Summary
Malignancy can be suppressed by the immune system in a process termed immunosurveillance. However, to what extent immunosurveillance occurs in spontaneous cancers and the composition of participating cell types remain obscure. Here we show that cell transformation triggers a tissue-resident lymphocyte response in oncogene-induced murine cancer models. Non-circulating cytotoxic lymphocytes, derived from innate, TCRαβ and TCRγδ lineages, expand in early tumors. Characterized by high expression of NK1.1, CD49a and CD103, these cells share a gene expression signature distinct from those of conventional NK cells, T cells and invariant NKT cells. Generation of these lymphocytes is dependent on the cytokine IL-15, but not the transcription factor Nfil3 that is required for the differentiation of tumor-infiltrating NK cells, and IL-15, but not Nfil3, deficiency results in accelerated tumor growth. These findings reveal a tumor-elicited immunosurveillance mechanism that engages unconventional type 1-like innate lymphoid cells and type 1 innate-like T cells.
Introduction
Understanding how the immune system impacts the process of tumorigenesis has captivated some of the greatest minds in immunology for more than a century. In the 1860s, following the observation that cancer arises at sites of chronic inflammation, Rudolf Virchow proposed a tumor-promoting function for leukocytes. However, at the turn of the last century, Paul Ehrlich reasoned that protective immune responses were likely required to suppress cancer in long-lived organisms (Ehrlich, 1909), and by the 1950s, the cancer immunosurveillance hypothesis was formally postulated to ascribe a plausible function of adaptive cellular immunity in eliminating transformed cells (Burnet, 1957; Thomas, 1959). Indeed, studies in the past two decades have revealed both tumor-promoting inflammation and protective tumor immunity in mouse models of cancer (Grivennikov et al., 2010). Such apparently opposing activities of inflammatory responses can be integrated into the framework of cancer immunoediting which, in its most complete manifestation, is composed of three sequential phases of tumor “elimination”, “equilibrium” and “escape” (Schreiber et al., 2011).
The original cancer immunosurveillance hypothesis attributed the role of protective tumor immunity to antigen-specific lymphocytes (Burnet, 1957; Thomas, 1959). Studies utilizing recombination-activating gene (Rag)-deficient mice or T lymphocyte depletion antibodies have revealed increased tumor incidence or tumor outgrowth in a carcinogen-induced sarcoma model (Koebel et al., 2007; Shankaran et al., 2001). In addition, sarcomas that develop under conditions of immunodeficiency are more immunogenic than tumors from wild-type mice (Koebel et al., 2007; Shankaran et al., 2001), and the dominant rejection antigen in one such tumor encodes a mutated neoepitope for CD8+ T cells (Matsushita et al., 2012). In a genetic mouse model of sarcoma, introduction of immunogenic peptides by lentivirus infection suppresses tumor development, and the loss of antigen expression or presentation on major histocompatibility complex (MHC) I results in tumor escape from T cell attack (DuPage et al., 2012). These findings demonstrate that cytotoxic T cells play a critical role in restraining tumor development in response to tumor-associated foreign antigens accompanied with viral infections or mutated antigens induced by carcinogens.
Yet, tumor development does not always generate neoantigens that mediate rejection, or induce host-protective antigen-specific T cell responses. In a transgenic model of sporadic cancer, the oncogenic simian virus 40 T antigen (SV40 Tag) is somatically induced, and functions as a tumor-associated neoantigen (Willimsky and Blankenstein, 2005). However, SV40 Tag triggers CD8+ T cell tolerance, and fails to reject nascent transformed cells (Willimsky and Blankenstein, 2005). In a transgenic adenocarcinoma of mouse prostate (Tramp) model, CD8+ T cells reactive to the unmutated histone H4 peptide as a tumor-associated antigen arise spontaneously in tumor-bearing mice (Savage et al., 2008). Adoptive transfer of H4 antigen-reactive T cells into Tramp mice does not result in effector T cell differentiation (Savage et al., 2008), which is in part due to immune repression by the regulatory cytokine transforming growth factor-β (TGF-β) (Donkor et al., 2011). These findings reveal that although tumor antigen-specific CD8+ T cell responses are induced in oncogene-induced cancers, their activities are restrained from inducing effective cancer immunosurveillance.
The lack of host-protective antigen-specific T cell responses implies that oncogene-induced tumors bypass the “elimination” and “equilibrium” phases of cancer immunoediting, and thus may, by default, display an “escaped” phenotype. Since tumors are derived from normal cells, it has been postulated that tumors may not be “foreign” or “dangerous” enough to induce a protective immune response (Matzinger, 2002; Pardoll, 2003). However, studies involving mice deficient in several immune effector molecules have revealed signs of immunosurveillance in genetic mouse models of cancer. For instance, deficiency of the activating receptor NKG2D results in earlier tumor onset in Tramp mice (Guerra et al., 2008). In addition, mice devoid of the cytotoxic molecule perforin (Smyth et al., 2000; Street et al., 2007), or the death receptor TNF-related apoptosis-inducing ligand (TRAIL) (Finnberg et al., 2008; Zerafa et al., 2005) manifest accelerated tumor growth in models of mammary carcinoma and B cell lymphoma. These observations suggest that cytotoxic immune responses are involved in repressing oncogene-induced cell transformation, although they may not engage tumor antigen-specific CD8+ T cells. Nevertheless, immunodeficiency could impair host elimination of infections (Enzler et al., 2003), which may result in chronic inflammation and secondarily affect tumor development (Coussens and Werb, 2002; Grivennikov et al., 2010; Mantovani et al., 2008). Whether cell transformation elicits a specific protective immune response in oncogene-induced tumors and the nature of the potential response have not been defined.
In this report, we set forth by characterizing lymphocytes with cytotoxic potential in transgenic models of murine cancer. We found that cell transformation expands a distinct group of non-circulating innate, TCRαβ and TCRγδ lymphocytes that express high levels of the cytolytic molecule granzyme B, and display potent cytotoxic activities against tumor cells. Generation of these lymphocytes is dependent on the cytokine IL-15, and IL-15 deficiency or overexpression results in accelerated or delayed tumor growth, respectively. These findings demonstrate that cell transformation triggers a distinct class of protective immune response by engaging unconventional tissue-resident type 1-like innate lymphoid cells and type 1 innate-like T cells.
Results
Tumor growth elicits a granzyme B response in genetic models of murine cancer
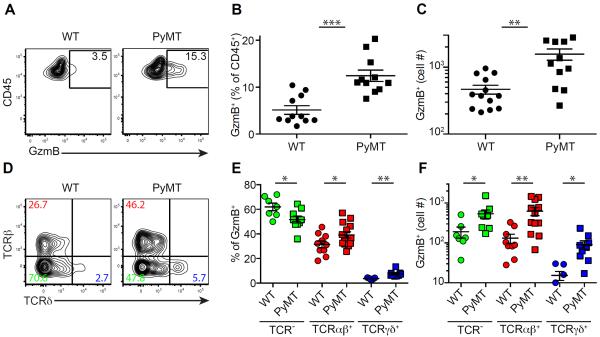
To investigate whether immunosurveillance is induced in spontaneous cancers, we utilized the murine MMTV-PyMT (PyMT) mammary tumor model on the C57BL/6 background (Franklin et al., 2014). Mammary glands from 8-week-old PyMT mice had early stage tumors, while palpable carcinomas were detected at older ages (Figure S1A–B). To determine whether lymphocytes with cytotoxic potential were induced in precancerous lesions, we examined expression of the cytolytic molecule granzyme B (GzmB) in leukocyte populations from the mammary glands of 8-week-old wild-type (WT) or PyMT mice. A higher percentage and number of GzmB-expressing cells were observed in PyMT mice (Figure 1A–C). GzmB-producing cells in mammary tissues further expanded as tumor grew (Figure S1C), but were barely detectable in the spleen or mammary tissue-draining lymph nodes of PyMT mice (Figure S1D), revealing that the lasting GzmB response is restricted to the transformed tissue.
