Abstract
We previously reported that tacrolimus (TAC) trough blood concentrations for African American (AA) kidney allograft recipients were lower than those observed in white patients. Subtherapeutic TAC troughs may be associated with acute rejection (AR) and AR-associated allograft failure. This variation in TAC troughs is due, in part, to differences in the frequency of the cytochrome P450 CYP3A5*3 allele (rs776746, expresses nonfunctional enzyme) between white and AA recipients; however, even after accounting for this variant, variability in AA-associated troughs is significant. We conducted a genomewide association study of TAC troughs in AA kidney allograft recipients to search for additional genetic variation. We identified two additional CYP3A5 variants in AA recipients independently associated with TAC troughs: CYP3A5*6 (rs10264272) and CYP3A5*7 (rs41303343). All three variants and clinical factors account for 53.9% of the observed variance in troughs, with 19.8% of the variance coming from demographic and clinical factors including recipient age, glomerular filtration rate, anti-cytomegalovirus drug use, simultaneous pancreas-kidney transplant and antibody induction. There was no evidence of common genetic variants in AA recipients significantly influencing TAC troughs aside from the CYP3A gene. These results reveal that additional and possibly rare functional variants exist that account for the additional variation.
Introduction
Tacrolimus (TAC) is a common immunosuppressant used in solid organ transplantation. TAC is metabolized by CYP3A4 and CYP3A5 enzymes to active and inactive metabolites (1). CYP3A5, however, has twice the intrinsic catalytic activity of CYP3A4 for TAC, and up to 60% of the hepatic metabolism of TAC is through CYP3A5 in patients who carry at least one CYP3A5* 1 allele (2). A common loss-of-function (LoF) allele of CYP3A5 (*3, rs776746), significantly affects TAC concentrations in whole blood due to reduced TAC metabolism, resulting in higher concentrations compared with persons with the *1 functional allele (2–5). The formation rate of the primary TAC metabolites is also significantly higher in liver microsomes from persons who have at least one functional CYP3A5* 1 allele compared with those who are homozygous for the *3 LoF allele. We created a genotype-based TAC dosing equation including both clinical variables and the CYP3A5*3 genotype (6,7). Although this equation provides valuable guidance for optimizing TAC dosing, a significant amount of variation is not accounted for, especially in African American (AA) kidney transplant recipients.
It has been reported previously that AA recipients have a higher incidence of acute rejection (AR) and reduced allograft survival compared with white recipients (8–10). We and others have reported that TAC trough concentrations in blood (TAC troughs) for AA recipients are much lower than those observed in white recipients, and thus AA recipients require higher doses of TAC to meet immunosuppression targets (3,11). In addition, subtherapeutic immunosuppression concentrations may be associated with the increase of AR observed in AA recipients (12). These lower concentrations are thought to be due to the functional CYP3A5* 1 allele that is much more frequent within the sub-Saharan African population (allele frequency 0.85) than the white population (allele frequency 0.25), resulting in greater TAC metabolism in persons associated carrying one or two CYP3A5* 1 alleles. Even after accounting for this variant, variability in AA TAC troughs is still significant. We hypothesized that additional genetic variants must be present to account for the unexplained variability. Inan effort to identify genetic variants associated with variation in TAC troughs, we evaluated 644 224single-nucleotide polymorphisms (SNPs) in a total of 357 AA recipients with available TAC trough concentrations in this genomewide association study (GWAS).
Materials and Methods
Study design and population
A discovery cohort of 197 adult AA kidney transplant recipients enrolled in the Deterioration of Kidney Allograft Function (DeKAF) Genomics study was used in the GWAS (13). Kidney allograft recipients who self-reported as AA were from three centers of a seven-center prospective study of recipients undergoing kidney or simultaneous pancreas–kidney (SPK) transplantation. An additional 160 AA participants from the same centers and two additional centers were used as a validation cohort. Participants were selected for this analysis if they were aged ≥18 years, received TAC for maintenance immunosuppression and had TAC troughs available in the first 6 months after transplant. High-and low-risk participants were included, although each center used slightly different criteria to attribute risk. This study is registered at www.ClinicalTrials.gov (NCT01714440). Participants were enrolled at time of transplant and signed informed consents approved by the institutional review boards of the enrolling centers.
Clinical information was obtained through the DeKAF Genomics study (13). Participants received oral TAC therapy with mycophenolate maintenance with varying durations of steroid according to transplant standard of care protocols. Induction therapy was administered based on transplant center preference but consisted mainly of Thymoglobulin (Genzyme, Cambridge, MA), Simulect (Novartis, Basel, Switzerland), or Campath (Genzyme). High-risk patients were more likely to receive Thymoglobulin. Donor and recipient characteristics, race, serum creatinine and estimated creatinine clearance, and concomitant medications at time of each TAC trough measurement were obtained from the respective medical records. TAC troughs were measured from whole blood by liquid chromatography-mass spectrometry and were obtained as part of routine clinical care. This was an observational trial, and troughs were not measured in a central laboratory; however, all TAC measurements were done in a Clinical Laboratory Improvement Amendments (CLIA)-approved laboratory. When available, two measurements were obtained in weeks 1 and 8 and in months 3, 4, 5 and 6 for a maximum of 24 trough concentrations per patient. TAC doses were adjusted based on trough concentrations to reach institution-specific trough goals based on time after transplant (generally 8–12 ng/mL in months 0–3 and 6–10 ng/mL in months 4–6). Additional dose adjustments were performed for toxicity using center-specific preferences. Trough values were normalized for dose (nanograms per milliliter per total daily dose in milligrams) prior to statistical analysis.
