Abstract
For successful gene delivery, plasmid DNA must be able to access the nucleus in order to be transcribed. Numerous studies have shown that gene delivery occurs more readily in dividing cells, which is attributed to increased nuclear access when the nuclear envelope disassembles during mitosis; however, nonviral carriers continue to have low transfection efficiencies and require large quantities of DNA per cell to achieve reasonable gene transfer, even in dividing cells. Therefore, we hypothesized that using histone-derived nuclear localization sequences (NLS)s to target polyplexes might enhance nuclear delivery by facilitating interactions with histone effectors that mediate nuclear partitioning and retention during mitosis. We discovered a novel interaction between polyplexes linked to histone 3 (H3) N-terminal tail peptides and the histone nuclear import protein importin-4, as evidenced by strong spatial colocalization as well as significantly decreased transfection when importin-4 expression was reduced. A fraction of the histone-targeted polyplexes was also found to colocalize with the retrotranslocon of the endoplasmic reticulum, Sec61. Super resolution microscopy demonstrated a high level of polyplex binding to chromatin post-mitosis, and there also was a significant decrease in the amount of chromatin binding following importin-4 knockdown. These results provide evidence that natural histone effectors mediate both nuclear entry and deposition on chromatin by histone-targeted polyplexes, and a translocation event from the endoplasmic reticulum into the cytosol may occur before mitosis to enable the polyplexes to interact with these essential cytoplasmic proteins.
Keywords: nuclear delivery, importin-4, histone polyplexes, non-viral gene delivery, chromatin
Introduction
Gene therapy has the potential to treat a variety of inherited and acquired diseases. Nonviral vehicles have attracted particular interest because of their relative safety and tailorability. However, nonviral vehicles such as polyplexes and lipoplexes have significant efficacy issues, with a limited capacity to reach the nucleus. In particular, gene delivery in non-dividing cells is typically highly limited, and requires a carrier able to provide active transport through the nuclear pore complex (NPC). Transfections work significantly better in dividing populations of cells in which the nuclear envelope disassembles during mitosis, thus largely eliminating the barrier.1-2 However, nonviral vectors continue to be less efficient compared to viral vectors, and the amount of DNA that gains access to the nucleus is small, regardless of how it is delivered or whether the cells are actively dividing. It is therefore of critical importance to study the nanostructure design features governing transport of DNA into the nucleus during mitosis, and an understanding of these mechanisms is key to improving non-viral delivery strategies.3
In nature, NLSs shuttle proteins and RNA into and out of the nucleus by binding to nuclear transport proteins such as importins, which engage complementary NLSs in order to shuttle bound cargoes through the NPC.4 Two of the best characterized NLSs are the NLS from SV40 large T antigen (PKKKRKV) and the bipartite NLS (KKKX5-20RK).5 Several groups have reported that the attachment of these sequences, or related localization signals, can enhance nuclear uptake of DNA carriers and other nanostructures.6-9 In non-dividing cells, DNA nuclear import requires an NLS.10 However, there have been conflicting reports about whether such signals are necessary for transfection in dividing cells. Multiple studies have been performed using nuclear chaperones,11 transcription factors,12 or chromatin interacting proteins13 as vehicles or ligands to improve DNA delivery in mitotic cells, and many of these reports indicate that NLSs can enhance gene transfer in these cell populations, potentially by engaging the importin-mediated import machinery. There also have been studies where NLSs were added to nanostructures and the addition of the NLSs did not increase transfection in dividing cells. For example, the localization sequences derived from the yeast transcriptional activator GAL4 and the SV40 large T antigen, which normally function by interacting with importins, were unable to enhance NPC-mediated nuclear import by DNA plasmids or polyplexes, respectively.14-15 This lack of activity may be due to the fact that the nanocarriers were unable to present their NLSs in an orientation accessible to importins for nuclear delivery, or that other aspects of nanocarrier size, shape, or structure prevented effective interactions with the nuclear import machinery. Alternatively, some investigators have proposed that NLSs are unnecessary for nuclear delivery in dividing populations of cells, and that nanocarriers are retained in the nucleus following mitosis in a more passive or probabilistic manner.16-17
Our previous studies examined the effects of incorporating a specific NLS, derived from the N-terminal tail of histone H3, into polyethylenimine (PEI) polyplexes to determine whether H3-targeting enhanced nuclear accumulation and improved gene transfer efficiency by engaging H3 effectors responsible for vesicular transport and nuclear processing. The H3-targeted polyplexes were biocompatible and improved post-mitotic nuclear retention2 and gene expression18 significantly as compared with untargeted polyplexes. In particular, we demonstrated that incorporation of the H3 peptides improved gene transfection by engaging the histone effector H3K4 methyltransferase to enhance endocytic trafficking via caveolae-mediated endocytosis and retrograde routing through the Golgi and endoplasmic reticulum (ER).19 The H3-targeted polyplexes accumulated in the ER prior to mitosis, and the ER-localized population of polyplexes redistributed into the nucleus following cell division. In nature, histones are also transferred through the ER and cytoplasm during synthesis and post-translational processing as well as during cellular division.20 Therefore, we questioned whether the H3-targeted polyplexes were interacting with histone chaperones or cytoplasmic proteins to gain access to the nucleus.
Core histones, consisting of histones H2A, H2B, H3, and H4, comprise the primary protein component of chromatin. These proteins come together to form the nucleosome when combined with chromosomal DNA. Histones H3 and H4 each contain an NLS in their amino-terminal domains, and these N-terminal domains were found to be essential for the nuclear accumulation of these proteins.21 After their synthesis in the cytoplasm, histones are bound by different histone chaperones, subjected to a series of posttranslational modifications, and imported into the nucleus. The last step in this assembly line of H3 is the association with importin-4 and importin-5 for translocation into the nucleus.22 H3 also associates with importins-4 and -5 during post-mitotic redistribution, when it is deposited onto chromatin. Importin proteins and other histone chaperones shield histones from nonspecific interactions until they are assembled into chromatin.23 The transfer of histones onto DNA involves various histone binding proteins, such as nucleoplasmin, N1/N2, and NAP-1.24
In this work, we sought to determine the mechanisms of nuclear access for H3-targeted polyplexes during mitosis, and specifically, to probe the role for the histone importin proteins in regulating the nuclear transport and retention of these polyplexes. Using confocal microscopy, short interfering RNA (siRNA) knockdown experiments, and super resolution imaging experiments, we showed that H3-targeted polyplexes required importin-4 for post-mitotic nuclear retention in CHO cells, and we also showed that regulation and potential interactions with importin-4 occurred before mitosis. Silencing of importin-4 affected post-mitotic nuclear retention as well as transfection efficiency by reducing polyplex co-deposition with chromatin inside the nucleus. We questioned the intracellular trafficking routes that permitted polyplexes to gain access to importin-4, since the histone-targeted polyplexes were ER-localized prior to mitosis, whereas importin-4 is known to shuttle between the cytoplasm and nucleus. Previous studies determined that retrotranslocation of proteins from the ER into the cytoplasm can occur,25 and that this retrotranslocation is often regulated by Sec61, an ER membrane protein translocon located exclusively in the ER and ER/Golgi transitional region.26-27 Accordingly, we studied polyplex regulation by Sec61 and showed that H3-targeted polyplexes had a significantly decreased transfection efficiency when Sec61 was inhibited, suggesting that Sec61 may facilitate polyplex/importin-4 interactions by regulating polyplex retrotranslocation across the ER membrane. Our overall findings identified the specific mechanisms by which H3-targeted polyplexes utilize components from the nuclear import machinery to gain access to the nucleus, and demonstrate the more general possibility that NLSs may affect gene delivery in dividing cells by altering interactions with chromatin to enhance nuclear retention. Previous studies have found NLSs to be important in nuclear pore import; however, our results show an alternative function in chromatin retention. These novel findings on mechanisms of polyplex nuclear import and retention can be utilized for future design and development of more efficient drug delivery vehicles.
