Abstract
Deeper understanding of the bacteriostatic and bactericidal mechanisms of antimicrobial peptides (AMPs) should help in the design of new antibacterial agents. Over several decades, a variety of biochemical assays have been applied to bulk bacterial cultures. While some of these bulk assays provide time resolution on the order of 1 min, they do not capture faster mechanistic events. Nor can they provide subcellular spatial information or discern cell-to-cell heterogeneity within the bacterial population. Single-cell, time-resolved imaging assays bring a completely new spatiotemporal dimension to AMP mechanistic studies. We review recent work that provides new insights into the timing, sequence, and spatial distribution of AMP-induced effects on bacterial cells.
Keywords: Antimicrobial peptide mechanisms, single-cell fluorescence microscopy, membrane permeabilization, oxidative stress
Mechanistic Studies of Antimicrobial Peptides
In this era of multi-drug resistant bacterial infections, new antibacterial treatments are badly needed. Natural antimicrobial peptides (AMPs, also called host-defense peptides) serve as templates for the design of new antibacterial agents [1–3]. Deeper understanding of how AMPs kill bacterial targets should facilitate this design effort. Decades of intensive study have shown that the underlying mechanisms by which specific AMPs kill specific bacterial species exhibit remarkable variety from peptide to peptide and remarkable specificity for particular AMP-bacteria pairings. Clear relationships between AMP structure and killing mechanisms have not yet emerged.
AMPs comprise an ancient class of short polypeptides (typically < 40 aa) that exhibit broad-spectrum antibacterial activity against both Gram-negative and Gram-positive bacteria [1]. They are secreted constitutively by epithelial cells within tissue repeatedly exposed to new microbes, such as in the lungs and the digestive tract. They are also found in vesicles within neutrophils and macrophages, cells that envelop and kill invading bacteria as part of the innate human immune response. One well studied class of cationic AMPs adopts amphipathic, α-helical conformations on binding to a membrane, often with a Pro-induced kink [4]. A second cationic class, the defensins, uses three disulfide bonds to enforce the three-β-strand defensin fold, a globular amphipathic structure [5, 6]. Many other categories occur in nature. In addition, bacteria synthesize a wide variety of peptide-like agents that attack competing bacteria—the bacteriocins [7].
For a long time it was widely believed that most AMPs halt growth and kill bacterial cells by permeabilizing the cytoplasmic membrane (CM), thus destroying the proton-motive force (pmf) that drives ATP production [1]. Biophysical studies of AMP interactions with synthetic lipid bilayers have provided models for the mechanisms of bilayer disruption. These include ‘barrel stave pores’ made of contiguous amphiphilic AMPs inserted perpendicular to the bilayer surface; larger ‘toroidal pores’ lined by inserted AMPs and highly curved lipid surfaces; and ‘carpet mechanisms’ involving large-scale ‘micellization’ of the bilayer [1]. Molecular dynamics simulations of pore formation support a ‘chaotic pore’ model—a fluxional, localized permeabilization site comprising a time-varying number of peptide and lipid molecules [8]. The absence of apparent structure-function relationships has led to the suggestion that interfacial activity determines the ability of a peptide to permeabilize membranes [9]. This refers to a balance of hydrophobic and hydrophilic components with sufficiently imperfect amphiphilicity to facilitate disruption of normal lipid packing. The strength of the connection between AMP permeabilization of lipid vesicles and the killing mechanisms for real bacterial cells remains to be seen [10, 11]. There is a correlation between an AMP’s tendency to induce highly negatively curved phases in mixtures with lipids and its antibacterial activity [12].
Most mechanistic studies of the bacteriostatic and bactericidal effects of AMPs have focused on bulk, planktonic cultures. These bulk assays provide great insight into a variety of specific biophysical and biochemical mechanistic events, sometimes with time resolution on the order of several minutes. For example, they can distinguish disruption of the outer membrane (OM) from disruption of the cytoplasmic membrane using fluorogenic dyes; measure real-time release of K+ from the cytoplasm [13]; and monitor dissipation of the pmf [14]. A remarkably thorough 2014 study of the effects of the synthetic hexapeptide RWRWRW-NH2 on Bacillus subtilis by Bandow and colleagues [15] applied some 15 different assays to the same system! However, the onset of damage mechanisms can occur within seconds, much faster than the response time of most bulk measurements. For example, a 2010 study from the Belcher lab used time-resolved atomic force microscopy (AFM) to image the attack of CM15 on Escherichia coli [16]. Corrugation of the outer surface of live bacteria began within 13 s of AMP addition. In addition, the bulk assays provide no subcellular spatial information.
Recent work employs imaging methodologies such as immuno-transmission electron microscopy (TEM), immuno-fluorescence, and AFM to directly observe the effects of AMPs on single cells. Most of these studies involve fixation and permeabilization of the cells, and they are typically carried out at a single time point after addition of the AMP. In a 2012 review, Munoz and Read surveyed studies that imaged the effects of AMPs on single, live microbes [17]. Most of the work reviewed involved yeast. Only two studies involved bacteria. The 2009 study of the interactions of the Sushi 1 peptide with E. coli used nanoparticle labeling of the peptide in both TEM and single-particle tracking methods [18].
The present review highlights a handful of recent studies in which spatial or temporal information gleaned from single-cell imaging experiments provides new insights into mechanisms of AMP attack on bacteria. The most common tool is fluorescence microscopy, either confocal or widefield, in conjunction with phase contrast microscopy (Box 1). A variety of one- and two-color fluorescent labeling schemes have proven useful, including labeling of the peptide itself. Single-cell imaging methods can provide a remarkably direct view of when and where key mechanistic events occur (Box 2). In our opinion, the most powerful mechanistic studies of the near future will augment time-resolved, live-cell imaging with increasingly powerful biochemical and genetic assays to provide a comprehensive picture of how AMPs halt growth and ultimately kill bacteria.
Box 1. Time-resolved Widefield Microscopy.