Genotyping
Pretransplant recipient DNA was isolated at time of transplant from peripheral blood lymphocytes. Lymphocytes were isolated by centrifugation after red blood cell lysis, and the DNA was isolated. Genotypes of the DNA from the discovery cohort (n = 197) were determined with the AFR-AMR Axiom chip (Affymetrix, Santa Clara, CA) (14), which contains 837 930 variants. Genotype calling was performed in one batch on the Affymetrix Genotyping Console v4.0 using the GT1 algorithm, which is based on BRLMM-P (15).
Genotyping of the validation cohort (n = 160) was performed on a custom exome-plus Affymetrix TxArray SNP chip (16). The CYP3A5*3 (rs776746), CYP3A5*6 (rs10264272) and CYP3A5*7 (rs41303343) genotypes were taken from this SNP chip and used for the analysis.
GWAS genotyping data quality control
Data quality control was carried out with PLINK software (version 1.90b1a) (17). Genotypes were subjected to a 95% call rate threshold. Samples with very high heterozygosity and suspected contamination were reassayed and removed if high heterozygosity could not be resolved. Unrelated samples with pairwise identity by descent >0.3 were excluded from the study. Individual SNPs were excluded if they were monomorphic or had low minor allele frequency (<0.5%). SNPs were not excluded based on divergence from the Hardy-Weinberg equilibrium, given the admixed nature of the cohort genotyped. The final number of variants analyzed from the AFR-AMR Axiom chip was 644 224. Genomewide significance was declared with association p-value <5 × 10−8.
Statistical analysis
Linear mixed-effects models (LMMs) were used to test for associations between natural log (ln)–transformed dose-normalized TAC troughs and genotypes. A log transformation was used to ensure that the outcome was normally distributed. Visual inspection showed that dose-normalized trough concentrations initially started low, rose quickly until day 9 after transplant and then plateaued in the early weeks after transplant (3,18). A simple spline method was used to model the effect of time on all trough concentrations, with the change in slope occurring at day 9. The longitudinal LMMs included a random intercept and random slopes for days after transplant and days after posttransplant day 9.
Confounding fixed clinical factors were identified by backward selection with a retention p-value of 0.10 in the discovery cohort. Tested clinical factors included transplant center; donor age and sex; and recipient factors such as age, sex, diabetes at baseline, donor type (living or deceased), antibody induction and SPK transplantation. Time-varying covariates considered for the backward selection, defined at each TAC trough observation, were steroid use, closest creatinine clearance to the trough (linear and quadratic), calcium channel blocker use, angiotensin-converting enzyme inhibitor use and antiviral drug use. The multivariable model determined with the discovery cohort data was applied to the validation cohort analyses.
To improve computational efficiency, we carried out the GWAS of TAC troughs in two steps. In step one, we estimated the day 9 posttransplant ln-transformed dose-normalized TAC trough concentration for each recipient by using an LMM with no clinical variables except days after transplant and days after posttransplant day 9, using the longitudinal trough measures in the discovery cohort. A GWAS was then run between each SNP and the estimated day 9 ln-transformed dose-normalized TAC trough using linear regression (PLINK, version 1.90b1a). This approach is slightly less powerful than running the full longitudinal LMM genomewide but is much faster (>10 times). To overcome the power lost in this approach, we identified SNPs in step one with a p-value <1 × 10−4. In the second step, we ran the multivariable LMM on the SNPs found in step one but applied a stringent genomewide significance level of p-value <5 × 10−8. SNPs were then confirmed in the validation cohort with the multivariable LMM.
We also analyzed the association of the most significant SNPs with the estimated glomerular filtration rate (eGFR) at 12 months after transplant and clinical AR up to 6 months after transplant using linear regression. The eGFR (in milliliters per minute per 1.73 m2) was calculated using the four-level MDRD equation (19). Analyses were conducted with SAS version 9.2 software (SAS Institute, Cary, NC).
Results
Recipient characteristics for the discovery cohort of 197 AA participants and the validation cohort of 160 AA participants are shown in Table 1. The participants in the discovery cohort were aged 35–64 years (77.7%) and primarily male (65.5%). Characteristics of the validation cohort were similar. The TAC troughs, total daily dose and dose-normalized TAC troughs were similar in the discovery and validation cohorts. There was a median of 18 troughs (interquartile range [IQR]: 14–21 troughs) per participant in the discovery cohort and 17 troughs (IQR: 13–21 troughs) per participant in the validation cohort.
