Abstract
EndoBI-1 is a recently isolated endo-β-N-acetylglucosaminidase, which cleaves the N-N′-diacetyl chitobiose moiety found in the N-glycan core of high mannose, hybrid and complex N-glycans. These N-glycans have selective prebiotic activity for a key infant gut microbe, Bifidobacterium longum subsp. infantis. The broad specificity of EndoBI-1 suggests the enzyme may be useful for many applications, particularly for deglycosylating milk glycoproteins in dairy processing. To facilitate its commercial use, we determined kinetic parameters for EndoBI-1 on the model substrates ribonuclease B and bovine lactoferrin, as well as on concentrated bovine colostrum whey. Km values ranging from 0.25 to 0.49, 0.43 to 1.00 and 0.90 to 3.18 mg/mL and Vmax values ranging from 3.5 × 10−3 to 5.09 × 10−3, 4.5 × 10−3 to 7.75 × 10−3 and 1.9 × 10−2 to 5.2 × 10−2 mg/mL × min were determined for ribonuclease B, lactoferrin and whey, respectively. In general, EndoBI-1 showed the highest apparent affinity for ribonuclease B, while the maximum reaction rate was the highest for concentrated whey. EndoBI-1-released N-glycans were quantified by a phenol-sulphuric total carbohydrate assay and the resultant N-glycan structures monitored by nano-LC-Chip-Q–TOF MS. The kinetic parameters and structural characterization of glycans released suggest EndoBI-1 can facilitate large-scale release of complex, bioactive glycans from a variety of glycoprotein substrates. Moreover, these results suggest that whey, often considered as a waste product, can be used effectively as a source of prebiotic N-glycans.
Keywords: Deglycosylation, N-glycans, Endo-β-N-acetylglucosaminidase
1. Introduction
Glycosylation of human and bovine milk proteins is a posttranslational modification that results from enzymatic processing of polypeptide chains in mammary epithelial cells. Protein glycosylation influences the biological and physicochemical properties of the protein including targeting, folding and stability [1]. Glycans are linked to proteins through O-glycosidic or N-glycosidic bonds. O-linked glycans (O-glycans) are frequently attached to the polypeptide via N-acetylgalactosamine to a hydroxyl group of a serine or threonine residue, and can be extended into a variety of different structural core classes. N-linked glycans (N-glycans) are linked via N-acetylglucosamines (HexNAc) to an asparagine residue of proteins in the specific amino acid sequence AsN-X-Ser/Thr (where X can be any amino acid except proline) [2].
Human milk oligosaccharides (HMO) are known to guide the development of the infant intestinal microbiota, in particular enriching a beneficial bifidobacterial population [3,4], which has recently been implicated in a range of health benefits including anti-adhesive antimicrobial activity, positive modulation of intestinal epithelial cell responses, immune modulation, protection against necrotizing enterocolitis and stimulation of brain development [5–9]. Unfortunately, many infants do not have access to human milk and, consequently, there have been numerous efforts to identify functionally equivalent molecules for infant formula and other prebiotic, therapeutic uses. Bovine milk represents one possible source of structurally similar oligosaccharides [10], however the concentration of these oligosaccharides in fluid milk is 20-fold lower than HMOs in human milk—a fact that limits their usage commercially [11,12]. Alternatively, bovine milk glyco-conjugates such as glycoproteins might be considered as an additional source of bovine glycans with desirable bioactivities. In fact, we have recently shown that a key infant gut microbe, Bifidobacterium longum subsp. infantis, consumes these conjugated N-glycans as a carbon source while other less desirable microbes do not (unpublished results). Whey, a by-product of cheese-making, contains ~4.5 g of glycoproteins per L [13]. Because only half of the total whey (190,000,000 ton/year) is currently utilized by the food industry, it is still considered a waste product [14]. Therefore, the use of whey as a source of bioactive glycans may also improve the sustainability of the dairy industry. However, robust enzymes with the appropriate activity to release the conjugated glycans are necessary for this process to become a commercial reality.
Peptidyl-N-glycosidases (PNGases) are widely used for deglycosylation of glycoproteins for analytical research purposes [15]. A PNGase (peptide- N4- (N-acetyl-β-glucosaminyl), glycopeptidase, N-glycanase, glycoaminidase or N-glycosidase) cleaves asparagine-linked oligosaccharides from glycoproteins by hydrolyzing the amide side chain [16,17]. Commercially available PNGases (PNGase A from sweet almond and PNGase F from Falvobacterium meningosepticum) release several kinds of N-linked glycans irrespective of their size or charge [18–20]. However, if fucose is α1,3-linked to N-acetlyglucosamine, the N-glycans are resistant to hydrolysis by PNGase F [21]. We recently showed that an endo-β-N-acetylglucosaminidase (EndoBI-1) isolated from B. longum subsp. infantis ATCC 15697 has the ability to cleave the N-N′-diacetyl chitobiose moiety found in the N-glycan core of high mannose, hybrid and complex N-glycans [1]. EndoBI-1 activity is not affected by fucosylation of the N-glycan core, and it is heat resistant making it attractive for use in various industrial processes. Despite the wide potential use of this enzyme, data are not yet available on its kinetics and substrate specificity in terms of initial maximum rates and Km-values.
In this study, we examine the kinetic parameters of EndoBI-1 for three purified, glycosylated substrates: model proteins ribonuclease B (RNase B) and bovine milk lactoferrin (bLF), as well as bovine colostrum whey protein concentrate. A goal was to investigate the potential use of EndoBI-1 for large-scale N-glycan production of released glycans from whey. Different direct and linearized plotting methods [22–24] were used to facilitate the prediction of kinetics parameters of the EndoBI-1. The values obtained enable comparison to other enzymes and estimations of optimal combinations of enzyme concentration and incubation time to yield complete N-glycan release from whey at a commercial-scale. These well determined kinetic parameters will enable us to produce a new prebiotic source efficiently. Finally, the structures of the released N-glycans were elucidated for each substrate by advanced mass spectrometry methods.