Results
H3-targeted polyplexes colocalize with importin-4
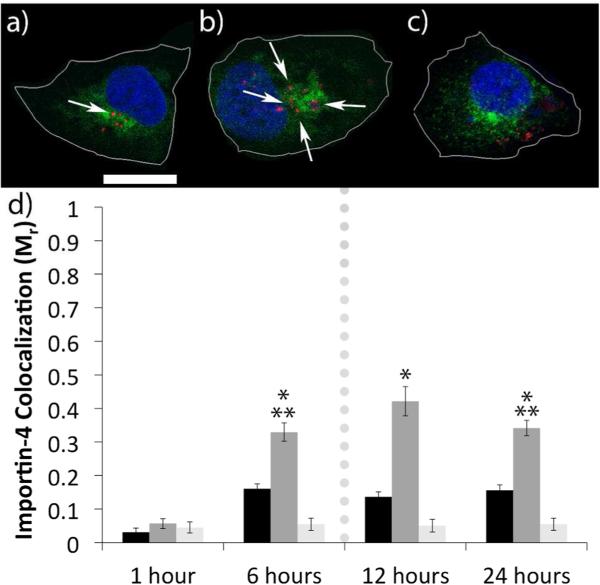
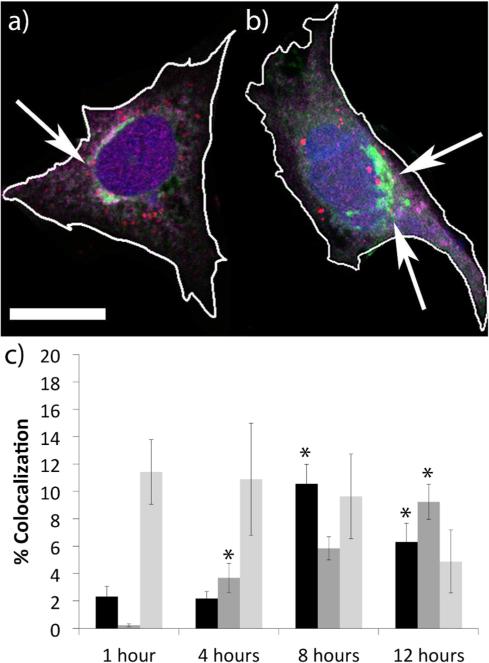
Importins regulate the nuclear import of a variety of macromolecules such as proteins and RNA, which they bind within the cytoplasm and then shuttle into the nucleus via interactions with the NPC;28-29 importin assisted transport is therefore crucial for a variety of cellular processes, including viral disease and oncogenesis.30-31 We asked whether importins might be involved in mediating post-mitotic nuclear accumulation by H3-targeted polyplexes, and hence we used immunostaining and confocal microscopy in CHO cells to determine whether the H3-targeted polyplexes trafficked coincident with importin-4 and importin-5. To enable analysis of polyplex transport kinetics, the cells were briefly pulse transfected with polyplexes containing plasmids labeled with AlexaFluor 555-peptide nucleic acids (PNA555),32 and at various times following transfection, the cells were immunocytochemically (ICC) stained with antibodies targeting importin-4 and importin-5. We discovered evidence of coordinated transport and a potential interaction between the H3-targeted polyplexes and importin-4, as evidenced by strong (42%) colocalization at 12 h post-transfection (Figure 1). Colocalization increased until reaching a maximum at 12 h, with a slight decrease at 24 h. Regulation by importin-5 appeared more limited, as only 18% of the H3-targeted polyplexes colocalized with importin-5 at 6 h post-transfection (Figure S1). Importin-5 colocalization levels reach a maximum at 6 h with no significant decrease through 24 h post-transfection. These experiments were also performed with untargeted PEI polyplexes, as well as with polyplexes containing a scrambled histone sequence (sH3). Both the untargeted polyplexes and the sH3-targeted polyplexes displayed substantially lower levels of colocalization with both importin-4 and importin-5 as compared with the H3-targeted polyplexes. These results suggest that importin-4 may play a key role in nuclear delivery of H3-targeted polyplexes, via a defined interaction with the H3 NLS; in contrast, importin-5 may only interact with polyplexes outside of the nucleus before mitosis, with little effect on delivery.
Figure 1.
(a-c) Representative confocal microscopy z-slice images of cells expressing importin-4 (green) with the nuclei stained with DAPI (blue) 6 h after a pulse-transfection with PEI polyplexes (a), H3-targeted polyplexes (b), or sH3 polyplexes (c). Polyplexes are in red; arrows indicate regions of colocalization between polyplexes and importin-4. The scale bar (shown in a) = 10 μm. The cell borders were outlined in white by comparison with the corresponding phase images by using Zen software. (d) Mander's coefficients quantifying colocalization between polyplexes and importin-4 from confocal microscopy images taken at different times post-transfection, performed by Volocity Image Analysis Software. Untargeted PEI polyplexes (black), H3-targeted polyplexes (dark gray), and sH3 polyplexes (light gray) were transfected in CHO cells and colocalization was analyzed at various times post-transfection. Each data point represents the mean ± SE with a minimum of 80-100 polyplexes analyzed. The dotted line indicates mitosis. *Indicates a statistically significant difference from PEI polyplexes at the same time point (P < 0.05). **Indicates a statistically significant difference from the previous time point for the given polyplex (P < 0.05).
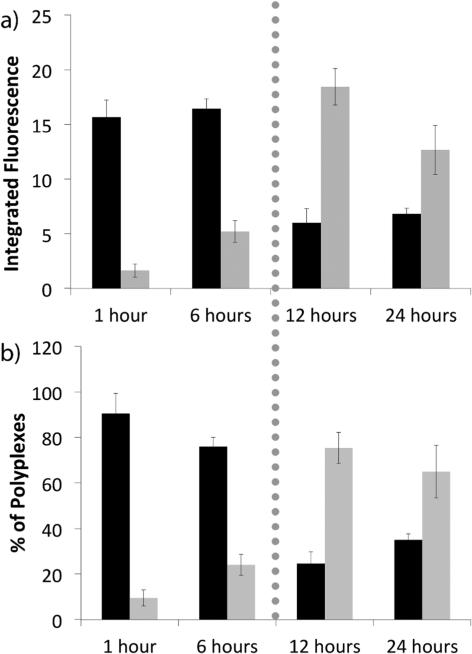
After verifying that the H3-targeted polyplexes were interacting with importin-4 both before and after cellular division, we sought to determine the intracellular locations of these interactions. Further analyses of the confocal images demonstrated that colocalization between the H3-targeted polyplexes and importin-4 occurred either inside the nucleus or in the nuclear periphery (Figure 2), with increased nuclear colocalization and decreased non-nuclear colocalization during and after mitosis. In these studies, we analyzed the subset of polyplexes that were colocalized with importin-4, and we determined which percentage were localized within the nucleus versus the nuclear periphery by using integrated density calculations to quantify the amount of polyplexes in each location. At one hour post-transfection, 90% of the H3-targeted, importin-4 colocalized polyplexes were in the non-nuclear region, whereas the other 10% of these polyplexes were found within the nucleus. At 6 h post-transfection, there was a decrease to 68% within the non-nuclear region. At the 12-hour time point, immediately following mitosis, there was a large shift from non-nuclear to nuclear colocalization, with 68% of polyplex-importin-4 colocalization within the nuclear region and only 32% in the non-nuclear regions. These data indicated that importin-4 may be shuttling H3-targeted polyplexes into the nucleus during mitosis.
Figure 2.
Nuclear vs. non-nuclear accumulation of H3-targeted polyplexes. (a) Integrated fluorescence calculations were used to determine the quantity of importin-4-colocalized polyplexes in the non-nuclear regions of the cell (black) or within the nucleus (grey). Integrated fluorescence was calculated using the area of the polyplex grouping and the mean intensity of the grouping, using ImageJ software. (b) Percentages of importin-4-colocalized polyplexes in the nucleus (grey) vs. non-nuclear regions of the cell (black). The dotted line represents the approximate time of mitosis. Each data bar represents the mean ± SE, with a minimum of 80-100 polyplex groupings analyzed per bar.