Figure 1. Precancerous lesions elicit an immune response characterized by GzmB B expression.
(A–C) Flow cytometric analysis of granzyme B (GzmB) expression in CD45+ leukocytes from pooled mammary glands of wild-type (WT) and 8-week-old PyMT mice with percentage (A–B) and absolute number (C) of CD45+GzmB+ cells. Data are representative of ten independent experiments.
(D–F) Flow cytometric analysis of TCRβ and TCRδ expression in CD45+GzmB+ subsets isolated from pooled mammary glands of WT and 8-week-old PyMT mice with percentage (D–E) and absolute number (F) of GzmB+ cells that are TCR− (green), TCRβ+ (red) or TCRδ+ (blue). Data are representative of five independent experiments. Each symbol denotes an individual mouse. Error bars represent the mean ± SEM. Two-tailed unpaired t-test was used for statistical analysis. *P<0.05, **P<0.01, ***P<0.001.
See also Figure S1.
To determine whether the tumor-elicited GzmB response represents a general mechanism of tumor surveillance, we utilized the Tramp model of murine prostate cancer (Greenberg et al., 1995). Compared to WT mice, a higher percentage and number of GzmB-expressing cells were present in the prostates of Tramp mice, and the cell number increased as tumors progressed to later stages (Figure S1E–F). GzmB-expressing cells in mammary and prostate tumors were made of heterogeneous populations of lymphocytes that could be differentiated by their surface expression of TCRβ and TCRδ (Figure 1D and data not shown). Unlike GzmB-producing TCRαβ+ or TCRγδ+ T cells, TCR−GzmB+ cells were negative for intracellular CD3 (Figure S1G), and were classified as innate lymphocytes. Notably, GzmB-expressing cells from all three lineages were present in WT mice at low numbers (Figure 1D–F). Cell transformation expanded all populations, with TCRαβ and TCRγδ lineage cells proportionally more represented in PyMT mice (Figure 1D–F).
GzmB-expressing TCRαβ+ T cells are unconventional
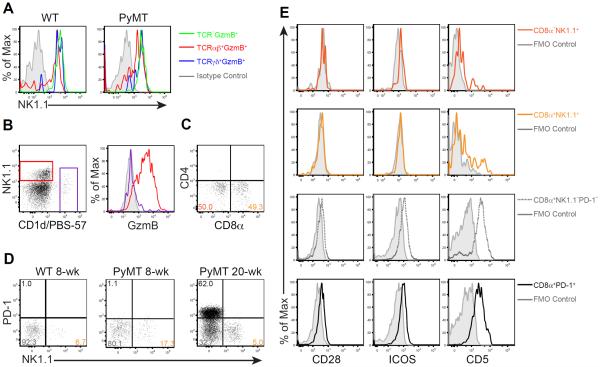
TCR−GzmB+ cells from WT and PyMT mice expressed NK1.1 (Figure 2A), a cell surface marker typically used to define conventional natural killer (cNK) cells. Notably, GzmB-producing TCRαβ and TCRγδ lineage cells also expressed high levels of NK1.1 (Figure 2A). To determine whether TCRαβ+NK1.1+GzmB+ cells belonged to the CD1d-restricted invariant natural killer T (NKT) cell lineage known to express NK1.1, we co-stained tumor-associated TCRαβ+ T cells with NK1.1 and a CD1d tetramer loaded with the NKT cell ligand PBS-57 (CD1d/PBS-57). CD1d/PBS-57+ NKT cells expressed low levels of NK1.1, but not GzmB (Figure 2B). In contrast, CD1d/PBS-57−NK1.1+ T cells expressed high levels of GzmB (Figure 2B). Moreover, unlike some CD1d/PBS-57+ NKT cells that expressed the co-receptor CD4, but not CD8α, nearly half of CD1d/PBS-57−NK1.1+ T cells expressed CD8α, but not CD4 (Figure 2C and Figure S2A). Importantly, tumor-associated TCRαβ+NK1.1+GzmB+ cells were unperturbed in the absence of CD1d (Figure S2B–C), demonstrating that they are distinct from CD1d-restricted NKT cells.
Figure 2. Tumor-associated GzmB-expressing T cells are unconventional.
(A) Flow cytometric analysis of NK1.1 expression in TCR−GzmB+ (green), TCRβ+GzmB+ (red) and TCRδ+GzmB+ (blue) cells in wild-type (WT) and 8-week-old PyMT mice.
(B) Flow cytometric analysis of NK1.1 expression and CD1d/PBS-57 tetramer reactivity in TCRβ+ lymphocytes (left), and GzmB expression in TCRβ+NK1.1+CD1d/PBS-57− cells (red) and TCRβ+CD1d/PBS-57+ cells (purple) from pooled mammary glands of 8-week-old PyMT mice. Grey histogram indicates fluorescence minus one (FMO) control.
(C) Flow cytometric analysis of CD4 and CD8α expression in TCRβ+NK1.1+CD1d/PBS-57− lymphocytes from pooled mammary glands of 8-week-old PyMT mice. Numbers in quadrants indicate percentage of cells.
(D) Flow cytometric analysis of PD-1 and NK1.1 expression in TCRβ+CD8α+ cells from pooled mammary glands of WT, 8- and 20-week-old PyMT mice. Numbers in quadrants indicate percentage of cells.
(E) Flow cytometric analysis of CD28, ICOS and CD5 expression among CD8α−NK1.1+ (dark orange), CD8α+NK1.1+ (light orange), CD8α+NK1.1−PD-1− (dotted grey) and CD8α+PD-1+ (black) T cell populations from pooled tumors of 20- to 24-week-old PyMT mice. Solid grey line in histograms indicates fluorescence minus one (FMO) control for each population.
See also Figure S2.
Activated conventional CD8+ T cells express cytolytic molecules including GzmB, and terminally differentiated cytotoxic T lymphocytes upregulate NK cell-associated markers such as KLRG1 (Zhang and Bevan, 2011). In PyMT mice, tumor growth induces exhaustion of CD8+ T cells characterized by high expression of the inhibitory co-receptor PD-1 (Franklin et al., 2014). To determine whether CD8α+NK1.1+ T cells might represent an activated conventional T cell population, we assessed the expression of NK1.1 and PD-1 in tumor-associated CD8α+ T cells. CD8α+NK1.1+ T cells were substantially expanded in 8-week-old PyMT mice compared to WT mice, while CD8α+PD-1+ T cells were undetectable at this stage (Figure 2D). At 20 weeks of age, PD-1 and NK1.1 remained mutually exclusive, although CD8α+PD-1+ T cells outpaced CD8α+NK1.1+ T cells to dominate the tumors (Figure 2D). Notably, compared to CD8α+NK1.1−PD-1− and CD8α+PD-1+ T cells, CD8α+NK1.1+ T cells maintained high GzmB expression (Figure S2D). Moreover, TCR profiling analyses showed that, unlike conventional CD8α+PD-1+ T cells that had substantially skewed TCR usage, CD8α+NK1.1+ T cells maintained a diverse TCR repertoire (Figure S2E). These observations suggest that CD8α+NK1.1+ T cells do not undergo clonal expansion or functional exhaustion, and thus are distinct from conventional cytotoxic T lymphocytes.
To further explore the lineage relationship between TCRαβ+NK1.1+GzmB+ T cells and conventional CD8+ T cells, we performed transcriptome analysis and included tumor-associated TCR−NK1.1+ cells as controls. Principal component analyses revealed that TCRαβ+CD8α+NK1.1+ and TCRαβ+CD8α−NK1.1+ cells formed a tight cluster, whereas CD8α+PD-1+ and CD8α+PD-1−NK1.1− T cells were closely associated (Figure S2F). Hierarchical clustering analyses showed that TCRαβ+NK1.1+ cells were more closely related to TCR−NK1.1+ cells than conventional CD8+ T cells (Figure S2G and Table S1). Notably, several conventional T cell lineage markers including the co-stimulatory molecules CD28 and ICOS, as well as the signal-tuning receptor CD5, were undetectable or expressed at very low levels in TCRαβ+NK1.1+ cells (Figure 2E and Table S1, cluster 4). Furthermore, inhibitory receptors such as CTLA4 and BTLA, as well as TCR signaling molecules such as LAT, were also downregulated in TCRαβ+NK1.1+ cells (Table S1, cluster 4). Together, these findings imply that TCRαβ+NK1.1+GzmB+ cells are unconventional T cells.