2. Materials methods
2.1. Gene cloning, expression and purification
A pEcoTM-T7-cHis, Eco Cloning Kit (GeneTarget Inc., San Diego, CA, USA) was used for gene cloning. The EndoBI-1 coding sequence was amplified from B. longum subsp. infantis ATCC 15697 genomic DNA with primers; 5′-TTTGTACAAAAAAGCAGGCACCATGAATGCGGACGCCGTTTCTCCGAC-3′ and 5′-TTTGTACAAGAAAGCTGGGTTGCCGGTCGCACTCAGTTGCTTCGG-3′ and then cloned into the pEco-T7-cHis vector. The vector was transformed into Escherichia coli Dh5α and E. coli BL21* for protein expression. The sequence was confirmed (IC-1006-forward; TAATACGACTCACTATAGG, IC-1006-reverse; TGCTAGTTATTGCTCAGCGG). Poly-histidine-tagged EndoBI-1 was produced in the E. coli host with a yield of ~1.7 mg/L on LB media under optimal induction conditions (0.5 mM IPTG, 37 °C for 4 h). The protein was purified following bacterial lysis using affinity chromatography with 5 mL prepacked Ni-charged columns (Bio-Rad, Hercules, CA, USA). The bound protein was eluted with 80 mM imidazole with high purity (data not shown).
2.2. Substrates
The concentration of bovine colostrum whey proteins was carried out using a pilot-scale cross-flow membrane system (Model L, GEA Filtration, Hudson, WI, USA). The system was composed of a 6.4 cm diameter spiral membrane housing (1–2 m2 area), a 95 L jacketed stainless steel hold tank, a Proline Promass 80 E flow-meter (Endress + Hauser, Reinach, Switzerland), a heat exchanger, and a 7.0HP feed pump (Hydra-Cell™ Pump, model D10EKSGSNECF, Minneapolis, MN, USA). After upstream lactose hydrolysis (0.1% lactase, 30 min, 40–43 °C), seventy-four liters of bovine colostrum whey were ultrafiltered using this system in single batch with a 10 kDa molecular weight cut-off polyethersulfone spiral-wound membrane (effective area of 1.86 m2) up to a 5.4 concentration factor (concentration factor = volume of feed/volume of retentate). Whey protein concentration was performed at 40–43 °C with a transmembrane pressure of 3.0 bars and a recirculation flow rate of 10 L/min. After a concentration factor of 5.4 was achieved, the protein-rich retentate was diluted back to its original volume with water. Two diafiltrations were performed to increase the removal of monosaccharides and oligosaccharides from the ultra-filtration retentate. RNase B from bovine pancreas and bLF were obtained from Sigma–Aldrich (St. Louis, MO, USA). To directly compare the pure glycoproteins (bLF and RNase B) to concentrated whey protein, five times the whey protein mass was used in reactions, to account for the mixed population of glycosylated (20%) and un-glycosylated (80%) proteins in whey [13,25].
2.3. Glycoprotein digestion by EndoBI-1 and glycan quantification
Enzyme and substrate concentration were determined by a Qubit Protein Assay Kit (Life Technologies, Grand Island, NY, USA). RNase B, bLF and concentrated bovine whey (0.1–0.8 mg/mL) were incubated for various times from 0 to 45 min at 37 °C with 0.025 mg/mL EndoBI-1 in a 0.02 M Na2HPO4 buffer solution at pH 5. The reactions were terminated by the addition of 1 M Na2CO3. Protein precipitation was carried out using a ratio of 4:1 cold pure ethanol added into the samples to precipitate proteins and collect the released N-glycans. Samples were dried overnight by vacuum centrifugation and were rehydrated in 100 µL of water, vortexed and sonicated. All experiments were performed in at least triplicate. A Carbohydrate Assay Kit, from Biovision, (Milpitas, CA, USA), was used to quantify released N-glycans with mannose as a standard. Aliquots of 30 µL of sample and 150 µL of concentrated sulfuric acid (98%) were added to each well. Samples were mixed on a shaker for ~1 min and incubated at 85 °C for 15 min. After incubation, 30 µL of developer (provided by Biovision) were added to each well. Samples were mixed on the shaker for 5 min and the OD of each sample was measured at 490 nm. Sample OD was converted to carbohydrate concentration using a mannose standard curve linear function.
2.4. Determination of kinetic constants
Because of the complex nature of glycosylated and non-glycosylated proteins in whey, we first tested the activity of the EndoBI-1 on the purified model glycoproteins RNaseB and Lactoferrin. This allowed us to calculate kinetic parameters that could be compared to those for the more complex substrate found in whey, as well as identify a concentration range for the whey protein that would facilitate measurement of kinetic parameters and the heterogeneous structures of glycans released.
In this manner, RNase B, bLF and bovine whey concentrate with initial concentrations ranging from 0.1 to 0.8 mg/mL were chosen to determine the reaction curves for a total time of 45 min. The initial rate of reaction was obtained from the slopes calculated from samples taken in the first 20 min of reaction for each of the three substrates. Kinetic values (Km and Vmax) were determined from the initial rates at various initial substrate concentrations by using Graph Prism 6.0 software with non-linear and linearized plotting techniques including Lineweawer–Burk, Hanes–Woolf and Eadie–Hofstie [22,23,26].
2.5. Glycan purification for nano-LC-Chip–Q-TOF MS
After enzymatic digestion and protein precipitation by ethanol, 100 µL of samples in water were loaded in duplicate on PGC SPE plates (Glygen, Columbia, MD, USA) that were conditioned using 3 × 100 µL of 80% ACN containing 0.1% TFA in water, followed by 3 × 100 µL of water. After sample loading, wells were washed using 6 × 200 µL of water and N-glycans were eluted using 3 × 200 µL of 40% ACN containing 0.1% TFA in water. The enriched N-glycans fractions were dried overnight in vacuum. Samples were rehydrated in 50 µL of water, vortexed, sonicated and diluted 50 times prior to mass spectrometry analysis. For each sample, duplicates were analyzed by mass spectrometry.
