Abstract
There is increasing evidence that regulators of the spindle checkpoint, kinetochore microtubule attachments and sister chromatid cohesion are part of an interconnected mitotic regulatory circuit with two positive feedback loops and the Chromosome Passenger Complex (CPC) at its center. If true, this conceptual breakthrough needs to be integrated into models of mitosis. In this review, we describe this circuit and point out how the double feedback loops could provide insights into the self-organization of some mitotic processes and the autonomy of every chromosome on the mitotic spindle. We will also provide working models for how mitotic events may be coordinated by this circuit.
Are mitotic events coordinated?
During mitosis the cell is dramatically reorganized and a number of events occur simultaneously (Figure 1). Traditionally, the activation of the spindle checkpoint, sister chromatid cohesion and the generation of kinetochore-microtubule attachments were thought to have distinct regulation. This simplification has been necessary to dissect these very complex cellular processes. However, these mitotic processes share kinase and phosphatase regulators. A number of recent papers suggest that these regulators also control each other and a regulatory circuit can now be drawn that connects the major regulators of these three seemingly distinct mitotic events. Moreover, employing this circuit can provide answers to paradoxical situations that arise during mitosis such as how the Aurora kinase phosphorylates kinetochores (where the kinase is low), while at the same time Aurora B activity must be kept in check on inner centromere cohesin substrates (where the kinase is high) to protect cohesion. While it is possible that isolated circuits independently regulate these events, we will explore the possibility that these interconnected circuits coordinate mitotic events to provide robust regulation of mitosis.
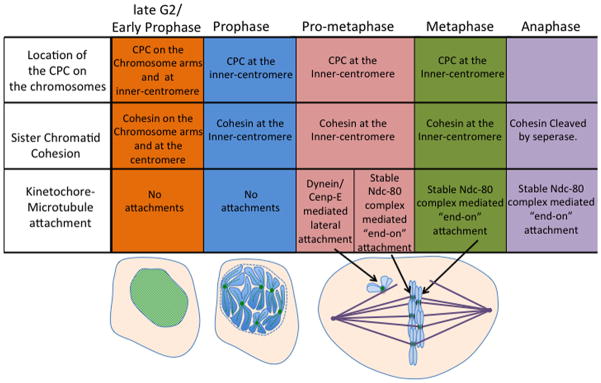
Figure 1. Table showing the temporal order of events during mitosis.
Spatial and temporal changes to the CPC location on the chromosome, Sister-chromatid cohesion and Kinetochore-microtubule attachment are described as the cell goes through different stages of mitosis. Below, morphology of the cells during each phase of mitosis with the CPC (green) localization on the chromosomes (blue) depicted at each stage of mitosis (mitotic spindle is represented in purple).
We propose to name this greater regulatory circuit the Centromere Signaling Network (CSN). The CSN is a kinase phosphatase signaling network that contains four kinases: Aurora B kinase, which is part of the CPC, MPS1 kinase, Bub1 kinase and Haspin kinase, as well as Sgo1, which binds Protein Phosphatase 2a (PP2A). Plk1 kinase is also involved [1,2,3,4], however we have limited our discussion of Plk1 because its kinetochore functions are poorly understood. One major reason to explore the concept that the CSN proteins coordinate mitotic events is that sets of CSN proteins regulate different events in mitosis (Figure 2). First, formation of proper kinetochore-microtubule attachments is regulated by Aurora B, Mps1, Sgo1 [5–15]. Second, the activation of the spindle checkpoint, which arrests the cell cycle until kinetochores make mature kinetochore attachments and is regulated by Aurora B, Mps1, Bub1 [15–17,5,18–22]. Third, cohesin is removed from chromosome arms while it is protected at the inner centromere region, which is regulated by Aurora B, Haspin and Sgo1 [23–25]. Fourth, the inner centromere is identified on each chromosome as a chromosome territory for CPC localization by the entire circuit [26–36].
Figure 2. Key Figure: The Centromere Signaling Network (CSN) contains sets of proteins that regulate multiple mitotic events.
Schematic representation of the CSN is shown with block arrows pointing at the process regulated by the proteins in the CSN.
Another reason to consider that the CSN may coordinate mitosis is that the four events occur with spatial and temporal regularity (Figure 1). For example, the spindle checkpoint is generated on chromosomes that are not aligned at the metaphase plate, while on the same spindle aligned kinetochores are not generating the signal. Since chromosomes are regulated differently, depending upon their location on the spindle, this is a form of spatial regulation. A second form of spatial regulation is the fact that cohesin is differentially regulated on chromosomes arms and centromeres. There is also temporal regulation. For example, kinetochores first generate “lateral” kinetochore-microtubule attachments, which then mature to “end-on” attachments (Figure 1). Finally, there is coordination between events as the kinetochore-microtubule attachment status is coupled to the generation of the spindle checkpoint signal.
The importance of linking the regulators of distinct events through a common circuit is that the CSN may act as an information processor that integrates information regarding the environment of each chromosome and produces outputs that ensure genomic stability. For example, it was recently shown that the microtubule plus end binding protein EB1 and microtubules regulate the CPC and this also controls Bub1 and Haspin activity to connect spindle status with kinetochore regulation (37). Because many of these events happen at distinct times on different chromosomes we also highlight how the CSN may underlie chromosome autonomy, wherein each chromosome regulates itself independently of adjacent chromosomes on the same spindle (Box 1).
Box 1. The kinetochore sub-network provides chromosome autonomy and regulation on the spindle.
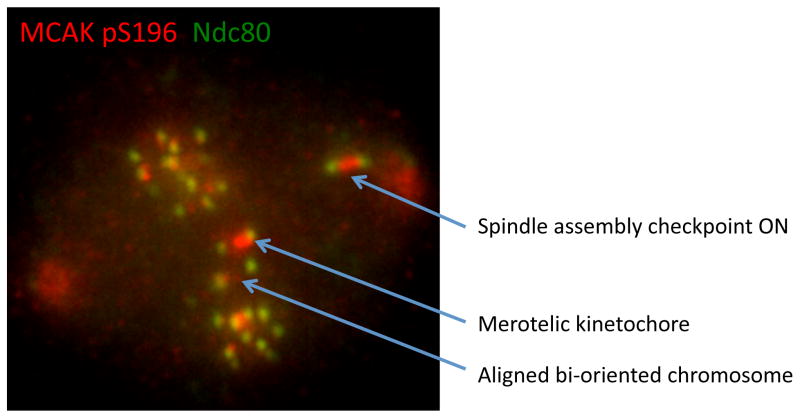
Unaligned chromosomes are able to recruit spindle checkpoint and outer kinetochore proteins, while aligned chromosomes on the same spindle have released these same proteins [16, 17]. This autonomy of chromosomes can at least in part be explained by the CSN. The chromosomal autonomy of centromere signaling can be visualized by following the phosphorylation of MCAK on S196 by Aurora B kinase (Figure I) [50]. MCAK phosphorylation is high on unaligned chromosomes but low on most aligned chromosomes. This autonomous signaling by the CPC is only partially due to the regulation of Aurora B kinase, which is only 2–4 fold higher on chromosomes that are not aligned to the metaphase plate in non-transformed cells (Figure I) [12, 37, 50]. The distinct outputs also require tight regulation of phosphatases and substrate localization to the inner centromere, thus they are products of an entire circuit rather than products of kinase regulation alone [23].