Importin-4 knockdown affects transfection and nuclear delivery
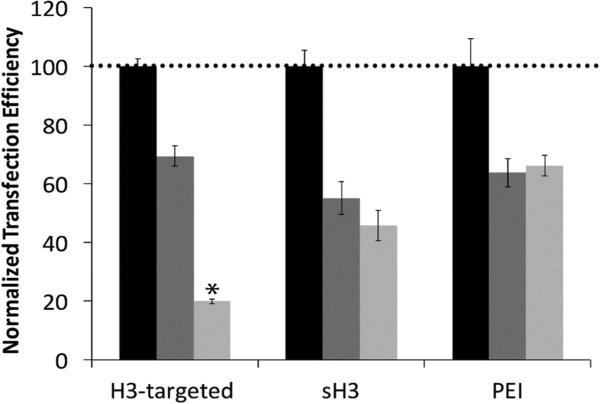
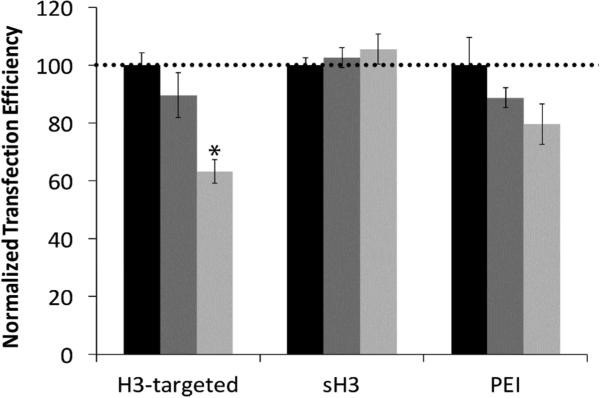
The high levels of colocalization between the H3-targeted polyplexes and importin-4 in both the nuclear periphery and the nucleus strongly suggested that importin-4 might mediate nuclear delivery and affect transfection efficiency. To further scrutinize this possibility, we pre-transfected CHO cells with siRNAs targeting importin-4 and subsequently quantified the reductions in transfection efficiency with the H3-targeted polyplexes, PEI polyplexes, or sH3 polyplexes by using flow cytometry. A scrambled siRNA was used to control for non-specific effects of siRNA transfection. Microscopy experiments and western blots were performed to confirm knockdown (Figures S2 & S3), and these experiments showed a 60% reduction in importin-4 expression following siRNA treatment as well as decreased levels of fluorescence in samples where ICC staining was used to detect importin-4. As compared to the scrambled control, siRNA induced importin-4 silencing produced an approximately 80% reduction in transfection efficiency, confirming a likely role for importin-4 in H3-targeted polyplex trafficking and delivery, consistent with imaging experiments (Figure 3). In contrast, there was no statistically significant difference between the transfection efficiency after importin-4 knockdown and the transfection efficiency following scrambled siRNA treatment when either PEI polyplexes or sH3 polyplexes were used for transfection, indicating that these polyplexes were not significantly transported by importin-4.
Figure 3.
Summary of flow cytometry analyses of CHO cell transfection following siRNA-mediated importin-4 knockdown. Transfection efficiencies of the indicated polyplexes were assessed 24 h post-transfection. Transfection with no treatment control (black), scrambled siRNA (dark gray), or importin-4 siRNA (light gray). Each data point represents the mean ± standard deviation for a total of at least four separately prepared samples. *Indicates a statistically significant difference from the scrambled siRNA treatment control (P < 0.05).
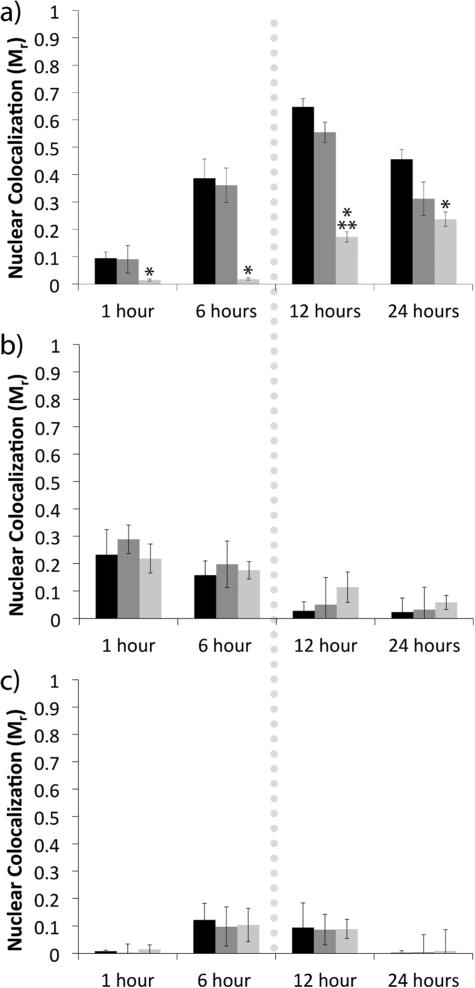
We expected that transfection was reduced because importin-4 was necessary for nuclear delivery of the H3-targeted polyplexes. Accordingly, we performed transfection experiments with PNA555-labeled polyplexes, and we used fluorescence microscopy experiments to quantify the reductions in the amount of nuclear-localized polyplexes following siRNA-mediated importin-4 silencing. These experiments demonstrated that four fold fewer H3-targeted polyplexes accessed the nucleus within 12 h (e.g. the time frame immediately following mitosis) when importin-4 was inhibited (Figure 4). When samples were treated with the scrambled siRNA for importin-4, there was no decrease in nuclear localization as compared with the untreated sample. Nuclear localization was undetectable until after mitosis in the siRNA treated sample, whereas the nuclear fraction of polyplexes increased significantly during mitosis in both the non-treated and scrambled siRNA samples. Nuclear localization peaked at 12 h post-transfection for all samples. In contrast, importin-4 silencing did not affect nuclear localization by the untargeted PEI polyplexes (Figure 4b) or by the sH3 polyplexes (Figure 4c), demonstrating that importin-4 was necessary only for nuclear localization of the H3-targeted polyplexes, and that the effect was most likely mediated by sequence-specific interactions between the H3 tail peptides and importin-4.
Figure 4.
Nuclear localization of H3-targeted polyplexes (a), PEI polyplexes (b), or sH3 polyplexes (c) following siRNA mediated importin-4 knockdown. Quantification of nuclear localization from confocal microscopy images was performed with Volocity Image Analysis software for polyplexes in CHO cells with no treatment (black), treatment with scrambled siRNA (dark gray), or treatment with importin-4 siRNA (light gray). The dotted line represents the approximate time of mitosis. Each data point represents the mean ± SE for a minimum of 100 polyplexes, with ~10 images analyzed per replicate. *Indicates a statistically significant difference from the no treatment control at the same time point (P < 0.05). **Indicates a statistically significant difference from the previous time point for the given sample (P < 0.05).