2.6. Nano-LC-Chip–Q-TOF mass spectrometry analysis
N-glycans were analyzed using the Agilent 6520 accurate-mass Q-TOF–LC/MS with a microfluidic nano-electrospray chip (Agilent Technologies, Santa Clara, CA, USA). N-glycans were separated using an HPLC-chip with a 40-nL enrichment column and a 43-mm × 75-µm analytical column, both packed with 5 µm porous graphitized carbon (PGC). The system was composed of a capillary and nanoflow pump, and both used binary solvents consisting of solvent A (3% ACN, 0.1% formic acid in water (v/v)) and solvent B (90% ACN, 0.1% formic acid in water (v/v)). Two microliters of sample were loaded with solvent A at a capillary pump flow rate of 4 µL/min. N-glycan separation was performed on a 65-min gradient delivered by the nanopump at a flow rate of 0.3 µL/min. The 65-min gradient followed this program: 0% B (0.0–2.5 min), 0 to 16% B (2.5–20.0 min), 16 to 44% B (20.0–30.0 min), 44 to 100% B (30.0–35.0 min) and 100% B (35.0–45.0 min). The gradient was followed by equilibration at 0% B (45.0–65.0 min). Data were acquired within the mass range of 450–3000 m/z for N-glycans in the positive ionization mode with an acquisition rate of 1 spectra/s. An internal calibrant ion of 922.010 m/z from the tuning mix (ESI–TOF Tuning Mix G1969–85000, Agilent Technologies) was used for continual mass calibration. For tandem MS analysis, N-glycans were fragmented with nitrogen as the collision gas. Spectra were acquired within the mass range of 100–3000 m/z. The collision energies correspond to voltages (Vcollision) that were based on the equation: Vcollision = m/z (1.5/100 Da) Volts – 3.6 Volts; where the slope and offset of the voltages were set at (1.5/100 Da) and (−3.6), respectively. Data acquisition was controlled by MassHunter Workstation Data Acquisition software (Agilent Technologies).
2.7. N-glycan identification
Compounds were identified using MassHunter Qualitative Analysis software (version B.06.00 SP2, Agilent Technologies) and the Find by Formula algorithm. The compounds were matched to a bovine milk N-glycan library using a mass error tolerance of 20 ppm [27]. The N-glycans from the library were composed of hexose (Hex), HexNAc, fucose, N-acetylneuraminic acid (NeuAc) and N-glycolylneuraminic acid (NeuGc). For compound identification, a minimum abundance of 1000 counts was used to filter out low-abundance compounds. Identified compounds were extracted allowing charge states of +1 to +3 and the isotope model was “glycans.” Extracted ion chromatogram peaks were smoothed with the Gaussian function, and the resulting peaks were integrated via the “agile” algorithm. The assignment of N-glycans was confirmed by tandem mass spectrometry.
3. Results
3.1. EndoBI-1 production and confirmation of its activity on RNase B
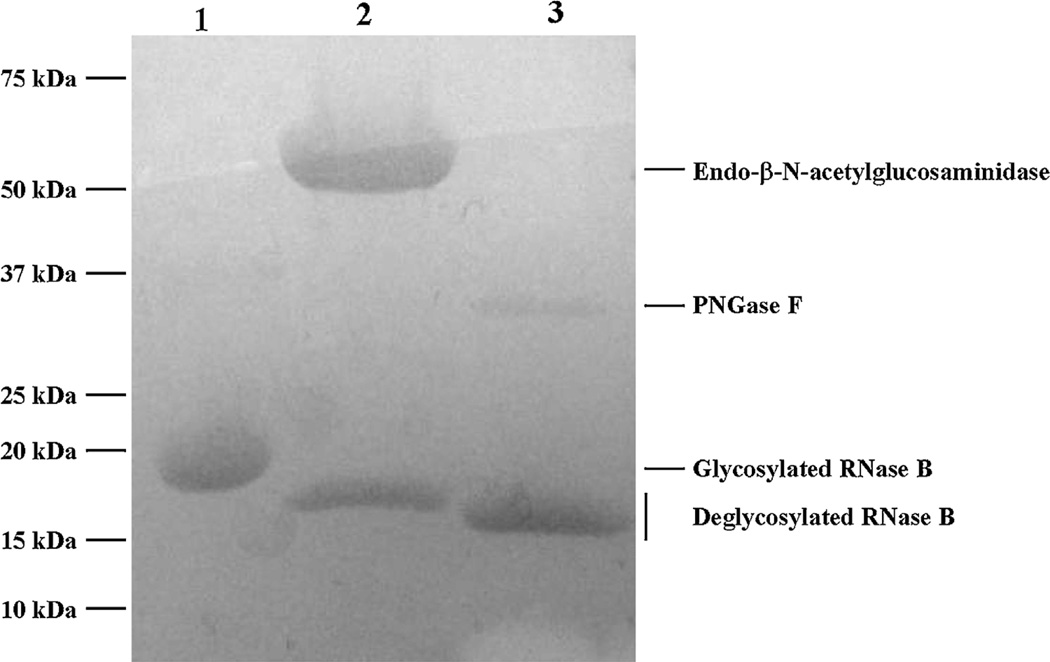
The activity of recombinant EndoB-1 was tested on model glycoprotein RNase B that contains only high mannose N-linked glycans. RNase B is a 17 kDa N-glycosylated protein and its deglycosylation results in a 14–15 kDa deglycosylated protein. Confirmation of the deglycosylation activity of RNase B by purified EndoB-1 and a control commercial enzyme PNGase F are shown in Fig. 1. Because EndoBI-1 cleaves one HexNAc residue less than PNGase F, the molecular weight of deglycosylated RNase B is slightly higher than 14 kDa for the EndoBI-1 enzyme. Substrates were denatured at 95 °C for 5 min in a final concentration of 10 mM DTT prior to the hydrolysis.
Fig. 1.
Enzymatic deglycosylation of RNase B by Endo-B-N-acetylglucosaminidase and PNGase F. on 12% SDS-PAGE gel. Lanes 1:glycosylated RNase B (17 kDa). Lane 2:deglycosylated Rnase B by Endo-β-N-acetylglucosaminidase (47 kDa). Lane 3: de glycosylated Rnase B by PNGase F (36 kDa).
3.2. Determination of Km and Vmax by non-linear and linearized plotting
Using the Michaelis–Menten model, the velocity of the reaction is obtained from the initial linear decrease of the substrate concentration or the increase in product concentration. Km, the substrate concentration when the reaction reaches half of Vmax, can also be calculated from the obtained curve [28]. The direct plot may be obtained by plotting the initial reaction rate (ν) against the corresponding initial substrate concentration (S) that yields a hyperbolic curve of saturation. However, it may be difficult to locate the asymptotes properly or potential deviations from the expected curve may be difficult to monitor. Thus, there are several linearized plotting methods that have been developed including Lineweawer–Burk, Hanes–Woolf and Eadie–Hofstman, each with their own advantages and disadvantages [22–24,29].