In a typical widefield fluorescence microscopy experiment, the microscope objective looks upward through a glass coverslip at the plated bacterial sample. In effect, the microscope squashes the fluorescence from three-dimensional objects (the cells) into a two-dimensional, magnified image at the camera plane. To enable imaging of two different species (e.g., GFP within a cell and red-labeled AMP attacking the cell), the two fluorescent species must have different excitation and emission spectra. The two colors are imaged in alternate frames by toggling the excitation laser wavelength to excite first the one, and then the other, fluorescent species. Phase contrast microscopy exploits the small phase shifts caused by variation in the refractive index of the sample to produce intensity modulations in the image on the camera.
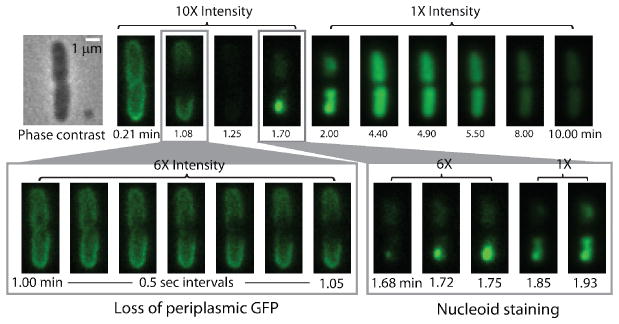
Time-lapse imaging creates a movie from a long sequence of images. In three-channel studies, shutters and the camera are synchronized to control the frame rate, the sequence of images acquired in each camera cycle, and the illumination time within each frame. In each cycle, we might obtain a sequence of three images, for example: red fluorescence, green fluorescence, and phase contrast repeated throughout the time-lapse movie. In the schematic example (Figure I), the red image shows the spatial distribution of a rhodamine-labeled AMP. The phase contrast image enables measurement of tip-to-tip cell length vs time, a proxy for cell growth or cell shrinkage due to osmotic effects. The green image monitors two different species, GFP and Sytox Green. GFP is initially located in the periplasm, resulting in a ‘halo’ image like the one shown at right of Figure I. If the outer membrane (OM) is permeabilized to GFP before the cytoplasmic membrane (CM), then the GFP halo will disappear. If the CM is permeabilized to GFP first, then GFP will fill the cytoplasm because its volume is much larger, resulting in a very different spatial distribution (Figure 8). The same green channel also detects Sytox Green, but only after it has traversed both membranes, bound to the chromosomal DNA, and thus become fluorescent. For survey work, the three-channel cycle might be carried out every 18 s, with each snapshot acquired in only 50 ms (time-lapse imaging). Concatenation of a time series of 200 such cycles then yields a one-hour long movie of four different variables, each depending on space and time.
Figure I.
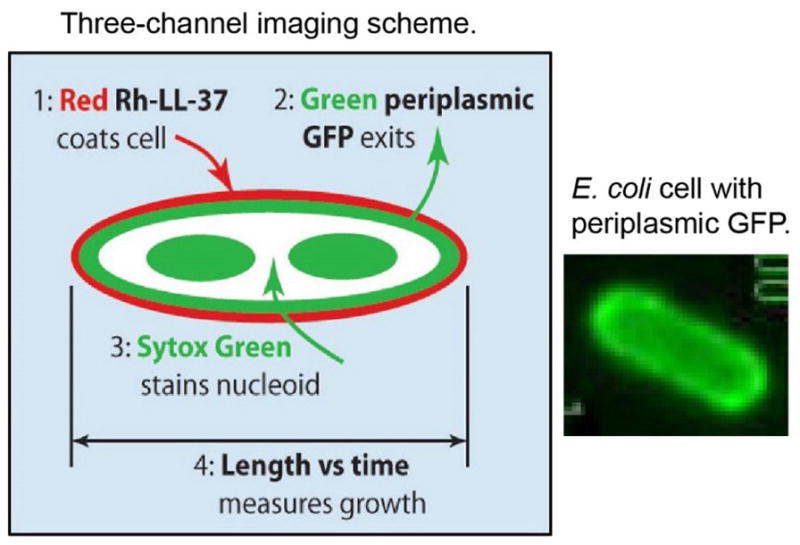
Three-channel Imaging Scheme and Image of Periplasmic GFP.
Box 2. Why Study Single Cells?
Bulk assays study AMP effects averaged over millions of cells in a culture. While bulk time resolution on the order of 1 min is sometimes achieved, there is no spatial information. In contrast, single-cell assays use microscopy to directly observe AMP effects on a field of well isolated cells, often using fluorescently labeled peptide or cellular content such as periplasmic GFP. The advantages are both spatial and temporal. If a red-fluorescent AMP binds selectively to a cellular sub-region, such as the septal region (illustrated as the red dots in Figure IA) or an endcap, this can be observed directly to provide mechanistic hints. In addition, single-cell methods routinely discern heterogeneity of the cellular response to an AMP. Perhaps septating cells are attacked preferentially over non-septating cells (as in the two cells with red bands in Figure IB). Growth of some cells, but not others, may be inhibited at a given AMP concentration. If a bulk assay detects reactive oxygen species after AMP addition, it cannot tell if 10%, 50%, or all of the cells are experiencing oxidative stress; single-cell imaging can distinguish these cases. Finally, ‘symptom’ development in each cell can be observed over time, by the methods described in Box 1. This enables determination of the time sequence of events and of the distribution of time intervals between successive events. That type of information is averaged out in time-resolved studies of bulk samples.
Figure I.
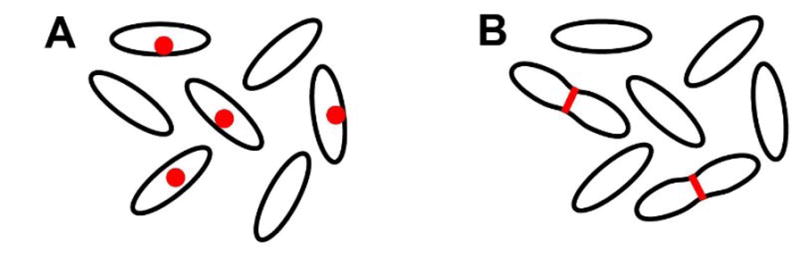
Schematic Examples of Heterogeneity among Cells.