Mechanism of nuclear import and importin-4 access
To further analyze the polyplex transport pathways leading to nuclear delivery, we questioned the mechanisms by which the H3-targeted polyplexes were gaining access to importin-4. Importin-4 is a known cytoplasmic protein, and in our previous studies, we showed that the H3-targeted polyplexes trafficked to the nucleus using an endomembrane transport pathway that resulted in polyplex accumulation in Rab6-linked ER vesicles prior to mitosis, followed by redistribution into the nucleus during mitosis.19 The Sec61 translocon functions to insert secretory and transmembrane proteins into the ER during protein synthesis, or to retrotranslocate misfolded proteins in the ER to the cytosol for degradation.26 It is also known to retrotranslocate certain toxins, such as cholera toxin and ricin, which are trafficked from the cell surface to the ER to the cytosol.33-34
Accordingly, we examined whether Sec61 might retrotranslocate the H3-targeted polyplexes to the cytoplasmic face of the ER membrane from the lumen of Rab6/ER vesicles. There was up to 10% triple colocalization between the H3-targeted polyplexes, Rab6, and Sec61, with a significant increase immediately prior to mitosis (~8 h post-transfection), at which point the colocalization values peaked (Figure 5). While the triple colocalization values were low overall as compared with the total amount of H3-targeted polyplex colocalization with Rab6 at these same time points (e.g. 70% at 8 h),19 the significant increase immediately prior to mitosis at 8 h suggested a transient interaction with Sec61 that might enable polyplex retrotranslocation to the cytoplasmic face of the vesicles; in the literature, the ability to capture interactions with Sec61 is typically relatively limited due to their characteristic transient nature.26 Triple colocalization between the H3-targeted polyplexes, Rab6, and Sec61 then decreased significantly after mitosis. We also analyzed triple colocalization between the H3-targeted polyplexes, Sec61, and importin-4, to determine whether the polyplexes might gain access to importin-4 on the cytoplasmic face of the ER vesicles, and we also found up to 10% triple colocalization, with maximal colocalization after mitosis (12 h post-transfection). These results demonstrate a delayed tri colocalization with importin-4 as compared with Rab6, which correlates with the trafficking pattern we identified with our H3-targeted polyplexes and Rab6.19 We note that in dual colocalization analyses of importin-4 and Sec61 alone, we observed a maximum of 12% colocalization, and in fact, there was less colocalization during mitosis (at the 12 h time point) as compared with the triple colocalization of these two proteins with the H3-targeted polyplexes. The higher coincidence of importin-4 and Sec-61 in the presence of the polyplexes suggested that binding between the H3-targeted polyplexes and importin-4 may drive interactions between importin-4 and Sec61.
Figure 5.
Triple colocalization experiments with H3-targeted polyplexes, importin-4, and Rab6, or H3-targeted polyplexes, importin-4 and Sec61. (a) Representative confocal z-stack slice of H3-targeted polyplexes (red), nucleus (blue), Rab6 (green), and importin-4 (magenta). (b) Representative confocal z-stack slice of H3-targeted polyplexes (red), nucleus (blue), Sec61 (green), and importin-4 (magenta). Arrows represent points of triple colocalization. The scale bar (shown in a) = 10 μm. (c) Quantification of colocalization with H3-targeted polyplexes, importin-4, and Rab6 (black); H3-targeted polyplexes, importin-4, and Sec61 (dark gray); or importin-4 and Sec61 alone (light gray). * Indicates a statistically significant difference between the previous time point for a given sample (P < 0.05).
To further probe the role for Sec61 in nuclear access by the H3-targeted polyplexes, we performed knockdown experiments employing siRNA coding for Sec61. A Western blot was performed to quantify Sec61 knockdown, which occurred at a level of approximately 70% (Figure S3). To determine whether Sec61 knockdown impacted H3-targeted polyplex transfection, cells were pre transfected with Sec61-encoding siRNAs and subsequently transfected with H3-targeted polyplexes, PEI polyplexes, or sH3 polyplexes. As shown in Figure 6, there was a 35% decrease in H3-targeted polyplex transfection following Sec61 knockdown, whereas there was no significant effect on transfection by the PEI polyplexes or sH3 polyplexes when Sec61 was silenced. These data suggest that Sec61 may trigger H3-targeted polyplex retrotranslocation for at least a fraction of the polyplexes, leading to cytoplasmic access and nuclear delivery.
Figure 6.
Summary of flow cytometry analyses of CHO cell transfection following siRNA-mediated Sec61 knockdown. Transfection efficiencies of the indicated polyplexes were analyzed 24 h post-transfection. Transfection with no treatment control (black), treatment with scrambled siRNA (dark gray), or treatment with Sec61 siRNA (light gray). Each data point represents the mean ± standard deviation for a total of at least four separately prepared samples. *Indicates a statistically significant difference from the no treatment control (P < 0.05).
Post-mitotic polyplex interaction with chromatin and importin-4 knockdown
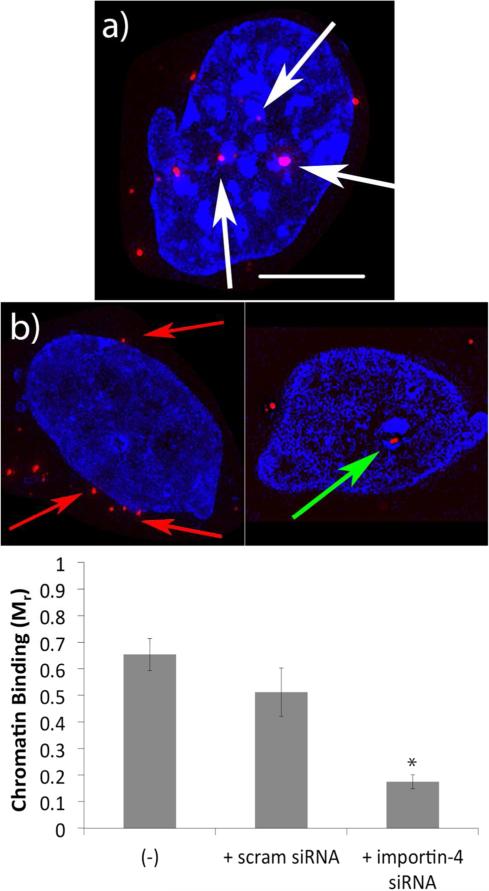
Importin-4 plays an instrumental role in post-mitotic redeposition of the H3 protein in chromatin, and we hypothesized that importin-4 might also deposit the polyplexes on chromatin in a similar manner, leading to improved nuclear retention. Accordingly, we sought to determine the extent to which the polyplexes bound to chromatin post-mitosis, and whether this effect required importin-4. Using super resolution microscopy, we assessed the interactions between the H3-targeted polyplexes and DAPI stained chromatin (Figure 7). Super resolution microscopy, combined with structured illumination microscopy (SIM), resolves objects beyond the diffraction limit by illumination with multiple interfering beams of light. The emitted light contains higher resolution image information, resulting in an image with twice the resolution of a conventional image taken on the same microscope, and the ability to distinguish features as small as 50 nm.35 Therefore, we used this technique to identify interactions between chromatin and the polyplexes according to procedures previous used in the literature.36 65% of the H3-targeted polyplexes interacted with chromatin following mitosis. We further explored this interaction by performing the same experiments after siRNA-mediated silencing of importin-4. There was an approximately 75% decrease in chromatin binding by the H3-targeted polyplexes when importin-4 was inhibited (Figure 7c), consistent with confocal microscopy analyses at 12 h post-transfection (Figure 4). From super resolution images, it was also evident that the majority of the polyplexes did not even enter the nucleus when importin-4 was reduced, and that the polyplexes were instead trapped around the nuclear periphery. Those polyplexes that did enter the nucleus did not interact with chromatin to a measurable extent when importin-4 was inhibited (Figure 7b). Therefore, these data corroborate the finding that importin-4 was necessary for the H3-targeted polyplexes to enter the nucleus, and the data also indicated that importin-4 drives interactions with chromatin to affect nuclear retention.
Figure 7.
Super resolution imaging experiments of H3-targeted polyplex binding to chromatin. Representative images of chromatin (blue) binding by the H3-targeted polyplexes (red) when cells were treated with scrambled importin-4 siRNA (a), and representative images showing the lack of H3-targeted polyplex binding to chromatin when cells were treated with importin-4 siRNA (b). The scale bar (shown in a) = 5 μm. Chromatin was stained with DAPI. White arrows indicate chromatin binding, green arrows represent the lack of chromatin binding by nuclear polyplexes when importin-4 was inhibited, and red arrows indicate those polyplexes that remained largely in the nuclear periphery with importin-4 inhibition. (c) Quantification of chromatin binding using Manders’ coefficients at 12 h post-transfection. Ten images containing 8–10 cells per image were analyzed for each sample.