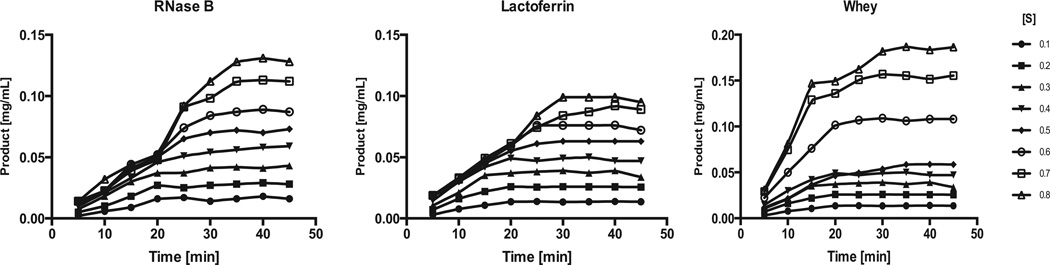
The kinetics of EndoBI-1 deglycosylating RNase B, bLF and whey glycoproteins were determined using various initial concentrations of each substrate over 45 min for each reaction (Fig. 2). The reaction rate for each substrate was constant (i.e., slope of concentration vs. time was constant) for the first 20 min, and initial rates of reaction were therefore calculated from the slopes of these data. The assumption of [S]» [E] is not valid after 20 min. Therefore, the enzyme concentration becomes limited and reaction rate gets slower.
Fig. 2.
Reaction curves of EndoBI-1 acting on RNase B, bLF and concentrated whey. The initial substrate concentration ranged from 0.1 to 0.8 mg/ml for and the enzyme concentration for each experiment was 0.025 mg/mL.
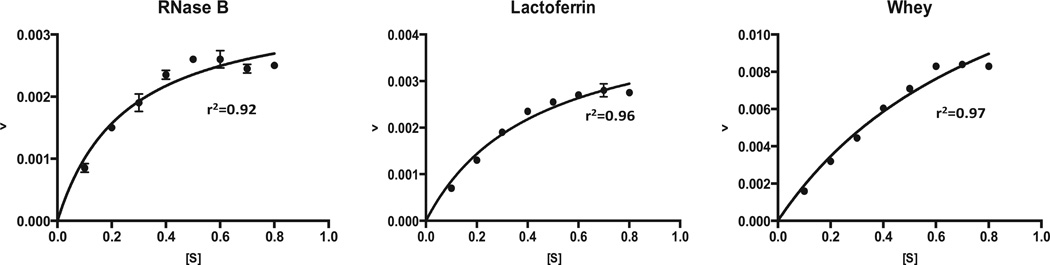
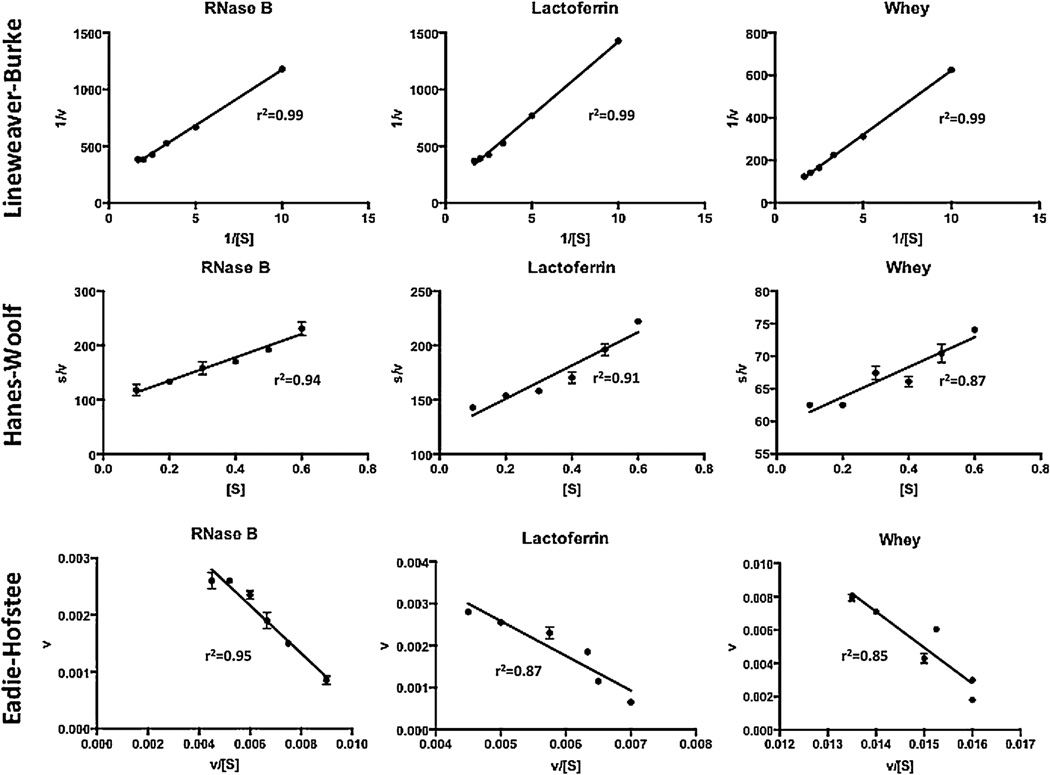
The direct plot was created by plotting the reaction rate (ν) against the initial substrate concentration (S). These plots all resulted in a hyperbolic or saturation relationship (Fig. 3). Km values of 0.25, 0.43 and 0.90 mg/mL and Vmax values of 5.09 × 10−3, 7.75 × 10−3 and 5.2 × 10−2 mg/mL × min were calculated for RNase B, bLF and bovine whey, respectively. The linearization methods, Lineweaver–Burk (1/S vs. 1/ν), Hanes-Woolf (S vs. S/ν) and Eadie-Hofstee (ν/S vs. ν), plotting techniques were also used (Fig. 4) to estimate the kinetic parameters for each substrate. Km and Vmax values estimated from these plotting techniques and a comparison with the non-linear curve fitting is shown in Table 1. In general, there is a significant difference in the parameters derived from the non-linear and linearized fitting techniques for each substrate. It was shown that Km values estimated from non-linear fitting are 1.77, 2 and 2.92 fold lower than the average of the three linearized fitting methods for RNase B, bLF and bovine whey, respectively, though the general trends are the same. Km refers to the minimum substrate concentration that achieves half of Vmax. Thus, the results demonstrate that the enzyme has a higher apparent affinity for RNase B than for the other substrates. Vmax values ranging from 3.5 × 10−3–5.09 × 10−3, 4.5 × 10−3–7.75 × 10−3 and 1.9 × 10−2–5.2 × 10−2 mg/mL × min were found for RNase B, bLF and bovine whey, respectively. These results indicate that the maximum rate of reaction is higher with the concentrated bovine colostrum whey as substrate compared with the model glycoprotein substrates and that the concentrated whey prepared at the UC Davis Milk Processing Lab can be used as N-glycan source at a rate of 2.08 mg N-glycan/ml × min per 1 mg/mL EndoBI-1 under optimal conditions.