GzmB-expressing innate lymphocytes are unconventional
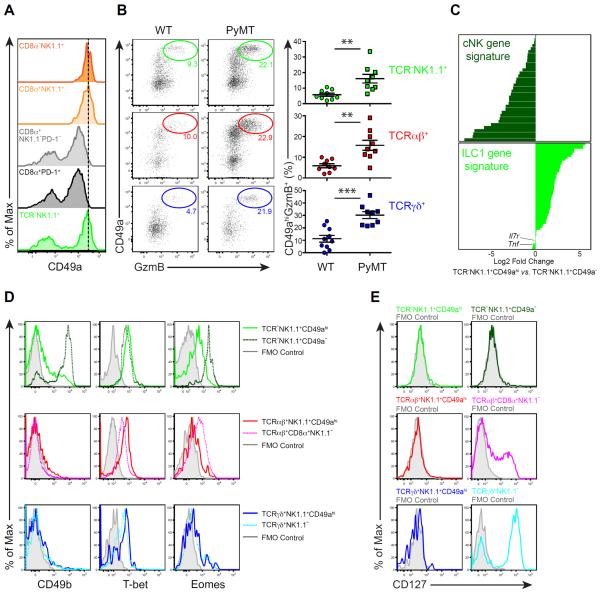
Among the transcripts enriched in TCRαβ+NK1.1+ T cells was Itga1 encoding the collagen-binding integrin CD49a (Table S1, cluster 1). Indeed, CD49a protein was expressed at higher levels in TCRαβ+CD8α+NK1.1+ or TCRαβ+CD8α−NK1.1+ cells than in CD8α+PD-1+ or CD8α+PD-1−NK1.1− T cells (Figure 3A). Surprisingly, TCR−NK1.1+ cells had mixed CD49ahi and CD49a− populations (Figure 3A). Notably, in addition to TCRαβ+ T cells, high CD49a expression marked GzmB-producing TCR−NK1.1+ innate lymphocytes as well as GzmB-producing TCRγδ+ T cells (Figure 3B). Compared to WT mice, PyMT mice had increased numbers of CD49ahi GzmB-expressing TCR−NK1.1+, TCRαβ+ and TCRγδ+ cells in precancerous mammary tissue (Figure 3B), further supporting that the early GzmB response engages all three lineages of lymphocytes.
Figure 3. Tumor-associated GzmB-expressing innate lymphocytes are type 1-like ILCs (ILC1ls).
(A) Flow cytometric analysis of CD49a expression in the indicated cell populations isolated from pooled tumors of 20- to 24-week-old PyMT mice. CD8α−NK1.1+, CD8α+NK1.1+, CD8α+NK1.1−PD-1− and CD8α+PD-1+ cell subsets were gated among TCRβ+ cells. Data are representative of ten independent experiments.
(B) Left panel: flow cytometric analysis of CD49a and GzmB expression in the indicated subsets isolated from pooled mammary glands of wild-type (WT) and 8-week-old PyMT mice. Data are representative of five independent experiments. Right panel: percentages of CD49ahiGzmB+ cells in the indicated subsets (determined by flow cytometric analysis as in the left panel). Each symbol denotes an individual mouse. Data are pooled from five independent experiments. Error bars represent the mean ± SEM. Two-tailed unpaired t-test was used for statistical analysis. **P<0.01, ***P<0.001.
(C) Log2 mean fold change of TCR−NK1.1+CD49ahi vs. TCR−NK1.1+CD49a− gene expression corresponding to the cNK and ILC1 core gene signatures. See also Table S2.
(D–E) Flow cytometric analysis of CD49b, T-bet and Eomes (D) as well as CD127 (E) expression in the indicated cell populations isolated from pooled tumors of 20- to 24-week-old PyMT mice. Grey histogram indicates fluorescence minus one (FMO) control. Data are representative of three independent experiments.
See also Figure S3.
In addition to cNK cells, innate lymphoid cells (ILCs) function as early effectors during immune challenge (Artis and Spits, 2015; Eberl et al., 2015). Both cNK cells and type 1 ILCs (ILC1s) participate in type 1 immune responses, but their classification has been controversial because of overlapping phenotypes (Diefenbach et al., 2014; Serafini et al., 2015). A recent study has identified gene expression signatures for cNK cells and ILC1s by cross-referencing differentially expressed transcripts between cNK cells and ILC1s from spleen and liver (Robinette et al., 2015). We sought to determine the identity of tumor-associated GzmB-expressing innate lymphocytes, and performed transcriptome analysis of sorted TCR−NK1.1+CD49ahi and TCR−NK1.1+CD49a− cells. To ensure that TCR−NK1.1+ cells were innate lymphocytes, we validated our gating strategy by confirming that these cells expressed the Nkp46 receptor (Figure S3A), which was considered a reliable marker for cNK cells and ILC1s (Diefenbach et al., 2014). Transcriptome analyses showed that the signature genes of cNK cells were expressed at high levels in TCR−NK1.1+CD49a− cells, whereas most transcripts used to define the ILC1 gene signature were enriched in TCR−NK1.1+CD49ahi cells (Figure 3C and Table S2). In line with these observations, TCR−NK1.1+CD49a− expressed high levels of Itga2 encoding the cNK cell marker CD49b compared to TCR−NK1.1+CD49ahi cells (Figure 3D and Table S2). Furthermore, while both cell populations expressed comparable amounts of the transcription factor T-bet, Eomes was expressed at substantially higher levels in TCR−NK1.1+CD49a− cells (Figure 3D and Table S2). These findings demonstrate that the GzmB-expressing innate lymphocytes are distinct from cNK cells.
One of the characteristics of conventional ILC1s is their lack of Eomes expression (Diefenbach et al., 2014; Eberl et al., 2015; Klose et al., 2014). However, Eomes was expressed at low but detectable levels in TCR−NK1.1+CD49ahi cells (Figure 3D). In addition, 10 out of 54 ILC1 signature genes were not upregulated in TCR−NK1.1+CD49ahi cells (Figure 3C and Table S2), suggesting that they are distinct from conventional ILC1s. Indeed, CD127, encoded by the Il7r gene and considered one of the best markers for ILCs (Diefenbach et al., 2014; Serafini et al., 2015; Spits et al., 2013), was not expressed in these cells (Figure 3C and 3E). Furthermore, Tnf, another ILC1 signature gene, was expressed at lower levels in TCR−NK1.1+CD49ahi cells (Figure 3C and Table S2), and TCR−NK1.1+CD49ahi cells poorly produced TNF-α, IFN-γ or both cytokines (Figure S3B–C). These characteristics, which are distinct from those of conventional ILC1s, prompted us to name TCR−NK1.1+CD49ahi cells type 1-like ILCs (ILC1ls), while the TCR−NK1.1+CD49a− cells were classified as cNK cells.
A distinct gene expression signature is shared among GzmB-expressing cells
Similar to ILC1ls, CD49ahi GzmB-expressing TCRαβ+ and TCRγδ+ T cells expressed high levels of T-bet, but low to undetectable amounts of Eomes or CD49b (Figure 3D). While CD127 was expressed in a fraction of TCRαβ+CD8α+NK1.1− and TCRγδ+NK1.1− T cells, it was not detectable on TCRαβ+NK1.1+CD49ahi or TCRγδ+NK1.1+CD49ahi cells (Figure 3E). Likewise, expression of TNF-α was substantially lower in TCRαβ+NK1.1+CD49ahi and TCRγδ+NK1.1+CD49ahi cells (Figure S3B–C). Furthermore, similar to TCRαβ+ T cells (Figure 2D), NK1.1 and PD-1 expression was mutually exclusive in TCRγδ+ T cells (Figure S3D), and only TCRγδ+NK1.1+ T cells expressed high levels of GzmB (Figure S3E).
