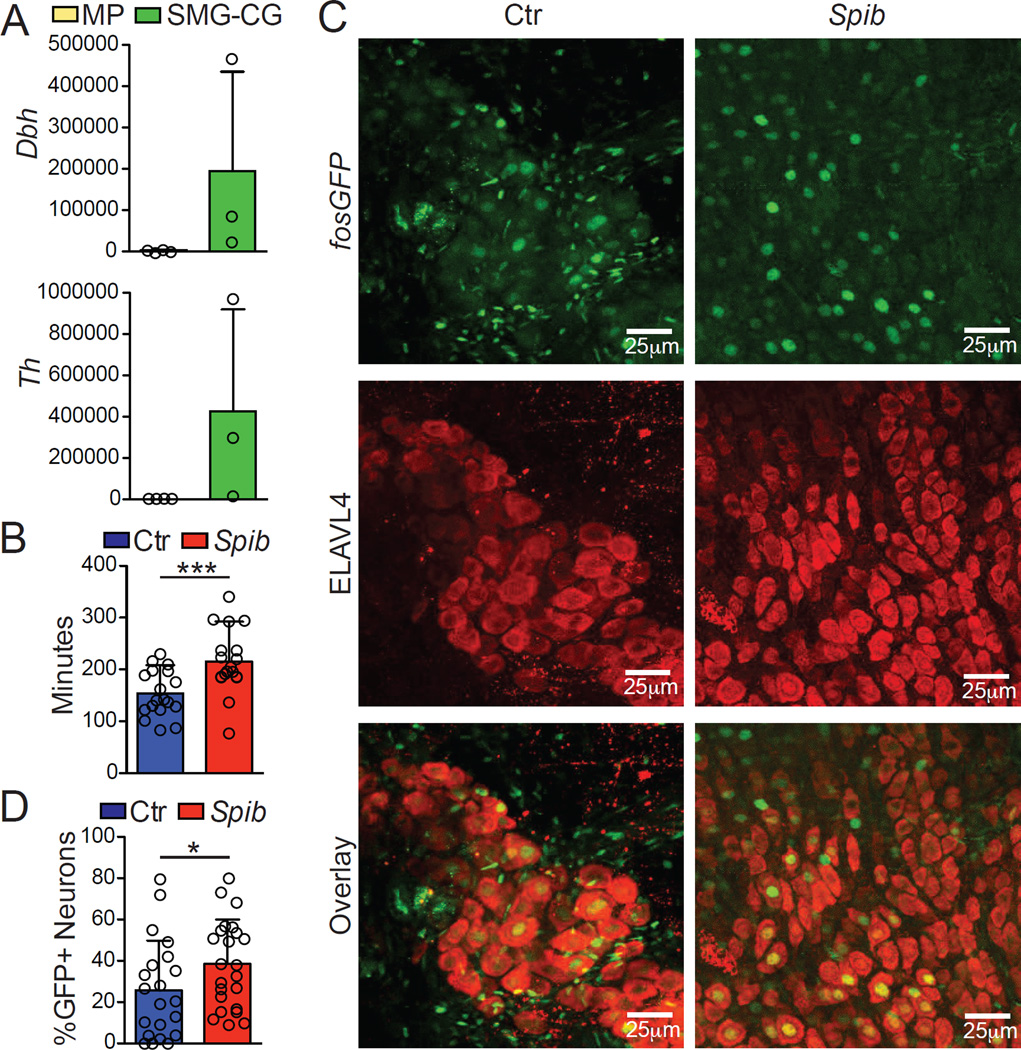
To address whether intrinsic innervation also contributed to the catecholaminergic response in the muscle, we generated a broad neuronal RiboTag strain by crossing Rpl22lsl-HA with Snap25Cre (Snap25RiboTag) mice. Analysis of HA-bound fractions from the muscularis and the SMG-CG revealed high expression of both Th and Dbh by SMG-CG but a lack of expression in the myenteric plexus (Figure 6A). We therefore concluded that a majority of the NE release in the gut muscularis microenvironment is derived from extrinsic sympathetic innervation.
Figure 6. Extrinsic Sympathetic Innervation is Activated During Enteric Infection.