Discussion
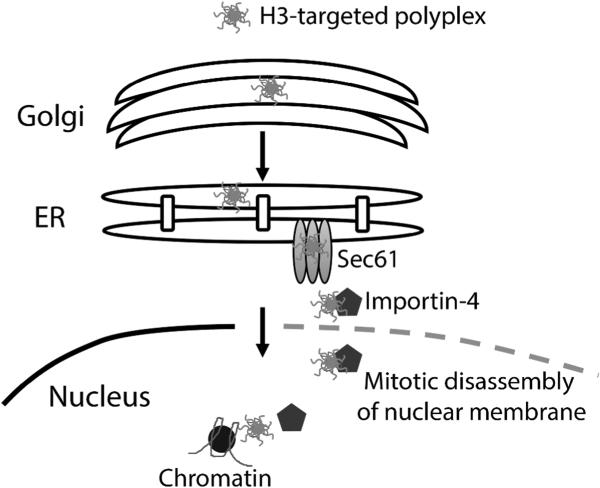
A critical limitation of many nonviral gene delivery vehicles is their low transfection efficiency, which is partially due to their limited ability to cross the nuclear membrane. Multiple studies have explored whether the addition of NLS sequences to nanostructures can enhance nuclear delivery, yet effective strategies to exploit NLSs in mitotic cells have not been fully elucidated. Accordingly, this study documents the first comprehensive examination of the mechanisms involved in NLS-mediated transfer of polyplexes from endomembrane vesicles to the nucleus. Our studies not only identify novel steps involved in gene delivery employing the histone H3 NLS, but also suggest general nuclear delivery mechanisms involving retrotranslocation from the ER into the cytosol as well as NLS-mediated chromatin deposition via interaction with cytoplasmic chaperones (Figure 8). These pathways may represent new targets to increase the efficacy of nanocarrier-mediated gene transfer.
Figure 8.
Proposed mechanisms for H3-targeted polyplex transfer into the nucleus. Key regulators include importin-4, Sec61, as well as ER membrane mediated nuclear entry during mitosis.19
In our previous studies, we determined that H3 targeting peptides enhance the utilization of caveolar endocytic routes and improve transfection by transferring polyplexes through compartments that localize to the ER and nucleus, similar to the trafficking behavior of several types of native proteins37 and pathogens.19 In these prior studies, we specifically showed that the transport behavior of the H3-targeted polyplexes was conferred in part by interactions with histone H3 effectors, such as H3K4 methyltransferase subunits involved in regulating vesicular transport between late endosomes and the Golgi.19 These findings motivated the analyses herein to determine whether other H3 effectors, such as the H3 importins, might be involved in shuttling H3-targeted polyplexes into the nucleus during mitosis.
Our work identified several key similarities with the importin mediated post-mitotic nuclear deposition process for the histone H3 protein. In particular, core histones are synthesized and imported during S phase, when they are needed for the assembly of newly replicated DNA, as well as for chromatin remodeling that takes place during transcription.38 H3 and H4 proteins form a tetramer within the cytosol after synthesis, and are actively transported into the nucleus through the NPC.21 Early studies have shown that nuclear import is typically facilitated by importins 4 and 5, which bind to either H3 or H4 NLS in the positively charged core domain of a H3 H4 tetramer.21, 39-40 However, Baake and coworkers found that core histones can also be transported by any of several members of the importin β superfamily, and H3 can be imported into the nucleus in the absence of H4 via interactions with importin-4 and subsequent binding to the import receptors.41 Also, recent studies have demonstrated that only importin-4 is needed for nuclear deposition.20, 22 Therefore, we focused on importin-4 as a nuclear chaperone for our H3-targeted polyplexes.
Our results demonstrate that the same importins are also critical for polyplex import, but the detailed processes appear to be distinct, likely because our polyplexes are too large for import through the NPC. During mitosis, the nucleus disassembles and the NPCs disassociate. At the end of mitosis, the nuclear envelope reassembles on the surface of chromatin, while NPCs begin to reassemble and actively reimport proteins that contain NLSs. Some viruses are known to access the nucleus during mitosis when the nuclear envelope is disassembled. For example, the retrovirus murine leukemia virus can only access the nucleus during mitosis because the viral complex is too large for the NPC.42 These viruses wait for the dispersion of the nuclear membrane, and become included within the nucleus during membrane reformation. This leads us to believe that our polyplexes may also be utilizing the reassembly of the nuclear envelope as a means to enter the nucleus during cell division. In fact, some studies have shown that the ER membrane forms the source of the newly forming nuclear membrane,43 which wraps around chromosomes until the nuclear envelope is reformed. Our studies demonstrated strong colocalization with importin-4, which increased within the nucleus post-mitosis, indicating that the NLS on the H3 peptide is functional and may be used in docking our polyplexes with the NPC. It is possible that the polyplexes associate with importin-4 in the cytoplasm, and this interaction may stabilize an association with chromatin during ER-mediated nuclear envelope reassembly. This hypothesis is consistent with the role for importin-4 in shuttling histone proteins onto chromatin, via coordinated interactions with the histone chaperone Asf1 and the chromatin assembly factor Caf1.23
The capacity to interact with importins requires access to the cytoplasm or nucleus, and hence we also questioned the mechanisms by which the H3-targeted polyplexes might exit ER/Rab6 vesicles. Pathogens are highly efficient at exploiting nature's machinery for this purpose; in particular, Shiga toxin B has evolved mechanisms to travel from the ER to cytosol via the ER-associated protein degradation pathway, ultimately escaping degradation and retrotranslocating into the cytosol.44 This process depends upon the Sec61 translocon, which was also implicated in localization of the epidermal growth factor receptor to the nucleus through a process involving retrotranslocation from the ER to the cytosol followed by importin β-mediated nuclear import.26 Small peptides have also been observed to exit the ER by this route.45 Accordingly, a reasonable suggestion was that the same channel used for the ER associated protein degradation (ERAD) pathway, Sec61 and its partner subunits, was also employed for translocating polyplexes out of the ER. Our results indicated that at least some of the H3-targeted polyplexes associate with Sec61 and rely on this association to mediate transfection, presumably by using an ERAD like pathway to access the cytoplasm and interact with importin-4. In particular, partial (~70%) suppression of Sec61 expression reduced H3-targeted gene transfer levels substantially, by approximately 35% (Fig. 6), despite the relatively low levels of detectable colocalization between the H3-targeted polyplexes and Sec61. Studies of Sec61 in protein transport have similarly provided mixed results. Membrane proteins show Sec61-dependence for degradation, which is evidence for Sec61-mediated retrotranslocation.46 Some experiments show strong dependence of the ERAD on Sec61,47-48 yet in other ERAD assays, there was no apparent role for Sec61.49-50 These studies may indicate that another, unidentified mechanism exists for protein translocation in some contexts. This alternate mechanism, or a similar process, may also be responsible for releasing a portion of the H3-targeted polyplexes into the cytosol.
In contrast to H3-targeted polyplexes, untargeted and sH3 polyplexes exhibited fundamentally different results, with significantly decreased localization with importin-4 and sec61, and a lack of dependence upon mitosis. These results are consistent with our previous findings where untargeted polyplexes were trafficked through a different pathway towards the nucleus.19 This significant difference in colocalization with importin-4, whose natural role is the import of the H3 protein, strongly indicates that it interacts sequence specifically with the H3-targeted polyplexes during import.
In this study, we observed the identification of specific mechanisms by which H3-targeted polyplexes reach the nucleus by retrotranslocating from ER vesicles and subsequently entering the nucleus during postmitotic redistribution of ER membranes, utilizing the natural import machinery of histone protein chaperones. These findings demonstrate a need for fundamentally different approaches in nonviral design. Additionally, as gene transfer within dividing cells continues to require unreasonable excesses of DNA, improved methods to target the nucleus are essential to advance gene therapy.