Survey of Recent Spatiotemporal Studies
AMPs That Bind Locally Near Cell Division Sites
Two recent studies illustrate how the distribution of binding sites of a fluorescently labeled AMP can provide clues to the bacterial targets of the attack, suggesting remarkably specific mechanisms of growth disruption. In 2013, Hultgren, Kline and co-workers [19] investigated the hypothesis that human β-defensins (hBDs) interfere with the protein translocation and secretion machinery of Gram-positive bacteria. A combination of immunofluorescence microscopy (IFM) and immunoelectron microscopy (IEM) had previously shown that in Enterococcus faecalis the ATP-binding translocase SecA and the sortases SrtA and SrtC colocalize into one or a few membrane-bound foci [20]. These puncta are located at the septum or at nascent cell division sites, depending on the phase within the cell cycle. In studies of live cells observed 1–5 min after treatment, both hBD2 (Figure 1) and hBD3 labeled with Cy3 also concentrate in a few foci that preferentially locate in similar fashion [19]. Subsequent immunolabeling experiments then showed that the hBD treatment disrupted the SecA and sortase foci, presumably interfering with proper secretion. As stained by nonyl acridine orange (NAO), the anionic lipids phosphatidylglycerol (PG) and cardiolipin (CL), also concentrated in similarly located foci. Such anionic lipid domains may attract cationic hBDs to the vicinity of protein secretory foci.
Figure 1.
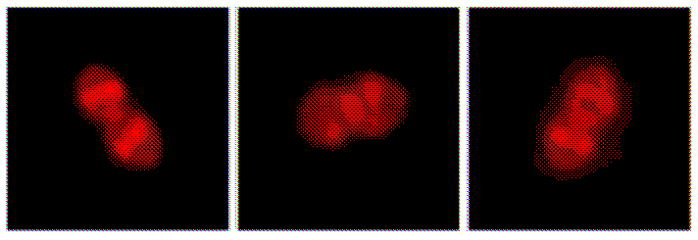
Pattern of Binding of hBD2 to E. faecalis Cells. Adapted from [19] with permission.
In 2015, Tomita and co-workers studied the interactions of BacL1, a subunit of Bacteriocin 41, with E. faecalis as well [21]. Once again the protein preferentially located at sites of cell division, either ongoing or nascent. The BacL1 pattern of binding was banded, not punctal. The bacteria subsequently suffered lysis. Removal of the triple-repeat SH3 domain abolished the specific spatial targeting. The specific localization and the lytic activity only occurred for growing cells, not for stationary phase cells. From comparisons with other Gram-positive bacteria, the authors suggested that BacL1 specifically targets peptidoglycan layers that use L-Ala-L-Ala cross-bridges, eventually leading to lysis during cell division.
Human α-defensin HD5 Induces Blebbing and Enters the Cytoplasm of E. coli
The Nolan lab recently published a thorough time-resolved, single-cell study of E. coli under attack by HD5ox, the folded form of the human defensin with all three Cys-Cys bonds intact [22]. This study provides a further example of the diversity of defensin-induced growth-halting mechanisms. Phase contrast imaging after attack by unlabeled HD5 revealed bacterial elongation and the formation of blebs, typically at the mid-plane region of septating cells (Figure 2). These are similar to the blebs observed after treatment with β-lactam antibiotics. As the concentration of HD5ox increases, the fraction of cells forming blebs and the log attenuation of cell viability both increased. Stationary phase E. coli were much less susceptible to HD5ox. The spatial distribution of cytoplasmic GFP expands to fill the blebs, suggesting they are caused by breaches in the peptidoglycan layer. Studies using periplasmic GFP (Box 1) showed that at least some of the blebs are surrounded by both membranes. Similar blebbing was observed in a variety of Gram-negative bacteria after treatment with HD5ox. HD5 analogues lacking the three disulfide bonds did not cause blebbing.
Figure 2.
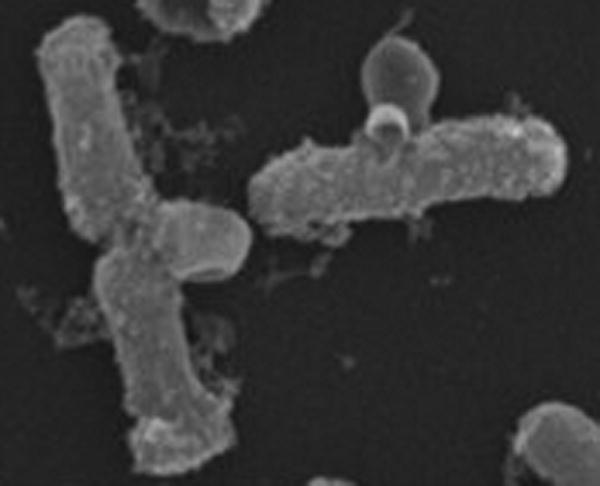
Scanning Electron Microscopy (SEM) Image Shows Blebbing of E. coli Induced by HD5ox. Adapted from [22] with permission.
Studies of rhodamine-labeled HD5 (Rh–HD5ox) showed that the peptide reaches the cytoplasm and exhibits a non-uniform spatial profile, concentrating at the two poles and at mid-cell. The specific target of the peptide within the cytoplasm remains uncertain. We note in passing that the Rh–HD5ox cytoplasmic distribution is reminiscent of that of ribosomes in normal growth conditions [23]. In rapidly growing cells, ribosomes concentrate in three ribosome-rich regions interleaved with two nucleoid lobes.
