Abstract
Osteosarcoma (OSA) is the most common primary bone tumor in humans. However, the cell of origin of OSA is not clearly defined although there is evidence that osteoblasts may serve as OSA progenitors. The role of osteocytes, terminally differentiated osteoblasts, as OSA progenitors has yet to be described. Analysis of patient cDNA from publicly available microarray data revealed that patients with OSA have increased expression of dentin matrix phosphoprotein 1 (DMP1), a marker of osteocytes. Analysis of multiple murine, human, and canine OSA cell lines revealed DMP1 expression. To test the tumorigenic potential of osteocytes, MLO-Y4, an SV-40 immortalized murine osteocyte cell line, was injected into subcutaneous and orthotopic (intratibial) sites of mice. Tumor growth occurred in both locations. Orthotopic MLO-Y4 tumors produced mixed osteoblastic/osteolytic radiographic lesions; a hallmark of OSA. Together, these data demonstrate for the first time that osteocytes can serve as OSA progenitors.
Keywords: DMP1, cancer, bone, osteocyte, osteosarcoma
Introduction
Osteosarcoma (OSA), which mainly impacts adolescents, is the most common primary bone tumor of humans [Bielack et al., 2008]. OSA only has a 60% 5-year survival rate due to a high incidence of lung metastasis at the time of diagnosis [Bielack et al., 2008]. Despite numerous studies, there have been minimal therapeutic gains in patients presenting with metastatic disease [Zhu et al., 2013]. Even though chemotherapy is an important component of therapy, few changes to standardized care have been implemented over recent decades. The deficit in therapeutic gains for OSA is due, in part, to the fact that little is known concerning the tumorigenic process and cell of origin for OSA. Defining the cells that contribute to the development of OSA will enhance the ability to identify therapeutic targets.
Bone is a complex tissue composed of numerous cell types. The mineralized portion of bone is primarily composed of osteoblasts, which promote new bone growth, osteoclasts, which resorb old and damaged bone, and osteocytes, which orchestrate the activities of osteoblasts and osteoclasts. Osteocytes (OCy) are terminally differentiated osteoblasts that become sequestered in mineralized bone [Bonewald, 2011]. OCy are also the most abundant cell type in mineralized bone, comprising over 90% of cells [Bonewald, 2011]. OCy are responsible for maintaining bone homeostasis through mechanosensation, the sensation of physical forces in the bone environment, and mechanotransduction, the translation of physical forces into biochemical signals that can be communicated to surrounding cells. These processes are necessary to measure physical forces and relate them to osteoblasts and osteoclasts, the effector cells. OCy are long lived cells, in that a single OCy may have a lifespan that measure decades [Franz-Odendaal et al., 2006]. The ability of OCy to coordinate bone formation and lysis in the presence or absence of mechanical stimuli makes them a unique cellular component of bone.
OSA is typically described as producing mixed osteoblastic/osteolytic regions within the tumor. This is unique as many cancers, such as breast and prostatic carcinoma, metastasize to bone and produce predominantly osteolytic or osteoblastic lesions, respectively. Osteoblasts have been implicated as OSA progenitors [Cao et al., 2005; Patane et al., 2006]. Due to the potential for OSA to be derived from osteoblasts, OSA cell lines, such as Saos2, are frequently utilized in many bone biology laboratories for exploring osteoblast biology [Czekanska et al., 2012; Fogh et al., 1977; Rodan et al., 1987]. However, due to their presence in the bone and bone-remodeling ability, it is plausible that OCy may also contribute to the development of OSA. Accordingly, the goal of this study was to determine if OCy could serve as an OSA precursor cell.
Materials and Methods
Cell Lines
The MLO-Y4 murine osteocyte cell line was maintained as previously described on collagen coated plates [Kato et al., 1997]. Plates were collagen coated by incubating 0.02 M acetic acid (Sigma Aldrich, St. Louis, MO) and 0.15 mg/ml rat tail type I collagen (BD Biosciences, Bedford, MA) in PBS for 1 hr at room temperature before being washed with PBS. MLO-Y4 was maintained in α-MEM (Cellgro, Henderson, VA) supplemented with 2.5% heat inactivated fetal bovine serum (FBS; Gibco, Grand Island, NY), 2.5% heat inactivated fetal calf serum (Hyclone, Logan, UT), and 1× penicillin/streptomycin (Gibco). DLM8, K12, K7M2 (murine); Saos2 (human); and Abrams, D17, Gracie, Moresco, OSCA40, Vogel (canine) cell lines were maintained in high-glucose DMEM with 10% FBS and 1× penicillin/streptomycin. The HMPOS, HOS, OOS, and POS (canine) cell lines were maintained in RPMI supplemented with 10% FBS and 1× penicillin/streptomycin. The MG63 and SJSA1 (human) cell lines were maintained in MEM with 10% FBS and 1× penicillin/streptomycin. All cells were serially passaged by trypsinization and maintained at 37°C and 5% CO2 in a humidified atmosphere (standard conditions).
Animals
All animal studies were performed in an AALAC-approved facility, with approval of the University Committee on Use and Care of Animals (UCUCA) of the University of Michigan. Male severe combined immunodeficiency disorder (SCID) mice 8–10 weeks of age were used for all experiments, and were bred at the University of Michigan. Five mice per group were used for all in vivo experiments.
In vivo tumor modeling
Subcutaneous tumors were established by injecting 1×106 MLO-Y4 cells suspended in 1× Hank’s balanced salt solution (HBSS; Gibco) into the right hind-flank of the mouse. Tumors were measured twice weekly using calipers, and tumor volume was calculated using the formula V = (S2 · L)/2, where L is the longest dimension measured and S is the perpendicular measurement.
Orthotopic tumors were established as previously described [Sottnik et al., 2010]. The proximal tibia was implanted with 1×106 MLO-Y4 cells while mice were under isoflurane anesthesia. Weekly radiographs were obtained using a Faxitron MX-20 (Wheeling, IL) at 4× magnification. Orthotopic tumor growth was assessed using a modified protocol previously described [Yin et al., 1999]. Briefly, radiographs were scanned at 600 dpi using a UMAX Powerlook 1000 and Magic Scan V4.71 software (Techville, Inc, Dallas, TX). It was determined that 600 dpi is equivalent to 55,800 pixels/cm2. Using Photoshop CS3 extended (Adobe Systems Inc, USA) a region of interest was created encompassing the radio-opaque area of the tibia between the growth plates. The number of pixels within this area was recorded. Pixel area (PA) was converted to geometric area using the following formula: A = (PA ÷ 55800) ÷ 4; where 55,800 = pixels/cm2 and 4 is the radiograph magnification.
Immunohistochemistry
Following euthanasia, primary tumor, lung, liver, and spleen were isolated from all animals, formalin fixed, and paraffin embedded. Organs were sectioned on 3 separate and distinct planes before undergoing hematoxylin and eosin (H&E) staining. Each organ section was then reviewed by a board-certified pathologist (R.M.) for the presence of metastases.