The differences in CPC activity on aligned and unaligned chromosomes are partially due to regulation of kinase localization by the kinetochore sub-network. Since kinetochores make mature attachments at different times means they each must down-regulate the CPC independently of other chromosomes on the spindle. In non-transformed cells, the level of MPS1, Bub1, H2ApT120 and the CPC is decreased on aligned chromosomes relative to chromosomes off the metaphase plate [12]. This suggests that this regulatory loop is dynamic and responsive to either position on the spindle or, more likely, the status of kinetochore-attachment, which allows movements of chromosomes to the metaphase plate. A recent finding suggests that these changes may be triggered by displacement of MPS1 from kinetochores after they make mature microtubule attachments [45, 46]. However, we note that the down-regulation of this loop on aligned chromosomes is often lost in transformed cells, and the misregulation of proteins in the CSN network have been recently shown to underlie Chromosomal instability in cancers [13].
There are many kinetochores in the same cytoplasm and it is not obvious how a signaling cascade could amplify a signal that is contained to one kinetochore. The requirement for histone phosphorylation may confine CSN signals to one chromosome by ensuring that the signals only amplify on chromatin and preventing signals from migrating from one chromosome to another. In addition, Aurora B and MPS1 share a common form of regulation. These kinases auto-phosphorylate their regulatory sites [107–109], which suggest that a major form of regulation is the concentration of these proteins at centromeres or kinetochores. Moreover, each upstream kinase in the pathway is required for localization of the downstream kinase [29–32,41–47], which serves to both concentrate and activate the downstream kinase. We suggest that this interesting form of kinase regulation is a key to chromosome autonomy; because local concentration of the downstream kinase is required to activate each subsequent step, the loop would only be achievable at a short distance.
The Centromere Signaling Network
Recent work suggests that CSN proteins can regulate each other and pathways can be drawn that are composed of two positive feedback loops that are interdependent because they share the CPC (Figure 3A). A central feature of these loops is that they recruit the CPC to inner centromeres through two histone phosphorylation events (Figure 3B, C). The haspin kinase phosphorylates histone H3 on Thr-3 (H3-pT3), which is directly bound by the survivin subunit of the CPC (Figure 3C) [26–28]. A second loop contains the Bub1 kinase, which phosphorylates Histone H2A on T120 [29–31] (Figure 3C). H2A-pT120 recruits Sgo1, which can bind the Borealin subunit of the CPC (Figure 3A) [30,31,33]. We will describe below the recent data from many groups that allow one to draw the circuit in this manner and the interesting regulatory properties that may be a function of the double positive feedback nature of the circuit.
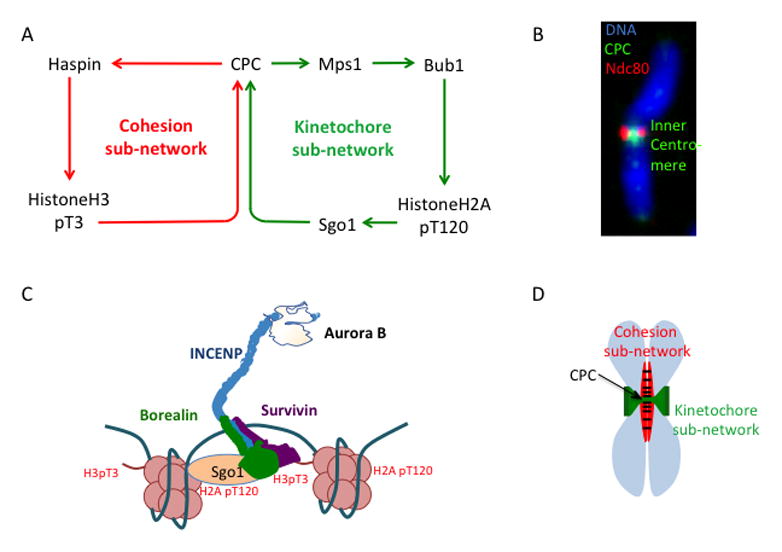
Figure 3. CPC localization pathways and epigenetic determination of inner-centromere.
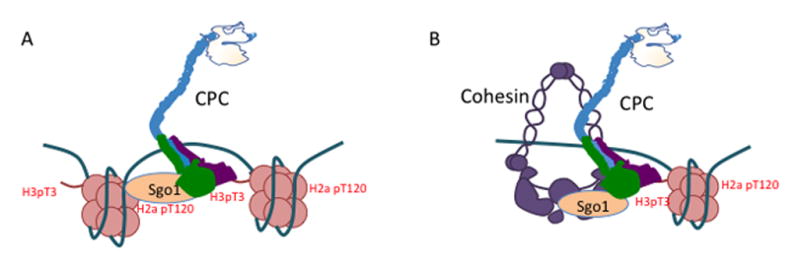
(A) Current model of the Chromosome Passenger Complex (CPC) localization to the inner-centromere by binding to two phospho-histone marks (in red). (B) Image showing the CPC (green) localized to the inner-centromere on a mitotic chromosome (original image from [119]). (C) Representation of the centromere signaling network (CSN). (D) Representation of the location of the cohesion and the kinetochore sub-network of the CSN on the mitotic chromosome.
Signaling network regulating the H3-pT3 histone mark (cohesion sub-network)
The Haspin kinase both recruits the CPC and it is thought to be recruited to chromosomes in a CPC dependent manner. Haspin kinase phosphorylates histone H3 on T3 during mitosis [26–28, 34]. The Survivin subunit of the CPC directly binds H3-pT3 to recruit the CPC to the inner-centromere (Figure 3A) [26–28,35]. The CPC in turn can stimulate haspin recruitment through phosphorylation, generating a positive feedback loop [36]. Haspin kinase is auto-inhibited by a domain known as the Haspin basic inhibitory segment (HBIS) and multisite phosphorylation of HBIS by Plk1 and CDK1 during mitosis neutralizes the HBIS to activate Haspin [2]. Aurora has been shown to activate Plk1 at kinetochores and thus may also indirectly activate Haspin [38].
The inner centromere localization of the CPC is highest in the center of the chromosomes between the kinetochores (Figure 3C) suggesting that there are mechanisms that allow the CPC to locate the central axis of mitotic chromosomes. The cohesin complex, which physically holds sister chromosomes together, is found in the central axis between the sister chromatids (39). Haspin is not abundant and difficult to localize on chromosomes so the localization is implied from H3pT3 patterns and biochemical interactions. Current models suggest that the spatial pattern of H3-T3 phosphorylation on the chromosome is controlled by the localization of Haspin by the Cohesin complex [27]. Haspin binds to the cohesin regulator Pds5 in yeast [27]. A similar mechanism is thought to work in mammals, because knockout of the Pds5 homologue Pds5B (but not Pds5A) results in low H3-T3 phosphorylation and low Aurora B at the inner-centromere [40]. This suggests that the H3pT3 loop directs the CPC to the central axis of the mitotic chromosome and we will refer to this part of the CSN as the cohesion sub-network (Figure 3C).