Potent Cyclic Hexapeptide Kills Bacteria Without Permeabilizing the Cytoplasmic Membrane
The Dathe lab has carried out a series of studies of cyclic hexapeptide interactions with single, live E. coli and B. subtilis cells [24]. These peptides are attractive drug candidates due to their resistance to proteolysis. The particular peptide c-(RRRWFW) (named c-WFW) is a potent killer of both Gram-negative and Gram-positive bacteria. In early work, a standard assay using the β-galactosidase substrate ONPG showed that lethal dosages of c-WFW never permeabilize the CM of E. coli. A recent imaging study using an analogue labeled with the fluorescent dye NBD (4-chloro-7-nitrobenzofurazan), called c-W(NBD)W (Figure 3) found strong accumulation of the peptide at the CM and no evidence that the peptide ever accesses the bulk of the cytoplasm [24]. In B. subtilis, both c-W(NBD)W (Figure 3) and the CL stain NAO showed similar patterns, a thin staining of the CM plus strong accumulation at the septal and polar regions. Treatment with unlabeled c-WFW abolished the domains in which CL accumulates strongly. The imaging studies augmented with evidence from vesicle-binding assays suggest a mechanism in which the peptide reaches the CM and disrupts lipid organization there. The ultimate consequences may include improper cell division, which relies on localization of the negatively curved lipids cardiolipin (CL) and phosphatidyl-ethanolamine (PE), or improper targeting of key membrane proteins to the polar and septal regions.
Figure 3.
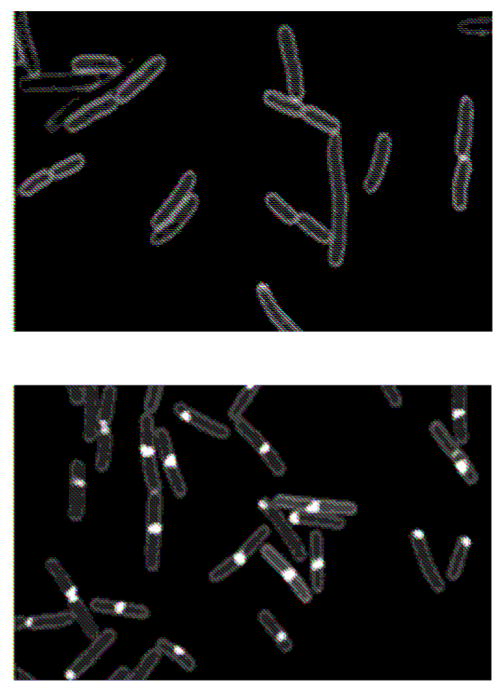
Membrane Binding Pattern of c-W(NBD)W on E. coli (A) and on B. subtilis (B). Adapted from [24] with permission.
Labeling and Imaging of Lantibiotics
New methods for labeling natural products with fluorescence probes have great potential for functional studies. Lanthipeptides are ribosomally synthesized, post-translationally modified bacteriocins, many of which have antibiotic properties (e.g., lantibiotics). The most well studied example is nisin. In 2013 Bindman and van der Donk [25] used bacterial overexpression to produce designed N-terminal mutants and developed organic reactions that provide a general method for preparing fluorescently labeled lanthipeptides. This enabled a two-color fluorescence imaging study of the attack of the synergistic, two-component system haloduracin-α (Halα and haloduracin-β (Halβ) on B. subtilis. The pattern of localization of rhodamine-labeled Halα alone (Figure 4) was consistent with binding to Lipid II. It comprised strong staining a thet mid-cell plus a series of puncta and transverse bands along the cell periphery. When rhodamine-labeled Halα and Cy5-labeled Halβ were added together, they colocalized. A series of controls showed that the mechanism is consistent with preferential binding of Halα to Lipid II and selective binding of Halβ to the Halα-Lipid II complex. The presence of Halβ greatly enhances the potency of Halα, suggesting that this ternary complex is important for optimal bioactivity.
Figure 4.
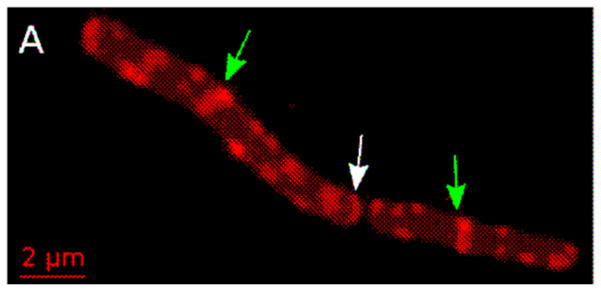
Membrane Binding pattern of Rh-Halα on B. subtilis. Arrows denote new (green) and old (white) cell division sites. Adapted from [25] with permission.
Multi-channel Imaging of AMP Effects with Sub-minute Time Resolution
Our own lab has been developing methods that enable direct, simultaneous observation of multiple AMP-induced effects on single cells as a function of time after injection of the peptide. The time resolution can be sub-minute or sub-second, as the phenomena require. We have used this approach to study the interactions of several AMPs with E. coli (Gram-negative) [26–29] and B. subtilis (Gram-positive) [30]. Each cell exhibits a series of ‘symptoms’ on its own peculiar timescale (Figure 5, Key Figure). Quantitative analysis of the images is time-consuming and because of this we call these ‘low throughput, high information content’ assays.
Figure 5 (Key Figure).
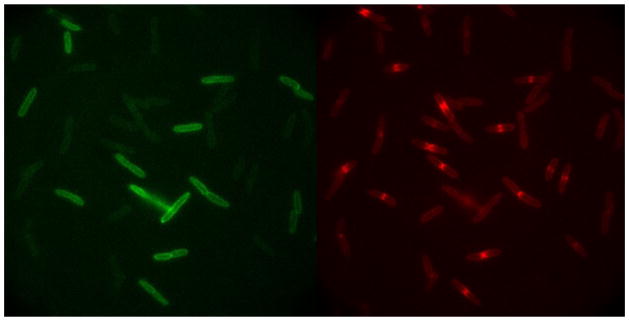
[0][0] Heterogeneity of E. coli Response to LL-37. Snapshot in time of the attack of Rhodamine-labeled LL-37 (Rh-LL-37; right panel) on live E. coli expressing GFP that is exported to the periplasm (left panel). Each cell tells its own story in real time. Rh-LL-37 permeabilizes the outer membrane of septating cells earlier than non-septating cells, leading to loss of periplasmic GFP (left panel). At the time point shown, most of the remaining GFP halos occur for non-septating cells. A red band at the septal region (right panel) signals passage of Rh-LL-37 into the periplasm, where it binds to an immobile species, possible peptidoglycan.
