Abstract
Killer cell immunoglobulin-like receptors (KIRs) regulate NK cell function. KIRs and their HLA ligands are highly polymorphic in nature with substantial allelic polymorphism. At present, there is a lack of an expedient method for KIR and HLA allele typing with relevant functional information. Here, we developed a single-nucleotide polymorphism (SNP) assay to type various allele groups of KIR2DL1 with distinct functional properties based on polymorphism at position 245. We also established a SNP assay to type different KIR ligands based on polymorphism at position 77 in HLA-C and position 83 in HLA-B and -A. Our SNP assays for KIR and KIR ligand typing are much cheaper and faster than existing high-resolution typing. Importantly, our high-throughput methods provide readouts that are informative in predicting NK cell activity in health, disease, and transplantation.
Keywords: Natural Killer cell, Killer cell immunoglobulin-like receptor, Human leukocyte antigen, Single-nucleotide polymorphism
Introduction
Natural killer (NK) cells are part of the innate immune system and are important for infection control [1; 2], cancer surveillance [3; 4; 5] and successful pregnancy [6; 7]. Their functions are regulated by various activating and inhibitory receptors present on the cell surface [8]. The primary inhibitory receptors are the class of killer immunoglobulin-like receptors (KIRs) [9; 10], which are highly polymorphic in nature [11]. Their polymorphism generates functional heterogeneity among alleles of the same KIR gene. For example, we have previously shown that KIR2DL1 alleles having arginine at amino acid position 245 have higher inhibitory activity and more durable surface expression upon identical ligand engagement than alleles that have cysteine at the same position [12]. Different alleles of KIR3DL1 are also reported to have different inhibitory capacity and levels of steady-state cell surface expression [13]. Some other KIRs also exhibit distinguishable functional differences among their alleles [14; 15; 16], although the exact molecular determinants of these differences have not been elucidated. Differences in KIR gene content are associated with many human diseases, including autoimmune diseases, inflammatory disorders, infectious diseases, immunodeficiency, cancer, and reproductive disorders [17], The relationship between these diseases and functional heterogeneity among the alleles of KIR is not yet known due to the lack of expedient methods for high-throughput typing of different functional groups of KIR alleles.
KIRs recognize the highly polymorphic human leukocyte antigen (HLA) class 1 protein with unique specificity [9; 18], NK cell functions are inhibited when the inhibitory KIRs recognize their specific ligands on target cells. For example, KIR2DL1 and KIR2DL2/2DL3 recognize HLA-C allotypes, whereas KIR3DL1 recognizes allotypes of HLA-B and -A. HLA-C ligands are divided into two groups, HLA-C1 and HLA-C2, based on the presence of asparagine or lysine at amino acid position 80 in the mature protein.[19] Furthermore, HLA-C1 contains a conserved serine residue at amino acid position 77, while an asparagine is in that position in HLA-C2. KIR2DL1 recognizes HLA-C2, and KIR2DL2/2DL3 recognizes HLA-C1 [20]. HLA-B is also divided into two groups, HLA-Bw4 and HLA-Bw6, based on their differences in amino acid positions 77–83. HLA-B and HLA-A (A*23, A*24, and A*32) alleles carrying the HLA-Bw4 epitopes are recognized by KIR3DL1 [21]. HLA-Bw6 is not a ligand for KIRs. Disease susceptibility has been associated with various KIR ligand constellations.
Biologically, NK cell reactivity toward target cells is based in part on the presence of KIRs and their cognate ligands. The functional competency of NK cells depends on a HLA class I dependant process known as ‘licensing’ [22]. The capacity of an individual NK cell expressing a certain KIR in response toward target cell missing its ligand depends on the education it received through prior interaction with self ligand. For example, if a NK cell expresses KIR2DL1 but lacks self HLA-C2, it is hyporesponsive against target cells missing HLA-C2 expression [23]. In contrary, a NK cell expressing KIR2DL1 in the presence of self HLA-C2 is reactive to target cells lacking the cognate ligand. Because not all alleles of a certain inhibitory KIR interact to the same degree with various HLA alleles of the same ligand group [24], allelic polymorphism makes the prediction of eventual NK cell reactivity difficult. The precise prediction of NK activity requires knowledge of the molecular determinants of both KIRs and their ligands. Here, we used a novel approach to develop a real-time polymerase chain reaction (PCR)–based method to assess the different functional groups of KIR2DL1, built upon our knowledge of the unique molecular determinants involved. We also established a parallel method for functional KIR ligand typing. These methods are easy, rapid, cost-effective, and informative both biologically and clinically.