Primary tumors from subcutaneous and orthotopically challenged mice were stained for H&E, dentin matrix phosphoprotein 1 (DMP1; Abcam, Cambridge, MA; 1:100), osteocalcin (OC; Millipore, Billerica, MA; 1:500), vimentin (Bioss, Woburn, MA; 1:200), and pan-cytokeratin (Bioss; 1:200). Antibody staining was accomplished using an intelliPATH FLX (Biocare Medical; Concord, CA) automatic staining system. Reveal decloaker (Biocare Medical) was used for antibody retrieval followed by rodent block M (Biocare Medical) to block non-specific binding. Primary antibodies were applied at dilutions described above. Secondary anti-rabbit antibody conjugated to horse radish peroxidase (HRP) was utilized and followed by staining with DAB followed by hematoxylin counterstaining (Biocare Medical). Images were captured by an Olympus BX41 microscope with matching software.
Real-Time qPCR methods
For each cell line, near-confluent cells were washed with 1× PBS, RNA extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and purified using an RNeasy Mini-Kit (Qiagen, Valencia, CA, USA) according to manufacturer instructions. RNA concentrations were measured using a ND-1000 Spectrophotometer (ThermoScientific, Wilmington, DE, USA). To maximize PCR sensitivity 5 µg of prepared RNA from each cell line was reverse transcribed using the SuperScript III Reverse Transcriptase Kit (Invitrogen). Quantitative real time PCR was performed in triplicate using SYBR Green qPCR MasterMix (Qiagen) in a 10 µL reaction volume on a Roche LightCycler 480 (Roche, Indianapolis, IN, USA). Primers were purchased from SABiosciences (Qiagen) or designed using NCBI RefSeq mRNA sequences and NCBI Primer-BLAST software [Ye et al., 2012]. All primers utilized, forward/reverse sequences, and amplicon length can be found in Supplementary Tables 1 (Mouse), 2 (Human), and 3 (Dog). Measurements from triplicate Ct values were averaged. Mean Ct values of >30 cycles were considered negative (−), 25–30 (+), 20–24.99 (++), and <20 (+++). Beta-actin, GAPDH, and β2-microglobulin were used as controls to ensure reaction activity. All samples were positive (+++) for control gene expression.
Western Blot
Cells were grown to near confluence before whole cell lysis using a Triton X-100 based lysis buffer [1% Triton X-100, 20 mM Tris-HCl, 137 mM NaCl, 10% glycerol, 2mM EDTA, 1mM PMSF, 1 mM sodium orthovanadate, and protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO)]. Protein quantification was performed using a CBA kit (Thermo-Scientific, Rockford, IL) with a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA). Equal amounts of protein were loaded on a 4–12% Bis-Tris gel (Invitrogen, Eugene, OR) and electrophoresed in 1× MES/SDS running buffer (Invitrogen, Carlsbad, CA). Bands were transferred to a PVDF membrane (Bio-Rad Laboratories, Hercules, CA) and blocked using 5% instant milk (Biorad) or 5% bovine serum albumin (BSA; Celliance, Kankakee, IL) in tris-buffered saline with Tween 20 (TBST; 50 mM Tris, 150 mM NaCl, and 0.05% Tween 20). Primary antibodies used were: rabbit polyclonal anti-SV40 T-Antigen (Santa Cruz Biotechnology, Dallas, TX), mouse monoclonal anti-p53 (Cell Signaling Technology, Danvers, MA), syrian hamster monoclonal anti-podoplanin (Santa Cruz Biotechnology), rabbit polyclonal anti-DMP-1 (Abcam) and rabbit monoclonal anti-HSP90 (Cell Signaling). Primary antibodies were diluted 1:1000 and incubated overnight at 4° C. Donkey anti-rabbit (Jackson Immunoresearch, West Grove, PA), donkey anti-mouse (Jackson Immunoresearch), and goat anti-syrian hamster (Santa Cruz Biotechnology) secondary antibodies conjugated to HRP were used for detection. Detection was accomplished using ECL reagent (Thermo-Scientific, Rockford, IL) and developed using radiographic film (Research Products International Corp., Mt. Prospect, IL).
ONCOMINE Study
The ONCOMINE repository (www.oncomine.com) is a repository of cDNA microarrays. Searching ONCOMINE for the term “osteosarcoma,” we identified 15 studies. Of the 15 studies identified, two did not differentiate OSA specifically from sarcoma, 9 were composed primarily of cell lines, and 1 only had a single OSA patient thus these studies were excluded from further analysis and the remaining 3 were analyzed [Baird et al., 2005; Henderson et al., 2005; Kobayashi et al., 2010]. The Kobayashi Sarcoma data set was determine to have the greatest number of patient samples (n = 27) for analysis and differentiated OSA into subtypes [Kobayashi et al., 2010]. Baird (n = 5; [Baird et al., 2005]) and Henderson (n = 11; [Henderson et al., 2005]) data sets did not differentiate OSA subtypes but were also analyzed. ONCOMINE was queried for genes associated with OCy and OSA. Statistical analyses were performed using ONCOMINE algorithms to adjust for multiple comparisons as previously described [Rhodes et al., 2004; Sottnik et al., 2013].
Statistics
Statistical analysis was performed using Prism 5 (GraphPad Software, La Jolla, CA). Doubling time for in vivo tumor growth was determined by non-linear regression of an exponential growth curve for tumor growth. ONCOMINE data was analyzed as previously described utilizing ONCOMINES algorithms [Rhodes et al., 2004; Sottnik et al., 2013]. Supplemental analyses of the Kobayashi dataset were performed using a two-tailed t-test comparing osteoblastic OSA to all other OSA subtypes. For all analyses, p-values of less than 0.05 were considered statistically significant.
Results
ONCOMINE cDNA microarray analysis
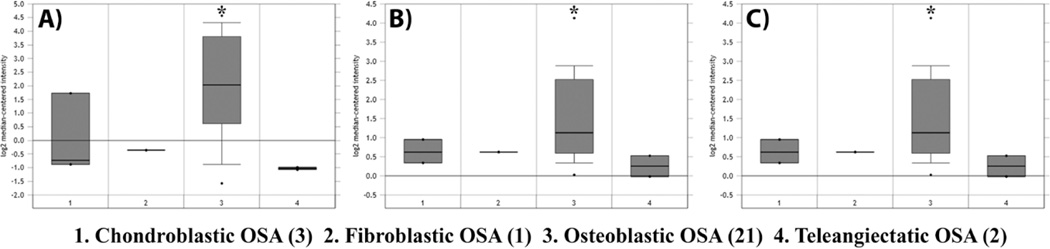
The OCy specific gene DMP1 has been previously reported to be expressed by OSA, suggesting that OCy may contribute to the development of OSA [Kashima et al., 2013]. DMP1 expression is characteristic of OCy [Bonewald, 2011]. Accordingly, the ONCOMINE microarray depository was queried for previous studies with sufficient data encompassing OSA patients. The Kobayashi sarcoma data set had the greatest number of patients for analysis (n = 27) and was investigated for significant expression differences in OCy markers [Kobayashi et al., 2010]. DMP1 was expressed in 0/6 non-osteoblastic OSA tumor samples, whereas 10/21 osteoblastic OSA had DMP1 overexpression (Figure 1; p<0.001). Osteoblastic OSA is the most common subtype of OSA, composing approximately 60% of all cases [Mutsaers et al., 2013]. DMP1 was found to have a gene rank of 17, signifying that there were only 16 other genes with more significant p-values in the data set (Figure 1A; Supplemental Figure 1). Significant overexpression of the OCy-associated genes matrix extracellular phosphoglycoprotein (MEPE), involved in integrin association; and phosphate-regulating neutral endopeptidase homolog x-linked (PHEX), involved in mineralization, were also observed (Figure 1; Supplemental Figure 1). Interestingly, alkaline phosphatase (ALPL), which has been a controversial prognostic factor in OSA biology [Bielack et al., 2009; Schmidt et al., 2013], was not significantly associated with osteoblastic OSA in this data set. When the dataset was analyzed for gene expression differences in the above noted genes based on age, sex, primary tumor location, metastasis at the time of diagnosis, or response to chemotherapy, there was no significant difference (p>0.05) associated with expression of OCy marker expression (data not shown).