(A) Expression of polysome-associated mRNA from small intestine muscularis and SMG-CG of naïve Snap25RiboTag mice determined by qPCR, presented relative to Hprt. (B) Total GI transit time (time required to expel feces containing carmine red dye) in mice 30 min post intragastric exposure to Spib or PBS (control). (C) Confocal images from the SMG-CG isolated from FosGFP mice 2 h post Spib intragastric exposure and stained using anti-GFP (upper panels) and anti- ELAVL4 (middle panels) antibodies. (D) Quantification of the number of GFP+ nuclei amongst ELAVL4+-TH+ neurons within SMG-CG. (A), pooled data shown are from 3 independent experiments, n = 3; (B), pooled data shown are from 4 independent experiments, n = 18; (C), images are representative of 2 independent experiments, n=6; (D), pooled data shown are from 2 independent experiments, n = 6 (each dot represents one image of sympathetic ganglion neurons). Data were analyzed by unpaired T-test and are shown as average±SD, *p ≤ 0.05,***p ≤ 0.001. See also Figure S4.
One of the consequences of NE in the muscularis region is the relaxation of intestinal smooth muscle (Pullinger et al., 2010). Consistent with this effect, we observed a significant impairment of gastrointestinal motility following oral gavage with Salmonella Typhimurium mutant Spib (Figure 6B). Additionally, quantification of NE in the cecal contents also indicated increased NE release in the intestinal tissue upon Spib infection (Figure S4). Finally, to confirm catecholaminergic neuronal activation after luminal infection, we measured c-Fos activation using FosGFP reporter mice. Oral administration of Spib resulted in pronounced activation of SMG-CG ganglia neurons and undetectable (or non-neuronal) c-Fos expression in the intestinal muscularis (Figure S4 and Figures 6C and 6D). Collectively, these data suggest that enteric infections activate gut extrinsic sympathetic innervation resulting in NE release within the muscular region and signaling through β2ARs in MMs.
β2AR–mediates alternative activation of intestinal macrophages
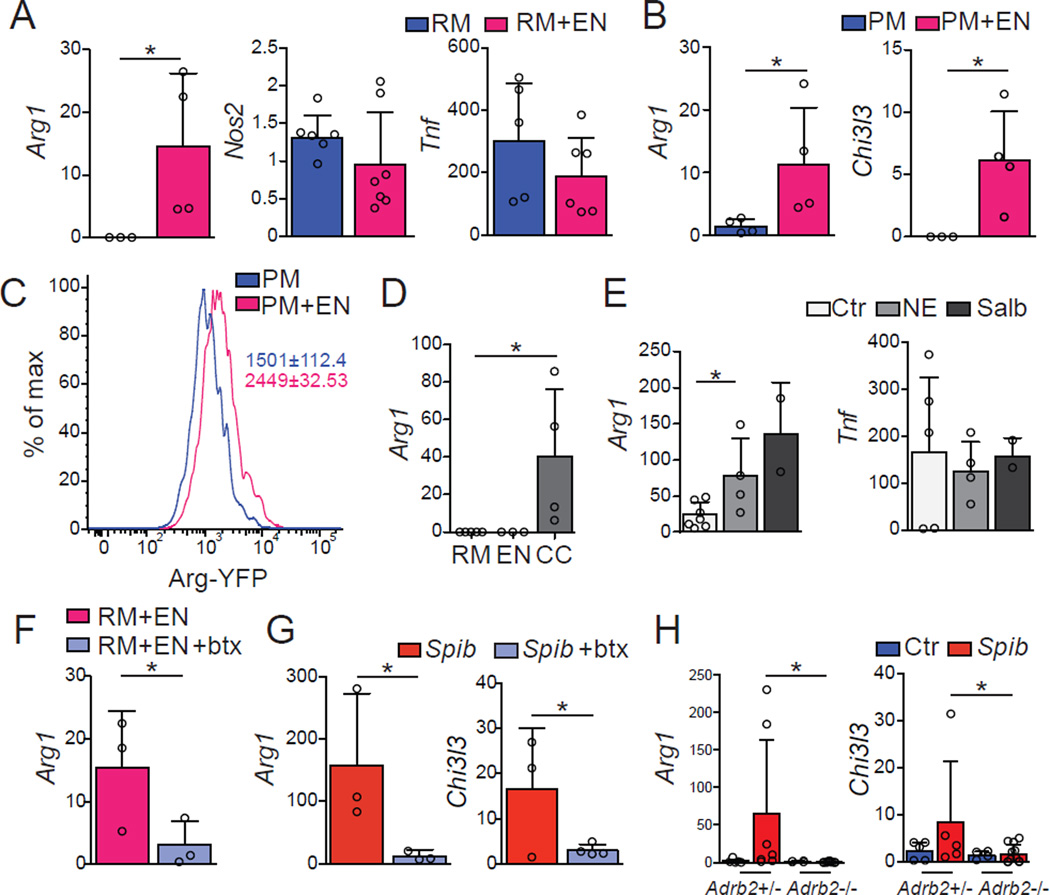
To investigate whether interaction with neurons is sufficient to directly modulate gene expression in macrophages, we first developed an in vitro co-culture system using neurospherederived primary enteric-associated neurons and either peritoneal macrophages or a macrophage cell line (RAW 264). Co-culture of RAW 264 macrophages or WT peritoneal macrophages with primary EANs resulted in increased levels of Arg1 and Chi3l3 expression with no change in Nos2 or Tnf expression (Figures 7A–7C). The supernatant of macrophage- EAN co-cultures (conditioned media), but not EAN or macrophage media alone, was also able to induce Arg1 in macrophages, indicating that, at least under these conditions, EANs may require cell-extrinsic signals in order to release soluble factors responsible for macrophage polarization (Figure 7D). Aptly, both NE and Salbutamol, a specific β2AR agonist, directly induced Arg1 