Materials and Methods
Cells, culture, and transduction
DNA samples used for KIR2DL1 allele and KIR ligand typing in this study were obtained from peripheral blood mononuclear cells (PBMCs) of healthy donors at St. Jude Children’s Research Hospital. The B-lymphoblastic cell line 721.221 was purchased from the International Histocompatibility Working Group and cultured in RPMI 1640 supplemented with 20% fetal bovine serum and 1 mM penicillin/streptomycin. The 721.221 cells were transduced with retroviral vector MMP-IC-GFP-W containing HLA-Cw6 and HLA-Cw7. High-expressing cells were sorted by flow cytometric cell sorting using green fluorescent protein (GFP) expression. The human chronic myelogenous leukemia cell line K562 was purchased from American Type Culture Collection (ATCC) and cultured in RPMI 1640 supplemented with 10% fetal bovine serum and 1 mM penicillin/streptomycin.
Detection of effector NK cell reactivity toward target cells
To separate the effector cells from target cells, effector cells were labeled with pan-leukocyte marker CD45. The labeled effector cells were washed extensively and then mixed with the target cells. The CD107 cytotoxicity assay was performed as described previously [12]. Briefly, labeled effector PBMCs were mixed with target K562 or 721.221 cells and incubated at 37°C in the presence of anti-CD107a monoclonal antibody H4A3 (BD Pharmingen). After 1 hour of coculturing, Golgi stop (BD Biosciences) was added to a final concentration of 5 mM, followed by 3 hours of incubation in a cell culture incubator. Cells were then washed and labeled with an antibody cocktail containing appropriate antibodies (monoclonal antibody against KIR2DL1, KIR2DL2/2DL3, KIR3DL1, and NKG2a). Single KIR+ NK cell populations were gated from the CD45+ cell population, and cytotoxicity was measured based on CD107 surface expression by flow cytometry (LSR II cytometer, Becton Dickinson). The absolute number of CD107+ cells in single KIR+ NK cell subsets was calculated using the following formula:
Single-nucleotide polymorphism assay
The single-nucleotide polymorphism (SNP) assay was performed on an HT7900 Sequence Detection System (Applied Biosystems) following the allelic discrimination assay protocol provided by the manufacturer. Primers for the assay were designed in such a way that they amplified all the alleles of a particular HLA type (HLA-B or HLA-C) as well as the amplicon containing the polymorphic region of interest. Two probes were designed with a single mismatch between them. Each probe bound only one group of alleles and was labeled with either 6FAM or VIC fluorescent dye at its 5′ end. The 3′ end of the probes contained a quencher. The universal primer pair that was designed to amplify all the alleles of HLA-B was: forward primer 5′-GAGGGGCCGGAGTATTGGGA-3′ and reverse primer 5′-TGTAATCCTTGCCGTCGTAGG-3′. The probe for HLA-Bw4–associated HLA-B and -A was 6FAM-CCGCTACTACAACCAG-MGBNFQ and for HLA-Bw6 was VIC-CGGCTACTACAACCAG-MGBNFQ. For HLA-C, forward primer 5′-TTGGGACCGGGAGACACAG-3′ and reverse primer 5′-CGATGTAATCCTTGCCGTC-3′ were used. The probes used for HLA-C1 and HLA-C2 were 6FAM-CCGAGTGAG CCTGC-MGBNFQ and VIC-CCGAGTGAA CCTGC-MGBNFQ, respectively. Each assay reaction mix contained a 250 nM probe concentration and 20 ng of genomic DNA in 1× TaqMan genotyping master mix (Applied Biosystems). For KIR2DL1 functional allele typing, the probe was designed based on a single-nucleotide mismatch at amino acid position 245 in the mature protein. The sequence for the probes used to distinguish the two functional groups of KIR2DL1 alleles were: 6FAM-CATCGCTGGTGCTC-MGBNFQ and VIC-CATTGCTGGTGCTCC-MGBNFQ. A universal primer was designed that could specifically amplify all the alleles of KIR2DL1. The sequence of the primer pair used was: forward primer 5′-CTCTTCATCCTCCTCTTCTTTC-3′ and reverse primer 5′-GAAAACGCAGTGATTCAACTG-3′. The SNP assay was run on the HT7900 using the same protocol as described for KIR ligand typing. The only exception was that the amount of DNA used to amplify the KIR2DL1 alleles was 50 ng instead of 20 ng.