Figure 1. Human patients with osteoblastic OSA have increased expression of osteocyte-specific genes.
The ONCOMINE database was searched for the term “osteosarcoma.” The Kobayashi sarcoma dataset was identified as having sufficient information for further analysis. OSA subtypes were analyzed for comparison of the OCy specific markers: (A) dentin matrix protein-1 (DMP1), (B) matrix extracellular phosphoglycoprotein (MEPE), and (C) phosphate-regulating neutral endopeptidase homolog x-linked (PHEX). (Heatmap of data present in Supplemental figure 1). Results are reported as log2 median-centered intensity as reported by ONCOMINE. *p < 0.05.
We subsequently screened the Baird (n = 5; [Baird et al., 2005]) and Henderson (n = 11; [Henderson et al., 2005]) data sets for expression of OCy markers in OSA patients (Supplementary Figure 1). The Henderson data set showed that DMP1 was significantly (p = 0.014) increased in OSA patients. Even though the Baird data set only had 5 OSA patients present, DMP1 was increased, though not significantly (p = 0.054). The Henderson dataset showed a significant increased expression of MEPE (p = 0.005) in OSA patients, but not PHEX. MEPE and PHEX were not present in the Baird dataset and thus could not be analyzed. OSA subtypes were not defined in either the Baird or Henderson datasets. Together, these data provide evidence of DMP1 overexpression in clinical OSA.
Expression of osteocyte markers in OSA cell lines
We next sought to determine if established OSA cell lines express markers known to be associated with OCy. RNA was isolated from murine, human, and canine OSA cell lines. cDNA was prepared and qPCR performed for OCy-specific markers and other proteins important in OSA biology (Supplemental Tables 1–3). β-actin, GAPDH, and β2-microglobulin were used as controls to ensure reaction activity. As a control for OCy, we utilized the well characterized murine OCy cell line MLO-Y4. DMP1 and podoplanin (Pdpn or e11/gp38) have been used to characterize OCy previously [Bonewald, 2011; Franz-Odendaal et al., 2006; Kashima et al., 2013; Woo et al., 2011; Zhang et al., 2006]. Murine OSA cell lines showed low expression of the OCy marker Dmp1 (Table 1). Pdpn (e11/gp38) is the earliest expressed gene in OCys and high expression was observed in all cell lines [Zhang et al., 2006]. The OCy related gene Phex had robust expression in all cell lines; however, Mepe was not observed in any of the cell lines. All cell lines expressed osteocalcin (Bglap) to various degrees yet K12 and K7M2 did not express Alpl. Osteocalcin expression has been shown to be associated with late osteoblast and early OCy differentiation [Lee et al., 2007]. Sclerostin (Sost) and receptor activator of nuclear factor kappa-B ligand (RANKL) are the primary mediators by which OCy mediate bone lysis [Bonewald, 2011]. RANKL activity is opposed by osteoprotegerin (OPG), a soluble inhibitor and decoy receptor for RANKL. MLO-Y4 is a known low expresser of Sost [Yu et al., 2011] even though mature OCy express Sost regularly [Wijenayaka et al., 2011]. Low expression of Sost by MLO-Y4 has been attributed to the early differentiated state of MLO-Y4 [Bonewald, 2011]. Sost was not expressed in MLO-Y4, as previously described, or DLM8. However, Sost was expressed in K12 and its more metastatic variant K7M2. Conversely, RANKL was expressed by to a greater degree by the Sost negative cell MLO-Y4 and DLM8, with lower expression in K12 and K7M2 cell lines. Bone sialoprotein (integrin binding sialoprotein; Ibsp) is a component of mineralized tissues and indicative of the differentiation of mature osteoblasts into OCy. DLM8 had the greatest expression of Ibsp while no other cell expressed Ibsp. Together, these data show the complex expression profile of OSA and the expression of various OCy markers by established murine OSA cell lines.
Table 1. Expression of osteocyte markers from murine OSA cell lines.
RNA was isolated from murine OSA cell lines, reverse transcribed, and assayed using qPCR. OSA marker expression was scored based on mean qPCR Ct values as follows: - >30; +, 25– 30; ++, 20–25; +++, <20.
| MLO-Y4 | DLM8 | K12 | K7M2 | |
|---|---|---|---|---|
| alpl | + + | + + | − | − |
| bglap | + + | + + + | + | + + |
| ibsp | − | + + | − | − |
| dmp1 | + | + | + | + |
| fgf23 | − | + | + | + |
| mepe | − | − | − | − |
| opg | + | + + | + + | + + |
| pdpn | + + + | + + | + + + | + + + |
| phex | + + | + | + | + + |
| sost | − | − | + | + |
| tnfsf11 | + + + | + | − | − |
| β-actin | + + + | + + + | + + + | + + + |
| gapdh | + + + | + + + | + + + | + + + |
We investigated the expression of OCy markers in the human OSA cell lines Saos2, MG63, and SJSA1 (Table 2). Saos2 has been previously described as more representative of osteoblastic OSA than MG63 and SJSA1, which are described as more fibroblastic, by ATCC (the American Type Culture Collection) and independent investigators [Mohseny et al., 2011; Pautke et al., 2004]. Saos2 was the only cell line to express DMP1. Since Saos2 is the only human cell line with an osteoblastic phenotype, these data are similar to the observations from the ONOCOMINE study. Saos2 expressed both MEPE and PHEX, similar to the results obtained from the murine OSA cell lines. However, MG63 and SJSA1 only expressed one of these markers each. PDPN was expressed by MG63 and SJSA1. Moderate expression of ALPL was present across all cell lines, but only low expression of BGLAP was observed in Saos2. IBSP was expressed in MG63 and SJSA1 but not in Saos2. The expression of ALPL, BGLAP, and IBSP, paint a complex picture concerning the derivation and OSA subtype as previously described [Pautke et al., 2004]. We observed that only Saos2 expressed RANKL, with none of the cell lines expressing SOST, differing from the mouse cell lines were cells predominantly expressed one or the other. Interestingly, OPG was highly expressed in MG63 and to a lesser degree in Saos2 and SJSA1.
Table 2. Expression of osteocyte markers from human OSA cell lines.
RNA was isolated from human OSA cell lines, reverse transcribed, and assayed using qPCR. OSA marker expression was scored based on mean qPCR Ct values as follows: - >30; +, 25– 30; ++, 20–25; +++, <20.
| MG63 | Saos2 | SJSA1 | |
|---|---|---|---|
| ALPL | + | + + | + + |
| BGLAP | − | + | − |
| IBSP | + | − | + |
| DMP1 | − | + | − |
| FGF23 | − | − | − |
| MEPE | − | + | − |
| OPG | + + + | + + | + |
| PDPN | + + | − | + + |
| PHEX | + | + | − |
| SOST | − | − | − |
| TNFSF11 | − | + | − |
| β-ACTIN | + + + | + + + | + + + |
| GAPDH | + + + | + + + | + + + |
Dogs have a relatively high incidence of OSA compared to humans, and in many ways are uniquely suited for the study of OSA due to high incidence of disease and similar pathology [Fan, 2010; Schmidt et al., 2013]. Screening of numerous canine OSA cell lines show similar trends to those observed in the murine and human OSA cell lines; expression of Dmp1, Pdpn, and Bglap with mixed expression of Alpl (Table 3). Expression of Fgf23, Phex, and RANKL was relatively low across the cell lines queried. Taken together, these results suggest that OCy are an additional cell type for the origination of OSA across species.