upregulation but did not change Tnf expression in peritoneal macrophages (Figure 7E). Furthermore, addition of butaxamine, a selective β2AR blocker, prevented Arg1 upregulation in macrophages co-cultured with EANs (Figure 7F). In these settings, however, we were unable to detect gene changes in additional M2-genes associated with the MM profile (data not shown). These findings reveal that enteric neurons and β2AR ligands can induce macrophage polarization in vitro resembling MMs.
Figure 7. β2ARs–Mediate Polarization of Macrophages.
(A–F) WT mice neuro-sphere–derived primary enteric-associated neurons (EANs) were cocultured with RAW (RM) or peritoneal macrophages (PM) from WT mice. (A, B) Expression of mRNA for Arg1, Nos2, Tnf and Chi3l3 by sorted (A) RM or (B) PM 18 h post co-culture with EANs. (C) Representative flow cytometry histogram for YFP expression by sorted PM isolated from Arg1YFP mice cultured with EANs as in A. (D) Expression of mRNA for Arg1 by sorted RM 18 h post exposure to conditioned media from RM, EANs or RM-EANs co-cultures (CC). (E) Expression of mRNA for Arg1 and Tnf by sorted PM 1 h post exposure to NE or Salbutamol (β2AR agonists). (F) Expression of mRNA for Arg1 by sorted RM 18 h post co-culture with EANs with or without butaxamine (β2AR-selective blocker). (G) Expression of mRNA for Arg1 and Chi3l3 by sorted small intestine MMs isolated 2 h post intragastric exposure to Spib in mice treated with vehicle or butaxamine. (H) Expression of mRNA for Arg1 and Chi3l3 by sorted small intestine MMs isolated from Adrb1+/−Adrb2−/− and Adrb1+/−Adrb2+/− littermate control mice 2 h post intragastric exposure to Spib. qPCR results are presented relative to Rpl32 expression. (A-H), pooled data of at least 2 independent experiments (A, n = 3–7; B-D, n = 3–4; E, n = 2–4; F and G, n = 3–4;H, n = 4–11. Data were analyzed by unpaired T-test and are shown as average±SD, *p ≤ 0.05. See also Figure S2.
We next asked whether β2AR-mediated signaling contributes to in vivo MM polarization after intestinal infection. Previous studies have reported anti-inflammatory effects of β2AR signaling on macrophages, possibly mediated by upregulation of M2-related genes upon exposure to β2AR agonists (Spengler et al., 1994). In vivo administration of butaxamine significantly impaired upregulation of Arg1 and Chi3I3 (Figure 7G). To further address the role of β2ARs in this pathway, we also analyzed beta-adrenergic receptor null mice. MMs isolated from Adrb1+/− Adrb2−/− exhibited markedly reduced expression of Arg1 and Chi3I3 relative to MMs isolated from Adrb1/2+/− littermate controls upon Spib infection (Figure 7H). The above data provide evidence that enteric infection triggers changes in MM gene expression via neuron-derived adrenergic signaling. Altogether, our results reveal communication between enteric-associated neurons and macrophages in the muscularis microenvironment that serves to induce a tissueprotective gene-expression program in resident macrophages (Graphical Abstract).