Results
Molecular determinant-based KIR2DL1 allele typing
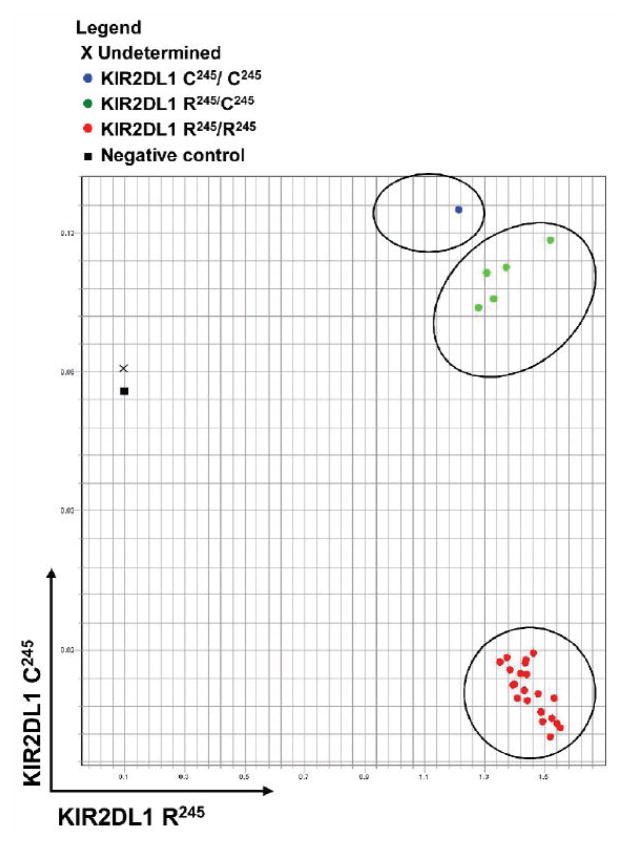
We tested the SNP assay using DNA from 27 donors (Figure 1). Results showed that 20 individuals had only arginine at amino acid position 245 of KIR2DL1, 5 were heterozygous for arginine and cysteine, and 1 had only cysteine in that position. We sequenced all 26 PCR products and confirmed the accuracy of the SNP assay. For the one individual with an “undetermined” result (Figure 1), there was no amplification of PCR products with repeated testings, indicating the absence of genomic KIR2DL1. Thus, our assay approach was useful not only for the identification of functional groups of KIR alleles, but also for determining the presence of the KIR gene itself.
Figure 1. Functional group typing of KIR2DL1.
DNA was extracted from donor peripheral blood mononuclear cells. Probes were designed for different functional groups of KIR2DL1 alleles based on single-nucleotide polymorphisms. Primers were designed that can specifically amplify all the alleles of KIR2DL1.
Functional relevance of molecular determinant-based KIR typing
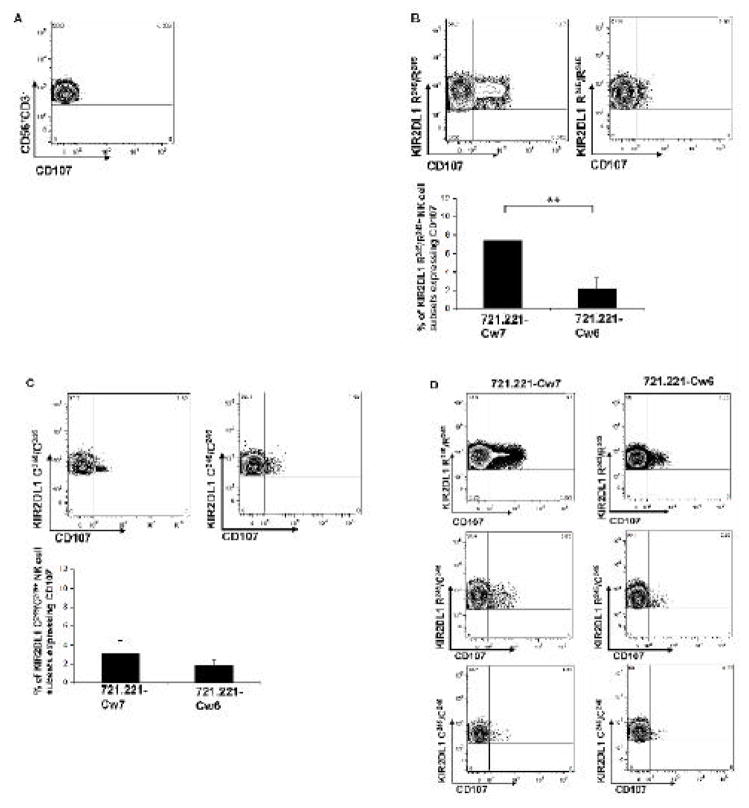
In our next experiment, we sought to validate the accuracy of our KIR typing method in discerning NK cell activity. NK cells were isolated from donor PBMCs using an AutoMACS automated magnetic cell sorter (Miltenyi) with CD56 microbeads. DNA was extracted from the same donor PBMCs and typed for the presence of KIR ligands (HLA-C1 and HLA-C2) and different functional allelic groups of KIR2DL1 (KIR2DL1 R245 and KIR2DL1 C245). NK cells from donors with HLA-C2 were chosen and mixed with 721.221 target cells expressing HLA-Cw6 (HLA-C2) or HLA-Cw7 (HLA-C1). As expected, NK cells showed no CD107 expression on their surface in the absence of the target cells (Figure 2A). NK cells having KIR2DL1 R245 were more reactive than KIR2DL1 C245 cells against target cells expressing non-ligand Cw7 [7.5% ± 2.6% and 2.8% ± 