Cells harvested in mid-log phase are plated on a glass coverslip for widefield microscopy (Box 1), enabling the study of dozens of cells in one field of view. For observations under aerobic conditions, fresh, aerated medium flows over the plated cells for the entire observation period, typically 60 min after initiation of flow of the AMP. Observations under anaerobic conditions are also feasible [26]. Use of a small microfluidics chamber enables us to flow growth medium plus AMP without undue expense. A steady concentration of nutrients, oxygen, and external AMP is established in seconds and maintained for hours as needed. This contrasts with the time-varying chemical conditions experienced by cells plated on agarose pads.
For each cell, the sequence of images provides detailed information about the timing of different AMP-induced events (Box 1). Three-channel imaging (phase contrast plus red and green fluorescence channels) provides quantitative, time-dependent information about cell length, AMP spatial distribution, and the permeabilization of membranes. Most recently, redox-sensitive dyes have been used to develop time-resolved, single-cell assays for the formation of reactive oxygen species (ROS) during AMP attack (Box 3).
Box 3. Single-cell Detection of Reactive Oxygen Species (ROS) in Live E. coli.
We use two different schemes to detect oxidative stress in real time within the cytoplasm of E. coli [26]. CellROX Green is a cell permeable profluorophore which converts to the green fluorescent species we call CellROX* on reaction with either superoxide (•O2−) or hydroxide (•OH) radicals in the presence of DNA. To detect those species, we add CellROX green to the growth medium prior to initiation of the flow of AMP. In separate experiments aimed at detection of hydrogen peroxide (H2O2), we use a cell line that expresses the non-native peroxidase APEX2 (mutated ascorbate peroxidase) from a plasmid and include the permeable dye Amplex Red in the flow of medium. APEX2 catalyzes the reaction of Amplex Red with H2O2 to form the red-fluorescent species resorufin.
In our study of CM15 attacking E. coli, we have observed a strong, transient CellROX* signal (Figure I, top panel) and a strong resorufin signal (Figure I, bottom panel) that rise within 30 s of permeabilization of the cytoplasmic membrane to GFP. It seems likely that CM15 is interfering with the electron transport chain in a way that abruptly produces •O2−, which is known to release Fe2+ from Fe-containing enzymes. Subsequent reactions of •O2− with superoxide dismutase would produce H2O2, whose reaction with free Fe2+ then produces the deadly •OH by the Fenton reaction. It is possible that many AMPs induce oxidative damage on reaching the cytoplasmic membrane and that such damage is an important growth-halting effect. Further mechanistic work is required.
Figure I.
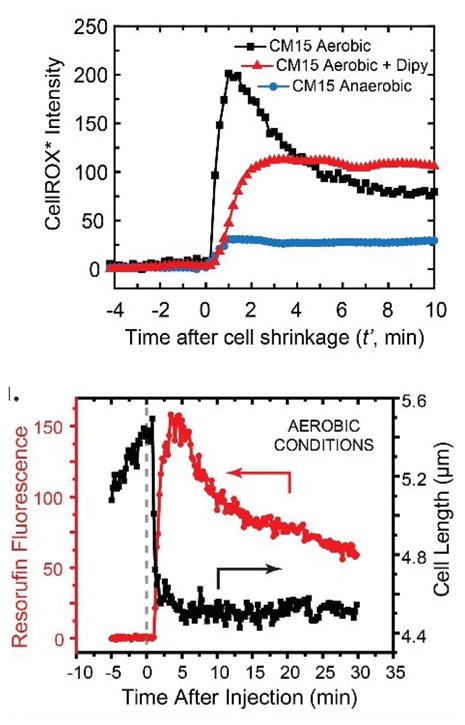
Time-resolved Reporters of Oxidative Stress. Top: CellROX* fluorescence intensity vs time after addition of CM15. Aerobic and anaerobic conditions as shown. Dipy is 2,2′-dipyridyl, a permeable scavenger of •OH radicals. Bottom: Resorufin fluorescence intensity vs time after addition of CM15 under aerobic conditions. Signal rises at the same time as cell shrinks. Images adapted from [26] with permission.
Comparison of the Attack of LL-37 and of Cecropin A on E. coli
Our initial study of time-dependent effects used rhodamine-labeled LL-37 (Rh-LL-37) and K-12 E. coli that expresses GFP from a plasmid and exports it to the periplasm [29]. The types of information gleaned from one septating cell are illustrated in Figure 6. We monitored cell length and the time-dependent intensity and spatial distributions of Rh-LL-37, periplasmic GFP, and Sytox Green over 60 min with an 18-s imaging cycle time. All cells become uniformly coated with Rh-LL-37 over the first minute after addition. Layer-by-layer penetration then occurs. Septating cells are attacked much earlier than non-septating cells. Rh-LL-37 gains access to the periplasm at curved regions of the OM, either at the septum (in septating cells) or at one endcap (non-septating cells). Seconds later, the OM becomes permeabilized to periplasmic GFP, which leaves the cell envelope. Cell length shrinks gradually as Rh-LL-37 enters the periplasm. We believe this is an osmotic effect. The influx of a large concentration of LL-37 and its counterions to the periplasmic space extracts water from the cytoplasm and reduces the turgor pressure, shrinking the cells. Importantly, growth halts significantly before the CM is permeabilized to Sytox Green. We speculate that the growth-halting mechanism is interference with peptidoglycan biosynthesis, not permeabilization of the OM and destruction of the pmf. A very similar sequence of events was observed for Cecropin A [28].
Figure 6.