Discussion
Dynamics of enteric neuron-macrophage interaction
Tissue-resident macrophages adapt to distinct niches to perform protective, reparative, or pro-inflammatory functions depending on their environmental cues and epigenetic state (Lavin et al., 2014; Okabe and Medzhitov, 2014). In the intestinal tissue, tissue macrophages are constantly exposed to both exogenous stimuli, such as bacteria and viruses, and endogenous stimuli, which can be membrane-bound or secreted stress molecules from diverse neighboring hematopoietic and stromal cells (Diehl et al., 2013; Farache et al., 2013). Several lines of evidence suggest that the enteric nervous system can directly communicate with MMs. Electron microscopy and immune staining-based studies have suggested that MMs form synapses with enteric neurons (Kinoshita et al., 2007; Phillips and Powley, 2012). Recent work by the Merad and Bogunovic groups has indicated that MMs directly regulate the activity of enteric neurons and peristalsis via secretion of BMP-2 in a microbiota-dependent manner (Muller et al., 2014). We provide evidence for reciprocal neuron-dependent macrophage adaptation to environmental perturbations in the intestinal muscularis, where macrophages represent the main hematopoietic lineage (Bogunovic et al., 2009) and share a niche with a dense network of neurons. Our analyses of the in vivo dynamics of neuron-macrophage “structural coupling” under inflammatory conditions provides insights into this complex interaction, with possible implications for disease tolerance (Medzhitov et al., 2012).
While lamina propria macrophages are motile and can directly sense signals derived from invading bacteria or epithelial cell stress responses (Diehl et al., 2013; Farache et al., 2013; Mazzini et al., 2014), macrophages residing in the submucosal layer, and in particular the intestinal muscularis, are less likely to directly sense perturbations from the lumen. At the same time, gastrointestinal infections may lead to long-lasting tissue damage induced by inflammatory cells (Rakoff-Nahoum et al., 2004). Hence, mechanisms that induce tissue-protective functions in resident macrophages are an essential component of responses to pathogens (Medzhitov et al., 2012). This might be of particular importance to tissues that include cells with reduced proliferative or regenerative capacity, such as neurons. The static position of MMs, primarily alongside neuronal cell bodies and nerve fibers, provides an optimal interface for such interaction. It is curious to note that the cell dynamics of the bipolar and stellate cells appear to match the structure to which they are most closely associated. For instance, bipolar cells, running parallel to nerve fibers, extend in only two directions and have small pseudopodial protrusions. Stellate MMs, which surround ganglia and neuronal cell bodies, can presumably completely embrace their surrounding area. These features suggest that MMs below the serosa are involved in the monitoring of neuronal status either through detection of electrical activity or secreted factors, such as CSF-1 (Muller et al., 2014). In these ways, the β2ARs-mediated MM response may represent an analogous activity to that described for microglia in the CNS; protecting closely associated neuronal processes from infection- or inflammation-induced tissue damage (Davalos et al., 2005; El Khoury et al., 2007; Nimmerjahn et al., 2005; Wang et al., 2015). Several of the genes modulated by the β2AR pathway are known to be involved in tissue-protective functions. For instance, Arginase 1 has been implicated in the development of the nervous system, axogenesis and neuro-regeneration, as well as anti-apoptotic effects on neurons (Cai et al., 2002; Estevez et al., 2006). Thereby, overt inflammation coupled with disruption of this pathway may lead to tissue damage. Indeed, clinical observations indicate that roughly 10% of irritable bowel syndrome (IBS) patients developed symptoms after episodes of gastrointestinal infection (Ohman and Simren, 2010). It seems plausible that severe or prolonged GI infection could overrun normal safeguards, such as the MM anti-inflammatory program, and damage EAN networks, permanently impacting GI physiology.