1.1%, respectively, p<0.05] (Figure 2B, 2C, left panels). The reactivity against 721.221 in the presence of the ligand Cw6 was suppressed in both KIR2DL1 R245 and KIR2DL1 C245 cells (Figure 2B, 2C, right panels). NK cell from one KIR2DL1 R245/C245 heterozygous individual had an intermediate cytolytic phenotype when compared with KIR2DL1 R245 or KIR2DL1 C245 homozygous cells (Figure 2D). These findings confirmed that our molecular determinant-based KIR typing method can accurately predict various degrees of NK cell activity against missing-self cells.
Figure 2. Molecular determinant-based KIR typing predicts NK cell activity.
CD56+ cells were isolated from donor peripheral blood mononuclear cells and mixed with target 721.221-Cw7 or 721.221-Cw6 cells. After a CD107 labeling procedure, cells were stained with KIR monoclonal antibodies. KIR2DL1+ NK cell subsets were gated, and surface expression of CD107 was detected using flow cytometry. A) Surface expression of CD107 on NK cells without any target cells. B) Degranulation of the KIR2DL1 R245/R245-positive NK cell subset against 721.221 cells expressing KIR2DL1 non-ligand HLA-Cw7 (left) and KIR2DL1 ligand HLA-Cw6 (right). Histogram shows average reactivity of 6 independent experiments; * p value <0.05. C) Degranulation of the KIR2DL1 C245/C245-positive NK cell subset against 721.221 cells expressing KIR2DL1 non-ligand HLA-Cw7 (left) and KIR2DL1 ligand HLA-Cw6 (right). Histogram shows average reactivity of 3 independent experiments; p value =0.05. D) Degranulation of the KIR2DL1 R245/R245 (upper panel), KIR2DL1 R245/C245 (middle panel) and KIR2DL1 C245/C245 (lower panel) -positive NK cell subset against 721.221 cells expressing non-ligand HLA-Cw7 (left) and ligand HLA-Cw6 (right).
SNP assay to detect KIR ligand groups
Once we successfully used the SNP assay to type different functional groups of KIR2DL1, we extended our method to type its ligand to be expedient for biological research and clinical applications.