Fig. 3.
Direct plotting of rate of reaction (ν) (mg/mL × min) and substrate concentration [S] (mg/mL).
Fig. 4.
Lineweaver–Burke (1/[S] (mL/mg) vs. 1/ν (mL × min/mg)), Hanes–Woolf ([S] (mg/mL) vs. s/ν (min)) and Eadie-Hofstee (ν/[S] (min−1) vs. ν (mg/mL × min)) linearized plotting techniques for the estimation of kinetic parameters of EndoBI-1 on Rnase B, bLF and concentrated whey.
Table 1.
Summary of the regressions shown in Figs. 3 and 4 (linearized equations, quantities plotted on the x and y axis and results for ν in mg/mL × min and Km in mg/mL.).
| Non-linearized plotting | Lineweaver–Burke | Eadie–Hofstee | Hanes–Woolf | ||||
|---|---|---|---|---|---|---|---|
| Linearized equation | – | ||||||
| x | S | 1/S | ν/S | S | |||
| y | ν | 1/ν | ν | S/ν | |||
| RNase B | |||||||
| Vmax | 3.5 × 10−3 | 5.09 × 10−3 | 4.6 × 10−3 | 4.6 × 10−3 | |||
| Km | 0.2528 | 0.497 | 0.42 | 0.426 | |||
| Lactoferrin | |||||||
| Vmax | 4.5 × 10−3 | 7.75 × 10−3 | 6.7 × 10−3 | 6.5 × 10−3 | |||
| Km | 0.4316 | 1.007 | 0.828 | 0.785 | |||
| Whey | |||||||
| Vmax | 1.9 × 10−2 | 5.2 × 10−2 | 3.7 × 10−2 | 4.3 × 10−2 | |||
| Km | 0.9026 | 3.18 | 2.16 | 2.575 |
3.3. N-glycan structures
Mass Spectrometry analysis of released N-glycan from RNase B, bLF and bovine colostrum whey protein concentrate after digestion by EndoBI-1 provides information on the glycosylation patterns for the three substrates as well as their potential bioactivity. Application of the bovine milk N-glycan library as a mass filter in Find by Formula allowed the identification of N-glycan compositions with high mass accuracy and retention time reproducibility [27]. Tandem MS analysis generated specific fragment ions that were common to all N-glycans allowing the confirmation of 5, 33 and 18 different oligosaccharide structures for RNase B, bLF and bovine colostrum whey protein concentrate, respectively. The details of all structures, including neutral and sialylated N-glycans, are described in the supporting information, (Tables S1–S3).
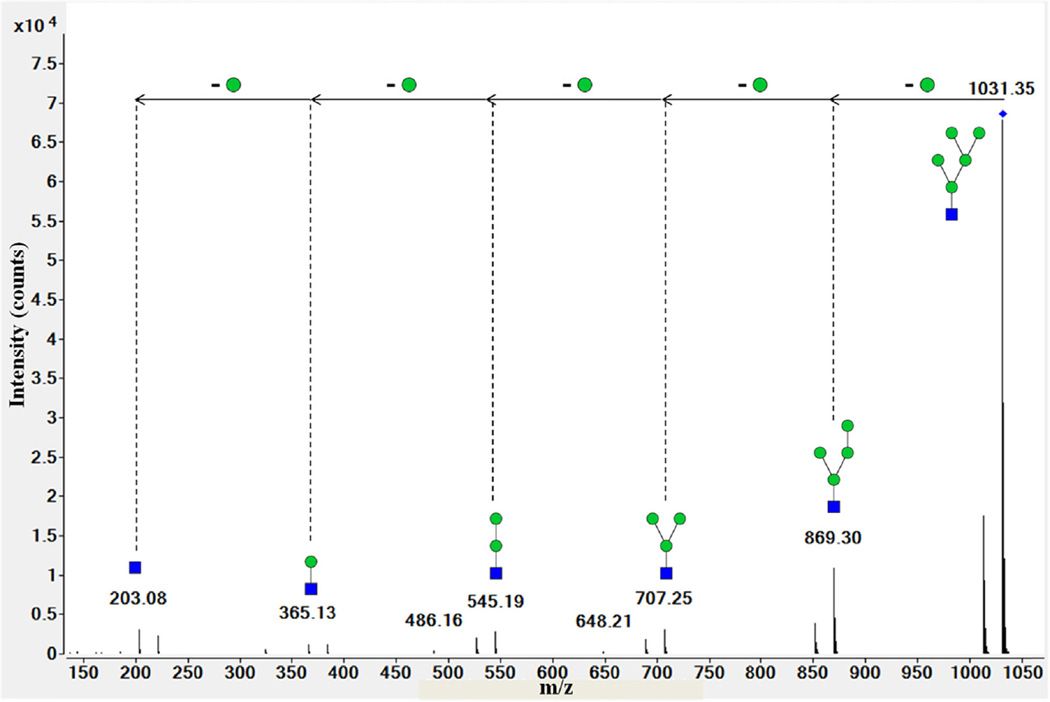
RNase B is a well-characterized bovine glycoprotein that contains high mannose glycans with five to nine mannoses. Nano-LC-Chip–Q-TOF MS confirmed the presence of only high mannose N-glycans that are eluted between 11 and 17 min and are linked to the unique N-glycosylation site of the protein, 60Asn [30]. The spectrum shown in Fig. 5 illustrates the fragment pattern of a high mannose N-glycan with 1032.36 m/z, z = 1. The spectrum shows a 162 Da difference between each peak that corresponds to the molecular weight of a mannose residue. Different high mannose isomers were resolved by nano-LC-Chip–Q-TOF MS for RNase B.