Sensing of intestinal insults at luminal sites
We observed that bacterial infection leads to activation of extrinsic sympathetic ganglia innervating the intestine. How do intrinsic or extrinsic neuronal pathways sense luminal bacteria? Attenuated strains (Tsolis et al., 1999), which already exhibit a reduced capacity to invade even with streptomycin pretreatment, are expected to have limited interaction with the epithelium. Many different pathways could operate in an epithelial-neuronal sensing mechanism. For instance, Wilson and coworkers have shown that epithelial cell-derived cytokine thymic stromal lymphopoietin (TSLP), upregulated during allergic responses, is sensed by afferent neurons to trigger itch behavior (Wilson et al., 2013). In the intestine, where epithelial cells are the source of more than 50% of the body’s dopamine and 90% of the body’s serotonin, it is very likely that enteroendocrine epithelial cells play an important role in communicating luminal perturbations to neuronal processes (Gershon and Tack, 2007). A recent report by Bohorquez and coworkers indicated that intestinal enteroendocrine cells directly connect with neurons innervating the small intestine and colon, possibly providing signals received from the lumen (Bohorquez et al., 2015). An alternative, albeit non-mutually exclusive, possibility is that neuronal processes reaching the intestinal villi directly sense microbial or stress signals, as recently shown in worms. Meisel et al. described a chemosensory mechanism used by Caenorhabditis elegans to detect bacterial metabolites and induce avoidance behavior. This pathway involves direct neuronal sensing of pathogenic stimuli, which in turn activates adjacent neurons (Meisel et al., 2014). In mammals, Chiu et al. showed that sensory neurons could directly respond to bacterial-derived N-formylated peptides or toxins, inducing pain (Chiu et al., 2013). In this study, ablation of nociceptors resulted in increased inflammatory infiltrate, suggesting an anti-inflammatory role for this pathway during bacterial infection (Chiu et al., 2013). An analogous pathway could be operational in the intestine, where potentially similar metabolites from commensal or invading bacteria would directly activate enteric neurons. Indeed, studies have suggested that enteric neurons express PRRs, such as TLR2 or TLR4 (Anitha et al., 2012) that can contribute to activation of nociceptive-associated channels in neurons such as TRPV1. Alternatively, TLR ligands may be able to directly activate these channels independent of TLRs (Meseguer et al., 2014). However, this sensing circuit still requires further studies specifically targeting PRR pathways or other potential direct sensing channels or receptors in EAN.
Regulation of inflammatory responses by the catecholamine-β2AR pathway
In addition to the well-described anti-inflammatory reflex, the involvement of catecholaminergic neurons in anti-inflammatory responses has many equivalents in the immune system (Tracey, 2009). However, catecholamines are also linked to enhanced inflammatory responses. It is thought that α-adrenergic signaling boosts inflammation while β-adrenergic signaling suppresses both innate and adaptive immunity (Guereschi et al., 2013; Nakai et al., 2014). Some of the broad anti-inflammatory effects of this pathway are attributed to widespread sympathetic innervation in peripheral lymph nodes associated with expression of β2AR in immune cells (del Rey and Besedovsky, 2008). The high expression of these receptors by MMs might represent a mechanism by which heavy norepinephrine innervation in the muscularis could preferentially target these cells, mitigating the pro-inflammatory state induced in LpMs close to the lumen. Plausibly, deep muscular and myenteric macrophages are in a privileged position to respond to neuronal signals, including NE, due to their contact with neuronal fibers, as opposed to their serosa-longitudinal muscle counterparts. Alternatively, lack of β2AR on neurons or another cell population may lead to a decrease in a necessary support factor in these areas. Similarly, subpopulations of B cells, macrophages and neutrophils preferentially express β2AR when compared to T cells (del Rey and Besedovsky, 2008), which could also explain how NE might regulate inflammatory responses without leading to immune suppression. Additionally, circulating levels of NE, as opposed to much higher concentrations around catecholaminergic nerve terminals comprise an additional layer of control of this MM-EAN-associated NE-β2AR axis (del Rey and Besedovsky, 2008). Although we found TH+ immunoreactivity within the intestinal muscularis as well as in the SMG-CG, we only observed activation of extrinsic innervation upon bacterial challenge. This is consistent with previous reports suggesting that the CG and SMG are the main sources of NE in the stomach and intestine (Li et al., 2010). Nevertheless, we cannot rule out the possibility that intrinsic innervation is also involved in this process since extrinsic innervation synapses directly onto EAN, and this could also be involved in sensing mechanisms (McVey Neufeld et al., 2015).