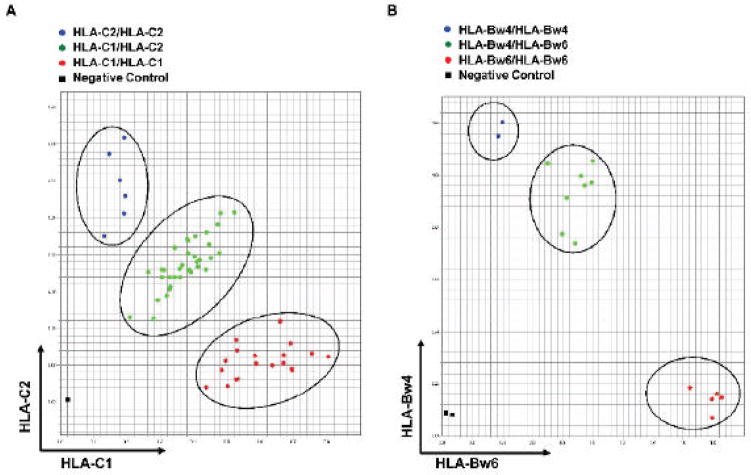
We tested 60 DNA samples that were HLA typed at the allelic level for HLA-A, -B, and -C. We ran the SNP assay initially without prior knowledge of the donor HLA type. We found that the SNP assay can distinguish HLA-C1 homozygous, HLA-C2 homozygous, and HLA-C1/HLA-C2 heterozygous cells (Figure 3A). Unblinding of the HLA typing results of these donor samples revealed 100% agreement in KIR ligand assignment. To confirm the accuracy of SNP typing, we performed HLA-C1 and HLA-C2 typing for an additional 47 individual donors using both high-resolution HLA typing and the KIR ligand SNP assay. Results from the two tests were again identical.
Figure 3. Functional group typing of KIR ligands.
DNA was extracted from donor peripheral blood mononuclear cells. Probes were designed to type KIR ligands based on single-nucleotide mismatches. A) Typing of KIR2DL1 and KIR2DL2/2DL3 ligand HLA-C. B) Typing of KIR3DL1 ligand HLA-Bw4.
After we successfully established the SNP assay to type HLA-C, we developed a similar method for HLA-Bw4 and HLA-Bw6 typing. We aligned HLA-B using the alignment tools in the IGMT/HLA database (http://www.ebi.ac.uk/imgt/hla/align.html), and a single-base-pair mismatch between HLA-Bw4 and HLA-Bw6 at nucleotide position 319 (amino acid position 83 in the mature protein) was chosen to design the probe for the Bw4/Bw6 SNP assay. Comparing the initial results from the SNP assay and high-resolution HLA typing, we found some discrepancies between these two tests if only HLA-B was considered, because HLA-Bw4–associated HLA-A alleles (A*23, A*24, and A*32) have the same sequence in the probe area as HLA-Bw4–associated HLA-B alleles. In the SNP assay, donors who were negative for the HLA-B–associated Bw4 epitope but positive for HLA-Bw4–associated HLA-A were shown to be HLA-Bw4–positive. Since both HLA-Bw4–associated HLA-B and HLA-A are ligands for KIR3DL1, our assay turns out to be ideal to rapidly detect both HLA-B– and -A–associated KIR3DL1 ligands. DNA samples from 94 donors were then tested for HLA-Bw4 and HLA-Bw6 (Figure 3B). Among the 94 samples, 93 SNP results matched those of high-resolution HLA typing. One sample was shown to be HLA-Bw4 homozygous in the SNP assay but HLA-Bw4/HLA-Bw6 heterozygous in HLA typing. The reason our SNP assay did not pick up the HLA-Bw6 signal in this sample was uncertain after extensive investigation. However, considering only the presence or absence of the clinically relevant HLA-Bw4 ligand (since HLA-Bw6 is not a KIR ligand), none of the SNP assays gave false-positive or -negative results. These findings suggest that the SNP assay can be used reliably for KIR ligand typing.