Signaling network regulating the pH2aT120 mark (kinetochore sub-network)
There must also be mechanisms to localize the CPC to the chromosome region between kinetochores (Figure 3C), which may be the function of the second positive feedback loop of the CSN. The kinase Bub1 recruits the CPC to the centromere by phosphorylating the histone H2A at T120 [29–31]. Sgo1 binds this phospho-histone mark and brings the CPC to the inner-centromere through its CDK1-dependent interaction with Bir1 (survivin) in Schizosaccharomyces pombe [32] (Figure 3A). A similar pathway exists in humans although the domain that binds Sgo1 has been transferred to the Borealin subunit [32] (Figure 3B). Bub1 is recruited to kinetochores by the MPS1 kinase, and MPS1 in turn is targeted to kinetochores by Aurora B phosphorylation, completing a second positive feedback loop [41–48]. Because both Bub1 and Mps1 are localized to kinetochores, this pathway will phosphorylate histones between the two kinetochores to direct the CPC to the inner centromere region. We will refer to this part of the CSN as the kinetochore sub-network (Figure 3A).
The CSN as drawn in Figure 3A shows a simple linear relationship between the proteins in the kinetochore sub-network. This model is based on recent experiments that combine cell biological observations and have strong biochemistry as support. However, we note that some older experiments suggest independence of Bub1 and Aurora B (15) and it is hard to reconcile all data in the literature. Thus, the network may be more of a web with some redundant pathways regulating proteins in the network.
It is unclear how proteins at kinetochores can affect Aurora B in the inner centromeres, which is hundreds of nanometers away. Bub1 was recently shown to activate RNA Polymerase II-dependent transcription at kinetochores and transcription is required to for the movement of Sgo1 from kinetochores to inner centromeres where it protects cohesion [33]. Sgo1 can also bind to cohesin [33]. Thus, the pool of Sgo1 that binds inner centromere Aurora B may be bound to cohesin and not H2ApT120 (Figure 4). Aurora B can bind RNA, which regulates its activity and localization (49) and an important area of future research is how transcription regulates the CPC and the entire CSN.
Figure 4. Two models of anchoring CPC to the inner-centromere.
A) Model 1: The CPC binds H3 pT3 and Cohesin bound Sgo1 at the same time. B) Model 2: The CPC binds H3 pT3 and H2a pT120 at the same time.
Functions of the CSN
It has been unclear how the CPC could regulate so many mitotic events. During pro-metaphase the CPC prevents or corrects improper kinetochore attachments [50,5–9], preserves centromeric cohesion [23–25], and generates the spindle assembly checkpoint [5,18–21]. We suggest that the CPC coordinates these events through its role within the CSN. To demonstrate this point we will describe how the network: 1) epigenetically defines the area between kinetochores to become the inner-centromere, 2) regulates centromeric cohesion protection and 3) allows proper coordination of kinetochore-microtubule attachments.
The inner centromere localization of the CPC is an emergent property of the CSN
Self-organizing systems underlie many biological processes by employing circuits with emergent properties to build resultant structures. Self-organizing systems are based on emergence, where a new property arises from the collective behavior of agents that themselves do not contain that property. Self-organization requires a positive feedback system to elicit dramatic changes to a system (51). We suggest that the CSN provides the emergent properties that drive the formation of the key aspects of inner centromere using self-organization principles including the localization of the CPC and the maintenance of cohesion (which we will discuss in another section).
In late G2 and early prophase the CPC is located throughout the nucleus. During prophase there is a dynamic reorganization of the CPC as it dissociates from the chromosome arms and then accumulates at the inner centromere (Figure 1) [52]. Histone H3 phosphorylation on T3 follows a similar pattern, providing a positive signal for these movements, but how H3T3 phosphorylation is spatially and temporally controlled is unclear. The CSN network may drive these dynamics to provide spatial information for CPC localization. To do this, the CSN must identify chromatin region between the kinetochores. However, if the signal was only derived from kinetochores then one would predict that the CPC would decrease as a function of distance from the kinetochores. This is not true; rather, it is highest in the center of chromosomes between the kinetochores, suggesting that there must be another mechanism to identify the central region.
The interaction between haspin and cohesin may identify the central axis of the chromosome. Like the CPC, cohesin is found throughout interphase chromatin (through its recruitment by CTCF) [53]. The bulk of cohesin is removed from chromatin during prophase by phosphorylation by mitotic kinases including CDK1, Plk1 and the CPC kinase Aurora B [54,55], and since the sisters remained cohesed along the central axis of chromosomes it is reasonable the cohesin remains high in this location (Figure 3D) [56]. The maintenance of cohesion in the central axis engages the cohesion loop of the CSN to spatially locate H3pT3 and the CPC to the central zone between the two sisters [26–28,35,40].
Mitotic chromosomes are bisected by a second axis of histone phosphorylation that is established by the second positive feedback loop (Figure 3D) [27, 29–32, 41–48]. Bub1 phosphorylates chromatin near kinetochores [27, 33]. This targets Sgo1 to chromatin between kinetochores, where it binds the CPC. Thus the CPC, which binds both H3pT3 and Sgo1, is localized by two orthogonal axes that are established on mitotic chromosomes: one axis between the sister chromosomes and one axis between the kinetochores (Figure 3C, D) [27]. The positive feedback nature of the two independent loops may reinforce the inner centromere location after initial recruitment of the CPC and the inner centromere chromosome region emerges as a function of the centromere-signaling network.
There is abundant evidence that the CSN may drive this rapid change of mitotic chromosomes and we will highlight the key findings that show that the inner centromere localization of the CPC is an emergent property of the two-loop circuit. In a pioneering paper the Dasso group showed that the CPC was distributed throughout chromatin after depletion of Bub1, even though kinetochores could form (29). Similarly, Aurora B remains localized to chromosome arms in cells depleted of haspin (27). In early prophase the CPC can localize along the inner chromatid axis as if this is an intermediate of the CPC moving to inner centromere (27). Thus the localization pattern of the CPC when one disrupts either loop suggests a dynamic process involving two feedback loops.
The network as written is dominated by kinases, although the Sgo1 protein can bind PP2A. Positive feedback systems must be limited to prevent them from dominating a system and there are likely additional phosphatase networks that need to be included to build robust models for mitotic regulation. For example, the protein Repo-Man recruits PP1 to dephosphorylate Histone H3 on T3 and limit Haspin activity. This may allow CPC to be released from chromosome arms so that it can be concentrated at the inner centromere during early mitosis [57]. Repo-Man-PP1 must be displaced from inner centromeres to allow the accumulation of the CPC during early mitosis. This may be achieved by Aurora B itself, which inhibits chromosome binding of PP1-Repo-Man by direct phosphorylation [58] and we suggest that there is only enough Aurora B to counter Repo-Man activity within the regions between kinetochores. However, there may be additional mechanisms to reverse Haspin phosphorylations on chromosome arms in prometaphase and metaphase when CDK inhibits PP1-Repo-Man interaction and Repo-Man binding to the chromatin (59–61). This may be through local reactivation of Repo-Man, which can bind PP2A to reactivate it [58].