Table 1. Recipient characteristics.
| Discovery cohort (n = 197) | Validation cohort (n = 160) | |
|---|---|---|
| Age group, years, n (%) | ||
| 18–34 | 33 (16.8) | 34 (21.3) |
| 35–64 | 153 (77.7) | 118 (73.8) |
| 65–84 | 11 (5.6) | 8 (5.0) |
| Donor age group, years, n (%) | ||
| 0–34 | 98 (49.8) | 71 (44.4) |
| 35–64 | 95 (48.2) | 85 (53.1) |
| 65–84 | 4 (2.0) | 4 (2.5) |
| Living donor status, n (%) | 65 (33.0) | 45 (28.1) |
| African American, n (%) | 197 (100) | 160 (100) |
| Female, n (%) | 68 (34.5) | 62 (38.8) |
| Diabetes at transplant, n (%) | 79 (40.1) | 52 (32.5) |
| SPK, n (%) | 9 (4.6) | 7 (4.4) |
| BMI, mean (SD) | 28.7 (5.2) | 29.2 (5.7) |
| Antibody induction, n (%) | ||
| Combination | 4 (2.0) | 3 (1.9) |
| Monoclonal/IL2RA | 93 (47.2) | 81 (50.6) |
| Polyclonal | 96 (48.7) | 74 (46.3) |
| None | 4 (2.0) | 2 (1.3) |
| Tacrolimus trough in the first 6 months in ng/mL, median (IQR) | 6.4 (4.5–8.7) | 6.5 (4.6–8.6) |
| Daily tacrolimus dose in first 6 months, mg, median (IQR) | 8.0 (6.0–10.0 | 8.0 (6.0–10.0) |
| Tacrolimus dose normalized tacrolimus trough, ng/mL/mg, median (IQR) | 0.80 (0.53–1.25) | 0.74 (0.50–1.13) |
IQR, interquartile range; SD, standard deviation, SPK, simultaneous pancreas–kidney.
In the initial genomewide unadjusted analysis of ln-transformed dose-normalized TAC troughs within the discovery cohort, a single region at the CYP3A5 locus on chromosome 7 was observed including the ZSCAN25 (overlaps CYP3A5), CYP3A5, CYP3A7, CYP3AP2, and CYP3A4 genes (Figure 1). Twenty-eight variants had p-values <1 × 10−5, and five were significant at the genomewide level (p<5 × 10−8) (Table S1). The most significant variants were within the pseudogene CY-P3AP2 (rs17161880, p = 9.26 × 10−14, and rs34880695, p = 1.03 × 10−12). Both variants are in linkage disequilibrium with rs776746 (D′ of 0.804 for rs17161880 and 0.834 for rs34880695), a well-known LoF allele (CYP3A5*3), which was the fifth top variant, with p = 2.28 × 10−9.
Figure 1. Manhattan plot of single-nucleotide polymorphisms (SNPs) associated with tacrolimus trough concentrations.
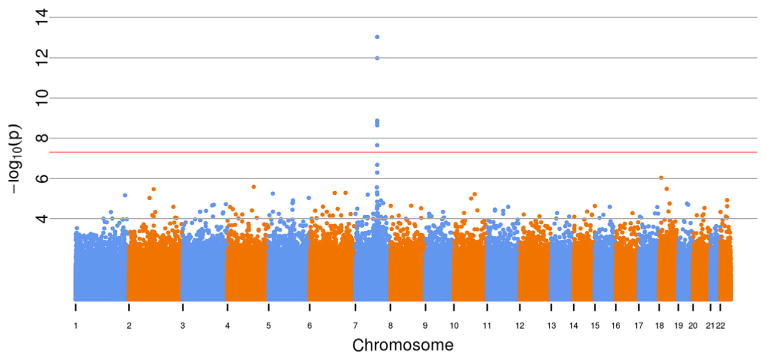
Overall, 644 224 SNPs were analyzed toward natural log of dose-normalized tacrolimus trough concentrations (nanograms per milliliter per total daily dose in milligrams) with no additional covariates. All SNPs are shown in order from chromosome 1 to 22. The red line is at the level of statistical significance (p < 5 × 10−8). The major peak is above the CYP3A locus.
After adjusting for rs776746 (CYP3A5*3) and important clinical covariates (enrolling center, time of trough after transplant, recipient age, anticytomegalovirus drug use, SPK and antibody induction), rs6956305, located within the ZSCAN25 gene, was the most significant variant (p = 2.9 × 10−15) (Table S2). After adjustment for the effect of rs6956305 and rs776746, the variant rs41303343 (CYP3A5*7), an LoF allele, was then found to be highly significant (p = 3.00 × 10−16) (Table S3) (20). We also performed a conditional analysis adjusting for rs776746 (CYP3A5*3), rs41303343 (CYP3A5*7), and rs10264272 (CYP3A5*6), which is another well-known LoF variant for persons of African origin (21). No other common SNPs in the GWAS were found to be significant including the two SNPs in the pseudogene CYP3AP2 and rs6956305.
The genotype frequencies in both cohorts for the three LoF alleles are shown in Table 2. Overall, 75% of the recipients had at least one LoF allele. Combining both cohorts, only 24% (84 of 357) were homozygous WT for all three alleles. Fifty percent (180 of 357) were heterozygous for one of the three LoF alleles (intermediate metabolizers), and 26% (93 of 357) were either homozygous or compound heterozygous for two LoF alleles (poor metabolizers). Analysis of ln-transformed dose-normalized TAC troughs with time showed that participants who were heterozygous for any of the three LoF alleles were intermediate metabolizers compared with homozygous participants for either the WT alleles (bottom line) or for two LoF alleles (top line) (Figure 2).