Functional relevance of KIR ligand group typing
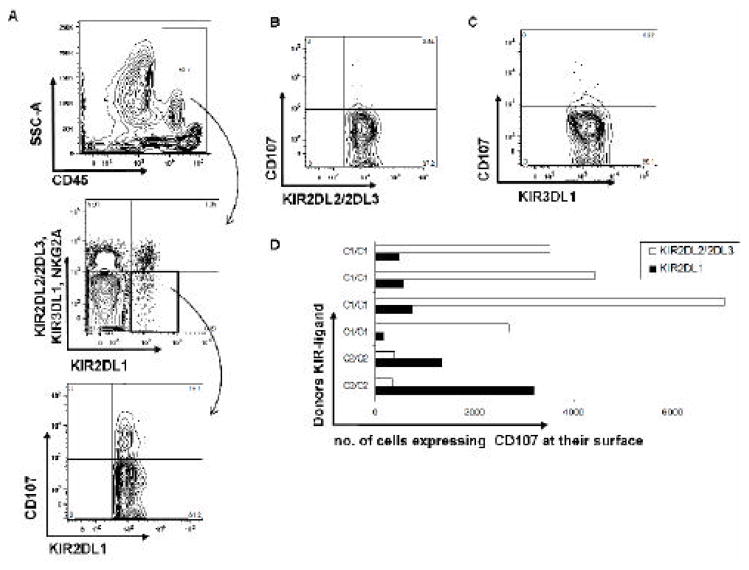
After we successfully typed KIR-ligand groups using the SNP assay, we sought to assess whether the typing results could predict NK cell reactivity. We labeled effector PBMCs with anti-CD45 monoclonal antibody to distinguish them from target K562 cells (which lack KIR ligand expression) after mixing. The gating strategies for the detection of CD107 surface expression in KIR2DL1+ subsets of donor NK cells are shown in Figure 4A. In one donor who was positive for HLA-C2 but not for HLA-C1 and -Bw4, the single KIR2DL1+ subset showed the highest degranulation (19%) (Figure 4A), whereas KIR2DL2/KIR2DL3+ and KIR3DL1+ subsets showed much lower degranulation (Figure 4B, 4C), indicating that NK cells could specifically recognize missing-self cells. These findings were then validated in samples from 6 other donors (Figure 4D), demonstrating the ability of our KIR ligand assay to predict NK cell activity through ligand participation in functional “licensing” or “education.”
Figure 4. KIR ligand group typing predicts NK cell activity.
Donor peripheral blood mononuclear cells were labeled with CD45 and mixed with K562 cells. Single KIR+ NK cell subsets were gated using monoclonal antibodies against KIRs. A) Gating strategy for single KIR+ cells (first two panels) and degranulation of the KIR2DL1+ NK cell subset (bottom panel). B) Degranulation of the KIR2DL2/2DL3+ subset. C) Degranulation of the KIR3DL1+ subset. D) Reactivity of 6 donors’ KIR2DL1+ and KIR2DL2/2DL3+ NK cell subsets toward K562 cells.
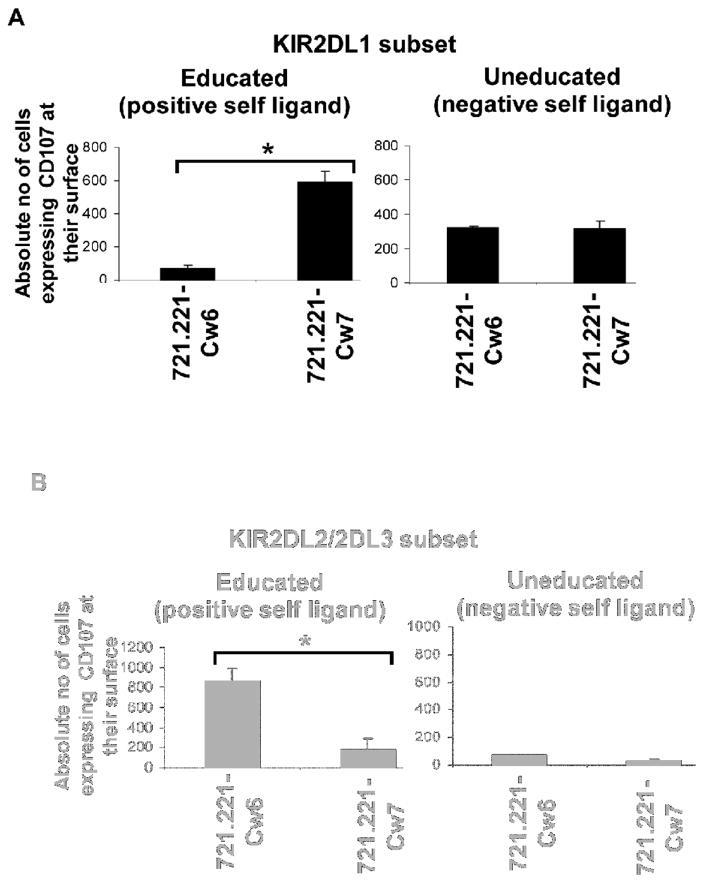
To further establish the accuracy of the prediction of NK cell licensing and recognition of missing-self cells based on our SNP results of the presence of a KIR ligand, we used 721.221 cells, which ectopically express a single KIR ligand. We stained the effector cells with CD45, gated on different single KIR+ NK cell subsets, and determined NK cell activation based on cell surface expression of CD107, as shown in Figure 4A. The predicted reactivity of educated versus uneducated KIR2DL1+ NK cell subsets based on the our SNP assay was highly accurate (Figure 5A, 5B). Educated KIR2DL1+ NK cell subsets were able to discern the presence or absence of ligands, but uneducated cells were not (Figure 5A). Similar results were also found in donor KIR2DL2/2DL3+ NK cell subsets (Figure 5B). Therefore, our SNP assay could be a useful method to type different functional groups of KIR ligands.