Fig. 5.
Deconvoluted tandem spectrum of high mannose N-glycans 5Hex-1HexNAc from RNase B. This glycan corresponded to m/z 1032.36 with z = +1. Green circles and blue squares represent mannose and HexNAc residues, respectively.
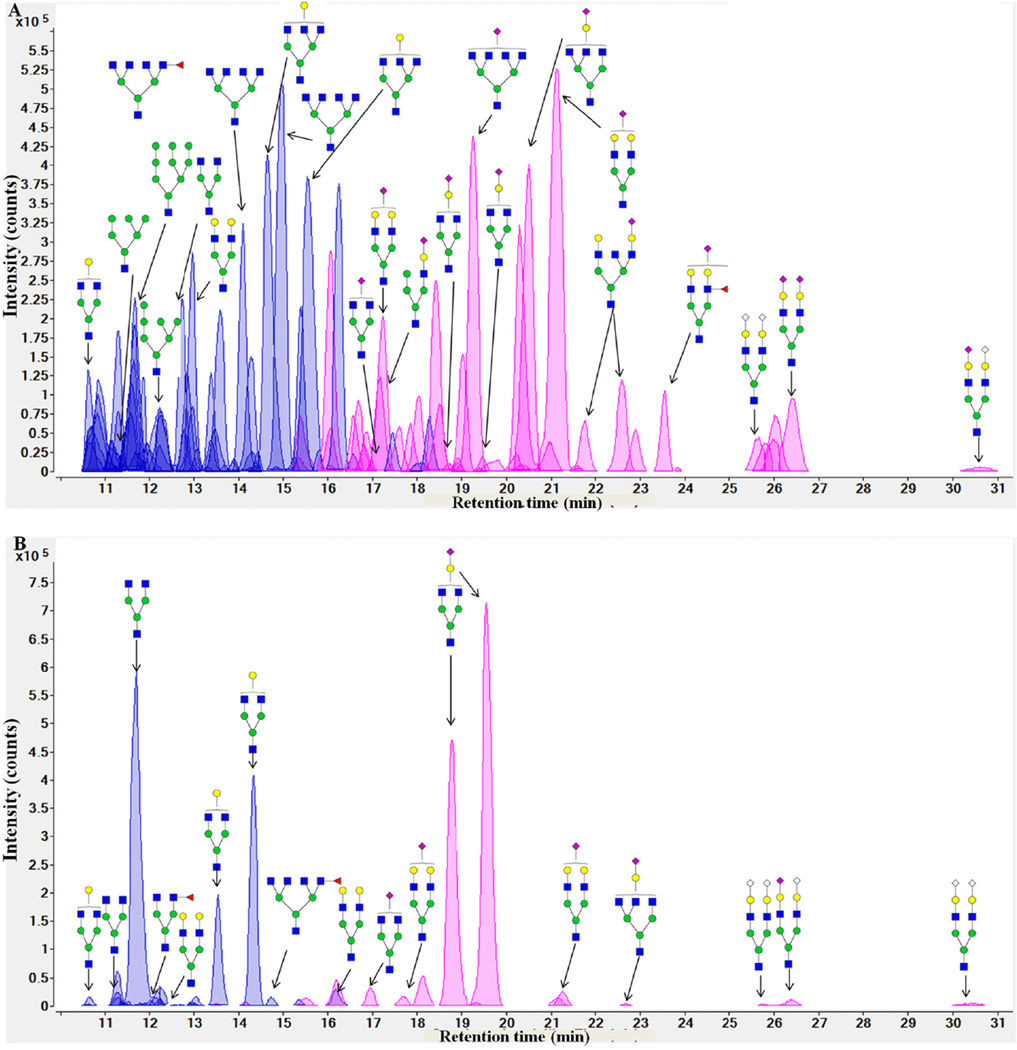
Fig. 6 presents extracted compound chromatograms (ECCs) of bLF and bovine colostrum whey protein concentrate. Glycomics profiling demonstrates different patterns for these two substrates. Whey glycoproteins and pure bLF exhibit high-mannose N-glycans and complex/hybrid N-glycans, including fucosylated and sialylated N-glycans.
Fig. 6.
Extracted compound chromatograms (ECCs) of N-glycans from bovine lactoferrin (A) and bovine colostrum whey concentrate (B). The glycan types are differentiated by color: neutral glycans are purple and sialylated glycans are pink. Green circles, yellow circles, blue squares, red triangles, purple diamonds and gray diamonds represent mannose, galactose, HexNAc, fucose, NeuAc and NeuGc residues, respectively.
4. Discussion
In the present study, we characterize the kinetics of glycan release from model glycoproteins using a novel endo-β-N-acetylglucosaminidase, EndoBI-1, obtained from B. longum subsp. infantis ATCC 15697. Importantly, we also evaluated the ability of this enzyme to catalyze the conversion of a dairy waste stream, concentrated bovine colostrum whey prepared at the pilot-scale (UC Davis Milk Processing Lab), into a potential bioactive N-glycan source. Using this enzyme, we demonstrated N-glycan release from glycoproteins in whey with a maximum rate of 5.2 × 10−2 mg/mL min using 0.025 mg enzyme/mL.
We first showed that N-glycans are, in fact, released from both the purified model glycoproteins and the bovine colostrum whey protein concentrate using this novel enzyme. This demonstrates that the EndoBI-1 enzyme has the potential to release bioactive glycans from bovine-milk-related glycoproteins. A wider variety of N-glycans, including anomers and isomers, was released from pure bLF than from glycoproteins in bovine colostrum whey protein concentrate. This difference can be explained by the fact that whey is a complex fluid that contains N-linked as well as O-linked and nonglycosylated proteins, including β-lactoglobulin, α-lactalbumin, serum albumin bovine immunoglobulin and lactoferrin. Among these proteins, lactoferrin and immunoglobulins are some of the most abundant glycoproteins in bovine milk [25,31]. Non-glycosylated whey proteins, including α-lactalbulmin (1.2 g/L) and β-lactoglobulin 3.2 g/L, are present at higher concentration than whey N-linked proteins like lactoferrin and immunoglobulins, [32]. These differences could explain why the N-glycan release by EndoBI-1 is more efficient on a pure glycoprotein than a complex mixture that contains lower relative amount of glycoproteins. It should be noted, however, that structure determinations were made using samples taken early on in the reaction corresponding to when the reactions were still at their initial rate. It is possible that samples taken at a later stage of the reactions could yield a different distribution of released glycans.