Table 2. Participants with CYP3A5 loss-of-function alleles observed.
| Discovery cohort (n = 197) | Validation cohort (n = 160) | |
|---|---|---|
| rs776746 (*3) | ||
| 0 | 98 (49.8) | 88 (55.0) |
| 1 | 77 (39.1) | 59 (36.9) |
| 2 | 22 (11.2) | 13 (8.1) |
| rs10264272 (*6) | ||
| 0 | 154 (78.2) | 119 (74.4) |
| 1 | 42 (21.3) | 39 (24.4) |
| 2 | 1 (0.5) | 2 (1.3) |
| rs41303343 (*7) | ||
| 0 | 162 (82.2) | 126 (78.8) |
| 1 | 31 (15.7) | 34 (21.3) |
| 2 | 4 (2.0) | 0 (0) |
| All 3 LoF alleles | ||
| 0 | 51 (25.9) | 33 (20.6) |
| 1 | 88 (44.7) | 92 (57.5) |
| 2 | 58 (29.4) | 35 (21.9) |
Data shown as n (%). LoF, loss of function.
Figure 2. Mean tacrolimus trough concentration by the number of loss-of-function (LoF) alleles.
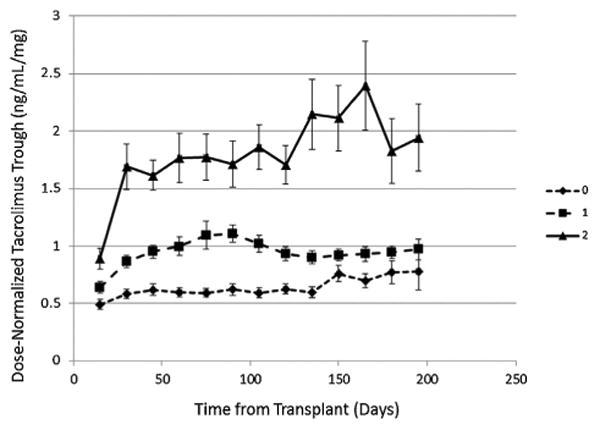
The effect of the genotype on dose-normalized tacrolimus trough concentrations (nanograms per milliliter per total daily dose in milligrams) with time is shown. Triangles and solid line indicate homozygous WT for all three alleles, square and dashed line indicate heterozygous for one wild-type allele and one LoF allele, and diamond and dashed line indicate either homozygous for an LoF allele or a compound heterozygote for two different LoF alleles.
The multivariable analysis of the clinical covariates for the discovery cohort is shown in Table 3. Time from transplant and anticytomegalovirus drug use were the most significant clinical variables affecting longitudinal trough measurements. Results were similar in the validation cohort (Table 4).
Table 3. Multivariable model for natural log–transformed dose-normalized tacrolimus trough concentrations in the discovery cohort.
| Variables | Effect (95% CI) | p-value |
|---|---|---|
| For each day after transplant | 0.10 (0.08, 0.12) | 8.0 × 10−18 |
| Additional effect for each day after day 9 after transplant | −0.10 (−0.12, −0.08) | 4.2 × 10−17 |
| Age, recipient, years | ||
| 18–34 vs. 65–84 | −0.35 (−0.70, −0.01) | 1.6 × 10−2 |
| 35–64 vs. 65–84 | −0.08 (−0.39, 0.23) | |
| GFR center | −0.002 (−0.003, 0.000) | 1.1 × 10−2 |
| Anti-CMV drug use | 0.08 (0.05, 0.12) | 6.7 × 10−7 |
| SPK | 0.48 (0.15, 0.81) | 4.6 × 10−3 |
| Induction immunosuppression | ||
| Combination vs. polyclonal | −0.24 (−0.80, 0.32) | 7.2 × 10−3 |
| Monoclonal/IL2RA vs. polyclonal | 0.18 (0.03, 0.32) | |
| None vs. polyclonal | 0.63 (0.15, 1.12) |
Data are adjusted for enrolling center. CI, confidence interval; CMV, cytomegalovirus; SPK, simultaneous pancreas–kidney.
Table 4. Multivariable model for natural log–transformed dose-normalized tacrolimus trough concentrations in the validation cohort.
| Variables | Effect (95% CI) | p-value |
|---|---|---|
| For each day after transplant | 0.06 (0.04, 0.08) | 3.0 × 10−6 |
| Additional effect for each day after day 9 after transplant | −0.06 (−0.08, −0.04) | 5.6 × 10−6 |
| Age, recipient, years | ||
| 18–34 vs. 65–84 | −0.31 (−0.66, 0.05) | 3.8 × 10−2 |
| 35–64 vs. 65–84 | −0.08 (−0.41, 0.24) | |
| GFR center | −0.001 (−0.003, 0.001) | 3.0 × 10−1 |
| Anti-CMV drug use | 0.08 (0.04, 0.13) | 1.4 × 10−4 |
| SPK | 0.49 (0.13, 0.85) | 7.0 × 10−3 |
| Induction immunosuppression | ||
| Combination vs. polyclonal | −0.07 (−0.60, 0.46) | 4.5 × 10−2 |
| Monoclonal/IL2RA vs. polyclonal | 0.19 (0.03, 0.35) | |
| None vs. polyclonal | −0.35 (−0.98, 0.28) |
Data are adjusted for enrolling center. CI, confidence interval; CMV, cytomegalovirus; SPK, simultaneous pancreas–kidney.