Figure 5. Reactivity of NK cell subsets can be predicted based on KIR ligand group typing.
KIR ligand typing of donor cells was done using a single-nucleotide polymorphism assay and followed by prediction of NK cell licensing. Single KIR+ NK cell subsets were gated using monoclonal antibodies against KIRs, and NK cell degranulation was measured using a CD107 marker. Reactivity of NK cell subsets toward target cells ectopically expressing their ligands was determined. Shown are the reactivity of the A) educated (left panel) and uneducated (right panel) KIR2DL1+ NK cell subsets toward 721.221 cells expressing the ligand Cw6 or non-ligand Cw7; * p value <0.05; and the B) educated (left) and uneducated (right) KIR2DL2/2DL3+ NK cell subsets toward 721.221 cells expressing the non-ligand Cw6 or ligand Cw7; * p value <0.05.
Discussion
Although numerous studies have established the importance of KIRs and their HLA ligands in health, disease, and transplant outcomes[10; 25; 26; 27], the significance of allele polymorphisms among these two sets of most highly polymorphic gene families has not yet been elucidated. The obstacles were two-fold: first, although it is certain that functional heterogeneity exists among different alleles, the molecular determinants are largely unknown; second, even when the molecular determinants are known, there is no expedient laboratory method for high-volume testing. Herein, we describe a novel approach for rapid and cost-effective typing of KIR alleles and KIR ligands once the molecular determinants are identified. Importantly, the readouts are informative biologically and will be useful in clinical diagnostics.
In the past, NK cell reactivity could be predicted in part based on the presence of KIRs and their ligands. However, allelic polymorphism generates another layer of functional heterogeneity. For the precise prediction of NK cell reactivity (e.g., in donor selection for NK cell transplantation), the knowledge of the presence of specific functional allelic groups of KIRs and their ligands are important. For example, using our functional KIR allele group typing based on the knowledge of molecular determinants, we demonstrated that we could successfully predict that KIR2DL1 R245/R245 NK cells were more reactive than KIR2DL1 C245/C245 cells toward missing-self cells. Thus, a clinical hypothesis might be that HLA-C2 positive bone marrow transplant donors with KIR2DL1 R245/R245 are better choices than those with KIR2DL1 C245/C245 for a patient lacking the HLA-C2 ligand. Another example is pregnancy. When pregnant mothers who lack most or all activating KIRs are carrying fetuses with the KIR2DL1 ligand HLA-C2 group, they are at a greater risk for preeclampsia [28]. This is true even if the mother herself also has HLA-C2. Since KIR2DL1 R245/R245 is more inhibitory than KIR2DL1 C245/C245, one may predict that if the mother has KIR2DL1 R245/R245, she may be at higher risk for preeclampsia when carrying an HLA-C2–positive baby. These mothers should then be monitored more closely during pregnancy.
Similar predictions are also possible for KIR2DL1-related diseases. For instance, individuals with KIR2DL1 R245/R245 alleles may have less risk for autoimmune diseases than those with the less inhibitory KIR2DL1 C245/C245. In contrast, individuals with KIR2DL1 C245/C245 alleles may be at lower risk for cancer or chronic infection. With the high-throughput KIR2DL1 functional group typing described here, we should now be able to study these hypotheses expeditiously.
Similar to typing KIR alleles, current HLA ligand typing requires high-resolution allele typing which is expensive and time-consuming. The SNP assay we developed in this study can type KIR ligands for 94 individuals within 4 hours, and the cost is about $4 per sample, whereas high-resolution HLA typing takes several weeks, and each sample costs about $200 [29]. In summary, we have developed a novel approach for KIR and HLA ligand typing that can be easily adopted in clinical diagnostic laboratories. Importantly, our readouts are biologically informative in the prediction of NK cell activity. This information will be valuable for future biological study of NK cells in health and disease and for clinical medicine in prognostication and transplant donor selection. Our functional molecular-determinant based approach will also be useful for the development of rapid SNP assays for many other highly polymorphic loci in the entire human genome.