Materials & Methods
Materials
H3 tail peptides comprised of the mammalian N-terminal H3 residues 1–25 (ARTKQTARKSTGGKAPRKQLATKAA-CONH2) were purchased from Anaspec (Fremont, CA) at ≥95% purity. The sH3 peptide sequence was designed to incorporate residues 1-25 of the N-terminal tail of the H3 protein in a randomized sequence (LSAATPRTAKGARQTKRQKAKGTAK-CONH2). The peptide was synthesized using previous protocols19. The peptide was synthesized by solid phase peptide synthesis with a Protein Technologies, Inc. Tribute series peptide synthesizer (Tucson, AZ) and a rink amide ChemMatrix resin from Pcas Biomatrix, Inc (Quebec, Canada). Cleavage of the peptide from the resin was performed using a cocktail consisting of 95 vol % trifluoroacetic acid (TFA), 2.5 vol % H2O, and 2.5 vol % triisopropylsaline. The gWIZ mammalian expression plasmid encoding GFP was obtained from Genlantis (San Diego, CA), amplified in DH5α Escherichia coli in Lysogeny Broth, and purified with a QIAGEN Plasmid Mega Kit (QIAGEN, Valencia, CA) following the manufacturer's protocols. Alexa Fluor 555-labeled PNA clamps were custom synthesized and purified to >90% by Panagene (Daejeon, Korea). Cell culture reagents were purchased from Fisher Scientific (Pittsburgh, PA). Branched PEI (25 kDa) and all other reagents were purchased at analytical grade from Sigma (St. Louis, MO). Rab-GFP plasmids were purchased from Addgene (ID#31734). Primary antibody reactive with importin-4 (ab28387) was obtained from Abcam (Cambridge, MA), and importin-5 (sc-11369) and actin (sc1616R) primary antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX). Secondary antibody (AlexaFluor488 anti-rabbit IgG) was obtained from Invitrogen (Carlsbad, CA). Sec61 and Rab6 –GFP plasmids were obtained from Addgene (15108, 31733).
Cells culture and synchronization
CHO-K1 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured according to ATCC protocols at 37°C and 5% CO2 in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were passaged when they reached approximately 80% confluency.
For cell synchronization, CHO cells were plated at approximately 7200 cells/cm2. Twenty-four h after plating, solutions of lovastatin (10μM) in growth medium (serum/antibiotic-supplemented DMEM) were added to cells, and the cells were incubated in the lovastatin solutions for 32–36 h51. Subsequently, the medium containing lovastatin was removed, the cells were washed with PBS, and fresh growth medium was added to cells to resume the cell division cycle. Cells were incubated for an additional 16 h so that transfection could take place during the S phase of cell division52.
Polyplex formation and transfection
For colocalization studies, pDNA was fluorescently labeled with PNA555 at a ratio of 1:20 (DNA:PNA) incubated overnight at 37 °C.32 H3-targeted PEI polyplexes, untargeted PEI polyplexes, or sH3 polyplexes were formed in 20 mmol/l 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) at a pH of 6 as previously described18. For all experiments, polyplexes were formed at an N:P ratio of 10. For polyplexes formed with a mixture of H3 (or sH3) and PEI, an N:P ratio of 6/4 was used where the total N:P ratio was 10, with N = 6 from H3 (or sH3) and N = 4 from PEI. This corresponds to ~90% (w/w) H3 and 10% PEI in the polycation solution used for pDNA complexation. For the formation of H3-targeted polyplexes and scrambled H3 polyplexes, the H3 (or sH3) peptide was added before the PEI peptide. The structures of the polyplexes were analyzed in our prior work by gel electrophoresis experiments as well as dynamic light scattering and zeta potential experiments.18, 53 The amount of peptide within the polyplex was also quantified previously53. Unless otherwise indicated, cells were pulse transfected for 15 minutes with polyplexes, and subsequently rinsed with phosphate-buffered saline (PBS) and fresh growth medium (serum/antibiotic supplemented DMEM) was added to the cells until a specified time point.
Immunocytochemistry analysis
For colocalization studies with importin-4 and Sec61, CHO cells were plated, synchronized, and 15 minute pulse transfected with H3-targeted polyplexes, untargeted PEI polyplexes, or sH3 polyplexes. Pulse/chase transfection approaches are necessary to accurately determine cellular transport kinetics, since endomembrane trafficking events occur on time scales that are much shorter than typical transfections.19, 54 At the specified time points, cells were rinsed with PBS, washed with 10 μg/ml heparin, washed again with PBS, and fixed with 4% paraformaldehyde in PBS for 15 minutes. Cells were subsequently permeabilized with 0.1% saponin in PBS (Sap) and blocked with 0.5% bovine serum albumin in 0.1% Sap. A 1 μg/ml solution of primary antibody was incubated with the cells overnight at 4 °C in the blocking buffer. Cells were subsequently rinsed three times with PBS and incubated with the secondary antibody in blocking buffer for 1 h at room temperature. Following secondary antibody treatment, cells were rinsed three more times with PBS and stored at 4 °C prior to imaging.
Confocal microscopy and quantification of polyplex colocalization
Cell imaging was performed with a 40× water immersion objective (NA = 0.55) on an LSM 710 microscope (Carl Zeiss, Thornwood, NY) equipped with appropriate lasers and filters for the selected fluorescent dyes. Volocity Imaging Software (PerkinElmer) was utilized for image analysis and quantification of colocalization, where the locations of polyplexes and importin-4, Sec61, Rab6, and the nucleus were determined from measurement statistics associated with individual voxel intensities. The fraction of polyplexes (red voxels) that colocalized with the stain of interest (green voxels) was analyzed by calculation of the Mr, which represents the sum of the colocalized red intensity divided by the sum of the total red intensity. Mr values range from 0 (no colocalization) to 1 (complete colocalization of red voxels with green voxels)55. Volocity software automatically determined minimum values for red and green intensities, and these minimum values were set as the threshold to distinguish signal from background. Statistical analyses of Mr values were performed using a two tailed Student's t-test and the SE reported represents the population of polyplexes analyzed. A range of 80–100 polyplexes per data point in each colocalization study was analyzed to obtain these values. For triple colocalization, we used an algorithm for determining colocalization by measuring groupings of polyplexes rather than individual voxels, as such a measure more likely represents the underlying functional relationships56.
Flow cytometry and transfection efficiency analysis
For cell transfection efficiency experiments, cells were transfected with polyplexes for 2 h, according to previously published procedures.18 GFP expression was quantified on an Accuri C6 flow cytometer. For cytometry analyses, cells were collected after imaging and prepared for analysis by standard trypsin-mediated collection protocols. Briefly, cells were rinsed with PBS, incubated with trypsin, collected in 15 ml centrifuge tubes. Subsequently, cells were collected by centrifugation, resuspended in 300 ml of cold PBS, filtered through a 35 μm nylon mesh to remove aggregates, and stored at 4 °C until analysis. Scattering plots were gated for quantification purposes, and a total of 10,000 cells were analyzed for each cell sample. Dead cells were excluded from the analyses of transfection efficiency.
siRNA silencing (Imp4, Sec61)
Importin-4 and Sec61 siRNAs were synthesized by Dharmacon (Boulder, CO). Cells were transfected using the DharmaFECT reagent using 10 μM pooled siRNA oligos for 48 hrs. Importin-4 target sequences: GCAUUUCGCUGUACAAGUU, AGUCAGAGGUGCCGGUCAU, CCUCGCAAGUUGUACGCAA, AUGGAGCACCUGCGGGAAU. Sec61 target sequences: CAGUAUUGGUUAUGAGUCU, GUUCGUAGAUUCAGUUACA, GCUCAAAGUUGGCCCUGUU, CUGUAAGCUUGCUGUUUUA
Super Resolution Microscopy
CHO cells were plated in eight-well plates and synchronized. Cells were pulse transfected for 30 min with polyplexes in Opti-MEM (1 μg of DNA/well), rinsed with PBS, and cultured in complete medium prior to imaging. Super resolution imaging was performed on an Elyra PS1 super resolution microscope (Carl Zeiss, Oberkochen, Germany) using a 561 nm laser for excitation. The objective was a 63×/1.4 oil immersion apochromat (Carl Zeiss). Ten images containing 80–100 cells in total were analyzed to collect the data for each sample.