The estimates of variance in the longitudinal trough measurements of the discovery cohort associated with the clinical variables and the CYP3A5 SNPs are shown in Table 5. Using multivariable analysis, the variance in TAC troughs declined with addition of the LoF alleles; the clinical variables alone captured 19.8% of the variance in troughs, and the three LoF alleles capturedan additional 34.1%, for a total of 53.9% explained variance. Similar results were shown in the analysis of the validation cohort (Table 6). A combined analysis of both cohorts showed that clinical variables and the 3 LoF alleles captured a total of 43.0% of the explained variance (data not shown). Figure 3 shows a box plot of participants with 0, 1 or 2 LoF alleles compared with dose-normalized tacrolimus trough concentrations.
Table 5. Variance in natural log–transformed dose-normalized tacrolimus troughs in the discovery cohort.
| Model | Variation of TAC troughs* | Variation explained by model**, % |
|---|---|---|
| Simple time-trend model | 0.3114 | – |
| Clinical variables | 0.2497 | 19.8 |
| Clinical variables + rs776746 | 0.1929 | 38.1 |
| Clinical variables + rs10264272 | 0.2495 | 19.9 |
| Clinical variables + rs41303343 | 0.231 | 25.8 |
| Clinical variables + rs776746 and rs10264272 | 0.1845 | 40.7 |
| Clinical variables + rs776746 and rs41303343 | 0.1553 | 50.1 |
| Clinical variables + rs776746, rs10264272, and rs41303343 | 0.1436 | 53.9 |
Variance estimated for day 9 posttransplant natural log–transformed dose-normalized TAC trough concentration.
Proportion of variation explained by each model compared with the simple time-trend model, namely, 1–var/0.3114, in which var is the estimated variance for the day 9 random variable in the previous column.
TAC, tacrolimus.
Table 6. Variance in natural log–transformed dose-normalized tacrolimus troughs in the validation cohort.
| Model | Variation of TAC troughs* | Variation explained by model**, % |
|---|---|---|
| Simple Time-trend Model | 0.2806 | – |
| Clinical Variables | 0.2283 | 18.6 |
| Clinical Variables + rs776746 | 0.213 | 24.1 |
| Clinical Variables + rs10264272 | 0.2267 | 19.2 |
| Clinical Variables + rs41303343 | 0.2224 | 20.7 |
| Clinical Variables + rs776746 and rs10264272 | 0.2049 | 27.0 |
| Clinical Variables + rs776746 and rs41303343 | 0.1976 | 29.6 |
| Clinical Variables + rs776746, rs10264272, and rs41303343 | 0.1842 | 34.3 |
Variance estimated for day 9 posttransplant natural log–transformed dose-normalized TAC trough concentration
Proportion of variation explained by each model compared with the simple time-trend model, namely, 1–var/0.3114, in which var is the estimated variance for the day 9 random variable in the previous column.
TAC, tacrolimus.
Figure 3. Box plot of dose-normalized tacrolimus trough concentration (nanograms per milliliter per total daily dose in milligrams) versus number of loss-of-function alleles.
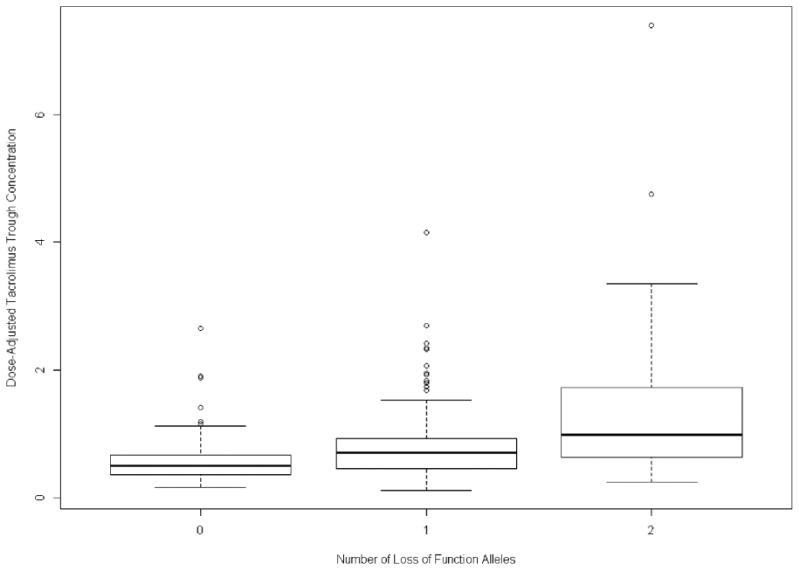
Horizontal bars represent 95% confidence levels.
The presence of LoF alleles with AR and 1-year eGFR was also analyzed. AR was not associated with the LoF alleles, but the number of LoF alleles was significantly associated with eGFR (p = 4.7 × 10−3). Each LoF allele increased the eGFR at 1 year by 9.1 mL/min (2.8–15.5 mL/min).
Discussion
It has been documented that AA recipients have reduced graft survival compared with white kidney allograft recipients (22). One-year AR is higher in AA than non-AA kidney allograft recipients. Reaching target immunosuppression concentrations is critical in reducing the risk for early AR events (10). There are significant differences in TAC troughs and dose requirements between AA and white recipients. Although we have shown that transplant center, days after transplant, age,concomitantmedications and SPK status are correlated with dose-normalized TAC trough concentrations, we cannot discern the specific causal covariates without extremely detailed clinical information on each participant.