Acknowledgments
This work was supported in part by research grants from NIH P30 CA-21765-24, CA-21765, the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities (ALSAC). We thank David Galloway for scientific editing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lodoen MB, Lanier LL. Natural killer cells as an initial defense against pathogens. Curr Opin Immunol. 2006;18:391–8. doi: 10.1016/j.coi.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001;1:41–9. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- 3.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–9. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008;27:5932–43. doi: 10.1038/onc.2008.267. [DOI] [PubMed] [Google Scholar]
- 5.Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology. 2007;121:1–14. doi: 10.1111/j.1365-2567.2007.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santoni A, Carlino C, Gismondi A. Uterine NK cell development, migration and function. Reprod Biomed Online. 2008;16:202–10. doi: 10.1016/s1472-6483(10)60575-5. [DOI] [PubMed] [Google Scholar]
- 7.Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol. 2002;2:656–63. doi: 10.1038/nri886. [DOI] [PubMed] [Google Scholar]
- 8.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long EO, Colonna M, Lanier LL. Inhibitory MHC class I receptors on NK and T cells: a standard nomenclature. Immunol Today. 1996;17:100. doi: 10.1016/0167-5699(96)80590-1. [DOI] [PubMed] [Google Scholar]
- 10.Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol. 2008;20:311–6. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson J, Marsh SG. IPD: the Immuno Polymorphism Database. Methods Mol Biol. 2007;409:61–74. doi: 10.1007/978-1-60327-118-9_4. [DOI] [PubMed] [Google Scholar]
- 12.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, Holladay M, Rooney B, Leung W. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114:5182–90. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–45. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steiner NK, Dakshanamurthy S, VandenBussche CJ, Hurley CK. Extracellular domain alterations impact surface expression of stimulatory natural killer cell receptor KIR2DS5. Immunogenetics. 2008;60:655–67. doi: 10.1007/s00251-008-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, Hurley CK, Christiansen FT, Witt CS. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- 16.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–9. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 17.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–52. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parham P. Taking license with natural killer cell maturation and repertoire development. Immunol Rev. 2006;214:155–60. doi: 10.1111/j.1600-065X.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 19.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–22. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colonna M, Brooks EG, Falco M, Ferrara GB, Strominger JL. Generation of allospecific natural killer cells by stimulation across a polymorphism of HLA-C. Science. 1993;260:1121–4. doi: 10.1126/science.8493555. [DOI] [PubMed] [Google Scholar]
- 21.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–44. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–13. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 23.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 24.De Santis D, Foley BA, John E, Senitzer D, Christiansen FT, Witt CS. Rapid, flow cytometric assay for NK alloreactivity reveals exceptions to rules governing alloreactivity. Biol Blood Marrow Transplant. 16:179–91. doi: 10.1016/j.bbmt.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 25.Leung W, Iyengar R, Turner V, Lang P, Bader P, Conn P, Niethammer D, Handgretinger R. Determinants of antileukemia effects of allogeneic NK cells. J Immunol. 2004;172:644–50. doi: 10.4049/jimmunol.172.1.644. [DOI] [PubMed] [Google Scholar]
- 26.Leung W, Iyengar R, Triplett B, Turner V, Behm FG, Holladay MS, Houston J, Handgretinger R. Comparison of killer Ig-like receptor genotyping and phenotyping for selection of allogeneic blood stem cell donors. J Immunol. 2005;174:6540–5. doi: 10.4049/jimmunol.174.10.6540. [DOI] [PubMed] [Google Scholar]
- 27.Triplett BM, Horwitz EM, Iyengar R, Turner V, Holladay MS, Gan K, Behm FG, Leung W. Effects of activating NK cell receptor expression and NK cell reconstitution on the outcomes of unrelated donor hematopoietic cell transplantation for hematologic malignancies. Leukemia. 2009;23:1278–87. doi: 10.1038/leu.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiby SE, Walker JJ, O’Shaughnessy MK, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957–65. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yun G, Tolar J, Yerich AK, Marsh SG, Robinson J, Noreen H, Blazar BR, Miller JS. A novel method for KIR-ligand typing by pyrosequencing to predict NK cell alloreactivity. Clin Immunol. 2007;123:272–80. doi: 10.1016/j.clim.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]