Table 3. Expression of osteocyte markers from canine OSA cell lines.
RNA was isolated from canine OSA cell lines, reverse transcribed, and assayed using qPCR. OSA marker expression was scored based on mean qPCR Ct values as follows: - >30; +, 25–30; ++, 20–25; +++, <20.
| Abrams | D17 | Gracie | HMPOS | HOS | Moresco | OOS | OSCA40 | POS | Vogel | |
|---|---|---|---|---|---|---|---|---|---|---|
| Alpl | − | − | − | + + | − | − | − | − | + + | − |
| Bglap | + | + | + | + | + | + | + | + | + | + |
| Dmp1 | + | + | + | − | + | + | + | + | − | + |
| Fgf23 | − | + | − | − | − | − | − | + | − | − |
| Pdpn | + | + | + + | + | + + | + + | − | + + | + | + + |
| Phex | + | − | − | − | + | + | − | + | − | − |
| Tnfsf11 | − | − | − | − | − | + | + | − | + | + |
| β-actin | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + |
| β2-microglobulin | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + | + + + |
Protein expression of osteocyte markers
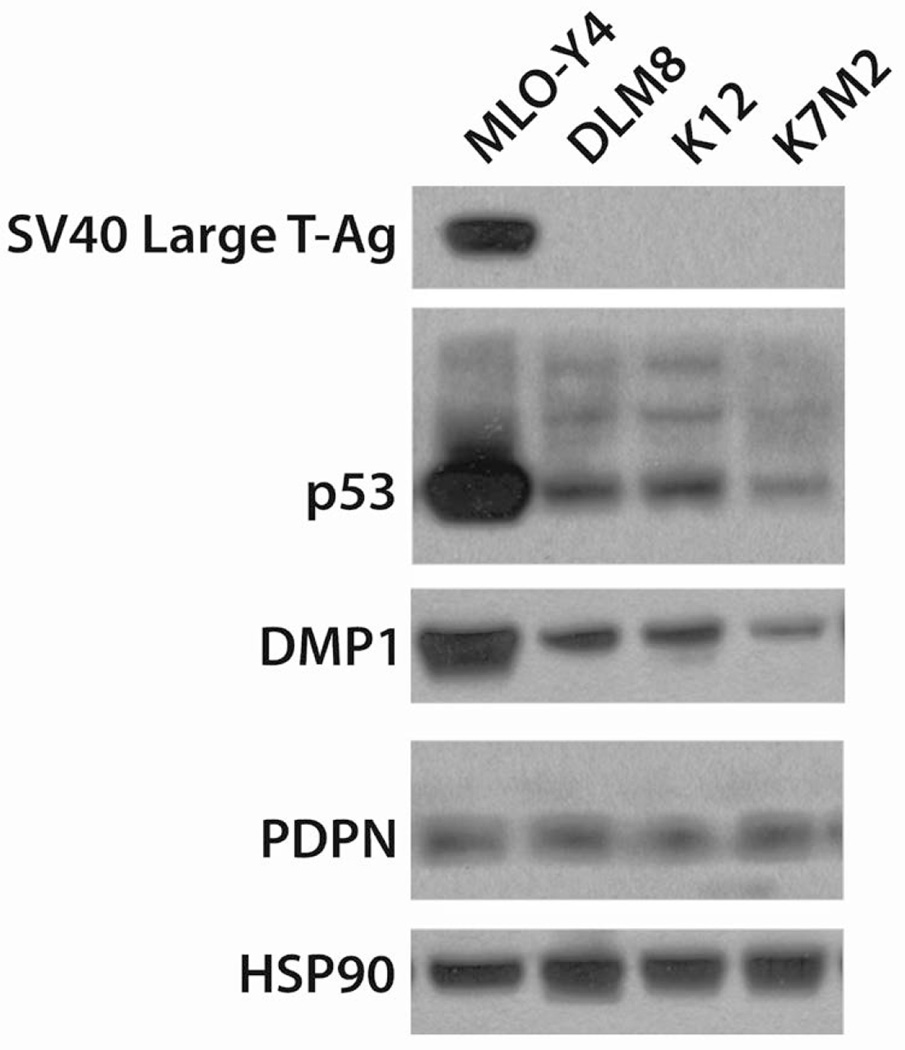
To confirm that the qPCR results for the OCy-specific markers reflected actual protein production, whole cell lysates were created from near confluent murine OSA cell lines. Since MLO-Y4 was created by expressing SV40-transduced large T antigen under a rat osteocalcin promoter, we investigated SV40 large T-antigen expression and observed that only MLO-Y4 expresses SV40 large T Antigen (Figure 2) as previously described [Kato et al., 1997]. MLO-Y4 had an overexpression of p53, likely due to SV40 expression and transformation. However, p53 is a known driver of OSA, and all murine cell lines were found to express p53 (Figure 2) [Berman et al., 2008].
Figure 2. Protein expression of osteocyte specific genes from murine OSA cell lines.
Whole cell lysates from murine OSA cell lines and MLO-Y4 OCys were examined by western blot. Since MLO-Y4 is SV40 large T-antigen transduced, we probed for SV40 large T antigen and p53. To determine the expression of OCy-specific markers, DMP1 and PDPN were examined. HSP90 was used as a loading control.
Next we sought to validate the expression of OCy markers by protein expression as we previously demonstrated by RNA expression. Indeed, all cell lines expressed PDPN to varying degrees confirming the qPCR results (Figure 2). DMP1 was expressed in all cell lines investigated, though with greater variability than observed by qPCR (Figure 2). Protein expression of DMP1 and PDPN is characteristic of OCy. HSP90 was used as a loading control. These data show that murine OSA cell lines express markers characteristic of the OCy phenotype.