Supplementary Material
Acknowledgments
This work was partially supported by the National Science Foundation under Grant No. DMR 0746458. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. This work was also partially supported by the National Institutes of Health under Grant No. R01 EB017766. We acknowledge the Delaware Biotechnology Institute Bioimaging Facility for use of their confocal microscopes. We also thank Jeffrey L Caplan and Michael Moore for training and continuing guidance with the equipment and software used for all cellular imaging and analysis.
Footnotes
Supplemental material:
Figure S1: Colocalization confocal images w/ importin-5
Figure S2: Colocalization confocal images & graph w/ imp4 siRNA
Figure S3: Western blots (importin-4 & sec61)
References
- 1.Fasbender A, Zabner J, Zeiher BG, Welsh MJ. A Low Rate of Cell Proliferation and Reduced DNA Uptake Limit Cationic Lipid-Mediated Gene Transfer to Primary Cultures of Ciliated Human Airway Epithelia. Gene Ther. 1997;4(11):1173–80. doi: 10.1038/sj.gt.3300524. [DOI] [PubMed] [Google Scholar]
- 2.Larsen JD, Ross NL, Sullivan MO. Requirements for the Nuclear Entry of Polyplexes and Nanoparticles During Mitosis. J. Gene Med. 2012;14(9-10):580–9. doi: 10.1002/jgm.2669. [DOI] [PubMed] [Google Scholar]
- 3.Dean DA, Strong DD, Zimmer WE. Nuclear Entry of Nonviral Vectors. Gene Ther. 2005;12(11):881–90. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorlich D, Kutay U. Transport between the Cell Nucleus and the Cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–60. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 5.McLane LM, Corbett AH. Nuclear Localization Signals and Human Disease. IUBMB Life. 2009;61(7):697–706. doi: 10.1002/iub.194. [DOI] [PubMed] [Google Scholar]
- 6.Ciolina C, Byk G, Blanche F, Thuillier V, Scherman D, Wils P. Coupling of Nuclear Localization Signals to Plasmid DNA and Specific Interaction of the Conjugates with Importin Alpha. Bioconjug. Chem. 1999;10(1):49–55. doi: 10.1021/bc980061a. [DOI] [PubMed] [Google Scholar]
- 7.Tkachenko AG, Xie H, Coleman D, Glomm W, Ryan J, Anderson MF, Franzen S, Feldheim DL. Multifunctional Gold Nanoparticle-Peptide Complexes for Nuclear Targeting. J. Am. Chem. Soc. 2003;125(16):4700–1. doi: 10.1021/ja0296935. [DOI] [PubMed] [Google Scholar]
- 8.Misra R, Sahoo SK. Intracellular Trafficking of Nuclear Localization Signal Conjugated Nanoparticles for Cancer Therapy. Eur. J. Pharm. Sci. 2010;39(1-3):152–63. doi: 10.1016/j.ejps.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Kang B, Mackey MA, El-Sayed MA. Nuclear Targeting of Gold Nanoparticles in Cancer Cells Induces DNA Damage, Causing Cytokinesis Arrest and Apoptosis. J. Am. Chem. Soc. 2010;132(5):1517–9. doi: 10.1021/ja9102698. [DOI] [PubMed] [Google Scholar]
- 10.Dean DA. Import of Plasmid DNA into the Nucleus Is Sequence Specific. Exp. Cell Res. 1997;230(2):293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Yu Z, Chen H, Gao J, Liang W. Transfection Efficiency and Intracellular Fate of Polycation Liposomes Combined with Protamine. Biomaterials. 2011;32(5):1412–8. doi: 10.1016/j.biomaterials.2010.09.074. [DOI] [PubMed] [Google Scholar]
- 12.Badding MA, Vaughan EE, Dean DA. Transcription Factor Plasmid Binding Modulates Microtubule Interactions and Intracellular Trafficking During Gene Transfer. Gene Ther. 2012;19(3):338–46. doi: 10.1038/gt.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagstaff KM, Fan JY, De Jesus MA, Tremethick DJ, Jans DA. Efficient Gene Delivery Using Reconstituted Chromatin Enhanced for Nuclear Targeting. FASEB J. 2008;22(7):2232–42. doi: 10.1096/fj.07-099911. [DOI] [PubMed] [Google Scholar]
- 14.Chan CK, Hubner S, Hu W, Jans DA. Mutual Exclusivity of DNA Binding and Nuclear Localization Signal Recognition by the Yeast Transcription Factor Gal4: Implications for Nonviral DNA Delivery. Gene Ther. 1998;5(9):1204–12. doi: 10.1038/sj.gt.3300708. [DOI] [PubMed] [Google Scholar]
- 15.van der Aa MA, Koning GA, d'Oliveira C, Oosting RS, Wilschut KJ, Hennink WE, Crommelin DJ. An Nls Peptide Covalently Linked to Linear DNA Does Not Enhance Transfection Efficiency of Cationic Polymer Based Gene Delivery Systems. J. Gene Med. 2005;7(2):208–17. doi: 10.1002/jgm.643. [DOI] [PubMed] [Google Scholar]
- 16.Tanimoto M, Kamiya H, Minakawa N, Matsuda A, Harashima H. No Enhancement of Nuclear Entry by Direct Conjugation of a Nuclear Localization Signal Peptide to Linearized DNA. Bioconjug. Chem. 2003;14(6):1197–202. doi: 10.1021/bc034075e. [DOI] [PubMed] [Google Scholar]
- 17.Grosse S, Thevenot G, Monsigny M, Fajac I. Which Mechanism for Nuclear Import of Plasmid DNA Complexed with Polyethylenimine Derivatives? J. Gene Med. 2006;8(7):845–51. doi: 10.1002/jgm.915. [DOI] [PubMed] [Google Scholar]
- 18.Reilly MJ, Larsen JD, Sullivan MO. Histone H3 Tail Peptides and Poly(Ethylenimine) Have Synergistic Effects for Gene Delivery. Mol. Pharm. 2012;9(5):1031–40. doi: 10.1021/mp200372s. [DOI] [PubMed] [Google Scholar]
- 19.Ross NL, Munsell EV, Sabanayagam C, Sullivan MO. Histone-Targeted Polyplexes Avoid Endosomal Escape and Enter the Nucleus During Postmitotic Redistribution of Er Membranes. Molecular Therapy—Nucleic Acids. 2015;4(2):e226. doi: 10.1038/mtna.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D. The Program for Processing Newly Synthesized Histones H3.1 and H4. Nat. Struct. Mol. Biol. 2010;17(11):1343–51. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosammaparast N, Guo Y, Shabanowitz J, Hunt DF, Pemberton LF. Pathways Mediating the Nuclear Import of Histones H3 and H4 in Yeast. J. Biol. Chem. 2002;277(1):862–8. doi: 10.1074/jbc.M106845200. [DOI] [PubMed] [Google Scholar]
- 22.Alvarez F, Munoz F, Schilcher P, Imhof A, Almouzni G, Loyola A. Sequential Establishment of Marks on Soluble Histones H3 and H4. J. Biol. Chem. 2011;286(20):17714–21. doi: 10.1074/jbc.M111.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamiche A, Shuaib M. Chaperoning the Histone H3 Family. Biochim. Biophys. Acta. 2012;1819(3-4):230–7. doi: 10.1016/j.bbagrm.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg RD, Lorch Y. Chromatin Structure and Transcription. Annu. Rev. Cell Biol. 1992;8:563–87. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 25.Tsai B, Ye Y, Rapoport TA. Retro-Translocation of Proteins from the Endoplasmic Reticulum into the Cytosol. Nature reviews. Molecular cell biology. 2002;3(4):246–55. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 26.Liao HJ, Carpenter G. Role of the Sec61 Translocon in Egf Receptor Trafficking to the Nucleus and Gene Expression. Mol. Biol. Cell. 2007;18(3):1064–72. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenfield JJ, High S. The Sec61 Complex Is Located in Both the Er and the Er-Golgi Intermediate Compartment. J. Cell Sci. 1999;112(Pt 10):1477–86. doi: 10.1242/jcs.112.10.1477. [DOI] [PubMed] [Google Scholar]