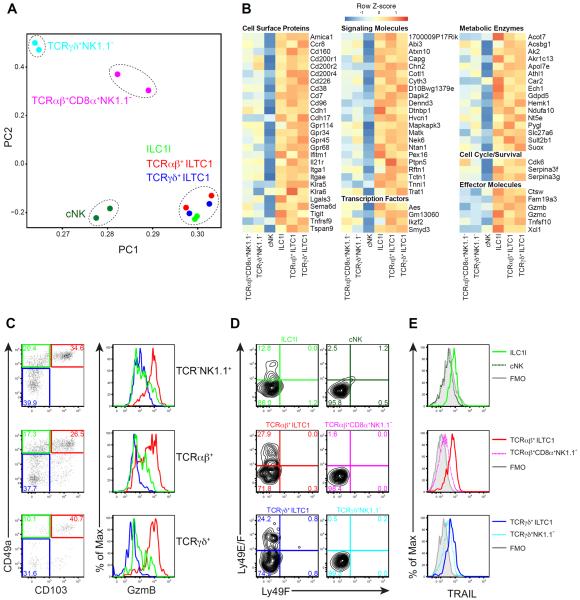
To further explore the likeness among the three lineages of GzmB-expressing lymphocytes, we sorted out ILC1ls, as well as cNK, TCRαβ+NK1.1+CD49ahi, TCRαβ+CD8α+NK1.1−, TCRγδ+NK1.1+CD49ahi and TCRγδ+NK1.1− cells from mammary tumors, and performed transcriptome analysis. As expected, all of these cell populations had classical lymphocyte morphology (Figure S4A). Principal component analyses revealed individual clusters for cNK, TCRαβ+CD8α+NK1.1− or TCRγδ+NK1.1− cells, but ILC1ls, TCRαβ+NK1.1+CD49ahi and TCRγδ+NK1.1+CD49ahi cells appeared indistinguishable as a single group (Figure 4A). Such striking similarity between ILC1ls and GzmB-expressing T cells, together with the finding that TCRαβ+NK1.1+CD49ahi and TCRγδ+NK1.1+CD49ahi cells did not display many features of conventional T cells, prompted us to name TCR+NK1.1+CD49ahi cells type 1 innate-like T cells (ILTC1s).
Figure 4. CD49ahiGzmB+ lymphocytes constitute a related group of cells distinct from conventional NK and T cells.
(A) Principal component analysis of gene expression in the indicated cell populations sorted from pooled tumors of 20- to 24-week-old PyMT mice. Expression data are biological replicates from two independent experiments.
(B) Gene expression signature of the indicated cell populations sorted from pooled tumors of 20- to 24-week-old PyMT mice. Genes were manually selected from those in cluster 3 (identified by hierarchical clustering shown in Figure S4B and Table S3). Heat map shows data pooled from two biological replicates.
(C) Flow cytometric analysis of CD49a and CD103 expression in the indicated cell populations isolated from pooled mammary glands of 8-week-old PyMT mice. For each population, the level of GzmB expression is shown in the histogram as follows: CD49ahiCD103+ (red), CD49a+CD103− (green), CD49a−CD103− (blue). Data are representative of four independent experiments.
(D–E) Flow cytometric analysis of Ly49E and Ly49F (D) or TRAIL (E) expression in the indicated cell populations isolated from pooled tumors of 20- to 24-week-old PyMT mice. Data are representative of two to three independent experiments. Grey histogram indicates fluorescence minus one (FMO) control.
The innateness of ILTC1s was further supported by hierarchical clustering analysis, which demonstrated closer association between ILTC1s and cNK cells than TCRαβ+CD8α+NK1.1− or TCRγδ+NK1.1− T cells (Figure S4B). Indeed, ILC1ls, TCRαβ+ ILTC1s and TCRγδ+ ILTC1s shared with cNK cells a set of transcripts that were substantially underrepresented in TCRαβ+CD8α+NK1.1− or TCRγδ+NK1.1− T cells (Figure S4B, S5A and Table S3, cluster1). Among these transcripts were genes encoding activating and inhibitory receptors such as Ly49 family molecules, NKG2A/C/E, NKG2D and CD244 (Figure S5A–C and Table S3, cluster1). In addition, transcripts of several signaling molecules including Tyrobp, transcription factors including Tbx21, and the effector molecule Prf1 were enriched in ILC1ls, ILTC1s, and cNK cells (Figure S5A and Table S3, cluster1).
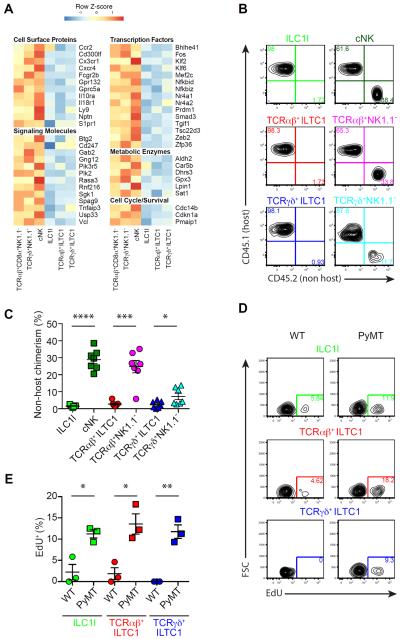
In line with principal component and hierarchical clustering analyses, a gene signature of 131 transcripts expressed at high levels in ILC1ls, TCRαβ+ ILTC1s and TCRγδ+ ILTC1s was identified (Figure 4B and Table S3, cluster 3). Gene Ontology and Pathway Annotation analyses showed that the gene signature was enriched for transcripts associated with cell adhesion (Figure 4B and Table S3, cluster 3). Indeed, several cell adhesion molecules including CD103, encoded by the Itgae gene, were upregulated in ILC1ls and ILTC1s, and GzmB-expressing lymphocytes from all three lineages were marked by high expression of both CD49a and CD103 (Figure 4C). Another shared feature of these cells was that a fraction of them expressed the inhibitory receptor Ly49E (Figure 4D and Figure S5D), encoded by the Klra5 gene (Figure 4B), previously reported to be expressed on fetal-liver-derived lymphocytes as well as intraepithelial lymphocytes (Denning et al., 2007; Stevenaert et al., 2003; Van Beneden et al., 2002). In addition to cell surface proteins, genes encoding several signaling molecules, transcription factors and metabolic enzymes were all substantially upregulated in ILC1ls and ILTC1s (Figure 4B). Furthermore, besides Gzmb, transcripts encoding several effector molecules including Gzmc and Tnfsf10 were enriched in ILC1ls and ILTC1s (Figure 4B), and the Tnfsf10-encoded cell death receptor TRAIL was expressed in all three lineages of GzmB-expressing lymphocytes (Figure 4E). These findings imply that ILC1ls and ILTC1s may engage multiple pathways to induce target cell cytotoxicity.