On chromosomes that have neocentromeres the CPC is found at the neocentromere but not the region of alpha satellite repeats that define the inactivated centromere arguing that it is localized by epigenetic mechanisms [62]. Centromeres are epigenetically identified by recruiting a histone H3 variant CENP-A, yet how this translates to epigenetic identification of the inner centromere is less well understood. Note that the CSN dependent mechanisms that we just discussed link the epigenetic mechanisms of CENP-A specification that localize kinetochores to specification of the inner centromere.
Regulation of sister chromatid cohesion by the CSN
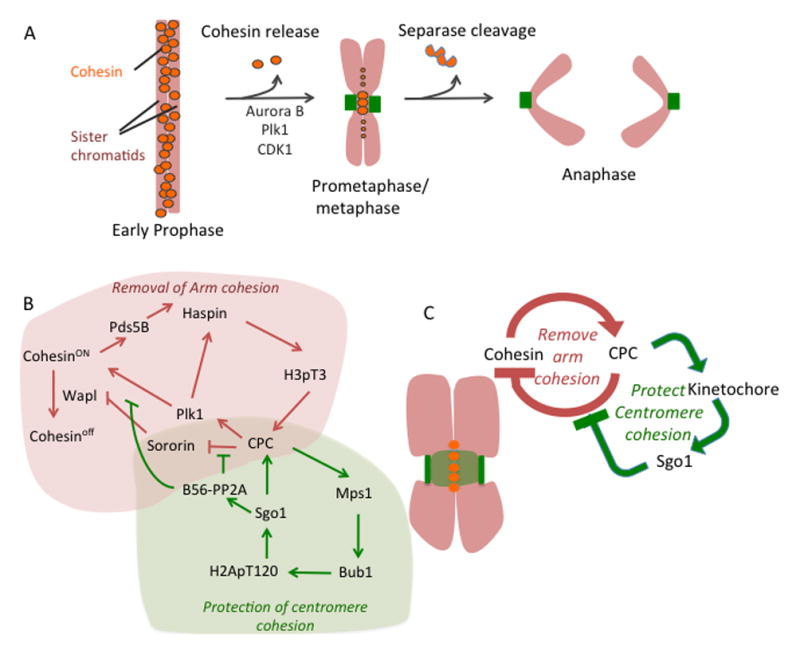
Sister chromatids must be held together until anaphase in order to faithfully segregate the genetic material to the daughter cells. This is accomplished by the ring-shaped cohesin complex, which physically pairs the sister chromatids [63,64]. Cohesion is established coincident with DNA replication, to enable faithful pairing of the two sister DNA strands [63,64]. Cohesin is removed from the chromatin in two steps during mitosis in higher eukaryotes [56]. The prophase pathway removes most of the cohesin from chromosome arms, but centromeric cohesin is protected from this pathway (Figure 5A) [56]. This generates a new state where mitotic chromosomes are paired by centromeric cohesin that is maintained until every chromosome obtains bipolar microtubule attachment (Figure 5A). At that point, centromeric cohesion is released by activation of the protease Separase, which cleaves chromatin-bound centromeric cohesin to drive the metaphase-to-anaphase transition (Figure 5A) [65–68].
Figure 5. Regulation of sister chromatid cohesion by the CSN.
(A) Two step cohesin removal from the chromosomes during mitosis. (B) Signaling network regulating sister chromatic cohesion during mitosis. Cohesion removing part of the network is represented by red arrows and green arrows represent cohesion-protecting network. (C) Simplified diagram of the cohesion regulation during the mitosis.
The release of cohesin from chromosome arms in prophase is controlled by three factors: Pds5 (Pds5 A/B in mammals), which directly binds cohesin; Wapl, which can release chromatin-bound cohesin [69,70], presumably by opening the ring; and Sororin, which competes for the Wapl binding site on Pds5 and therefore protects chromatin-bound cohesin [71–73]. The spatial segregation of cohesin on mitotic chromosomes is thus determined by the recruitment of Wapl: Wapl is recruited to cohesin on the chromosome arms, whereas cohesin at the centromere is protected from Wapl binding [69,70,72–74]. Proteins in the CSN control these events and we suggest that these dynamics can be explained by one branch of the CSN acting at chromosomes arms in prophase, while both positive feedback loops are engaged at centromeres. Aurora B and Cdk1 phosphorylate Sororin on multiple sites and reduce its interaction with Pds5, allowing Wapl to bind and remove cohesin from the chromosome arms (Figure 5A, B) [73] (Box 2). In addition, Plk1 can directly phosphorylate and release cohesin (Figure 5A, B) [75] (Box 2). However, we are left with a paradox: how is it that the CPC removes cohesin from chromosome arms, yet cohesin is protected in the centromere where the CPC is highest? We suggest the answer lies in the feedback loops, which can be redrawn as a spatially segregated negative feedback system (Figure 5B, C). That Pds5 recruits haspin suggests the cohesin complex indirectly recruits the CPC, which drives the dissociation of cohesin [27, 40]. This simple feedback system allows the CPC, via the cohesion sub-network of the CSN, to quickly bind and remove cohesin from the arms. However, the CPC recruits the Bub1 kinase to the region between kinetochores [5,20,47, 48], driving the association of Sgo1 preserves centromeric cohesion by recruiting the phosphatase PP2A, which removes the Aurora B and Plk1 phosphorylations to preserve cohesion [76–79]. Thus, on chromosome arms there is a simple feedback loop where cohesin recruits the CPC to release cohesin. However, between kinetochores the negative feedback system is negated because the CPC can also recruit the inhibitor of cohesion release. This is a spatially segregated negative feedback circuit that drives the self-organization of the mitotic chromosome by removing cohesion from all chromatin unless it is between kinetochores. Apart from recruiting the CPC to cohesin, Haspin may also play a positive role in protecting cohesin [24].
Box 2. Regulation by Multisite phosphorylation.
Mitotically regulated proteins often have domains that are phosphorylated at many independent sites and often by different kinases. For example, the cohesion regulator Sororin has 11 phosphorylation sites that regulate its association with Pds5, which protects cohesion. Phosphorylation of Sororin by CDK1 and Aurora B inhibits its binding to Pds5, driving cohesin release during prophase [73]. In addition, cohesin is released by the phosphorylation by Plk1 of 12 sites on the SA2 subunit of cohesin [75]. We do not know the exact number of sites that need to be phosphorylated for cohesin to release chromatin, but it may require up to 22 distinct phosphorylation events by three different mitotic kinases.
Why would such a complex mechanism evolve? We suggest that regulating a critical event by requiring the coincident phosphorylation of multiple sites by different kinases provides robustness to biological processes by four mechanisms that are not mutually exclusive.
Phosphorylation by multiple kinases can ensure that an event only happens at a specific cell cycle time. For example, the prophase cohesion release pathway only happens during mitosis. If it were only regulated by CDK1 then the cell risks the possibility that cohesin could be released by the activation of another kinase with a similar phosphorylation consensus site. In fact, CDK2 has a similar consensus site as CDK1 but is activate in S-phase. However, cohesin remains protected in S-phase as long as kinases that phosphorylate the same sites as both Plk1 and Aurora B are not activated.