The hardwiring of lymphoid and non-lymphoid tissues with neuronal processes allows for immediate regulation of closely-associated cells, and this could potentially synchronize immune responses in areas not directly in contact with the stimuli. We have uncovered a neuro-immune axis that reinforces a tissue-protective program in macrophages in response to a potentially pathogenic insult. Future studies are needed to dissect the exact (afferent and efferent) roles played by EAN in this process. Additionally, it remains unclear how luminal signals reach intrinsic or extrinsic innervation. Exploring these questions are the next steps to understanding how neuronal hardwiring in the intestine and other immune-rich tissues can modulate immune responses and how immune cells may prevent or cause neuronal damage.
Experimental Procedures
Animals
ArgYFP(Arg1tm1Lky/J), Adrb1tm1BkkAdrb2tm1Bkk, C57BL/6J, Cx3cr1GFP(Cx3cr1tm1Litt/LittJ), Rosa26lsl-GCaMP3, Rosa26lsl-tomato, Itgax(CD11c)eYFP, Lyz2Cre/Cre (Lyz2tm1(cre)Ifo/J), FosGFP(B6.Cg-Tg(Fos/EGFP)1-3Brth/J), Snap25Cre, Rpl22tm1.1Psam/J, Wnt1Cre and zDCDTR (Zbtb46DTR) mice were purchased from the Jackson Laboratories and maintained in our facilities. Hand2Cre and ThCre, mice were generously provided by D. Clouthier (UC Denver) and J. Friedman (RU), respectively. These lines were interbred in our facilities to obtain the final strains described in the text. Genotyping was performed according to the protocols established for the respective strains by Jackson Laboratories. Mice were maintained at the Rockefeller University animal facilities under specific pathogen-free conditions. Mice were used at 7–12 weeks of age for most experiments. Animal care and experimentation were consistent with the NIH guidelines and were approved by the Institutional Animal Care and Use Committee at the Rockefeller University.
iDISCO
The iDISCO protocol was followed as detailed on the continuously-updated website: http://idisco.info. For more detailed information, please see the Supplemental Experimental Procedures.
Intravital Two-Photon Imaging
Images were acquired as previously described (Farache et al., 2013). For more detailed information, please see the Supplemental Experimental Procedures.
RiboTag
Isolation of HA-tagged polysomes was performed as previously described (Sanz et al., 2009). For more detailed information, please see the Supplemental Experimental Procedures.
Single Cell Suspension of Intestinal Macrophages
After cleaning and washing in HBSS, the small intestine tissue was cut in two and the muscularis region was carefully dissected from the underlying mucosa. Each region was then finely cut and digested as previously described (Muller et al., 2014). For more detailed information, please see the Supplemental Experimental Procedures.
Salmonella Typhimurium Infections
Mice were intragastrically exposed to 109 of either mutant strain of Salmonella Typhimurium, Spib or invA. For more detailed information, please see the Supplemental Experimental Procedures.
Statistics
Statistical analyses were performed in GraphPad Prism software. Data were analyzed by applying one-way ANOVA or unpaired Student’s t-test where appropriate. A P value of less than 0.05 was considered significant.
For additional detailed information, please see the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We are indebted to K. Velinzon and N. Thomas for sorting cells, P. Ariel and the Rockefeller University BIRC for their invaluable help with IVM, N. Renier and R. Azevedo for their help with the iDISCO, D. Esterházy for kindly providing lyz2csfr1-DTRCD11ceYFP mice, M. Bogunovic and M. Merad (Mount Sinai) for their insightful discussions and help with the isolation of MMs and LpMs, J. Farache, G. Shakhar (Weizmann) and G. Victora (Whitehead, MIT) for their help in establishing live multiphoton microscopy of the gut, members of the Nussenzweig lab and The Rockefeller University employees for continuous assistance. We thank S. Tavazoie, V. Ruta, M. Nussenzweig (RU) and members of our laboratory, particularly V. Pedicord for discussions and critical reading and editing of the manuscript. D.M. is supported by a Kenneth Rainin Foundation Breakthrough Award, a National Institutes of Health NIH 5R21AI105047 grant. D.M. and P.M. are supported by the Leona M. and Harry B. Helmsley Charitable Trust. L.F. was supported by Crohn’s & Colitis Foundation of America. I.G. and F.A.C-P. were supported by FAPESP and CNPq (Brazil). The BIRC is supported the Empire State Stem Cell Fund through NYSDOH C023046. The authors have no conflicting financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.M. conceived and supervised this study. I.G., P.M., F. A.C-P and D.M. designed experiments. I.G., P.M., L.F. and F. A.C-P performed experiments. I.G. and P.M. prepared figures and helped with manuscript preparation (contributed equally to this manuscript, alphabetic order). T.Y.O. analyzed RNA-seq and helped with figure preparation. D.M. wrote the paper.