Characterization of the tumorogenicity of MLO-Y4 cells
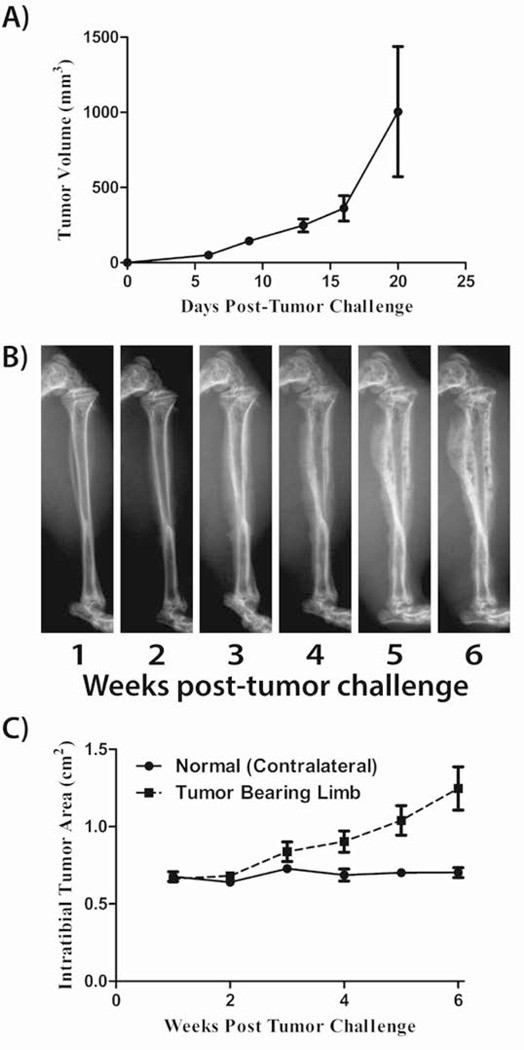
Transduction of cells with SV-40 large T antigen has been used to create multiple models of various tumor types, including OSA, breast, and prostate cancer, [Atkin et al., 2009; Pipas, 2009]. Thus we determined if large T antigen conferred an OSA phenotype on MLO-Y4 cells. Initially, to determine if MLO-Y4 cells are tumorigenic, 1×106 cells were implanted subcutaneously (SQ) in SCID mice. MLO-Y4 cells produced SQ tumors with a doubling time of 3.2 days (r2=0.523; Figure 3A). We subsequently injected mice intratibially (IT) with the MLO-Y4 cells to recapitulate the orthotopic environment of OSA. Orthotopic tumors developed and induced radiographically-defined mixed osteoblastic/osteolytic lesions similar to that observed in OSA patients (Figure 3B). Tumor burden was semi-quantified by measuring the radiographic area of the tumor from weekly radiographs as previously described for intraosseous tumors [Yin et al., 1999]. Orthotopic tumors were observed to have a doubling time of 36.3 days (r2=0.605; Figure 3C). Mice were euthanized 6 weeks after challenge due to primary tumor size. Gross metastases were not observed.
Figure 3. MLO-Y4 are tumorigenic in SCID mice.
MLO-Y4 were injected subcutaneously (SQ) and orthotopically (intratibial; IT). A) Caliper measurements of SQ tumors were used to determine tumor volume as measured twice weekly. B) Weekly radiographs of IT tumors of a representative mouse are shown. C) Measurement of cross-sectional tumor area measured from radiographs of IT tumors.
Histologic analysis of MLO-Y4 tumors
To examine the histologic features of the resulting MLO-Y4 tumors, SQ and orthotopic tumors were formalin fixed and paraffin embedded. Similar findings for SQ and orthotopic tumors were observed. Multiple organs from mice with orthotopic tumors were evaluated by conventional H&E histology to investigate metastatic tumor development; however no metastases were observed in the lung, liver, or spleen. H&E stained sections from primary tumors demonstrated a predominantly high grade spindle cell malignant neoplasm with cells demonstrating coarse chromatin and increased mitotic activity. However, no obvious osteoid production was identified in these tumors.
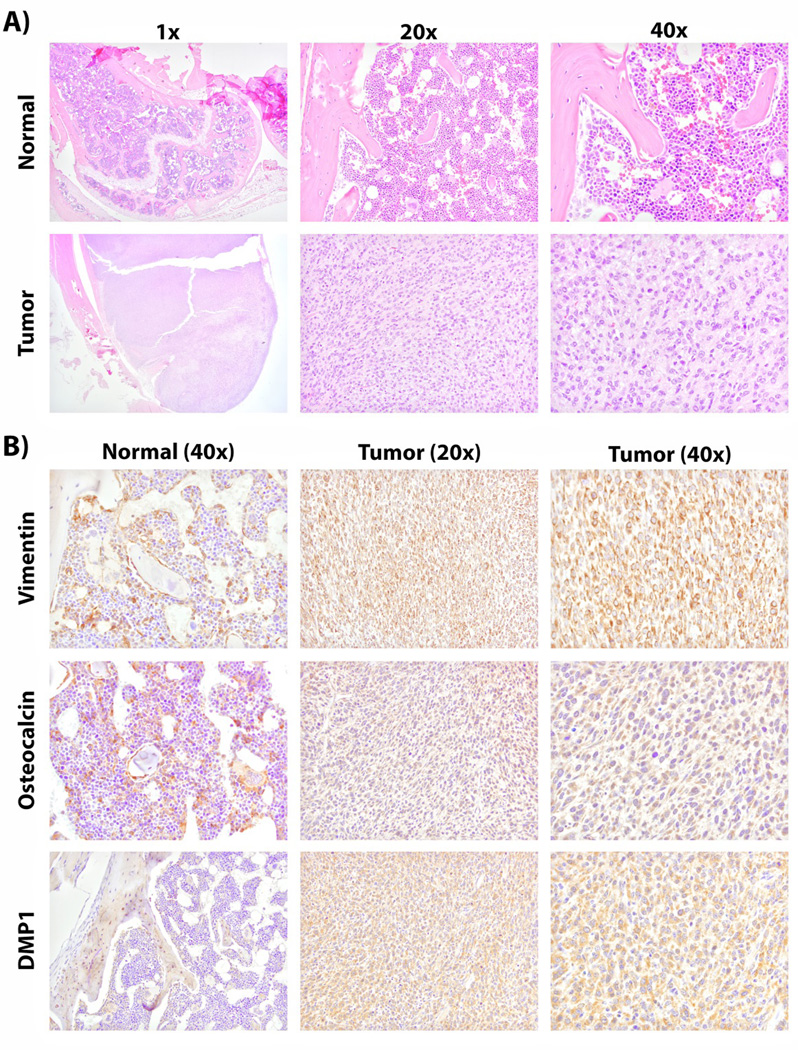
To further histologically characterize the tumor tissue of origin, immunohistochemistry for vimentin and pan-cytokeratin was performed. Vimentin is a marker of mesenchymal cells; whereas, pan-cytokeratin is attributed to epithelial-derived cells. Vimentin expression has previously been associated with OSA [Barger et al., 2005]. In this study, normal bone marrow and osteoblasts were found to positively express vimentin. Tumors resulting from MLO-Y4 injection were found to have strong cytoplasmic vimentin expression suggestive of their mesenchymal differentiation (Figure 4). However, pan-cytokeratin expression in tumor was negative (data not shown). These observations identify that MLO-Y4 tumors have a mesenchymal phenotype consistent with what is expected for OSA.
Figure 4. Immunohistochemistry of osteocyte specific markers in MLO-Y4 tumors.
Mice were injected with 1×106 MLO-Y4 cells orthotopically, in the tibia. After 6 weeks, mice were euthanized and tumor was formalin fixed and paraffin embedded. Immunohistochemical analysis of MLO-Y4 tumors and contralateral normal limbs was performed. A) Tumor was histologically assessed by H&E staining. B) Further characterization of the tumor was achieved by staining for vimentin (mesenchymal phenotype), osteocalcin (late osteoblast/early OCy) and DMP1 (OCy origin).
To characterize the bone cell of origin for the MLO-Y4 tumors immunohistochemistry for osteocalcin and DMP1 was performed. Osteocalcin staining was observed in cells of the osteoblast lineage of normal bone (Figure 4). Even though osteocalcin has been observed to be highly expressed in MLO-Y4 cells in culture [Qin et al., 2007], we observed only moderate staining in tumor sections (Figure 4). DMP1 was found to characteristically stain matrix surrounding OCys localized to the lacunar canalicular system of normal bone as previously described (Figure 4) [Kashima et al., 2013]. DMP1 was found to be strongly expressed in the cytosol of tumor cells. DMP1 is known to be secreted and act as a positive factor in mineralization [Qin et al., 2007]. Thus, these data demonstrate that MLO-Y4 tumors express (1) a mesenchymal phenotype (2) a late osteoblast/early OCy marker (osteocalcin) and (3) the OCy-specific marker DMP1. Taken together, these data indicate that OCy can serve as an OSA progenitor.