It is well documented that distributive phosphorylation on multiple sites can generate switch-like kinetics for cell cycle events. An outstanding example is the phoshphorylation of Clb-CDK inhibitor, Sic1, during the G1-to-S transition in budding yeast. Sic1 degradation activates the Clb-CDK kinases, allowing the cell to enter S-phase. Sic1 must be phosphorylated over six times by the G1-specific Cln kinases before it can be recognized by the F-box protein of SCF and marked for degradation[110, 111]. The distributive phosphorylation of multiple sites on Sic1 allows all of the Sic1 molecules to be degraded at approximately the same time, which drives a concerted entry into S-phase [112–114]. We suggest that, similarly, multiple phosphorylation events synchronize cohesin release in prophase. Only after CDK1, Aurora kinases and Plk1 are all robustly activated in prophase can sufficient phosphorylation begin to accumulate on Sororin and SA2, which would generate a lag phase after kinase activation followed by a relatively synchronous release of cohesin from chromosome arms.
-
Multi-site phosphorylation allows tight regulation by phosphatases. This type of multiple phosphorylation system might be crucial under conditions where the kinases are highly active, such as in the inner centromere in mitosis where Aurora B activity is probably at its highest but cohesion must be preserved. The phosphorylation of multiple sites establishes a lag between kinase activation and initiating an event, which provides time for a phosphatase to remove the phosphorylations. Thus, the recruitment of PP2A to centromeric cohesin by Sgo1 can protects cohesion in the centromere where both Aurora B and Plk1 are also targeted and active [77, 78]. We suggest that the requirement for these kinases to phosphorylate many sites is essential for PP2A/Sgo1 to protect cohesin in an environment where there are strong kinase activities.
During prometaphase when cohesion is protected by keeping Sororin dephosphorylated in inner centromeres, multisite phosphorylation generates robust spindle checkpoint activation at kinetochores. There are 19 MELT sequences on the human Knl1 protein that can be phosphorylated by MPS1 and Plk1 kinases to generate the SAC signal and control kinetochore-microtubule attachments [115]. It appears that the SAC signal requires phosphorylating more than a single site. The multisite requirement to generate the SAC probably prevents accidental activation of the SAC by transient increases in MPS1 activity in metaphase or silencing by transient increases of PP1 on unattached kinetochores.
In the case of Hec1 there is also evidence that the multisite phosphorylation also provides rheostat-like control (rather than a threshold) where the number of sites that are phosphorylated is proportional to the weakening of the interaction between Ndc80 and microtubules [116–118].
This insight provides important lessons about the system. First, the feedback loops contain regulators of the various processes (i.e., Sgo1) because they perform their functions as part of this whole centromere network. In other words, Sgo1 performs two functions in the circuit: to recruit more CPC through its interaction with Borealin, and to preserve cohesion [32, 33, 77]. The preservation of cohesin would also recruit more CPC through the CSN. Second, the system ensures robustness. When centromeric CPC activity increases the system will recruit more Sgo1/PP2A to ensure that the cohesin-releasing activity will never overwhelm the preserving activity. Third, the system approaches a steady state that can only be reversed by an external signal. In this case, centromeric cohesion is robustly maintained until the APC/C activates the Separase protease, which cleaves cohesin to drive the segregation of chromosomes [65–68]. Although this system is robust, it is not a true steady state because cohesin cannot be reattached and chromosomes will eventually lose their cohesion and exit from mitosis, which may contribute to cohesion fatigue [80].
CSN regulation of kinetochore-microtubule interaction
Human kinetochores bind approximately 17 microtubules and aneuploidy can develop if a kinetochore is pulled by microtubules attached to opposite poles (merotely) in anaphase [81, 82]. Thus, mitosis depends upon the sister kinetochores generating bipolar kinetochore microtubule attachments, meaning that one sister kinetochore binds microtubules from one pole, while the other sister kinetochore binds microtubules emanating from the opposite pole. It is likely that cells prevent merotely by orienting the two sister kinetochores toward the two opposites poles before they make stable attachments [82–84]. It is believed that the motors dynein and Centromere protein E (CENP-E) rotate chromosomes to achieve this orientation [85–87]. Based on these observations it is suggested that one of the central mechanisms to prevent merotely is to ensure these motors initially bind microtubules before kinetochores generate “end-on” Ndc80 complex-mediated attachments that segregate chromosomes in anaphase [83, 84].
We suggest that incorporating the CSN into the regulation of kinetochore microtubule attachments enable models with the attributes of a bistable switch (Figure 6A). First, the CSN may initially generate a stable system where lateral attachments dominate and end-on attachments are inhibited. Second, after end-on attachments form there is a new stable state where end-on attachments dominate. Third, the first state has mechanisms that ensure the transition to the second state. We will outline each of these states below and note that mathematical modeling is required to test if this is truly a bistable system.
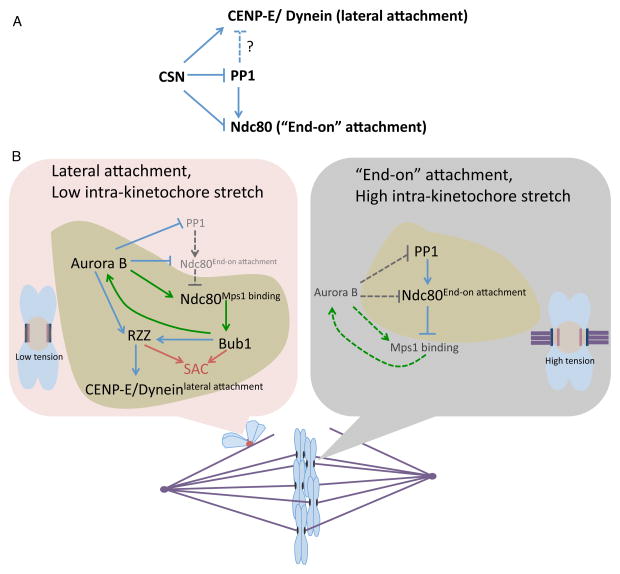
Figure 6. Regulation of kinetochore-microtubule interaction by the CSN.
(A) Network coordinating lateral and “end on” kinetochore-microtubule attachment. (B) Representation of the signaling network that is active on the laterally attached kinetochore (red) or on “end on” attached kinetochore (black). The signaling network represented by green arrows is part of the CSN; the dominant signaling network is represented in the yellow patch. Dotted lines represent inactive or weakened networks. Red lines indicate activation of spindle assembly checkpoint (SAC).
The CSN promotes lateral attachment by recruiting two microtubule motors, CENP-E and Dynein [88, 89]. The CSN plays a crucial role in the formation of motor-dependent attachments by both recruiting and regulating dynein and CENP-E [88, 89]. For example, current models suggest Mps1 and Bub1 recruit, and Aurora B regulates, CENP-E to properly align chromosomes [22, 90]. Thus, the entire CSN network may be needed to properly control CENP-E (Figure 6A).