To further characterize the reaction kinetics of EndoBI-1,wecalculated the Michaelis–Menten kinetic parameters, Vmax and Km for two purified glycoprotein substrates and bovine colostrum whey protein concentrate, an example of a potential commercial substrate for this novel enzyme. Interestingly, there are significant differences in the kinetic parameters derived from different fitting techniques for each substrate. These results are consistent with other studies that compared the non-linear and linearized plotting techniques [33–35]. In general, non-linear plotting resulted in the underestimation of the kinetic parameters compared to the linearized methods. It is possible that more sophisticated methods for non-linear fitting [36] would improve the agreement between methods. However, the general trends in kinetic parameters for the three substrates are, in themselves, instructive and consistent between methods.
The lowest Km was observed for RNase B while the highest Km value was determined for whey. The lower apparent affinity of the enzyme for whey glycoproteins is explained by the heterologous nature of the substrate as described above. Because whey includes not only glycoproteins but also non-glycosylated proteins, this may result in the overestimation of the Km value, or may also be a good indicator of the competitive inhibition that might be expected by non-substrate glycoproteins and non-glycosylated proteins [37,38].
Although, RNase B showed the highest enzyme-protein affinity, it has the lowest Vmax value compared to bLF and whey-derived glycoproteins. RNase B has a single high mannose N-glycosylation site that might result in low N-glycan release efficiency per amount of a given protein. While EndoBI-1 showed lower affinity for bLF than RNase B, it has a slightly higher Vmax for bLF than for RNase B. This might be the result of the multiple and more complex N-glycan content of bLF. The highest Vmax value was observed for whey. The enzyme exhibited a 5–10 fold higher Vmax on bovine colostrum whey protein concentrate than on RNase B and bLF depending on the fitting method used to calculate this parameter. Although, EndoBI-1 has lower apparent affinity value for whey glycoproteins, its high maximum reaction rate could be a result of a high biantennary N-glycan content of whey immunoglobulins. Immunoglobulins mainly contain biantennary N-glycans, while the model proteinsweused contain mostly tri-and tetra-antennary-type N-glycans (Fig. 6) Because most of endo-β-N-acetylglucosaminidases prefer the high mannose and biantennary N-glycans to tri- and tetra-antennary-type N-glycans, EndoBl-1 digests the N-glycans of immunoglobulin efficiently, which results in a higher observed activity on whey.
The kinetic parameters of PNGase F for various glycoproteins have been investigated by several groups [15,39,40]. In general, it was reported that the denaturation of glycoproteins is essential for PNGase F activity. The Km values of PNGase F for denatured glycoproteins are higher than EndoBI-1 values that we determined in this study. Moreover, lower Vmax values observed in these studies compared to our findings indicate that EndoBI-1 is an efficient alternative enzyme that can be used for large-scale glycan production. The low Vmax of PNGase F can be explained by its limited activity on core fucosylated N-glycans.
Successful release of these N-glycans from bovine milk glycoproteins, which were recently shown to be a new prebiotic source (submitted), by EndoBI-1, an enzyme active on all N-glycan cores, will enable further investigation of the biological and potential nutritional or therapeutic functions of bioactive glycans in bovine milk streams and lead to their larger-scale commercialization. However, a necessary step in this process is to fully characterize the enzymatic behavior and kinetics of the enzyme on a number of food substrates, including various dairy streams. Moreover, the use of these enzymes to release bioactive glycans from dairy processing waste streams like whey would also make dairy processing more economically and environmentally sustainable.
Supplementary Material
Acknowledgments
This work has been supported by University of California Discovery Grant Program, the UC Davis RISE program, the Bill and Melinda Gates Foundation, National Institutes of Health awards R21AT006180, R01AT007079 and the Peter J. Shields Endowed Chair in Dairy Food Science.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.enzmictec.2015.05.007
References
- 1.Garrido D, et al. Endo-beta-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol. Cell. Proteomics: MPC. 2012;11(9):775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Legrand D, et al. Lactoferrin: a modulator of immune and inflammatory responses. Cell. Mol. life Sci.: CMLS. 2005;62(22):2549–2559. doi: 10.1007/s00018-005-5370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sela DA, Mills DA. The marriage of nutrigenomics with the microbiome: the case of infant-associated bifidobacteria and milk. Am. J. Clin. Nutr. 2014;99(3):697S–703S. doi: 10.3945/ajcn.113.071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zivkovic A, et al. Establishment of a milk-oriented microbiota (mom) in early life: how babies meet their moms. Funct. Foods Rev. 2013;15:3–12. [Google Scholar]
- 5.Kunz C, et al. Oligosaccharides in human milk: structural: functional, and metabolic aspects. Annu. Rev. Nutr. 2000;20(1):699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 6.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 7.Rudloff S, et al. Urinary excretion of lactose and oligosaccharides in preterm infants fed human milk or infant formula. Acta Paediatr. 1996;85(5):598–603. doi: 10.1111/j.1651-2227.1996.tb14095.x. [DOI] [PubMed] [Google Scholar]
- 8.Patel BK, Shah JS. Necrotizing enterocolitis in very low birth weight infants: a systemic review. ISRN Gastroenterol. 2012;2012 doi: 10.5402/2012/562594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas A, et al. Breast milk and subsequent intelligence quotient in children born preterm. Lancet. 1992;339(8788):261–264. doi: 10.1016/0140-6736(92)91329-7. [DOI] [PubMed] [Google Scholar]
- 10.Zivkovic AM, Barile D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv. Nutr. 2011;2(3):284–289. doi: 10.3945/an.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao N, et al. Bovine milk glycome. J. Dairy Sci. 2008;91(10):3768–3778. doi: 10.3168/jds.2008-1305. [DOI] [PubMed] [Google Scholar]