- 28.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D. Importins Fulfil a Dual Function as Nuclear Import Receptors and Cytoplasmic Chaperones for Exposed Basic Domains. EMBO J. 2002;21(3):377–86. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chook YM, Blobel G. Karyopherins and Nuclear Import. Curr. Opin. Struct. Biol. 2001;11(6):703–15. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 30.Moseley GW, Filmer RP, DeJesus MA, Jans DA. Nucleocytoplasmic Distribution of Rabies Virus P-Protein Is Regulated by Phosphorylation Adjacent to C Terminal Nuclear Import and Export Signals. Biochemistry. 2007;46(43):12053–61. doi: 10.1021/bi700521m. [DOI] [PubMed] [Google Scholar]
- 31.Hogarth CA, Calanni S, Jans DA, Loveland KL. Importin Alpha Mrnas Have Distinct Expression Profiles During Spermatogenesis. Dev. Dyn. 2006;235(1):253–62. doi: 10.1002/dvdy.20569. [DOI] [PubMed] [Google Scholar]
- 32.Millili PG, Yin DH, Fan H, Naik UP, Sullivan MO. Formulation of a Peptide Nucleic Acid Based Nucleic Acid Delivery Construct. Bioconjug. Chem. 2010;21(3):445–55. doi: 10.1021/bc900328j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandvig K, Garred O, Prydz K, Kozlov JV, Hansen SH, van Deurs B. Retrograde Transport of Endocytosed Shiga Toxin to the Endoplasmic Reticulum. Nature. 1992;358(6386):510–2. doi: 10.1038/358510a0. [DOI] [PubMed] [Google Scholar]
- 34.Sandvig K, van Deurs B. Transport of Protein Toxins into Cells: Pathways Used by Ricin, Cholera Toxin and Shiga Toxin. FEBS Lett. 2002;529(1):49–53. doi: 10.1016/s0014-5793(02)03182-4. [DOI] [PubMed] [Google Scholar]
- 35.Gustafsson MG. Surpassing the Lateral Resolution Limit by a Factor of Two Using Structured Illumination Microscopy. J. Microsc. 2000;198(Pt 2):82–7. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 36.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, Leonhardt H, Sedat JW. Subdiffraction Multicolor Imaging of the Nuclear Periphery with 3d Structured Illumination Microscopy. Science. 2008;320(5881):1332–6. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang YN, Wang H, Yamaguchi H, Lee HJ, Lee HH, Hung MC. Copi-Mediated Retrograde Trafficking from the Golgi to the Er Regulates Egfr Nuclear Transport. Biochem. Biophys. Res. Commun. 2010;399(4):498–504. doi: 10.1016/j.bbrc.2010.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verreault A. De Novo Nucleosome Assembly: New Pieces in an Old Puzzle. Genes Dev. 2000;14(12):1430–8. [PubMed] [Google Scholar]
- 39.Muhlhausser P, Muller EC, Otto A, Kutay U. Multiple Pathways Contribute to Nuclear Import of Core Histones. EMBO reports. 2001;2(8):690–6. doi: 10.1093/embo-reports/kve168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blackwell JS, Jr., Wilkinson ST, Mosammaparast N, Pemberton LF. Mutational Analysis of H3 and H4 N Termini Reveals Distinct Roles in Nuclear Import. J. Biol. Chem. 2007;282(28):20142–50. doi: 10.1074/jbc.M701989200. [DOI] [PubMed] [Google Scholar]
- 41.Baake M, Bauerle M, Doenecke D, Albig W. Core Histones and Linker Histones Are Imported into the Nucleus by Different Pathways. Eur. J. Cell Biol. 2001;80(11):669–77. doi: 10.1078/0171-9335-00208. [DOI] [PubMed] [Google Scholar]
- 42.Goff SP. Host Factors Exploited by Retroviruses. Nat. Rev. Microbiology. 2007;5(4):253–63. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 43.Anderson DJ, Hetzer MW. Shaping the Endoplasmic Reticulum into the Nuclear Envelope. J. Cell Sci. 2008;121(Pt 2):137–42. doi: 10.1242/jcs.005777. [DOI] [PubMed] [Google Scholar]
- 44.Hazes B, Read RJ. Accumulating Evidence Suggests That Several Ab-Toxins Subvert the Endoplasmic Reticulum-Associated Protein Degradation Pathway to Enter Target Cells. Biochemistry. 1997;36(37):11051–4. doi: 10.1021/bi971383p. [DOI] [PubMed] [Google Scholar]
- 45.Gillece P, Pilon M, Romisch K. The Protein Translocation Channel Mediates Glycopeptide Export across the Endoplasmic Reticulum Membrane. Proc. Natl. Acad. Sci. U. S. A. 2000;97(9):4609–14. doi: 10.1073/pnas.090083497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott DC, Schekman R. Role of Sec61p in the Er-Associated Degradation of Short-Lived Transmembrane Proteins. J. Cell Biol. 2008;181(7):1095–105. doi: 10.1083/jcb.200804053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant Analysis Links the Translocon and Bip to Retrograde Protein Transport for Er Degradation. Nature. 1997;388(6645):891–5. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 48.Willer M, Forte GM, Stirling CJ. Sec61p Is Required for Erad-L: Genetic Dissection of the Translocation and Erad-L Functions of Sec61p Using Novel Derivatives of Cpy. J. Biol. Chem. 2008;283(49):33883–8. doi: 10.1074/jbc.M803054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE. Real-Time Fluorescence Detection of Erad Substrate Retrotranslocation in a Mammalian in Vitro System. Cell. 2007;129(5):943–55. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato BK, Hampton RY. Yeast Derlin Dfm1 Interacts with Cdc48 and Functions in Er Homeostasis. Yeast. 2006;23(14-15):1053–64. doi: 10.1002/yea.1407. [DOI] [PubMed] [Google Scholar]
- 51.Javanmoghadam-Kamrani S, Keyomarsi K. Synchronization of the Cell Cycle Using Lovastatin. Cell Cycle. 2008;7(15):2434–40. doi: 10.4161/cc.6364. [DOI] [PubMed] [Google Scholar]
- 52.Wu JR, Gilbert DM. Lovastatin Arrests Cho Cells between the Origin Decision Point and the Restriction Point. FEBS Lett. 2000;484(2):108–12. doi: 10.1016/s0014-5793(00)02135-9. [DOI] [PubMed] [Google Scholar]
- 53.Larsen JD, Reilly MJ, Sullivan MO. Using the Epigenetic Code to Promote the Unpackaging and Transcriptional Activation of DNA Polyplexes for Gene Delivery. Mol. Pharm. 2012;9(5):1041–51. doi: 10.1021/mp200373p. [DOI] [PubMed] [Google Scholar]
- 54.Sandin P, Fitzpatrick LW, Simpson JC, Dawson KA. High-Speed Imaging of Rab Family Small Gtpases Reveals Rare Events in Nanoparticle Trafficking in Living Cells. ACS Nano. 2012;6(2):1513–21. doi: 10.1021/nn204448x. [DOI] [PubMed] [Google Scholar]
- 55.Manders E, Verbeek F, Aten J. Measurement of Colocalization of Objects in Dual-Color Confocal Images. Journal of Microscopy-Oxford. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 56.Fletcher PA, Scriven DR, Schulson MN, Moore ED. Multi-Image Colocalization and Its Statistical Significance. Biophys. J. 2010;99(6):1996–2005. doi: 10.1016/j.bpj.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.