At the same time CSN promotes lateral attachment it also inhibits “end-on” attachments (Figure 6B). Aurora B directly prevents pre-mature “end-on” attachment by phosphorylating the Ndc80 complex and inhibiting its interaction with microtubules [9, 22, 91–93]. Proteins involved in Dynein recruitment to the kinetochore might also inhibit “end-on” attachment [94–96] and this might also be ultimately controlled by Aurora B [97] (figure 6B). Since microtubules displace Mps1 bound to Ndc80 it is reasonable to assume that unattached Ndc80 complex would recruit more Mps1 to the kinetochore [44–46]. This increase in Mps1 would lead to an increase in the CPC levels at centromeres, due to the CSN, which may further inhibit “end-on” attachment [5, 20, 46, 47]. This positive feedback loop between Mps1 and the CPC may lead to robust inhibition of “end-on” attachment (Figure 6B).
This robust inhibition of end-on attachment through positive feedback by three kinases in the kinetochore sub-network of the CSN has to be controlled to allow the kinetochores to initiate “end-on” attachment (Figure 6B). This is accomplished through recruitment of phosphatases by the CSN to decrease and counteract kinase activity. The CSN recruits phosphatase PP2A at two locations: to the inner-centromere, through BUB1 dependent recruitment of Sgo1-PP2A [27, 29–32]; and to the kinetochore, through BUB1, Plk1 and Aurora B dependent recruitment of BUBR1-PP2A [98–100]. These two pools of phosphatases allow the formation of initial “end-on” attachments by countering CPC-dependent destabilization of the “end-on” attachment. Sgo1-PP2A reduces the activity of the CPC by dephosphorylating the T-loop of Aurora B thus reducing the overall activity of the CPC [10]. Plk1 mediated BUBR1-PP2A dephosphorylates the kinetochore substrates of the CPC and allows PP1 recruitment to Knl1, which also dephosphorylates the kinetochore substrates and promotes “end-on” attachments [98–101].
Once proper “end-on” attachments begin to form there are at least three events that down-regulate the CSN to stabilize the initial “end-on” attachment (Figure 6B). First, the microtubule binding to the Ndc80 complex competes off Mps1 and reduces its levels at kinetochore, which subsequently causes reduction in the CPC levels at the centromere to stabilize “end-on” attachments [5, 20, 44–48]. Second, microtubule dependent pulling forces generated by bipolar attachment physically pulls the outer kinetochore away from the inner-centromere localized CPC [11, 102, 103]. This physical separation of the outer kinetochore from Aurora B would reduce the phosphorylation of the Ndc80 complex [11, 45, 46, 102, 103]. Third, recruitment of PP1 to the “end-on” attached kinetochores leads to further stabilization of the attachments [104, 105]. Multiple pools of PP1 are recruited to the kinetochore that stabilizes “end-on” attachments and localization of most of these pools of PP1 are inhibited by Aurora B activity [104, 105].
Coordination of Kinetochore-microtubule attachment formation and the Spindle assembly checkpoint (SAC)
The formation of kinetochore microtubule attachments and the spindle assembly checkpoint must be coordinated to ensure faithful genome segregation. The CSN may allow this coordination. Mps1, BUB1 and the CPC, which inhibit end-on attachment and promote lateral attachments, also activate and maintain the spindle assembly checkpoint [16, 17] (Figure 6B). This could ensure that the checkpoint is activated at unattached kinetochores. Similarly, the proteins or events involved in promoting end-on attachment (BubR1-PP2A, Knl1-PP1 and Ndc80-microtubule interaction) are also involved in silencing the spindle assembly checkpoint [16, 17, 101]. In fact, the most important step for spindle assembly checkpoint silencing is Ndc80-mediated end-on attachment itself. Microtubule binding to Ndc80 displaces Mps1 from the kinetochores, which leads to silencing of the spindle assembly checkpoint [45, 46]. The displacement of MPS1 would also down regulate the CSN to lower Bub1 and Aurora B coupling end-on attachment to dramatic changes to the kinetochore.
Since MPS1 and Bub1 are displaced from metaphase kinetochores, one could imagine that it is difficult to restart the SAC once chromosomes are aligned. Yet, the addition of taxol to metaphase cells quickly reinitiates spindle checkpoint signaling [106]. It is tempting to speculate that the reversibility of this system is ensured because the cohesion sub-network maintains some CPC at inner-centromeres even when kinetochore sub-network is down regulated, which would enable rapid binding of MPS1 to unattached molecules of Ndc80 (44–46).
The generation of the SAC signal involves tens to hundreds of phosphorylations on numerous substrates at each kinetochore by Aurora B, Plk1, Mps1 and Bub1 thus it is an emergent property of the network. The generation of so many phosphorylations can ensure that, once generated, the signal is robustly maintained until there is both active recruitment of phosphatases and down regulation of the kinases that accompanies the transition to mature kinetochore microtubule attachments (Box 2).
Concluding Remarks
It has recently become clear that proteins involved in regulating the spindle checkpoint, cohesion and microtubule attachments work together and may exist in a circuit. We have attempted to demonstrate how grouping the regulators into a circuit with two positive feedback loops may explain how some of the dynamic processes of mitosis may be coordinated.
Figure I. Chromosome autonomy during mitosis.
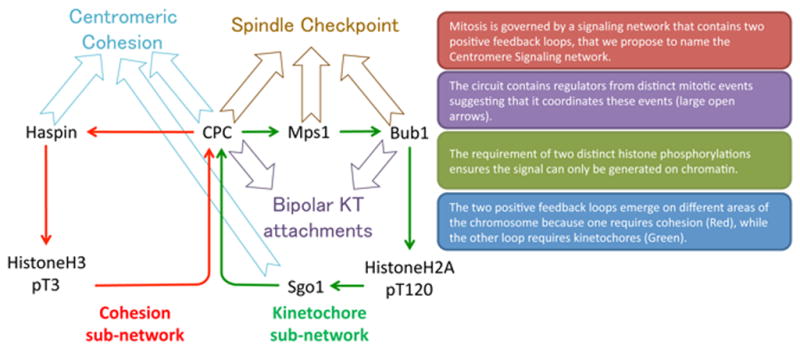
Image of a late pro-metaphase Xenopus S3 cell probed for Aurora B substrate MCAK phospho-S196 (pS196) and Ndc80. Kinetochores have different levels of Aurora B activity, as inferred from MCAK pS196, depending on their location along the spindle and attachment status. Original image from [50].
OUTSTANDING QUESTIONS BOX.
How does the inner-centromeric chromosome passenger complex (CPC) regulate proteins at the outer kinetochore? An important part of the centromere signaling network (CSN) depends on the regulation of outer-kinetochore proteins (such as Ndc80 and Mps1) by the inner-centromeric CPC. The distance between the inner-centromere and outer kinetochore is 500–600nm; it is unclear how the signaling takes place across these large length scales.
What are the missing details of the CSN network? For example, the most mysterious part of the circuit remains how Bub1 recruits the CPC. Does this work only through H2A-T120 phosphorylation? In addition, chromosome arm cohesion can be lost without Haspin, so it is unclear if it participates in the feedback loop to allow rapid removal of cohesin from arms.
What are the network properties of the CSN? Mathematical modeling studies are required to understand the emergent properties of the CSN and how they contribute to faithful mitosis.
CSN is the central circuit and it can be controlled by inputs from the spindle. Multiple inputs are known that feed into the CSN, such as microtubules, RNA, kinases, phosphatases and guanine nucleotide exchange factors; these inputs can change the properties of the network and influence its outcome. Are there more inputs to the CSN and how does each input regulate the CSN to control mitotic processes?