It has been reported that the LoF allele CYP3A5*3 is the major genetic cause of variation in TAC metabolism. This allele encodes for a mutation in intron 3, resulting in a cryptic splice site encoding for a mRNA with a premature stop codon (21). In this study, we showed that two other LoF alleles, CYP3A5*6 and CYP3A5* 7, also play significant roles in TAC metabolism in AA recipients. In CYP3A5*1/*6 heterozygotes, two mRNAs were identified, one normal 3A5 mRNA and one mRNA with a skipped exon 7 (21). Deletion of exon 7 results in a frameshift mutation encoding a truncated protein of 184 amino acids. The CYP3A5*7 variant also produces a frameshift mutation due to an insertion of a thymine in codon 346, leading to premature termination at codon 348 (23). Within our study population, only 24% were homozygous WT for all three WT alleles, showing that substantially more AA recipients carry LoF variants than previously presumed. Consequently, the lack of LoF variants, thought to be present at much higher frequency in white versus AA recipients, is not a satisfactory explanation of why AA recipients have higher TAC metabolism. We previously showed that there is a heterozygous effect for the CYP3A5*3 allele, and we showed in this study that this was the case for participants who were also heterozygous for the other two LoF alleles (Figure 2) (6,7). Variants within the CYP3A4 locus (CYP3A4*22, rs35599367) have also been implicated in affecting TAC troughs (4,24), but neither this variant nor any other variants within CYP3A4 were found to contribute to the observed variation in TAC troughs in this analysis of AA recipients. In addition, a known functional variant within the transporter ABCB1 (previously MDR1; rs1045642, c.3435T>C), which has also been associated with variation in TAC concentrations, was not found to contribute to the observed variation in TAC troughs in this analysis of AA recipients (25).
Determining accurate initial dosing of immunosuppresants at the time of transplantation is critical to reduce early AR events. The therapeutic range for TAC is narrow, and initial genotyping should provide for maximal efficacy of TAC concentrations. Although the CYP3A5*3 allele is a major LoF allele that affects TAC trough concentrations, additional LoF alleles need to be considered to accurately estimate TAC metabolism. In this report, we identified two additional common LoF variants that affect TAC trough concentrations in the AA kidney recipients, CYP3A5*6 and CYP3A5*7, both of which have been found in the African population (26). In Figure 3, several participants in the 0 or 1 LoF plots are outside of the 95% confidence range. It is possible that these participants have an LoF allele that is not the *3, *6, or *7 allele, resulting in higher dose-normalized TAC troughs. The variants genotyped in this study were limited to those on the GWAS chip. Analysis of the 1000 Genomes project data identifies many more putative LoF alleles within CYP3A5 including 15 germline stop-gained variants (nonsense variants) and 21 germline frameshift variants. Additional potentially functional variants include 56 donor or acceptor splice site variants and 216 missense variants (27). Each variant may have the same LoF effect as the three variants analyzed in this report, making DNA sequencing of CYP3A5 necessary to capture all genetic variation associated with this gene.
We also found that eGFR increases with an increased number of LoF alleles, making reduced metabolism of TAC protective. Although these alleles were not associated with AR, subclinical AR may be occurring when TAC concentrations are low, due to increased TAC metabolism resulting in lower GFR levels.
Our study has several limitations. TAC troughs were measured from whole blood by each institution and not at a central laboratory; however, all assays were CLIA certified. TAC area under the curve was not used because it was not obtained under the standard of care and thus was not available. In addition, nonadherence to medications has also been shown to be an important risk factor for AR and may influence results. Although we have shown that transplant center, days after transplant, age, concomitant medications, and SPK status are correlated with dose-normalized TAC trough concentrations, we cannot discern the specific causal covariates without extremely detailed clinical information on each participant. We were also not able to categorize calcium channel blockers as dihydropyridines versus nondihydropyridines. In future studies, we intend to gather more detailed concomitant medication information because it is likely that drug–drug interactions play a role for some patients. Nonetheless, this does not negate our findings.
We conclude that the AA-specific variants CYP3A5*3, CYP3A5*6, and CYP3A5* 7 explain a great proportion of the observed TAC trough variability in AA recipients. Better understanding of the impact of population-specific genomic variants is a critical step toward a personalized medicine approach to transplant immunosuppression.
DeKAF Investigators
Arthur Matas, MD, Department of Surgery, University of Minnesota, Minneapolis, MN; J. Michael Cecka, MD, UCLA Immunogenetics Center, Los Angeles, CA; John Connett, PhD, Division of Biostatistics, University of Minnesota, Minneapolis, MN; Fernando G. Cosio, MD, Division of Nephrology, Mayo Clinic, Rochester, MN; Robert Gaston, MD, Division of Nephrology, University of Alabama, Birmingham, AL; Rosalyn Mannon, MD, Division of Nephrology, University of Alabama, Birmingham, AL; Sita Gourishankar,MD, Division of Nephrology and Immunology, University of Alberta, Edmonton, Alberta, Canada; Joseph P. Grande, MD, PhD, Mayo Clinic College of Medicine, Rochester, MN; Lawrence Hunsicker, MD, Nephrology Division, Iowa City, IA; Bertram Kasiske, MD, Division of Nephrology, Hennepin County Medical Center, Minneapolis, MN; and David Rush, MD, Health Sciences Center, Winnipeg MB, Canada.