Discussion
In this study, we provide both expression and functional evidence that OCys can serve as a cell of origin of OSA. Additionally we have shown that a transformed OCy cell line, MLO-Y4, is tumorigenic in mice. The transition from osteoblast to OCy is a complex process with ill-defined endpoints [Woo et al., 2011]. For example, the expression of ALP and OC are common to both cell types, with ALP declining during the transition and OC increasing. It is during this time that markers characteristic of osteocytes, such as DMP1, PDPN, and MEPE begin to be expressed. Expression of PDPN and DMP1 have been described as the earliest markers associated with OCy [Bonewald, 2011; Franz-Odendaal et al., 2006; Kashima et al., 2013; Woo et al., 2011; Zhang et al., 2006]. OCy further differentiate leading to expression of markers such as RANKL, FGF23, and SOST [Bonewald, 2011; Franz-Odendaal et al., 2006]. The OCy cell line MLO-Y4 expresses DMP1, PDPN, Phex, and RANKL, consistent with previous reports describing these cells as OCy [Bonewald, 2011; Franz-Odendaal et al., 2006; Kashima et al., 2013; Woo et al., 2011; Zhang et al., 2006] Many established OSA cell lines investigated also express these markers. Data presented herein support OCy as a progenitor cell of OSA in addition to osteoblasts.
It is likely that transformation of MLO-Y4 occurred due to the introduction of SV40 large T-antigen during development of the cell line. SV40 large T antigen can transform cells by inactivating p53 and retinoblastoma (pRb); both molecules are critical in promoting apoptosis [Lane and Crawford, 1979; Levine and Oren, 2009]. SV40 based transformation has not only been used as a mechanism to promote murine models of OSA [Jones, 2011; Knowles et al., 1990; Marton et al., 2000], but SV40 DNA has also been observed in human patients with OSA [Lednicky et al., 1997]. Mutations and inactivations of p53 and pRb are known drivers of OSA [Berman et al., 2008; Walkley et al., 2008]. This does not diminish the usefulness of MLO-Y4 as a tool for the study of OCy biology, but does provide an additional caveat for its use. It is not uncommon for tumor cell lines to provide the basis for understanding physiologic processes. For example, the human OSA cell line Saos2 is widely utilized for the study osteoblast biology with MG63 to a lesser degree [Czekanska et al., 2012].
A majority of patients diagnosed with OSA have metastases at the time of diagnosis, with the majority being lung metastases. We investigated if MLO-Y4 is capable of producing spontaneous metastases from an orthotopic location. However, no metastases were observed in the lung, liver, or spleen of these mice. Even though metastases significantly increase the usefulness of a model, many OSA cell lines have required selection of metastatic variants to increase metastatic ability of weakly metastatic cell lines [Asai et al., 1998; Barroga et al., 1999; Khanna et al., 2000; Kimura et al., 2002; Su et al., 2009]. Furthermore, it is possible in the murine model, that the primary tumor grows rapidly, not allowing time for metastases to manifest as the mouse must be euthanized due to the large primary tumor. It may also be possible to develop metastatic variants of MLO-Y4 by selecting lung metastases after intravenous challenge similarly to the murine OSA cell lines DLM8 and K7M2 [Asai et al., 1998; Khanna et al., 2000]. We observed expression of PDPN, an early OCy marker, in a majority of the OSA cell lines studied confirming previous reports that PDPN is aberrantly expressed in OSA [Kunita et al., 2011]. PDPN is known to also play a role in platelet aggregation and has been proposed as a plausible target to inhibit OSA metastases [Kunita et al., 2011].
To investigate if the OCy markers DMP1, MEPE, and PHEX were associated with metastasis, we performed a post-hoc analysis of the Kobayashi sarcoma dataset from ONCOMINE [Kobayashi et al., 2010]. No significant association was present. Even though the Kobayashi dataset was the largest OSA dataset present, it is still relatively small with only 27 patients analyzed. A lack of statistical power may have led to the non-significant result. It is also plausible that the OCy factors DMP1, MEPE, and PHEX are not prognostic or associated with the development of metastases. Expression of these proteins by OSA tumors and cell lines suggests OCy as an ancestral cell to the resulting tumor. Investigation of human cell lines also described osteoblastic OSA (Saos2) as expressing DMP1; whereas, those described as fibroblastic OSA (MG63 and SJSA1) did not express DMP1. Results obtained from the Baird and Henderson data sets provide further evidence of DMP1 overexpression in OSA [Baird et al., 2005; Henderson et al., 2005]. These data correlate with the patient data suggesting DMP1 is expressed predominantly by osteoblastic OSA and are important in describing the etiology of OSA. DMP1, MEPE, and PHEX belong to the SIBLING (small integrin-binding ligand N-linked glycoproteins) family of proteins. The SIBLING family members osteopontin (OP) and IBSP have previously associated with a metastatic phenotype [Bellahcene et al., 2008]. Association of SIBLING family members in epithelial derived tumors and osteotropic metastatic disease has also been observed [Bellahcene et al., 2008]. Therefore, further investigation is necessary to determine if DMP1, MEPE, and PHEX play a role in OSA metastasis or may be of use as prognostic factors.
OSA typically forms mixed osteoblastic/osteolytic lesions. We observed these changes on radiographs but did not observe osteoid production in subcutaneous or orthotopic tumors. It is unclear why these differences persist, but it is plausible that these are primarily lytic lesions masked by the normal bone architecture. OCys do not secrete components of extracellular matrix, but do secrete molecules to mineralize ECM. It is possible that OCy are capable of mineralizing ECM, but other cells must be present to produce the more immature bone matrix. There is little evidence, most of it contradictory, concerning the ability of OCys to create or resorb bone on their own [Wysolmerski, 2013]. OCys are unique bone cells whose primary role is the sensation of physical forces in the bone microenvironment and conversion of those signals into biochemical signals. OCys coordinate osteoblastic and osteolytic responses. Expression of OPG, RANKL, and SOST are the primary mediators used to activate osteoblasts or osteoclasts, respectively. We observed mixed expression of RANKL, OPG, and SOST from OSA cell lines, but observed cells expressing either RANKL or SOST. All murine OSA cell lines expressed OPG. Therefore, the potential for antagonistic responses from OCy are plausible dependent on the microenvironment. Nevertheless, it is plausible that the mixed lesions characteristic of OSA are due to the underlying OCy nature of the cells driving these tumors. OCys provide a mechanism by which local alterations in the microenvironment lead to osteoblastic or osteolytic regions. Since OCy coordinate osteoblastic/osteolytic responses physiologically, it is plausible that a tumor composed of, or derived from, OCy would be capable of employing both arms of bone maintenance resulting in characteristically mixed lesions.