Are mitotic events coordinated by the CSN? The CSN is composed of regulators common to distinct mitotic events, thus it makes sense that the CSN allows coordination these events. However, this central hypothesis needs to be experimentally tested.
How does deregulation of the CSN affect genomic stability and how does it contribute to the progression of chromosomal instability (CIN) in cancers? CIN cancers have poor prognosis; transformed cells have been shown to deregulate CSN, which may underlie the CIN phenotype.
Trends Box.
Mitotic processes were recently connected in a signaling circuit composed of two positive feedback loops, which we name the Centromere Signaling Network (CSN).
The CSN was first identified for its ability to localize the Chromosome Passenger Complex (CPC) to inner centromeres. The circuit contains two histone phosphorylation events that generate two axes on mitotic chromosomes. The intersection of these axes defines the location of the CPC and preserves centromeric cohesion.
Recent observations suggest that the network configuration of the circuit may coordinate the spindle assembly checkpoint, kinetochore-microtubule attachment, and inner-centromeric cohesion protection.
The circuitry of the CSN self-contains signals from mitotic regulators within chromosome, and thus may underlie the autonomous behavior of chromosomes during mitosis.
Glossary
- Neocentromere
Ectopic centromeres that occasionally arise at the non-centromeric location of a chromosome. Because they functionally replace normal centromeres at chromosome regions lacking the alpha-satellite DNA sequences that underlie most centromeres, they demonstrate that centromeres are specified by epigenetic rather than genetic mechanisms
- Kinetochore
A specialized protein structure that is assembled on the centromeres during mitosis that generates the spindle checkpoint signal and attaches spindle microtubules to chromatids
- Chromosome Passenger Complex
Four-protein complex composed of kinase Aurora-B and three regulatory subunits INCENP, Borealin and Survivin that regulates numerous mitotic events
- Inner-centromere
Is a chromatin region between two sister centromeres. This is also the site on the mitotic chromosome where the CPC is localized and cohesin is protected until anaphase
- Centromere
Region of the chromosome where kinetochore is assembled in mitosis. This region is specified epigenetically in most of the organisms by the presence of a histone H3 variant CENP-A
- Spindle Assembly Checkpoint
A cell cycle feedback-system initiated at the improperly attached kinetochore, which inhibits the onset of anaphase until all the kinetochores are properly attached with the mitotic spindle
- “Lateral” kinetochore-microtubule attachment
A type of kinetochore-microtubule attachment that precedes end-on kinetochore-microtubule attachment, and is formed through interaction of the, kinetochore localized, motor proteins with lateral walls of spindle microtubules
- “End-on” kinetochore-microtubule attachment
A type of kinetochore-microtubule attachment, which is mediated through the interaction of the kinetochore localized Ndc80 complex with the plus end of spindle microtubules
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhou L, et al. Polo-like kinase-1 triggers histone phosphorylation by Haspin in mitosis. EMBO Rep. 2014;3:273–81. doi: 10.1002/embr.201338080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghenoiu C, et al. Autoinhibition and Polo-Dependent Multisite Phosphorylation Restrict Activity of the Histone H3 Kinase Haspin to Mitosis. Molecular Cell. 2013;52:734–745. doi: 10.1016/j.molcel.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Schubert C, et al. Plk1 and Mps1 Cooperatively Regulate the Spindle Assembly Checkpoint in Human Cells. Cell Reports. 2015;12:66–78. doi: 10.1016/j.celrep.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Espeut J, et al. Natural Loss of Mps1 Kinase in Nematodes Uncovers a Role for Polo-like Kinase 1 in Spindle Checkpoint Initiation. Cell Rep. 2015;12(1):58–65. doi: 10.1016/j.celrep.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161(2):281–94. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimini D, et al. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16(17):1711–8. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 7.Knowlton AL, et al. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 8.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLuca KF, et al. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124(4):622–34. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meppelink A, et al. Shugoshin-1 balances Aurora B kinase activity via PP2A to promote chromosome bi-orientation. Cell Rep. 2015;11(4):508–15. doi: 10.1016/j.celrep.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D, et al. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salimian KJ, et al. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol. 2011;21(13):1158–65. doi: 10.1016/j.cub.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanno, et al. The inner centromere-shugoshin network prevents chromosomal instability. Science. 2015;349(6253):1237–40. doi: 10.1126/science.aaa2655. [DOI] [PubMed] [Google Scholar]
- 14.Jelluma N, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132(2):233–46. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 15.Meraldi P, Sorger PK. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24(8):1621–33. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacristan C, Kops GJ. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol. 2015;25(1):21–8. doi: 10.1016/j.tcb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Stukenberg PT, Burke DJ. Connecting the microtubule attachment status of each kinetochore to cell cycle arrest through the spindle assembly checkpoint. Chromosoma. 2015 doi: 10.1007/s00412-015-0515-z. [DOI] [PubMed] [Google Scholar]
- 18.Kallio MJ, et al. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol. 2002;12(11):900–5. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- 19.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15(23):3118–29. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santaguida S, et al. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 2011;30(8):1508–19. doi: 10.1038/emboj.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matson DR, et al. A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes Dev. 2012;26(6):542–7. doi: 10.1101/gad.184184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrieu A, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106(1):83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 23.Tanno Y, et al. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 2010;24(19):2169–79. doi: 10.1101/gad.1945310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai J, et al. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell. 2006;11(5):741–50. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Resnick, et al. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev Cell. 2006;11(1):57–68. doi: 10.1016/j.devcel.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;303(6001):231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamagishi Y, et al. Two Histone Marks Establish the Inner Centromere and Chromosome Bi-Orientation. Science. 2010;330(6001):239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- 28.Kelly AE, et al. Survivin reads phosphorylated Histone H3 Threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330(6001):235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyarchuk Y, et al. Bub1 is essential for assembly of the functional inner centromere. J Cell Biol. 2007;176(7):919–928. doi: 10.1083/jcb.200609044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawashima SA, et al. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing Shugoshin. Science. 2010;327(5962):172–7. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 31.Ricke RM, et al. Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J Cell Biol. 2012;199(6):931–949. doi: 10.1083/jcb.201205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukahara T, et al. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 2010;467(7316):719–23. doi: 10.1038/nature09390. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, et al. Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Mol Cell. 2015;59(3):426–36. doi: 10.1016/j.molcel.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 34.Dai, et al. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19(4):472–88. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niedzialkowska E, et al. Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres. Mol Biol Cell. 2012;23:1457–1466. doi: 10.1091/mbc.E11-11-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, et al. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr Biol. 2011;21(12):1061–1069. doi: 10.1016/j.cub.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banerjee B, et al. EB1 enables spindle microtubules to regulate centromeric recruitment of Aurora B. J Cell Biol. 2014;204(6):947–63. doi: 10.1083/jcb.201307119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carmena M, et al. The Chromosomal Passenger Complex Activates Polo Kinase at Centromeres. PLoS Biol. 2012;10(1):e1001250. doi: 10.1371/journal.pbio.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giménez-Abián, et al. Regulation of sister chromatid cohesion between chromosome arms. Curr Biol. 2004;14(13):1187–93. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 40.Carretero M, et al. Pds5B is required for cohesion establishment and Aurora B accumulation at centromeres. EMBO J. 2013;32(22):2938–49. doi: 10.1038/emboj.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shepperd LA, et al. Phosphodependent Recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 Kinase Maintains the Spindle Checkpoint. Curr Biol. 2012;22(10):891–9. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.London N, et al. Phosphoregulation of Spc105 by Mps1 and PP1 Regulates Bub1 Localization to Kinetochores. Curr Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamagishi Y, et al. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol. 2012;14(7):746–52. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- 44.Nijenhuis W, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol. 2013;201(2):217–31. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiruma Y, et al. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 2015;348(6240):1264–7. doi: 10.1126/science.aaa4055. [DOI] [PubMed] [Google Scholar]