Accession Numbers
All RNA-seq data are submitted to NCBI’s Gene Expression Omnibus, with the accession number GEO: GSE74131.
References
- Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology. 2012;143:1006–1016. e1004. doi: 10.1053/j.gastro.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron F, Woods C, Kuhn K, Bishop J, Howard MJ, Clouthier DE. Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development. 2011;138:2249–2259. doi: 10.1242/dev.056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohorquez DV, Shahid RA, Erdmann A, Kreger AM, Wang Y, Calakos N, Wang F, Liddle RA. Neuroepithelial circuit formed by innervation of sensory enteroendocrine cells. J Clin Invest. 2015 doi: 10.1172/JCI78361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Deng K, Mellado W, Lee J, Ratan RR, Filbin MT. Arginase I and polyamines act downstream from cyclic AMP in overcoming inhibition of axonal growth MAG and myelin in vitro. Neuron. 2002;35:711–719. doi: 10.1016/s0896-6273(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Chiu IM, Heesters BA, Ghasemlou N, Von Hehn CA, Zhao F, Tran J, Wainger B, Strominger A, Muralidharan S, Horswill AR, et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature. 2013;501:52–57. doi: 10.1038/nature12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- del Rey A, Besedovsky HO. Sympathetic nervous system-immune interactions in autoimmune lymphoproliferative diseases. Neuroimmunomodulation. 2008;15:29–36. doi: 10.1159/000135621. [DOI] [PubMed] [Google Scholar]
- Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khoury J, Toft M, Hickman SE, Means TK, Terada K, Geula C, Luster AD. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat Med. 2007;13:432–438. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- Estevez AG, Sahawneh MA, Lange PS, Bae N, Egea M, Ratan RR. Arginase 1 regulation of nitric oxide production is key to survival of trophic factor-deprived motor neurons. J Neurosci. 2006;26:8512–8516. doi: 10.1523/JNEUROSCI.0728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farache J, Koren I, Milo I, Gurevich I, Kim KW, Zigmond E, Furtado GC, Lira SA, Shakhar G. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity. 2013;38:581–595. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness JB, Kunze WA, Clerc N. Nutrient tasting and signaling mechanisms in the gut. II. The intestine as a sensory organ: neural, endocrine, and immune responses. Am J Physiol. 1999;277:G922–G928. doi: 10.1152/ajpgi.1999.277.5.G922. [DOI] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397–414. doi: 10.1053/j.gastro.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Guereschi MG, Araujo LP, Maricato JT, Takenaka MC, Nascimento VM, Vivanco BC, Reis VO, Keller AC, Brum PC, Basso AS. Beta2-adrenergic receptor signaling in CD4+ Foxp3+ regulatory T cells enhances their suppressive function in a PKA-dependent manner. Eur J Immunol. 2013;43:1001–1012. doi: 10.1002/eji.201243005. [DOI] [PubMed] [Google Scholar]
- Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, Muller W, Sparwasser T, Forster R, Pabst O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, Bohn P, Mortrud M, Ouellette B, Kidney J, et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 2014;8:76. doi: 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Sarkar AA, Giovannucci DR, Clouthier DE, Abe M, Howard MJ. Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev Dyn. 2007;236:93–105. doi: 10.1002/dvdy.20989. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Horiguchi K, Fujisawa M, Kobirumaki F, Yamato S, Hori M, Ozaki H. Possible involvement of muscularis resident macrophages in impairment of interstitial cells of Cajal and myenteric nerve systems in rat models of TNBS-induced colitis. Histochem Cell Biol. 2007;127:41–53. doi: 10.1007/s00418-006-0223-0. [DOI] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Caron MG, Blakely RD, Margolis KG, Gershon MD. Dependence of serotonergic and other nonadrenergic enteric neurons on norepinephrine transporter expression. J Neurosci. 2010;30:16730–16740. doi: 10.1523/JNEUROSCI.2276-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1(+) macrophages to CD103(+) dendritic cells. Immunity. 