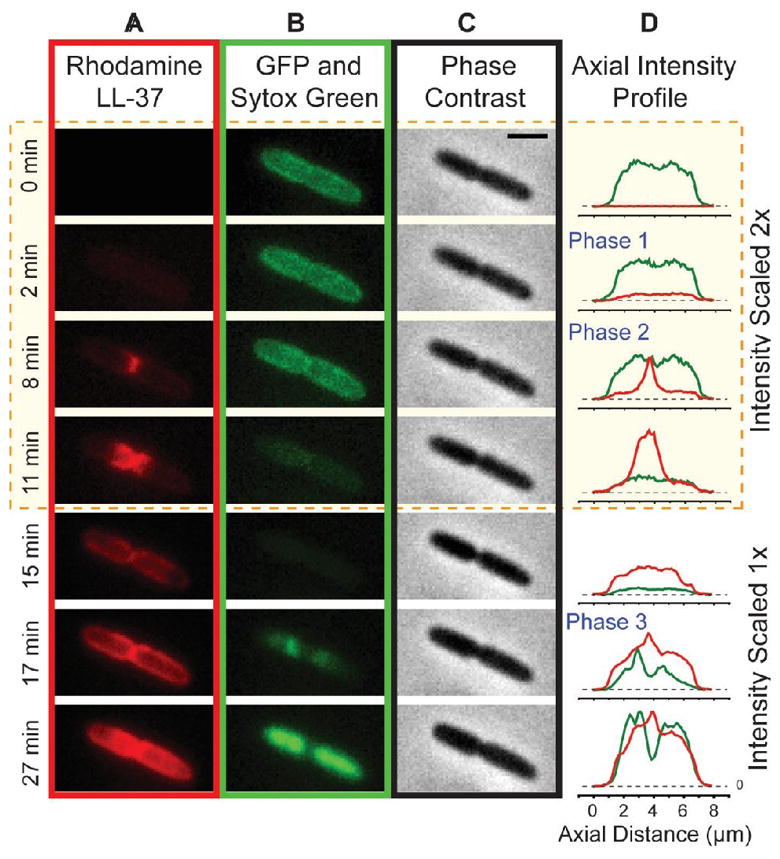
Attack of Rh-LL-37 on a Septating E. coli Cell. Montage at left shows time sequences of red (A; Rh-LL-37), green (B; first periplasmic GFP, then Sytox Green), and phase contrast images (C). (D) Axial projections of red and green intensity distributions. Reproduced from [29] with permission.
For both LL-37 and Cecropin A, there is a significant time lag of several minutes between addition of the peptide and permeabilization of the OM to GFP [28]. For both AMPs, there is strong heterogeneity in the time lag between peptide addition and permeabilization of the OM to GFP. Cells may differ in the transit time of peptide through the lipopolysaccharide layer. Alternatively, the permeabilization event itself may involve some type of nucleation – the cooperative, concerted action of multiple peptide molecules.
After permeabilization of the OM, there is a shorter, more homogeneous, sub-minute time lag before CM permeabilization, signaled by fluorescence of Sytox Green. For both AMPs, fast movies reveal that the OM and CM permeabilization events are localized and persistent (Figure 7), not global and transient [28]. The limited spatial resolution of fluorescence microscopy does not enable us to distinguish pore formation from a ‘localized carpet’ mechanism [31].
Figure 7.
Attack of Cecropin A on a Septating E. coli Cell. Initial phase contrast image plus montage of green fluorescence images. Periplasmic GFP leaves the cell, then Sytox Green crosses both membranes and stains DNA. Expanded timescale shows localized permeabilization pattern for both the OM and CM. Reproduced from [28] with permission.
One curious difference between Cecropin A and LL-37 is the pattern of attack sites for septating cells [28]. LL-37 permeabilizes the OM to periplasmic GFP at the septal region, and then permeabilizes the CM to Sytox Green at the septal region as well. In contrast (Figure 6), Cecropin A permeabilizes the OM to GFP at the septum but then ‘goes around the block’ to permeabilize the CM to Sytox Green at one endcap. Perhaps Cecropin A has a strong propensity to bind to the anionic lipids cardiolipin and phophatidylglycerol, which are known to sequester at curved membrane surfaces [32].
CM15 Attack on E. coli: Evidence for the Abrupt Onset of Oxidative Stress
The synthetic antimicrobial peptide CM15 is a hybrid that combines residues 1–7 of Cecropin A (from moths) with residues 2–9 of melittin (bee venom) [33]. It is highly charged, +5 at pH = 7. Within 24 s of addition of the peptide, most cells abruptly shrink and their periplasmic GFP moves inward to fill the cytoplasm, rather than leaving the cell envelope (Figure 8) [26]. Evidently this highly cationic peptide crosses the OM rapidly and permeabilizes the CM to GFP some 30 min before it permeabilizes the OM to GFP. In work under review, we have observed similarly rapid passage of highly cationic β-peptide random copolymers across the OM [34]. The detailed mechanism of facile translocation of a highly cationic peptide across the OM is not well understood yet.
Figure 8.
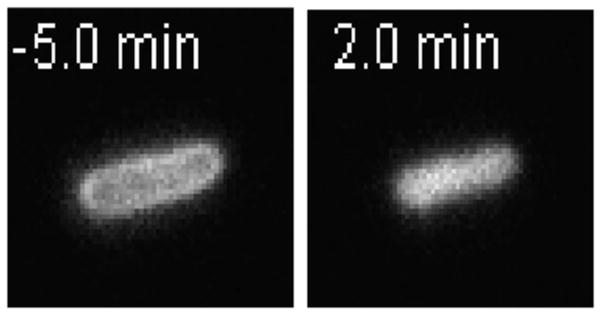
GFP Images Before and After Addition of CM15. CM15 induces periplasmic GFP to move inward and fill the cytoplasm. Adapted from [26] with permission.