ILC1ls and ILTC1s are tissue-resident and expand in precancerous lesions
In addition to the upregulated gene signature, hierarchical clustering analyses revealed 122 transcripts downregulated in ILC1ls, TCRαβ+ ILTC1s and TCRγδ+ ILTC1s (Figure 5A, Figure S4B and Table S3, cluster 2). Gene Ontology and Pathway Annotation analyses showed that proteins encoded by this gene cluster were associated with signaling and cell mobility (Figure 5A and Table S3, cluster 2). Indeed, genes encoding several chemokine receptors including Ccr2, Cx3cr1, Cxcr4, the cell trafficking receptor S1pr1, and the transcriptional regulator of cell migration Klf2, were all repressed in ILC1ls and ILTC1s (Figure 5A and Figure S6A–B). In line with these observations, parabiosis experiments revealed that the degree of non-host chimerism was much lower for ILC1ls, TCRαβ+ ILTC1s and TCRγδ+ ILTC1s when compared to cNK, TCRαβ+CD8α+NK1.1− and TCRγδ+NK1.1− cells, respectively (Figure 5B–C). These findings demonstrate that ILC1ls and ILTC1s are tissue-resident lymphocytes, a property likely further promoted by their high expression of tissue retention molecules such as CD103 and CD49a (Figure 4C).
Figure 5. Tissue-resident ILC1ls and ILTC1s expand in precancerous lesions.
(A) Gene expression signature of the indicated cell populations sorted from pooled tumors of 20- to 24-week-old PyMT mice. Genes were manually selected from those in cluster 2 (identified by hierarchical clustering shown in Figure S4B and Table S3). Heat map shows data pooled from two biological replicates.
(B–C) Flow cytometric analysis of CD45.1 (host) and CD45.2 (non-host) expression in the indicated cell populations isolated from the parabiotic PyMT mice 2 weeks after surgery (connected at 6 weeks of age). Data are representative of three independent experiments (B), and percentage of non-host chimerism was compiled from three independent experiments (C).
(D–E) Flow cytometric analysis of EdU incorporation in the indicated cell populations isolated from pooled mammary glands of 8-week-old wild-type (WT) and PyMT mice. Cells were first gated on CD49ahiGzmB+ cells, and then on TCR− (ILC1l), TCRβ+ (TCRαβ+ ILTC1) and TCRδ+ (TCRγδ+ ILTC1). Data are representative of two independent experiments (D). Percentage of EdU+ cells was compiled from two independent experiments (E). (C–E) Each symbol denotes an individual mouse and error bars represent the mean ± SEM. Two-tailed unpaired t-test was used for statistical analysis. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
See also Figure S6.
Notably, ILC1ls and ILTC1s from PyMT mice were enriched for the transcripts of Cdk6 (Figure 4B), encoding a cyclin-dependent kinase, but had low expression of Cdkn1a (Figure 5A), encoding the cyclin-dependent kinase inhibitor p21. We considered that cell transformation might trigger a higher proliferative capacity of ILC1ls and ILTC1s resulting in their increased numbers in PyMT mice. Indeed, ILC1ls and ILTC1s from the mammary tissue of 8-week-old PyMT mice incorporated more EdU, and expressed higher levels of the cell proliferation marker Ki67 than cells from WT mice (Figure 5D–E and Figure S6C). Together, these findings reveal that tissue-resident ILC1ls and ILTC1s selectively expand in tumors.
ILC1ls and ILTC1s exhibit innate cytotoxicity towards tumor cells
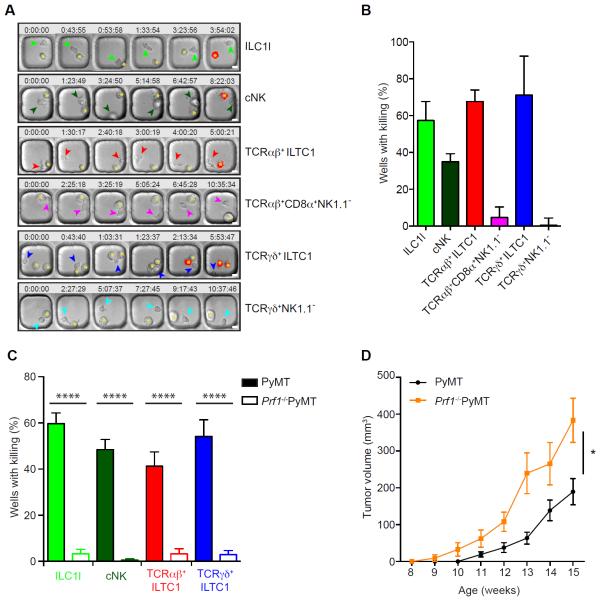
ILC1ls and ILTC1s expressed high levels of cytolytic molecules (Figure 4B). To investigate the cytotoxic potential of these cells, we performed single-cell killing assays using PyMT-derived AT3 cells as targets. Similar to cNK cells, ILC1ls as well as TCRαβ+ ILTC1s and TCRγδ+ ILTC1s were highly efficient in killing tumor cells compared to TCRαβ+CD8α+NK1.1− and TCRγδ+NK1.1− cells (Figure 6A–B and Supplemental Movies). Notably, the killing triggered by ILC1ls and ILTC1s proceeded with multiple short contacts (Supplemental Movies), and appeared distinct from the stable synapse formed between antigen-specific cytotoxic T cells and their targets (Deguine et al., 2010). These observations support that ILTC1s behave like innate lymphocytes and may not recognize peptide antigens to kill target cells. To test this hypothesis, we performed cytotoxic assays using the tumor cell line RMA-S, which lacks the transporter associated with antigen processing, a critical component of the conventional MHC Ia antigen-processing pathway. Indeed, ILC1ls as well as TCRαβ+ ILTC1s and TCRγδ+ ILTC1s potently induced cytotoxicity of RMA-S tumor cells (Figure S7A), supporting an innate mode of effector activities of ILTC1s.
Figure 6. ILC1ls and ILTC1s exhibit potent cytotoxic activities against tumor cells.
(A–C) Lymphocyte populations were purified and cultured from pooled tumors of 20- to 24-week-old PyMT (A–C) and Prf1−/−PyMT mice (C), incubated at a 1:1 ratio with AT-3 tumor cells and imaged in polydimethylsiloxane wells in the presence of propidium iodide (PI).
(A) Time-lapse montage showing the killing (marked by the red signal indicative of PI staining) of AT3 target cells (indicated by an asterisk) by ILC1l, TCRαβ+ ILTC1, TCRγδ+ ILTC1, cNK, TCRαβ+CD8α+NK1.1− and TCRγδ+NK1.1− cells (shown by arrowheads). Time (hours:minutes:seconds) is shown at the top of each image with the their initial co-appearance of target and effector cells in the well set at 0. Scale bar = 10 μm.
(B–C) Quantification of killing efficiency defined by number of wells with PI signal over total number of wells with background cell death subtracted. Wells containing one effector lymphocyte were included for quantification and only a single killing event per well was counted (n = 20–100). Filled and open bars represent killing efficiency of cells sorted from wild-type PyMT and Prf1−/−PyMT mice, respectively (C). Data are pooled from two to three independent experiments. Error bars represent the mean ± SEM. Oneway ANOVA was used for statistical analysis. ****P<0.0001.
(D) Total tumor burden of Prf1−/− PyMT and wild-type PyMT mice monitored between 8 and 15 weeks of age (n = 7–11). Error bars represent the mean ± SEM. Two-way ANOVA was used for statistical analysis. *P<0.05.
See also Figure S7, Movie S1, Movie S2, Movie S3, Movie S4, Movie S5 and Movie S6.
ILC1ls and ILTC1s expressed high levels of GzmB and GzmC as well as TRAIL (Figure 4B), suggesting that they may utilize several effector molecules to induce target cell cytotoxicity. To investigate a role of the lytic granule pathway in target cell killing, we crossed PyMT mice to the perforin-deficient background. Prf1−/− PyMT mice had comparable numbers of ILC1ls and ILTC1s to WT PyMT mice (Figure S7B and data not shown). Nonetheless, perforin deficiency resulted in diminished cytolytic activities of ILC1ls and ILTC1s as well as cNKs (Figure 6C). These findings demonstrate that ILC1ls, ILTC1s and cNKs predominantly engage the lytic granule pathway to kill tumor cells. Notably, Prf1−/− PyMT mice exhibited accelerated tumor growth (Figure 6D), suggesting a critical function for ILC1ls, ILTC1s and/or cNKs in cancer immunosurveillance.