Supplementary Material
Table S1: Association of top single-nucleotide polymorphisms with tacrolimus longitudinal troughs and no additional covariates in discovery cohort (adjusted for enrolling center).
Table S2: Association of top single-nucleotide polymorphisms with longitudinal tacrolimus troughs adjusted for CYP3A5*3 (rs776746) in the discovery cohort.
Table S3: Tacrolimus (TAC) model in African Americans in the discovery cohort: natural log of dose-normalized TAC trough concentrations with clinical factors and rs776746 and rs41303343 (CYP3A5*3 and *7) loss-of-function alleles*.
Table S4: Tacrolimus (TAC) levels model in African Americans in validation cohort: natural log of dose-normalized TAC trough concentration with clinical factors*.
Table S5: Tacrolimus (TAC) model in African Americans in the validation cohort: natural log of dose-normalized TAC trough concentration with clinical factors and CYP3A5*3, *6, and*7 (rs776746, rs10264272, and rs41303343) loss-of-function alleles*.
Table S6: Tacrolimus (TAC) model in African Americans in the validation cohort: natural log of dose-normalized TAC trough concentration with clinical factors and number of CYP3A5*3, *6, and *7 (rs776746, rs10264272, and rs41303343) loss-of-function alleles.
Acknowledgments
The authors wish to thank the research participants for their participation in this study. We acknowledge the dedication and hard work of our coordinators at each of the DeKAF Genomics clinical sites: University of Alberta, Nicoleta Bobocea, Tina Wong, Adrian Geambasu and Alyssa Sader; University of Manitoba, Myrna Ross and Kathy Peters; University of Minnesota, Mandi DeGrote and Danielle Berglund; Hennepin County Medical Center, Lisa Berndt; Mayo Clinic, Tom DeLeeuw; University of Iowa, Wendy Wallace and Tammy Lowe; University of Alabama, Jacquelin Vaughn and Tena Hilario. We also acknowledge the dedicated work of our research scientists Marcia Brott and Amutha Muthusamy. This study was supported in part by NIH/NIAID grants 5U19-AI070119 and 5U01-AI058013.
Abbreviations
- AA
African American
- AR
acute rejection
- CI
confidence interval
- DeKAF
Deterioration of Kidney Allograft Function
- eGFR
estimated glomerular filtration rate
- GWAS
genomewide association study
- LMM
linear mixed-effects models
- LoF
loss of function
- ln
natural log
- SD
standard deviation
- SNPs
single-nucleotide polymorphisms
- SPK
simultaneous pancreas-kidney
- TAC
tacrolimus
Footnotes
Disclosure: The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting Information: Additional Supporting Information may be found in the online version of this article.
References
- 1.Iwasaki K. Metabolism of tacrolimus (FK506) and recent topics in clinical pharmacokinetics. Drug Metab Pharmacokinet. 2007;22:328–335. doi: 10.2133/dmpk.22.328. [DOI] [PubMed] [Google Scholar]
- 2.Dai Y, Hebert MF, Isoherrannen N, et al. Effect of CYP3A5 polymorphism on tacrolimus metabolic clearance in vitro. Drug Metab Dispos. 2006;34:836–847. doi: 10.1124/dmd.105.008680. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson PA, Oetting WS, Brearley AM, et al. Novel polymorphisms associated with tacrolimus trough concentrations: Results from a multicenter kidney transplant consortium. Transplantation. 2011;91:300–308. doi: 10.1097/TP.0b013e318200e991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruckmueller H, Werk AN, Renders L, et al. Which genetic determinants should be considered for tacrolimus dose optimization in kidney transplantation? A combined analysis of genes affecting the CYP3A locus Ther Drug Monit. 2015;37:288–295. doi: 10.1097/FTD.0000000000000142. [DOI] [PubMed] [Google Scholar]
- 5.Rojas L, Neumann I, Herrero MJ, et al. Effect of CYP3A5*3 on kidney transplant recipients treated with tacrolimus: A systematic review and meta-analysis of observational studies. Pharmacogenomics J. 2015;15:38–48. doi: 10.1038/tpj.2014.38. [DOI] [PubMed] [Google Scholar]
- 6.Passey C, Birnbaum AK, Brundage RC, Oetting WS, Israni AK, Jacobson PA. Dosing equation for tacrolimus using genetic variants and clinical factors. Br J Clin Pharmacol. 2011;72:948–957. doi: 10.1111/j.1365-2125.2011.04039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passey C, Birnbaum AK, Brundage RC, et al. Validation of tacrolimus equation to predict troughs using genetic and clinica factors. Pharmacogenomics. 2012;13:1141–1147. doi: 10.2217/pgs.12.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neylan JF. Immunosuppressive therapy in high-risk transplant patients: Dose-dependent efficacy of mycophenolate mofetil in African-American renal allograft recipients. U.S. Renal Transplant Mycophenolate Mofetil Study Group. Transplantation. 1997;64:1277–1282. doi: 10.1097/00007890-199711150-00008. [DOI] [PubMed] [Google Scholar]