Essentially simultaneously with the CM permeabilization event, we observe abrupt enhancement of three different fluorescence signals which report on oxidative stress in the cytoplasm [26]. The CellROX* and resorufin signals (Box 3) suggest that CM15 induces a burst of superoxide (•O2−) and/or hydroxyl radicals (•OH) and also hydrogen peroxide (H2O2) [35]. Active respiration is a prerequisite for these signals. Importantly, the MIC for CM15 is 20-fold larger in anaerobic conditions than in aerobic conditions. This strongly implicates oxidative stress [36] as a key bacteriostatic mechanism of CM15 against E. coli in aerobic conditions. The oxidative damage evidently occurs within 30 s of CM permeabilization.
We have observed similar CellROX* signals following the attack on E. coli of the natural AMPs LL-37 and Cecropin A [26] and of random β-peptide copolymers [34]. Oxidative damage may prove to be a fairly commonplace downstream effect of AMPs. Recent work also implicates oxidative damage as a key bactericidal mechanism for standard antibiotics [37].
Concluding Remarks
Single-cell imaging provides unique, time-resolved spatial information about the mechanisms of action of AMPs on live bacterial cells. The new data complements well established bulk biochemical assays. At present the resulting spatiotemporal information is more biophysical (e.g., the sequence of membrane permeabilization events, osmotic effects, and halting of growth) than biochemical (e.g., interference with cell wall biogenesis, DNA replication, or transcription profiles). Development of new single-cell assays that measure specific downstream biochemical events vs time is an important goal (see Outstanding Questions). The oxidative stress assays (Box 3) provide one example [26]. In the future, bacteriologists might well transfer single-cell imaging methods proven useful in studies of yeast [17] and of mitochondria [38] to bacterial studies. There are also opportunities to convert bulk fluorescence assays into single-cell assays. In a different vein, well established single-particle tracking methods would enable studies of AMP effects on the spatial distribution and movement of key cytoplasmic actors. Possibilities include DNA morphology and local diffusive motion [26, 27], and the spatial distribution and biochemical activity of ribosomes and RNA polymerase [23, 39, 40], among many others.
Outstanding Questions.
How do short, highly cationic peptides traverse the outer membrane so readily? This is an important question for Gram-negative bacteria, because once a highly cationic peptide reaches the periplasm in sufficient quantity, multiple growth-halting effects occur rapidly.
How can we adapt fluorescence-based assays of specific biochemical events to the study of AMP effects on live bacteria in real time with subcellular spatial resolution? There are many opportunities for methods development.
How widespread is oxidative damage as a growth-halting mechanism? It is important to routinely measure minimum inhibitory concentrations in both aerobic and anaerobic media and to correlate the results with assays that detect oxidative stress in real time.
What is the complete range of bacteriostatic and bactericidal effects of any one AMP on any one bacterial species? It seems that the more thoroughly the community looks for effects, the more damage mechanisms we find.
Is there a discernible connection between AMP sequence and structure and the range of growth-halting effects? This has been a challenging question for decades, and remains so today.
Looking further ahead, we anticipate time-resolved, single-cell mechanistic studies of bacteria in more realistic contexts than planktonic growth. The effects of AMPs on stationary phase cells [22] are not well explored. Studies of AMP effects on single cells within model biofilms would represent a larger step towards realistic environments [41, 42]. It should also be possible to adapt the methods outlined here to direct, time-resolved imaging of the fate of bacterial cells as they are attacked by neutrophils or macrophages [43].
Trends.
Single-cell, real-time observations provide a remarkably detailed picture of the timing, sequence, and subcellular location of specific events during the attack of antimicrobial peptides (AMPs) on live bacteria.
In addition to permeabilizing membranes, AMPs induce a variety of ‘downstream effects’. Specific peptides may interfere with cell wall synthesis; induce osmotic shock; disrupt synthesis of DNA, RNA, or proteins; destroy the proton-motive force; or induce oxidative stress.
Environmental factors can modulate potency by enabling specific bacteriostatic mechanisms or by altering the AMP structure. LL-37 is more effective against E. coli in aerobic metabolism than in anaerobic conditions. In 2011, Schroeder and co-workers found that the reduced, unfolded form of human β-defensin-1 is much more potent against some intestinal bacteria that live in a naturally reducing environment.
Acknowledgments
We thank our colleagues Dr. Robert Bucki and Prof. Bill Wimley for encouragement and advice in the early stages of our AMP studies. The Weisshaar lab research described in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Awards Number R01GM094510 (to JCW as PI) and R01GM093265 (to JCW and Samuel Gellman as co-PIs). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 2.Afacan NJ, Yeung AT, Pena OM, Hancock RE. Therapeutic potential of host defense peptides in antibiotic-resistant infections. Curr Pharm Design. 2012;18:807–819. doi: 10.2174/138161212799277617. [DOI] [PubMed] [Google Scholar]
- 3.Wimley WC, Hristova K. Antimicrobial peptides: Successes, challenges and unanswered questions. J Membr Biol. 2011;239:27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tossi A, Sandri L, Giangaspero A. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Wehkamp J. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 7.Allen HK, Trachsel J, Looft T, Casey TA. Finding alternatives to antibiotics. Ann N Y Acad Sci. 2014;1323:91–100. doi: 10.1111/nyas.12468. [DOI] [PubMed] [Google Scholar]