IL-15, but not Nfil3, regulates ILC1l and ILTC1 generation and tumor growth
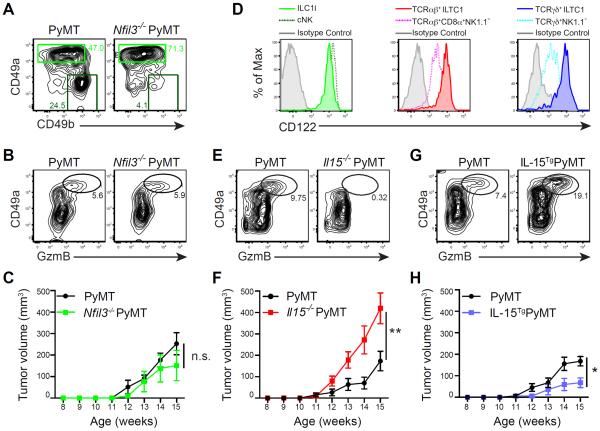
We next sought to define a specific function for ILC1ls and ILTC1s in control of tumor development. The transcription factor Nfil3 regulates the differentiation of cNK cells (Gascoyne et al., 2009; Kamizono et al., 2009) and conventional ILCs (Fuchs et al., 2013; Geiger et al., 2014; Klose et al., 2014; Seillet et al., 2014). To investigate a potential role for Nfil3 in ILC1l and ILTC1 differentiation, we first analyzed the mammary glands of Nfil3−/− mice in the steady state. As expected, cNK cell number was greatly reduced in the absence of Nfil3 (Figure S7C). Surprisingly, ILC1ls were unperturbed (Figure S7C), suggesting that differentiation of ILC1ls diverges from that of cNK cells or conventional ILCs. We further crossed PyMT mice to the Nfil3-deficient background and found that the tumor-infiltrating cNK cells were also greatly diminished (Figure 7A), while GzmB-expressing ILC1ls or ILTC1s from both TCRαβ and TCRγδ lineages remained unperturbed in the tumor (Figure 7A–B and data not shown). Importantly, Nfil3 deficiency did not affect tumor growth (Figure 7C), suggesting that cNK cells are dispensable for the control of tumor development in PyMT mice.
Figure 7. IL-15 regulates ILC1l and ILTC1 generation impacting on tumor growth.
(A) Flow cytometric analyses of CD49a and CD49b expression in TCR−NK1.1+ cells from pooled tumors of Nfil3−/− PyMT and wild-type PyMT mice. Numbers in plots indicate percentage of CD49ahiCD49b− (ILC1l) and CD49a−CD49b+ (cNK) cells in their respective gates. Data are representative of three independent experiments.
(B) Flow cytometric analysis of CD49a and GzmB expression in CD45+ leukocytes from pooled tumors of Nfil3−/− PyMT and wild-type PyMT mice. Numbers in plots indicate percentage of CD49ahiGzmB+ cells. Data are representative of three independent experiments.
(C) Total tumor burden of Nfil3−/− PyMT and wild-type PyMT mice monitored between 8 and 15 weeks of age (n = 3–6).
(D) Flow cytometric analysis of CD122 expression in the indicated cell populations isolated from pooled mammary glands of 8-week-old PyMT mice. Data are representative of five independent experiments.
(E) Flow cytometric analysis of CD49a and GzmB expression in CD45+ leukocytes isolated from pooled mammary glands of 8-week-old Il15−/− PyMT and wild-type PyMT mice. Numbers in plots indicate percentage of CD49ahiGzmB+ cells.
(F) Total tumor burden of Il15−/− PyMT and wild-type PyMT mice monitored between 8 and 15 weeks of age (n = 14–17).
(G) Flow cytometric analysis of CD49a and GzmB expression in CD45+ leukocytes isolated from pooled mammary glands of 8-week-old IL-15TgPyMT and wild-type PyMT mice. Numbers in plots indicate percentage of CD49ahiGzmB+ cells.
(H) Total tumor burden of IL-15TgPyMT and wild-type PyMT mice monitored between 8 and 15 weeks of age (n = 11–15).
(C, F, H) Error bars represent the mean ± SEM. Two-way ANOVA was used for statistical analysis. n.s. = not significant, *P<0.05, **P<0.01.
See also Figure S7.
To further investigate the in vivo function of ILC1ls and ILTC1s, we followed the observation that these cells expressed high levels of the Il2rb gene encoding CD122, the β chain receptor for cytokines IL-2 and IL-15 (Figure 7D and Figure S5A). These findings suggest that ILC1ls and ILTC1s may depend on IL-15 for differentiation and/or homeostasis. To test this hypothesis, we crossed PyMT mice to the IL-15-deficient background. Strikingly, IL-15 deficiency depleted ILC1ls and ILTC1s in addition to cNKs (Figure 7E and data not shown). Compared to control PyMT mice, Il15−/− PyMT mice exhibited accelerated tumor growth (Figure 7F), developed more widespread carcinomas (Figure S7D), and had reduced survival (Figure 7E), which was in line with a recent report (Gillgrass et al., 2015). To investigate whether IL-15 expression represents a rate-limiting step of ILC1l and ILTC1 generation, we bred PyMT mice with a strain of IL-15 transgenic (IL-15Tg) mice. IL-15 overexpression expanded ILC1ls and ILTC1s (Figure 7G and Figure S7F–H), and inhibited tumor growth (Figure 7H). Together, these observations suggest a critical role for ILC1ls and ILTC1s, but not cNKs, in cancer immunosurveillance, although potentially redundant functions of ILC1ls, ILTC1s and cNKs cannot be excluded.
Discussion
The immune system has well defined functions in inducing immunity to foreign pathogens while maintaining tolerance to normal self-tissues. Its role in cancer, however, has been enigmatic, in part because tumors are pathogenic while at the same time, self-derived. In this study, we show that cell transformation induces expansion of tissue-resident ILC1ls and ILTC1s with potent cytolytic activities against tumor cells, thus defining an unprecedented class of immune response well suited for the surveillance of nascent transformed cells.
Tumor-associated ILC1ls appear to be transcriptionally distinct but functionally related to cNK cells, and at the same time, transcriptionally related but phenotypically different from conventional CD127+ ILC1s. Recent cell lineage tracing experiments have revealed that CD127+ ILCs and cNK cells originate from separate progenitors (Constantinides et al., 2014; Klose et al., 2014). In line with these observations, studies using an Eomes-reporter mouse strain have demonstrated that the liver-resident CD127+CD103−Eomes− ILC1s, also known as liver-resident NK cells (Peng et al., 2013; Sojka et al., 2014) or immature NK cells (Takeda et al., 2005), are a stable population and do not give rise to cNK cells in cell transfer experiments (Daussy et al., 2014). Generation of mammary tissue ILC1ls, but not cNK cells, is independent of Nfil3, a feature shared with the ILC1-like population from the salivary gland (Cortez et al., 2014), suggesting that ILC1ls are developmentally distinct from cNK cells as well. Intriguingly, two populations of ILC1s with differential expression of CD127 and CD103 have recently been identified in humans (Bernink et al., 2015; Bernink et al., 2013; Fuchs et al., 2013). Unlike CD127+ ILC1s that can be converted to ILC3s by polarization cytokines, CD103+ ILC1s are refractory to such conversion, implying that CD103+ ILC1s and CD127+ ILC1s are distinct cell populations (Bernink et al., 2015). Notwithstanding, whether mouse ILC1ls and human CD103+ ILC1s are related and represent the third lineage of type 1 innate lymphocytes, and whether CD103 is a stable cell surface marker and can be well used for cell classification await future characterization of their exact developmental pathways in mouse and human.