- 12.Tao N, et al. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. J. Dairy Sci. 2009;92(7):2991–3001. doi: 10.3168/jds.2008-1642. [DOI] [PubMed] [Google Scholar]
- 13.de Wit JN. Marschall rhone-poulenc award lecture. Nutritional and functional characteristics of whey proteins in food products. J. Dairy Sci. 1998;81(3):597–608. doi: 10.3168/jds.s0022-0302(98)75613-9. [DOI] [PubMed] [Google Scholar]
- 14.Galvao CM, et al. Controlled hydrolysis of cheese whey proteins using trypsin and alpha-chymotrypsin. Appl. Biochem. Biotechnol. 2001;91–93:761–776. doi: 10.1385/abab:91-93:1-9:761. [DOI] [PubMed] [Google Scholar]
- 15.Altmann F, Schweiszer Weber SC. Kinetic comparison of peptide: N-glycosidases F and A reveals several differences in substrate specificity. Glycoconjugate J. 1995;12(1):84–93. doi: 10.1007/BF00731873. [DOI] [PubMed] [Google Scholar]
- 16.Nuck R, et al. Optimized deglycosylation of glycoproteins by peptide-N4-(N-acetyl-beta-glucosaminyl)-asparagine amidase from Flavobacterium meningosepticum. Glycoconjugate J. 1990;7(4):279–286. doi: 10.1007/BF01073372. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi N. Demonstration of a new amidase acting on glycopeptides. Biochem. Biophys. Res. Commun. 1977;76(4):1194–1201. doi: 10.1016/0006-291x(77)90982-2. [DOI] [PubMed] [Google Scholar]
- 18.O’Neill RA. Enzymatic release of oligosaccharides from glycoproteins for chromatographic and electrophoretic analysis. J. Chromatogr. A. 1996;720(1–2):201–215. doi: 10.1016/0021-9673(95)00502-1. [DOI] [PubMed] [Google Scholar]
- 19.Szabo Z, Guttman Karger ABL. Rapid release of N-linked glycans from glycoproteins by pressure-cycling technology. Anal. Chem. 2010;82(6):2588–2593. doi: 10.1021/ac100098e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morelle W, et al. Analysis of N- and O-linked glycans from glycoproteins using maldi-tof mass spectrometry. Methods Mol. Biol. 2009;534:5–21. doi: 10.1007/978-1-59745-022-5_1. [DOI] [PubMed] [Google Scholar]
- 21.Tretter V, Altmann F, Marz L. Peptide-N4-(N-acetyl-beta-glucosaminyl) asparagine amidase F cannot release glycans with fucose attached alpha 1—3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem./FEBS. 1991;199(3):647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- 22.Woolf B. The addition compound theory of enzyme action. Biochem. J. 1931;25(1):342–348. doi: 10.1042/bj0250342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanes CS. Studies on plant amylases: the effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley. Biochem. J. 1932;26(5):1406–1421. doi: 10.1042/bj0261406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tummler K, et al. New types of experimental data shape the use of enzyme kinetics for dynamic network modeling. FEBS J. 2014;281(2):549–571. doi: 10.1111/febs.12525. [DOI] [PubMed] [Google Scholar]
- 25.Madureira AR, et al. Bovine whey proteins – overview on their main biological properties. Food Res. Int. 2007;40(10):1197–1211. [Google Scholar]
- 26.Burk D, Lineweaver H, Horner CK. The specific influence of acidity on the mechanism of nitrogen fixation by azotobacter. J. Bacteriol. 1934;27(4):325–340. doi: 10.1128/jb.27.4.325-340.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nwosu CC, et al. Comparison of the human and bovine milk n-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J. Proteome Res. 2012;11(5):2912–2924. doi: 10.1021/pr300008u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaelis L, et al. The original michaelis constant: translation of the 13 Michaelis–Menten paper. Biochemistry. 2011;50(39):8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowd JE, Riggs DS. A comparison of estimates of Michaelis–Menten kinetic constants from various linear transformations. J. Biol. Chem. 1965;240:863–869. [PubMed] [Google Scholar]
- 30.An HJ, et al. Determination of N-glycosylation sites and site heterogeneity in glycoproteins. Anal. Chem. 2003;75(20):5628–5637. doi: 10.1021/ac034414x. [DOI] [PubMed] [Google Scholar]
- 31.Sánchez L, et al. Concentration of lactoferrin and transferrin throughout lactation in cow’s colostrum and milk. Biol. Chem. Hoppe-Seyler. 1988;369(9):1005–1008. doi: 10.1515/bchm3.1988.369.2.1005. [DOI] [PubMed] [Google Scholar]
- 32.De Wit JN. Nutritional and functional characteristics of whey proteins in food products. J. Dairy Sci. 1998;81(3):597–608. doi: 10.3168/jds.s0022-0302(98)75613-9. [DOI] [PubMed] [Google Scholar]
- 33.Ritchie RJ, Prvan T. Current statistical methods for estimating the K-m and V-max of Michaelis–Menten kinetics. Biochem. Educ. 1996;24(4):196–206. [Google Scholar]
- 34.Kadam RS, Iyer KR. Isolation of liver aldehyde oxidase containing fractions from different animals and determination of kinetic parameters for benzaldehyde. Indian J. Pharm. Sci. 2008;70(1):85–88. doi: 10.4103/0250-474X.40337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tummler K, et al. New types of experimental data shape the use of enzyme kinetics for dynamic network modeling. FEBS J. 2014;281(2):549–571. doi: 10.1111/febs.12525. [DOI] [PubMed] [Google Scholar]
- 36.Coleman MC, Block DE. Nonlinear experimental design using bayesian regularized neural networks. Aiche J. 2007;53(6):1496–1509. [Google Scholar]
- 37.Ascenzi P, Ascenzi MG, Amiconi G. Enzyme competitive-inhibition - graphical determination of ki and presentation of data in comparative-studies. Biochem. Educ. 1987;15(3):134–135. [Google Scholar]
- 38.Lizama HM, Suzuki I. Synergistic competitive-inhibition of ferrous iron oxidation by thiobacillus-ferrooxidans by increasing concentrations of ferric iron and cells. Appl. Environ. Microbiol. 1989;55(10):2588–2591. doi: 10.1128/aem.55.10.2588-2591.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee KB, et al. A new approach to assay endo-type carbohydrases: bifluorescent-labeled substrates for glycoamidases and ceramide glycanases. Anal. Biochem. 1995;230(1):31–36. doi: 10.1006/abio.1995.1433. [DOI] [PubMed] [Google Scholar]
- 40.Mussar KJ, et al. Peptide: N-glycosidase F studies on the glycoprotein aminoglycan amidase from Flavobacterium meningosepticum. J. Biochem. Biophys. Methods. 1989;20(1):53–68. doi: 10.1016/0165-022x(89)90081-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.