2014;40:248–261. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- McVey Neufeld KA, Perez-Burgos A, Mao YK, Bienenstock J, Kunze WA. The gut microbiome restores intrinsic and extrinsic nerve function in germ-free mice accompanied by changes in calbindin. Neurogastroenterol Motil. 2015;27:627–636. doi: 10.1111/nmo.12534. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell. 2014;159:267–280. doi: 10.1016/j.cell.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseguer V, Alpizar YA, Luis E, Tajada S, Denlinger B, Fajardo O, Manenschijn JA, Fernandez-Pena C, Talavera A, Kichko T, et al. TRPA1 channels mediate acute neurogenic inflammation and pain produced by bacterial endotoxins. Nat Commun. 2014;5:3125. doi: 10.1038/ncomms4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Koscso B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, Mortha A, Leboeuf M, Li XM, Mucida D, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Hayano Y, Furuta F, Noda M, Suzuki K. Control of lymphocyte egress from lymph nodes through beta2-adrenergic receptors. J Exp Med. 2014;211:2583–2598. doi: 10.1084/jem.20141132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KD, Qiu Y, Cui X, Goh YP, Mwangi J, David T, Mukundan L, Brombacher F, Locksley RM, Chawla A. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Ohman L, Simren M. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat Rev Gastroenterol Hepatol. 2010;7:163–173. doi: 10.1038/nrgastro.2010.4. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, 3rd, Lafaille JJ, Hempstead BL, Littman DR, Gan WB. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RJ, Powley TL. Macrophages associated with the intrinsic and extrinsic autonomic innervation of the rat gastrointestinal tract. Auton Neurosci. 2012;169:12–27. doi: 10.1016/j.autneu.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullinger GD, van Diemen PM, Carnell SC, Davies H, Lyte M, Stevens MP. 6-hydroxydopamine-mediated release of norepinephrine increases faecal excretion of Salmonella enterica serovar Typhimurium in pigs. Vet Res. 2010;41:68. doi: 10.1051/vetres/2010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber HA, Loschko J, Karssemeijer RA, Escolano A, Meredith MM, Mucida D, Guermonprez P, Nussenzweig MC. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J Exp Med. 2013;210:2025–2039. doi: 10.1084/jem.20130903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MP, Gozzelino R, Weis S. Tissue damage control in disease tolerance. Trends Immunol. 2014;35:483–494. doi: 10.1016/j.it.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Spengler RN, Chensue SW, Giacherio DA, Blenk N, Kunkel SL. Endogenous norepinephrine regulates tumor necrosis factor-alpha production from macrophages in vitro. J Immunol. 1994;152:3024–3031. [PubMed] [Google Scholar]
- Tracey KJ. Reflex control of immunity. Nat Rev Immunol. 2009;9:418–428. doi: 10.1038/nri2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis RM, Townsend SM, Miao EA, Miller SI, Ficht TA, Adams LG, Baumler AJ. Identification of a putative Salmonella enterica serotype typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun. 1999;67:6385–6393. doi: 10.1128/iai.67.12.6385-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol. 2013;31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, Barrow AD, Diamond MS, Colonna M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SR, The L, Batia LM, Beattie K, Katibah GE, McClain SP, Pellegrino M, Estandian DM, Bautista DM. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell. 2013;155:285–295. doi: 10.1016/j.cell.2013.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala HA, Borghuis BG, Hoogland TM, Madisen L, Tian L, De Zeeuw CI, Zeng H, Looger LL, Svoboda K, Chen TW. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J Neurosci. 2012;32:3131–3141. doi: 10.1523/JNEUROSCI.4469-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim KW, Brenner O, Krauthgamer R, Varol C, Muller W, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40:720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.