ILTC1s are tissue-resident T lymphocytes with innate cytolytic activities against tumor cells. Based on the expression of co-receptors, TCRαβ+ ILTC1s can be divided into CD4−CD8α− and CD4−CD8α+ subsets. The varying expression of co-receptors along with high expression of CD103 and several NK cell receptors in TCRαβ+ ILTC1s was reminiscent of that of TCRαβ+ intestinal epithelial lymphocytes (IELs) (Cheroutre et al., 2011). Recent studies have revealed that TCRαβ+ IELs are selected by agonist antigens associated with diverse MHC specificities in the thymus (Mayans et al., 2014; McDonald et al., 2014). Several innate-like TCRγδ+ T cells subsets have also been shown to require a strong TCR signal for their thymic differentiation (Wencker et al., 2014). Yet, upon maturation, these cells undergo attrition of TCR signaling, concomitant with acquisition of responsiveness to innate immune signals (Wencker et al., 2014). Notably, compared to conventional tumor-associated TCRγδ+ and TCRαβ+CD8+ T cells, ILTC1s exhibit profound downregulation of several TCR signaling molecules and do not undergo T cell exhaustion. These observations raise the intriguing possibility that ILTC1s may deviate from conventional lineage of T cells following their agonistic selection in the thymus and attenuation of TCR signaling in the periphery. Defining the selection antigens for ILTC1s and the mechanisms of signal rewiring will clarify the ontogeny of ILTC1s and how they are related to IELs and other innate-like TCRγδ+ T cell subsets.
ILC1ls and ILTC1s share a common transcriptional signature, respond similarly to tumors, and are functionally alike. Their rapid response in precancerous lesions could be attributed to their ability to sense tumor-associated stress signals such as the NKG2D ligands that are induced in transformed cells (Raulet et al., 2013). Similar to cNK cells, ILC1ls and ILTC1s constitutively express NKG2D. Notably, the lack of cNK cells in Nfil3−/− deficient mice does not affect tumor growth, implying that the previously demonstrated tumor suppressor function of NKG2D (Guerra et al., 2008) may be attributed to ILC1ls and ILTC1s instead of cNK cells. In fact, although cNK cells are found in tumor tissues and exhibit cytotoxic activities against tumor cells in vitro, they do not expand in precancerous lesions. Furthermore, cNK cells are a recirculating population, which is in contrast to the tissue-resident ILC1ls and ILTC1s. Such distinct tissue localization likely makes ILC1ls and ILTC1s unique sentinels of transformed epithelium. It is interesting to note that one of the markers shared among these cell populations is the epithelial cadherin (E-cadherin) receptor CD103. CD103 interaction with E-cadherin promotes cytotoxic T cell killing of target cells (Le Floc'h et al., 2007). Future studies will determine whether CD103 is crucial for tumor sensing and killing mediated by ILC1ls and ILTC1s.
Generation of ILC1ls and ILTC1s is dependent on the cytokine IL-15. However, it remains unknown whether IL-15 promotes their differentiation, homeostasis and/or activation in transformed tissues. Constitutive IL-15 overexpression expands ILC1ls and ILTC1s, but the endogenous cellular sources of IL-15 involved in their regulation remain to be determined. IL-15 is expressed in epithelial cells as well as stromal cell populations, and requires trans-presentation to regulate target cells (Burkett et al., 2004). In celiac disease, intestinal epithelial cells produce excessive amounts of IL-15 to activate IELs (Jabri and Sollid, 2009). Interestingly, a recent study showed that deletion of the IL15 gene in colorectal cancer is associated with reduced lymphocyte proliferation concomitant with enhanced risk of tumor recurrence and poor patient outcome (Mlecnik et al., 2014). These findings imply that secretion of IL-15 by tumor cells may promote cancer immunosurveillance. In agreement with this hypothesis, IL-15 is induced in senescent tumor cells in a transplantable tumor model, and may cooperate with NKG2D to eliminate tumor cells (Iannello et al., 2013). Nevertheless, the definitive function of tumor-produced IL-15 in cancer immunosurveillance awaits the generation of tumor-specific IL-15-deficient mouse model.
ILC1l- and ILTC1-associated responses are observed in two oncogene-induced murine cancer models, suggesting that they may represent a general mechanism of cancer immunosurveillance. Future studies will reveal whether the ILC1l- and ILTC1-associated responses are induced and modulated in human cancers, and whether their activities can be manipulated for cancer immunotherapy.
Experimental Procedures
Mice
MMTV-PyMT mice were backcrossed to the C57BL/6 background for 10 generations as previously described (Franklin et al., 2014). Tramp, Cd1d−/−, Prf1−/− and CD45.1+ congenic mice were purchased from Jackson Laboratory. Il15−/− mice were purchased from Taconic Farms. Nfil3−/− mice were previously described (Kashiwada et al., 2010), and kindly provided to us by P. Rothman. IL-15 Transgenic mice were previously described (Marks-Konczalik et al., 2000), and kindly provided to us by T. Waldmann and Y. Tagaya. Littermate controls were used in all experiments when possible. All mice were bred and maintained in a specific pathogen-free facility at MSKCC and animal experimentation was conducted in accordance with institutional guidelines. For tumor measurement, pathology scoring and ex vivo immune cell analyses, see Supplementary Experimental Procedures for details.
RNAseq and transcriptome analysis
Complementary DNA (cDNA) libraries were generated from RNA purified from immune cell populations, amplified using the SMARTer RACE Amplification Kit (Clontech), and sequenced in replicate for 40 million reads using 50 bp paired-end at the Integrated Genomics Operation Core Facility at MSKCC. The raw output BAM files were converted to FASTQ using PICARD. All heatmaps and PCA plots were generated using the ggplot2 package. All analyses after count table generation were conducted in the R statistical environment. For detailed analyses, see Supplementary Experimental Procedures.
Statistic analysis
Two-tailed unpaired t-test, One-way ANOVA and Two-way ANOVA were used to calculate statistical significance using Prism 6 software (GraphPad). A P value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank P. Rothman for providing the Nfil3−/− mouse strain, Y. Tagaya and T. Waldmann for providing the IL-15 Transgenic mouse strain and N. Cheung for the IL-15/IL-15Rα complex. We also thank the M. Li lab for insightful discussions. MSKCC has filed a provisional patent application with the U.S. Patent and Trademark Office towards methods and compositions for cancer immunotherapy targeting innate lymphoid cells and innate-like T cells. S.D. and M.O.L. are listed as inventors on this patent application. This work was supported by the Ludwig Center for Cancer Immunology (M.O.L.), and the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests The authors declare no competing financial interests.
Accession number The RNA-seq datasets were deposited at GEO (accession number: GSE76362).
Authors Contributions S.D. and M.O.L. conceived the project, analyzed the data and prepared the manuscript. S.D., B.M.W., R.A.F., C.T.L., S.A.O. and A.T. performed the experiments. S.C., Y.P. and C.S.L. performed the bioinformatic analysis of the gene expression data. M.H. conceived the single cell-killing assay.