- 46.Ji Z, et al. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science. 2015;348(6240):1260–4. doi: 10.1126/science.aaa4029. [DOI] [PubMed] [Google Scholar]
- 47.Saurin AT, et al. Aurora B potentiates mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun. 2011;2:316. doi: 10.1038/ncomms1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van der Waal MS, et al. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 2012;13(9):847–54. doi: 10.1038/embor.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jambhekar, et al. RNA stimulates Aurora B kinase activity during mitosis. PLoS One. 2014;9(6):e100748. doi: 10.1371/journal.pone.0100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lan W, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14(4):273–86. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 51.Solé Ricard V, Bascompte Jordi. Self-organization in complex ecosystems. Princeton University Press; 2006. [Google Scholar]
- 52.Carmena M, et al. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 54.Nishiyama T, et al. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc Natl Acad Sci U S A. 2013;110(33):13404–9. doi: 10.1073/pnas.1305020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sumara I, et al. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9(3):515–25. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- 56.Waizenegger IC, et al. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103(3):399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 57.Qian J, et al. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr Biol. 2011;21(9):766–73. doi: 10.1016/j.cub.2011.03.047. [DOI] [PubMed] [Google Scholar]
- 58.Qian J, et al. Aurora B defines its own chromosomal targeting by opposing the recruitment of the phosphatase scaffold Repo-Man. Curr Biol. 2013;23(12):1136–43. doi: 10.1016/j.cub.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 59.Trinkle-Mulcahy, et al. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J Cell Biol. 2006;172(5):679–92. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vagnarelli, et al. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol. 2006;8(10):1133–42. doi: 10.1038/ncb1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vagnarelli, et al. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev Cell. 2011;21(2):328–42. doi: 10.1016/j.devcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassett EA, et al. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol. 2010;190(2):177–85. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 64.Peters JM, et al. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 65.Uhlmann F, et al. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 66.Buonomo SB, et al. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- 67.Uhlmann F, et al. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 68.Hauf S, et al. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 69.Gandhi R, et al. Human Wapl is a cohesion binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kueng S, et al. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 71.Dreier MR, et al. Regulation of sororin by Cdk1-mediated phosphorylation. J Cell Sci. 2011;124(17):2976–87. doi: 10.1242/jcs.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishiyama T, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143(5):737–49. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 73.Nishiyama T, et al. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc Natl Acad Sci U S A. 2013;110(33):13404–9. doi: 10.1073/pnas.1305020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hara K, et al. Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion. Nat Struct Mol Biol. 2014;21(10):864–70. doi: 10.1038/nsmb.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hauf S, et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3(3):e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang Z, et al. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10(5):575–85. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 77.McGuinness BE, et al. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3(3):e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kitajima TS, et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441(7089):46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 79.Shintomi K, Hirano T. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev. 2009;23(18):2224–36. doi: 10.1101/gad.1844309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daum JR, et al. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21(12):1018–24. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153(3):517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McEwen BF, et al. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12(9):2776–89. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stukenberg PT, Foltz DR. Kinetochores: orchestrating the chromosomal minuet. Curr Biol. 2010;20(12):R522–5. doi: 10.1016/j.cub.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Magidson, et al. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol. 2015;17(9):1134–44. doi: 10.1038/ncb3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alexander SP, Rieder CL. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol. 1999;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Magidson V, et al. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shrestha RL, Draviam VM. Lateral to end-on conversion of chromosome–microtubule attachment requires kinesins CENP-E and MCAK. Curr Biol. 2013;23:1514–1526. doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kasuboski JM, et al. Zwint-1 is a novel Aurora B substrate required for the assembly of a dynein-binding platform on kinetochores. Mol Biol Cell. 2011;22(18):3318–30. doi: 10.1091/mbc.E11-03-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson VL, et al. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117(8):1577–89. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 91.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111(2):163–72. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 92.Cheeseman IM, et al. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127(5):983–97. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 93.Sarangapani KK, et al. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc Natl Acad Sci U S A. 2013;110(18):7282–7. doi: 10.1073/pnas.1220700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheerambathur DK, et al. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 2013;342(6163):1239–42. doi: 10.1126/science.1246232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gassmann R, et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22(17):2385–99. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gassmann R, et al. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24(9):957–71. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Famulski JK, Chan GK. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr Biol. 2007;17(24):2143–9. doi: 10.1016/j.cub.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 98.Suijkerbuijk SJ, et al. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell. 2012;23(4):745–55. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 99.Kruse T, et al. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J Cell Sci. 2013;126(5):1086–92. doi: 10.1242/jcs.122481. [DOI] [PubMed] [Google Scholar]
- 100.Xu P, et al. BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biol Open. 2013;2(5):479–86. doi: 10.1242/bio.20134051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nijenhuis W, et al. Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat Cell Biol. 2014;16(12):1257–64. doi: 10.1038/ncb3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184(3):373–81. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Uchida KS, et al. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184(3):383–90. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim Y, et al. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hagting A, et al. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J Cell Biol. 2002;157(7):1125–37. doi: 10.1083/jcb.200111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jelluma N, et al. Chromosomal instability by inefficient Mps1 auto- activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS One. 2008;3(6):e2415. doi: 10.1371/journal.pone.0002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kelly AE, et al. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12(1):31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rosasco-Nitcher SE, et al. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319(5862):469–72. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- 110.Feldman RM, et al. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91(2):221–30. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 111.Tang X, et al. Composite low affinity interactions dictate recognition of the cyclin -dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc Natl Acad Sci U S A. 2012;109(9):3287–92. doi: 10.1073/pnas.1116455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Borg M, et al. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci U S A. 2007;104(23):9650–5. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Klein P, et al. Mathematical modeling suggests cooperative interactions between a disordered polyvalent ligand and a single receptor site. Curr Biol. 2003;13(19):1669–78. doi: 10.1016/j.cub.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 114.Mittag T, et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci U S A. 2008;105(46):17772–7. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vleugel M, et al. Sequential multisite phospho-regulation of KNL1-BUB3 interfaces at mitotic kinetochores. Mol Cell. 2015;57(5):824–35. doi: 10.1016/j.molcel.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 116.Zaytsev AV, et al. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol Biol Cell. 2015;26(10):1829–44. doi: 10.1091/mbc.E14-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127(5):969–82. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 118.Alushin GM, et al. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol. 2012;19(11):1161–7. doi: 10.1038/nsmb.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McCleland ML, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17(1):101–14. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]