- 8.Marrink SJ, Vries AHd, Tieleman DP. Lipids on the move: Simulations of membrane pores, domains, stalks and curves. Biochim Biophys Acta. 2009;1788:149–168. doi: 10.1016/j.bbamem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He J, Starr CG, Wimley WC. A lack of synergy between membrane-permeabilizing cationic antimicrobial peptides and conventional antibiotics. Biochim Biophys Acta. 2015;1848:8–15. doi: 10.1016/j.bbamem.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Krauson AJ, Wimley WC. Toward the de novo design of antimicrobial peptides: Lack of correlation between peptide permeabilization of lipid vesicles and antimicrobial, cytolytic, or cytotoxic activity in living cells. Biopolymers. 2014;102:1–6. doi: 10.1002/bip.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt NW, Wong GC. Antimicrobial peptides and induced membrane curvature: Geometry, coordination chemistry, and molecular engineering. Curr Opin Solid State Mater Sci. 2013;17:151–163. doi: 10.1016/j.cossms.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlov DS, Nguyen T, Lehrer RI. Potassium release, a useful tool for studying antimicrobial peptides. J Microbiol Methods. 2002;49:325–328. doi: 10.1016/s0167-7012(01)00383-9. [DOI] [PubMed] [Google Scholar]
- 14.Rathinakumar R, Walkenhorst WF, Wimley WC. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: The importance of interfacial activity. J Am Chem Soc. 2009;131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wenzel M, Chiriac AI, Otto A, Zweytick D, May C, Schumacher C, Bandow JE. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci U S A. 2014;111:E1409–1418. doi: 10.1073/pnas.1319900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fantner GE, Barbero RJ, Gray DS, Belcher AM. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat Nanotechnol. 2010;5:280–285. doi: 10.1038/nnano.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munoz A, Read ND. Live-cell imaging and analysis shed light on the complexity and dynamics of antimicrobial peptide action. Front Immunol. 2012;3:248. doi: 10.3389/fimmu.2012.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leptihn S, Har JY, Chen J, Ho B, Wohland T, Ding JL. Single molecule resolution of the antimicrobial action of quantum dot-labeled sushi peptide on live bacteria. BMC Biol. 2009;7:22. doi: 10.1186/1741-7007-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandaswamy K, Liew TH, Wang CY, Huston-Warren E, Meyer-Hoffert U, Hultenby K, Kline KA. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2013;110:20230–20235. doi: 10.1073/pnas.1319066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, Hultgren SJ. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol. 2009;191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurushima J, Nakane D, Nishizaka T, Tomita H. Bacteriocin protein Bacl1 of Enterococcus faecalis targets cell division loci and specifically recognizes L-Ala2-cross-bridged peptidoglycan. J Bacteriol. 2015;197:286–295. doi: 10.1128/JB.02203-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chileveru HR, Lim SA, Chairatana P, Wommack AJ, Chiang IL, Nolan EM. Visualizing attack of Escherichia coli by the antimicrobial peptide human defensin 5. Biochemistry. 2015;54:1767–1777. doi: 10.1021/bi501483q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol Microbiol. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheinpflug K, Krylova O, Nikolenko H, Thurm C, Dathe M. Evidence for a novel mechanism of antimicrobial action of a cyclic R-,W-rich hexapeptide. PLoS One. 2015;10:e0125056. doi: 10.1371/journal.pone.0125056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bindman NA, van der Donk WA. A general method for fluorescent labeling of the N-termini of lanthipeptides and its application to visualize their cellular localization. J Am Chem Soc. 2013;135:10362–10371. doi: 10.1021/ja4010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi H, Yang Z, Weisshaar JC. Single-cell, real-time detection of oxidative stress induced in Escherichia coli by the antimicrobial peptide cm15. Proc Natl Acad Sci U S A. 2015;112:E303–310. doi: 10.1073/pnas.1417703112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakshi S, Choi H, Rangarajan N, Barns KJ, Bratton BP, Weisshaar JC. Nonperturbative imaging of nucleoid morphology in live bacterial cells during an antimicrobial peptide attack. Appl Environ Microbiol. 2014;80:4977–4986. doi: 10.1128/AEM.00989-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rangarajan N, Bakshi S, Weisshaar JC. Localized permeabilization of E. coli membranes by the antimicrobial peptide Cecropin A. Biochemistry. 2013;52:6584–6594. doi: 10.1021/bi400785j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc Natl Acad Sci U S A. 2011;108:E77–81. doi: 10.1073/pnas.1101130108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barns KJ, Weisshaar JC. Real-time attack of LL-37 on single Bacillus subtilis cells. Biochim Biophys Acta. 2013;1828:1511–1520. doi: 10.1016/j.bbamem.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 32.Oliver PM, Crooks JA, Leidl M, Yoon EJ, Saghatelian A, Weibel DB. Localization of anionic phospholipids in Escherichia coli cells. J Bacteriol. 2014;196:3386–3398. doi: 10.1128/JB.01877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato H, Feix JB. Osmoprotection of bacterial cells from toxicity caused by antimicrobial hybrid peptide CM15. Biochemistry. 2006;45:9997–10007. doi: 10.1021/bi060979m. [DOI] [PubMed] [Google Scholar]
- 34.Choi H, Chakraborty S, Liu R, Gellman SH, Weisshaar JC. Single-cell, time-resolved antimicrobial effects of a highly cationic, random nylon-3 copolymer on live E. coli. ACS Chem Biol. 2015 doi: 10.1021/acschembio.5b00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dwyer DJ, Collins JJ, Walker GC. Unraveling the physiological complexities of antibiotic lethality. Annu Rev Pharmacol Toxicol. 2015;55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 38.Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakshi S, Choi H, Mondal J, Weisshaar JC. Time-dependent effects of transcription- and translation-halting drugs on the spatial distributions of the Escherichia coli chromosome and ribosomes. Mol Microbiol. 2014;94:871–887. doi: 10.1111/mmi.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakshi S, Dalrymple RM, Li W, Choi H, Weisshaar JC. Partitioning of RNA polymerase activity in live Escherichia coli from analysis of single-molecule diffusive trajectories. Biophys J. 2013;105:2676–2686. doi: 10.1016/j.bpj.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Wong GC. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–391. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lakins MA, Marrison JL, O’Toole PJ, van der Woude MW. Exploiting advances in imaging technology to study biofilms by applying multiphoton laser scanning microscopy as an imaging and manipulation tool. J Microsc. 2009;235:128–137. doi: 10.1111/j.1365-2818.2009.03190.x. [DOI] [PubMed] [Google Scholar]
- 43.Moller J, Luhmann T, Chabria M, Hall H, Vogel V. Macrophages lift off surface-bound bacteria using a filopodium-lamellipodium hook-and-shovel mechanism. Sci Rep. 2013;3:2884. doi: 10.1038/srep02884. [DOI] [PMC free article] [PubMed] [Google Scholar]