References
- Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, Munneke JM, Hazenberg MD, Villaudy J, Buskens CJ, et al. Interleukin-12 and -23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity. 2015;43:146–160. doi: 10.1016/j.immuni.2015.06.019. [DOI] [PubMed] [Google Scholar]
- Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nature immunology. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- Burkett PR, Koka R, Chien M, Chai S, Boone DL, Ma A. Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. The Journal of experimental medicine. 2004;200:825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnet M. Cancer: a biological approach. III. Viruses associated with neoplastic conditions. IV. Practical applications. British medical journal. 1957;1:841–847. doi: 10.1136/bmj.1.5023.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nature reviews Immunology. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. Journal of immunology. 2014;192:4487–4491. doi: 10.4049/jimmunol.1303469. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. The Journal of experimental medicine. 2014;211:563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguine J, Breart B, Lemaitre F, Di Santo JP, Bousso P. Intravital imaging reveals distinct dynamics for natural killer and CD8(+) T cells during tumor regression. Immunity. 2010;33:632–644. doi: 10.1016/j.immuni.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Denning TL, Granger SW, Mucida D, Graddy R, Leclercq G, Zhang W, Honey K, Rasmussen JP, Cheroutre H, Rudensky AY, et al. Mouse TCRalphabeta+CD8alphaalpha intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. Journal of immunology. 2007;178:4230–4239. doi: 10.4049/jimmunol.178.7.4230. [DOI] [PubMed] [Google Scholar]
- Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor MK, Sarkar A, Savage PA, Franklin RA, Johnson LK, Jungbluth AA, Allison JP, Li MO. T cell surveillance of oncogene-induced prostate cancer is impeded by T cell-derived TGF-beta1 cytokine. Immunity. 2011;35:123–134. doi: 10.1016/j.immuni.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348 doi: 10.1126/science.aaa6566. aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich P. Uber den jetzigen stand der karzinomforschung. Ned Tijdschr Geneeskd. 1909;5:273–290. [Google Scholar]
- Enzler T, Gillessen S, Manis JP, Ferguson D, Fleming J, Alt FW, Mihm M, Dranoff G. Deficiencies of GM-CSF and interferon gamma link inflammation and cancer. The Journal of experimental medicine. 2003;197:1213–1219. doi: 10.1084/jem.20021258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnberg N, Klein-Szanto AJ, El-Deiry WS. TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. The Journal of clinical investigation. 2008;118:111–123. doi: 10.1172/JCI29900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, Pamer EG, Li MO. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nature immunology. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- Geiger TL, Abt MC, Gasteiger G, Firth MA, O'Connor MH, Geary CD, O'Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. The Journal of experimental medicine. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillgrass AE, Chew MV, Krneta T, Ashkar AA. Overexpression of IL-15 promotes tumor destruction via NK1.1+ cells in a spontaneous breast cancer model. BMC cancer. 2015;15:293. doi: 10.1186/s12885-015-1264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. Prostate cancer in a transgenic mouse. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, Knoblaugh S, Cado D, Greenberg NM, Raulet DH. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannello A, Thompson TW, Ardolino M, Lowe SW, Raulet DH. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. The Journal of experimental medicine. 2013;210:2057–2069. doi: 10.1084/jem.20130783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri B, Sollid LM. Tissue-mediated control of immunopathology in coeliac disease. Nature reviews Immunology. 2009;9:858–870. doi: 10.1038/nri2670. [DOI] [PubMed] [Google Scholar]
- Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. The Journal of experimental medicine. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M, Levy DM, McKeag L, Murray K, Schroder AJ, Canfield SM, Traver G, Rothman PB. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:821–826. doi: 10.1073/pnas.0909235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- Le Floc'h A, Jalil A, Vergnon I, Le Maux Chansac B, Lazar V, Bismuth G, Chouaib S, Mami-Chouaib F. Alpha E beta 7 integrin interaction with E-cadherin promotes antitumor CTL activity by triggering lytic granule polarization and exocytosis. The Journal of experimental medicine. 2007;204:559–570. doi: 10.1084/jem.20061524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Marks-Konczalik J, Dubois S, Losi JM, Sabzevari H, Yamada N, Feigenbaum L, Waldmann TA, Tagaya Y. IL-2-induced activation-induced cell death is inhibited in IL-15 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11445–11450. doi: 10.1073/pnas.200363097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- Mayans S, Stepniak D, Palida SF, Larange A, Dreux J, Arlian BM, Shinnakasu R, Kronenberg M, Cheroutre H, Lambolez F. alphabetaT cell receptors expressed by CD4(−)CD8alphabeta(−) intraepithelial T cells drive their fate into a unique lineage with unusual MHC reactivities. Immunity. 2014;41:207–218. doi: 10.1016/j.immuni.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald BD, Bunker JJ, Ishizuka IE, Jabri B, Bendelac A. Elevated T cell receptor signaling identifies a thymic precursor to the TCRalphabeta(+)CD4(−)CD8beta(−) intraepithelial lymphocyte lineage. Immunity. 2014;41:219–229. doi: 10.1016/j.immuni.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlecnik B, Bindea G, Angell HK, Sasso MS, Obenauf AC, Fredriksen T, Lafontaine L, Bilocq AM, Kirilovsky A, Tosolini M, et al. Functional network pipeline reveals genetic determinants associated with in situ lymphocyte proliferation and survival of cancer patients. Science translational medicine. 2014;6 doi: 10.1126/scitranslmed.3007240. 228ra237. [DOI] [PubMed] [Google Scholar]
- Pardoll D. Does the immune system see tumors as foreign or self? Annual review of immunology. 2003;21:807–839. doi: 10.1146/annurev.immunol.21.120601.141135. [DOI] [PubMed] [Google Scholar]
- Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. The Journal of clinical investigation. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annual review of immunology. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, Immunological Genome C. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nature immunology. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage PA, Vosseller K, Kang C, Larimore K, Riedel E, Wojnoonski K, Jungbluth AA, Allison JP. Recognition of a ubiquitous self antigen by prostate cancer-infiltrating CD8+ T lymphocytes. Science. 2008;319:215–220. doi: 10.1126/science.1148886. [DOI] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz GT, Carotta S. Nfil3 is required for the development of all innate lymphoid cell subsets. The Journal of experimental medicine. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nature reviews Immunology. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Thia KY, Street SE, MacGregor D, Godfrey DI, Trapani JA. Perforin-mediated cytotoxicity is critical for surveillance of spontaneous lymphoma. The Journal of experimental medicine. 2000;192:755–760. doi: 10.1084/jem.192.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nature reviews Immunology. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- Stevenaert F, Van Beneden K, De Creus A, Debacker V, Plum J, Leclercq G. Ly49E expression points toward overlapping, but distinct, natural killer (NK) cell differentiation kinetics and potential of fetal versus adult lymphoid progenitors. Journal of leukocyte biology. 2003;73:731–738. doi: 10.1189/jlb.0902443. [DOI] [PubMed] [Google Scholar]
- Street SE, Zerafa N, Iezzi M, Westwood JA, Stagg J, Musiani P, Smyth MJ. Host perforin reduces tumor number but does not increase survival in oncogene-driven mammary adenocarcinoma. Cancer research. 2007;67:5454–5460. doi: 10.1158/0008-5472.CAN-06-4084. [DOI] [PubMed] [Google Scholar]
- Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, Yagita H, Kinoshita K, Okumura K, Smyth MJ. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood. 2005;105:2082–2089. doi: 10.1182/blood-2004-08-3262. [DOI] [PubMed] [Google Scholar]
- Thomas L. Delayed hypersensitivity in health and disease. In: Lawrence HS, editor. Cellular and Humoral Aspects of the Hypersensitive States. Hoeber-Harper; New York: 1959. pp. 529–532. [Google Scholar]
- Van Beneden K, De Creus A, Stevenaert F, Debacker V, Plum J, Leclercq G. Expression of inhibitory receptors Ly49E and CD94/NKG2 on fetal thymic and adult epidermal TCR V gamma 3 lymphocytes. Journal of immunology. 2002;168:3295–3302. doi: 10.4049/jimmunol.168.7.3295. [DOI] [PubMed] [Google Scholar]
- Wencker M, Turchinovich G, Di Marco Barros R, Deban L, Jandke A, Cope A, Hayday AC. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nature immunology. 2014;15:80–87. doi: 10.1038/ni.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willimsky G, Blankenstein T. Sporadic immunogenic tumours avoid destruction by inducing T-cell tolerance. Nature. 2005;437:141–146. doi: 10.1038/nature03954. [DOI] [PubMed] [Google Scholar]
- Zerafa N, Westwood JA, Cretney E, Mitchell S, Waring P, Iezzi M, Smyth MJ. Cutting edge: TRAIL deficiency accelerates hematological malignancies. Journal of immunology. 2005;175:5586–5590. doi: 10.4049/jimmunol.175.9.5586. [DOI] [PubMed] [Google Scholar]
- Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.