The observations made herein incite new questions concerning the etiology of OSA. The development of tumor from what are believed to be terminally differentiated cells is important as these cells should not be proliferative. Further investigation concerning the importance of OCy specific factors to OSA progression and metastasis must also be investigated. Recently, Kashima et al. published data showing that DMP1 is specifically expressed by OSA [Kashima et al., 2013]. Their immunohistochemical investigation of a large sample size of primary bone tumors supports our conclusion that osteoblastic OSA specifically expresses DMP1 and that OCy may serve as a progenitor cell for OSA. These observations suggest that OCy may act as a cell of origin for OSA in addition to osteoblast. Analysis of the Kobayashi data set depicted 10 of 21 osteoblastic OSA patients expressing DMP1 [Kobayashi et al., 2010]. Targeted inactivation of p53 and pRb has previously been shown to promote OSA [Berman et al., 2008; Walkley et al., 2008]. Therefore, it is plausible that OCy are a novel cell of origin for OSA in addition to osteoblasts. Further investigation is necessary to determine how the cell of origin alters metastasis and potentially long term survival.
The observation that OSA develops from OCy may provide novel methods for clinical intervention. By better understanding OCy, and how they react to their environment, we may be able to develop novel methods of inhibiting OSA growth and metastasis. OCy biology has not been as elucidated in the same detail as osteoblasts and osteoclasts even though OCy comprise a much larger percentage of bone. Ongoing studies to explore the functional relationship of OCy with other cells in the bone microenvironment may lead to novel pathways and targets for OSA. Understanding that OCys may give rise to OSA is of importance for understanding the underlying tumor biology of OSA, and discovering new methodologies than can be exploited to inhibit this aggressive bone disease.
Supplementary Material
Acknowledgments
We would like to thank Dr. Lynda Bonewald at the University of Missouri-Kansas City for providing the MLO-Y4 cell line. We would also like to thank Dr. Douglas Thamm at Colorado State University and Dr. Thomas Rosol at Ohio State University for kindly providing the canine osteosarcoma cell lines utilized in the study. Thank you to Dr. Ken Pienta (Johns Hopkins) for providing SCID mice for experiments.
Funding
This work was supported by a Department of Defense Prostate Cancer Research Program Training Award (W81XWH-12-1-0172) to J.S. and a prostate cancer program project at the University of Michigan (P01 CA093900/CA/NCI NIH HHS/United States).
List of abbreviations
- ALPL
Alkaline phosphatase
- DMP1
Dentin matrix phosphoprotein 1
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- HSP90
Heat shock protein 90
- IBSP
Integrin binding sialoprotein; bone sialoprotein
- IT
Intratibial
- MEPE
Matrix extracellular phosphoglycoprotein
- OC
Osteocalcin
- Bglap
bone gamma-carboxyglutamic acid-containing protein
- OCy
Osteocyte
- OP
Osteopontin
- OPG
Osteoprotegerin
- OSA
Osteosarcoma
- PDPN
Podoplanin; e11/gp38
- PHEX
Phosphate-regulating neutral endopeptidase homolog x-linked
- RANKL
Receptor activator of nuclear factor kappa-B ligand
- SCID
Severe combined immunodeficiency disorder
- SIBLING
Small integrin-binding ligand N-linked glycoproteins
- SOST
Sclerostin
- SQ
Subcutaneous
Footnotes
Statement of author contributions
JS, RM, and EK were involved in study design. All authors were involved in data analysis and/or data interpretation. All authors participated in writing the manuscript and had final approval of the submitted version.
References
- Asai T, Ueda T, Itoh K, Yoshioka K, Aoki Y, Mori S, Yoshikawa H. Establishment and characterization of a murine osteosarcoma cell line (LM8) with high metastatic potential to the lung. International journal of cancer. Journal international du cancer. 1998;76:418–422. doi: 10.1002/(sici)1097-0215(19980504)76:3<418::aid-ijc21>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Atkin SJ, Griffin BE, Dilworth SM. Polyoma virus and simian virus 40 as cancer models: history and perspectives. Seminars in cancer biology. 2009;19:211–217. doi: 10.1016/j.semcancer.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer research. 2005;65:9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- Barger A, Graca R, Bailey K, Messick J, de Lorimier LP, Fan T, Hoffmann W. Use of alkaline phosphatase staining to differentiate canine osteosarcoma from other vimentin-positive tumors. Veterinary pathology. 2005;42:161–165. doi: 10.1354/vp.42-2-161. [DOI] [PubMed] [Google Scholar]
- Barroga EF, Kadosawa T, Okumura M, Fujinaga T. Establishment and characterization of the growth and pulmonary metastasis of a highly lung metastasizing cell line from canine osteosarcoma in nude mice. The Journal of veterinary medical science / the Japanese Society of Veterinary Science. 1999;61:361–367. doi: 10.1292/jvms.61.361. [DOI] [PubMed] [Google Scholar]
- Bellahcene A, Castronovo V, Ogbureke KU, Fisher LW, Fedarko NS. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): multifunctional proteins in cancer. Nature reviews. Cancer. 2008;8:212–226. doi: 10.1038/nrc2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein ML, Mukherjee S, Lees JA. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielack S, Carrle D, Casali PG. Osteosarcoma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2009;4(20 Suppl):137–139. doi: 10.1093/annonc/mdp154. [DOI] [PubMed] [Google Scholar]
- Bielack SS, Carrle D, Hardes J, Schuck A, Paulussen M. Bone tumors in adolescents and young adults. Current treatment options in oncology. 2008;9:67–80. doi: 10.1007/s11864-008-0057-1. [DOI] [PubMed] [Google Scholar]
- Bonewald LF. The amazing osteocyte. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Zhou Z, de Crombrugghe B, Nakashima K, Guan H, Duan X, Jia SF, Kleinerman ES. Osterix, a transcription factor for osteoblast differentiation, mediates antitumor activity in murine osteosarcoma. Cancer research. 2005;65:1124–1128. doi: 10.1158/0008-5472.CAN-04-2128. [DOI] [PubMed] [Google Scholar]
- Czekanska EM, Stoddart MJ, Richards RG, Hayes JS. In search of an osteoblast cell model for in vitro research. European cells & materials. 2012;24:1–17. doi: 10.22203/ecm.v024a01. [DOI] [PubMed] [Google Scholar]
- Fan TM. Animal models of osteosarcoma. Expert review of anticancer therapy. 2010;10:1327–1338. doi: 10.1586/era.10.107. [DOI] [PubMed] [Google Scholar]
- Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. Journal of the National Cancer Institute. 1977;59:221–216. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- Franz-Odendaal TA, Hall BK, Witten PE. Buried alive: how osteoblasts become osteocytes. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:176–190. doi: 10.1002/dvdy.20603. [DOI] [PubMed] [Google Scholar]
- Henderson SR, Guiliano D, Presneau N, McLean S, Frow R, Vujovic S, Anderson J, Sebire N, Whelan J, Athanasou N, Flanagan AM, Boshoff C. A molecular map of mesenchymal tumors. Genome biology. 2005;6:R76. doi: 10.1186/gb-2005-6-9-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KB. Osteosarcomagenesis: modeling cancer initiation in the mouse. Sarcoma. 2011;2011:694136. doi: 10.1155/2011/694136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima TG, Dongre A, Oppermann U, Athanasou NA. Dentine matrix protein 1 (DMP-1) is a marker of bone-forming tumours. Virchows Archiv : an international journal of pathology. 2013;462:583–591. doi: 10.1007/s00428-013-1399-z. [DOI] [PubMed] [Google Scholar]
- Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an osteocyte-like cell line, MLO-Y4. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 1997;12:2014–2023. doi: 10.1359/jbmr.1997.12.12.2014. [DOI] [PubMed] [Google Scholar]
- Khanna C, Prehn J, Yeung C, Caylor J, Tsokos M, Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clinical & experimental metastasis. 2000;18:261–271. doi: 10.1023/a:1006767007547. [DOI] [PubMed] [Google Scholar]
- Kimura K, Nakano T, Park YB, Tani M, Tsuda H, Beppu Y, Moriya H, Yokota J. Establishment of human osteosarcoma cell lines with high metastatic potential to lungs and their utilities for therapeutic studies on metastatic osteosarcoma. Clinical & experimental metastasis. 2002;19:477–485. doi: 10.1023/a:1020395816633. [DOI] [PubMed] [Google Scholar]
- Knowles BB, McCarrick J, Fox N, Solter D, Damjanov I. Osteosarcomas in transgenic mice expressing an alpha-amylase-SV40 T-antigen hybrid gene. The American journal of pathology. 1990;137:259–262. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A, Tochigi N, Morioka H, Toyama Y, Hirohashi S, Kawai A, Yamada T. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Molecular cancer therapeutics. 2010;9:535–544. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- Kunita A, Kashima TG, Ohazama A, Grigoriadis AE, Fukayama M. Podoplanin is regulated by AP-1 and promotes platelet aggregation and cell migration in osteosarcoma. The American journal of pathology. 2011;179:1041–1049. doi: 10.1016/j.ajpath.2011.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Lednicky JA, Stewart AR, Jenkins JJ, 3rd, Finegold MJ, Butel JS. SV40 DNA in human osteosarcomas shows sequence variation among T-antigen genes. International journal of cancer. Journal international du cancer. 1997;72:791–800. doi: 10.1002/(sici)1097-0215(19970904)72:5<791::aid-ijc15>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nature reviews. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton I, Johnson SE, Damjanov I, Currier KS, Sundberg JP, Knowles BB. Expression and immune recognition of SV40 Tag in transgenic mice that develop metastatic osteosarcomas. Transgenic research. 2000;9:115–125. doi: 10.1023/a:1008939620322. [DOI] [PubMed] [Google Scholar]
- Mohseny AB, Machado I, Cai Y, Schaefer KL, Serra M, Hogendoorn PC, Llombart-Bosch A, Cleton-Jansen AM. Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Laboratory investigation; a journal of technical methods and pathology. 2011;91:1195–1205. doi: 10.1038/labinvest.2011.72. [DOI] [PubMed] [Google Scholar]
- Mutsaers AJ, Ng AJ, Baker EK, Russell MR, Chalk AM, Wall M, Liddicoat BJ, Ho PW, Slavin JL, Goradia A, Martin TJ, Purton LE, Dickins RA, Walkley CR. Modeling distinct osteosarcoma subtypes in vivo using Cre:lox and lineage-restricted transgenic shRNA. Bone. 2013;55:166–178. doi: 10.1016/j.bone.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Patane S, Avnet S, Coltella N, Costa B, Sponza S, Olivero M, Vigna E, Naldini L, Baldini N, Ferracini R, Corso S, Giordano S, Comoglio PM, Di Renzo MF. MET overexpression turns human primary osteoblasts into osteosarcomas. Cancer research. 2006;66:4750–4757. doi: 10.1158/0008-5472.CAN-05-4422. [DOI] [PubMed] [Google Scholar]
- Pautke C, Schieker M, Tischer T, Kolk A, Neth P, Mutschler W, Milz S. Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 OS in comparison to human osteoblasts. Anticancer research. 2004;24:3743–3748. [PubMed] [Google Scholar]
- Pipas JM. SV40: Cell transformation and tumorigenesis. Virology. 2009;384:294–303. doi: 10.1016/j.virol.2008.11.024. [DOI] [PubMed] [Google Scholar]
- Qin C, D'Souza R, Feng JQ. Dentin matrix protein 1 (DMP1): new and important roles for biomineralization and phosphate homeostasis. Journal of dental research. 2007;86:1134–1141. doi: 10.1177/154405910708601202. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodan SB, Imai Y, Thiede MA, Wesolowski G, Thompson D, Bar-Shavit Z, Shull S, Mann K, Rodan GA. Characterization of a human osteosarcoma cell line (Saos-2) with osteoblastic properties. Cancer research. 1987;47:4961–4966. [PubMed] [Google Scholar]
- Schmidt AF, Nielen M, Klungel OH, Hoes AW, de Boer A, Groenwold RH, Kirpensteijn J. Prognostic factors of early metastasis and mortality in dogs with appendicular osteosarcoma after receiving surgery: An individual patient data meta-analysis. Preventive veterinary medicine. 2013 doi: 10.1016/j.prevetmed.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Sottnik JL, Daignault-Newton S, Zhang X, Morrissey C, Hussain MH, Keller ET, Hall CL. Integrin alpha2beta 1 (alpha 2beta 1) promotes prostate cancer skeletal metastasis. Clinical & experimental metastasis. 2013;30:569–578. doi: 10.1007/s10585-012-9561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottnik JL, Duval DL, Ehrhart EJ, Thamm DH. An orthotopic, postsurgical model of luciferase transfected murine osteosarcoma with spontaneous metastasis. Clinical & experimental metastasis. 2010;27:151–160. doi: 10.1007/s10585-010-9318-z. [DOI] [PubMed] [Google Scholar]
- Su Y, Luo X, He BC, Wang Y, Chen L, Zuo GW, Liu B, Bi Y, Huang J, Zhu GH, He Y, Kang Q, Luo J, Shen J, Chen J, Jin X, Haydon RC, He TC, Luu HH. Establishment and characterization of a new highly metastatic human osteosarcoma cell line. Clinical & experimental metastasis. 2009;26:599–610. doi: 10.1007/s10585-009-9259-6. [DOI] [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, Alt FW, McMahon AP, Orkin SH. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes & development. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijenayaka AR, Kogawa M, Lim HP, Bonewald LF, Findlay DM, Atkins GJ. Sclerostin stimulates osteocyte support of osteoclast activity by a RANKL-dependent pathway. PloS one. 2011;6:e25900. doi: 10.1371/journal.pone.0025900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SM, Rosser J, Dusevich V, Kalajzic I, Bonewald LF. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2011;26:2634–2646. doi: 10.1002/jbmr.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysolmerski JJ. Osteocytes remove and replace perilacunar mineral during reproductive cycles. Bone. 2013;54:230–236. doi: 10.1016/j.bone.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. The Journal of clinical investigation. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, van der Valk M, Cao J, Han CY, Juan T, Bass MB, Deshpande C, Damore MA, Stanton R, Babij P. Sclerostin expression is induced by BMPs in human Saos-2 osteosarcoma cells but not via direct effects on the sclerostin gene promoter or ECR5 element. Bone. 2011;49:1131–1140. doi: 10.1016/j.bone.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Zhang K, Barragan-Adjemian C, Ye L, Kotha S, Dallas M, Lu Y, Zhao S, Harris M, Harris SE, Feng JQ, Bonewald LF. E11/gp38 selective expression in osteocytes: regulation by mechanical strain and role in dendrite elongation. Molecular and cellular biology. 2006;26:4539–4552. doi: 10.1128/MCB.02120-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, McManus MM, Hughes DP. Understanding the Biology of Bone Sarcoma from Early Initiating Events through Late Events in Metastasis and Disease Progression. Frontiers in oncology. 2013;3:230. doi: 10.3389/fonc.2013.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.