- 9.Laftavi MR, Pankewycz O, Patel S, et al. African American rena transplant recipients (RTR) require higher tacrolimus doses to achieve target levels compared to white RTR: Does clotrimazole help? Transplant Proc. 2013;45:3498–3501. doi: 10.1016/j.transproceed.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Gralla J, Le CN, Cooper JE, Wiseman AC. The risk of acute rejection and the influence of induction agents in lower-risk African American kidney transplant recipients receiving modern immunosuppression. Clin Transplant. 2014;28:292–298. doi: 10.1111/ctr.12311. [DOI] [PubMed] [Google Scholar]
- 11.Neylan JF. Effect of race and immunosuppression in rena transplantation: Three-year survival results from a US multicenter, randomized trial. FK506 Kidney Transplant Study Group. Transplant Proc. 1998;30:1355–1358. doi: 10.1016/s0041-1345(98)00274-7. [DOI] [PubMed] [Google Scholar]
- 12.Taber DJ, Gebregziabher MG, Srinivas TR, Chavin KD, Baliga PK, Egede LE. African-American race modifies the influence of tacrolimus concentrations on acute rejection and toxicity in kidney transplant recipients. Pharmacotherapy. 2015;35:569–577. doi: 10.1002/phar.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Israni A, Leduc R, Holmes J, et al. Single-nucleotide polymorphisms, acute rejection, and severity of tubulitis in kidney transplantation, accounting for center-to-center variation. Transplantation. 2010;90:1401–1408. doi: 10.1097/TP.0b013e3182000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann TJ, Kvale MN, Hesselson SE, et al. Next generation genome-wide association tool: Design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vignettes: Genotype clustering for Axiom™ arrays. Affymetrix Web site. 2009 Oct 12; Available from: http://media.affymetrix.com/support/technical/whitepapers/brlmmp_whitepaper.pdf.
- 16.Li YR, van Setten J, Verma SS, et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med. 2015;7 doi: 10.1186/s13073-015-0211-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobson PA, Schladt D, Oetting WS, et al. Lower calcineurin inhibitor doses in older compared to younger kidney transplant recipients yield similar troughs. Am J Transplant. 2012;12:3326–3336. doi: 10.1111/j.1600-6143.2012.04232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Modification of Diet in Rena Disease Study Group Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 20.Hustert E, Haberl M, Burk O, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 22.Narayanan M, Pankewycz O, Shihab F, Wiland A, McCague K, Chan L. Long-term outcomes in African American kidney transplant recipients under contemporary immunosuppression: A four-yr analysis of the Mycophenolic acid Observational REna transplant (MORE) study. Clin Transplant. 2014;28:184–191. doi: 10.1111/ctr.12294. [DOI] [PubMed] [Google Scholar]
- 23.Hustert E, Haberl M, Burk O, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Pallet N, Jannot AS, El Bahri M, et al. Kidney transplant recipients carrying the CYP3A4* 22 allelic variant have reduced tacrolimus clearance and often reach supratherapeutic tacrolimus concentrations. Am J Transplant. 2015;15:800–805. doi: 10.1111/ajt.13059. [DOI] [PubMed] [Google Scholar]
- 25.Kurzawski M, Dabrowska J, Dziewanowski K, Domanski L, Peru(x0017C)yńska M, Droździk M. CYP3A5 and CYP3A4, but not ABCB1 polymorphisms affect tacrolimus dose-adjusted trough concentrations in kidney transplant recipients. Pharmacogenomics. 2014;15:179–188. doi: 10.2217/pgs.13.199. [DOI] [PubMed] [Google Scholar]
- 26.Bains RK, Kovacevic M, Plaster CA, et al. Molecular diversity and population structure at the cytochrome P450 3A5 gene in Africa. BMC Genet. 2013;14:34. doi: 10.1186/1471-2156-14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.1000 Genomes. Available from: http://browser.1000genomes.org/Homo_sapiens/Gene/Variation_Gene/Table?db=core;g=ENSG00000106258;r=7:99245817-99277621.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Association of top single-nucleotide polymorphisms with tacrolimus longitudinal troughs and no additional covariates in discovery cohort (adjusted for enrolling center).
Table S2: Association of top single-nucleotide polymorphisms with longitudinal tacrolimus troughs adjusted for CYP3A5*3 (rs776746) in the discovery cohort.
Table S3: Tacrolimus (TAC) model in African Americans in the discovery cohort: natural log of dose-normalized TAC trough concentrations with clinical factors and rs776746 and rs41303343 (CYP3A5*3 and *7) loss-of-function alleles*.
Table S4: Tacrolimus (TAC) levels model in African Americans in validation cohort: natural log of dose-normalized TAC trough concentration with clinical factors*.
Table S5: Tacrolimus (TAC) model in African Americans in the validation cohort: natural log of dose-normalized TAC trough concentration with clinical factors and CYP3A5*3, *6, and*7 (rs776746, rs10264272, and rs41303343) loss-of-function alleles*.
Table S6: Tacrolimus (TAC) model in African Americans in the validation cohort: natural log of dose-normalized TAC trough concentration with clinical factors and number of CYP3A5*3, *6, and *7 (rs776746, rs10264272, and rs41303343) loss-of-function alleles.


