Abstract
Ring1 and YY1 binding protein (RYBP) is a member of Polycomb group (PcG) proteins and regulates cell growth through both PcG-dependent and -independent mechanisms. Our initial study indicated that RYBP is down-regulated in human non-small cell lung cancer (NSCLC) tissues. The present study determined the molecular role of RYBP in the development of NSCLC. We systemically investigated the association between the RYBP expression and the survival of patients with NSCLC. We also carried out in vitro and in vivo studies to explore the molecular basis for the tumor suppressing role of RYBP in NSCLC. Our clinical results demonstrated that the RYBP mRNA and protein expressions were significantly down-regulated in NSCLC and significantly linked to the poor prognosis in NSCLC patients. The enforced expression of RYBP inhibited cell survival, induced apoptosis, and increase chemosensitivity in NSCLC cells; knockdown of RYBP showed the opposite effects. Moreover, adenoviral delivery of RYBP sensitized the NSCLC cells to chemotherapy in vivo. In addition, RYBP expression was induced by paclitaxel, the first-line chemotherapeutic agent for NSCLC. Our results reveal that RYBP inhibits the aggressiveness of NSCLC cells and downregulation of RYBP is associated with poor prognosis,, suggesting that RYBP could be developed as a biomarker and a novel target for therapy in patients with lung cancer.
Keywords: RYBP, Apoptosis, Chemotherapy, Non-small cell lung cancer, Patient survival
1. Introduction
Lung cancer is one of the most diagnosed human malignancies and leading cause of cancer-related deaths worldwide [1]. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancers, responds poorly to therapy, and resists chemotherapy [2]. Despite recent progress made in the diagnosis and treatment of NSCLC, the 5-year survival rate remains poor (~15%) [1]. Therefore, there is an urgent need for a better understanding the pathogenesis of NSCLC, identifying prognostic factors, and exploring novel therapeutic targets.
The polycomb group (PcG) genes, originally identified as developmental regulators of body segmentation in Drosophila melanogaster, have been shown to be crucial for many biological processes, including development and differentiation in mammals [3–5]. PcG proteins assemble in multi-subunit nuclear complexes that have various biochemical functions, including the recognition and modification of histone posttranslational modifications and chromatin compaction [3–5]. There are two major polycomb repressive complexes, PRC1 and PRC2 [4, 5]. RING1 and YY1-binding protein (RYBP) is a newly identified member of PRC1 [6]; it binds to PcG members RING1 [7] and YY1 [8, 9] and represses gene transcription in both Drosophila [10] and mammalian cells [9].
Recently, RYBP has been found to have PcG-independent functions. It binds to several apoptotic mediators and induces apoptosis [11–13]. Enforced expression of RYBP inhibits the growth of cancer cells, but has minimal effects on non-transformed cells [14]. Additionally, RYBP has been suggested to act as a negative regulator of cell invasion [15]. More recently, we have demonstrated that RYBP activates tumor suppressor p53 through blocking MDM2-mediated p53 ubiquitination and degradation [16]. Overexpression of RYBP inhibits cancer cell growth and enhances the sensitivity of cells to chemotherapy [17]. These results implicate that RYBP may be critical in cancer development, progression and therapy.
There is an increasing interest in exploring PcG in human cancer diagnosis and treatment. Aberrant expression, missense mutation, and chromosomal translocation of multiple PcG genes have been identified in various human cancer types [3–5, 18], suggesting that PcG deregulation may promote cancer development and progression through genetic and epigenetic mechanisms. In our initial study, we found that the RYBP level is reduced in human lung and liver cancer tissues compared to the paired non-cancerous tissues [16, 17]. However, the potential role of RYBP in NSCLC prognosis is unknown. The present study was designed to demonstrate the relationship between the RYBP expressions at both mRNA and protein levels and the survival of patients with NSCLC. To explore the molecular mechanisms by which RYBP regulates cancer cell survival and sensitivity to chemotherapy, we used in vitro and in vivo human NSCLC models to determine the effects of RYBP expression and knockdown on cancer cell growth and progression. It is hoped that these investigations would provide a basis for further development of RYBP as in prognostic biomarker and a valid therapeutic target in NSCLC.
2. Materials and Methods
2.1. Patients and tissue specimens
Archived tissue samples were obtained from NSCLC patients who underwent pulmonary surgery between April 1, 2006 and June 30, 2011 at the Tianjin Medical University Cancer Institute and Hospital, Tianjin, China. The study was approved by the Medical Ethics Committee of Tianjin Medical University and a written informed consent was obtained from each of the patients. Patient follow-up information was extracted by professional census enumerator according to a uniform guideline. The clinicopathological variables used in the present study are listed in Table 1. Further details can be found in the Supplementary Methods.
Table 1.
Clinical characteristics and RYBP expression in NSCLC patients
| Variables | RYBP mRNA expression (RT-PCR) | RYBP protein expression (TMA) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| n (%) | Low | High | High (%) | P value | n (%) | − | + | + (%) | P value | |
| Age at diagnosis | ||||||||||
| ≤ 60 | 199(45.1) | 90 | 109 | 54.8 | 0.409 | 82(43.2) | 44 | 38 | 46.3 | 0.12 |
| > 60 | 242(54.9) | 119 | 123 | 50.8 | 108(56.8) | 70 | 38 | 35.2 | ||
| Gender | ||||||||||
| Male | 267(60.5) | 162 | 105 | 39.3 | <0.001 | 122(64.2) | 83 | 39 | 32 | 0.002 |
| Female | 174(39.5) | 47 | 127 | 73 | 68(35.8) | 31 | 37 | 54.4 | ||
| Family history of cancer a | ||||||||||
| No | 357(81.7) | 166 | 191 | 53.5 | 0.715 | 153(80.5) | 66 | 153 | 69.9 | 0.113 |
| Yes | 80(18.3) | 39 | 41 | 51.3 | 35(18.4) | 10 | 35 | 77.8 | ||
| Smoking history | ||||||||||
| No | 165(37.4) | 39 | 126 | 76.4 | <0.001 | 59(31.1) | 26 | 33 | 55.9 | 0.003 |
| Yes | 276(62.6) | 170 | 106 | 38.4 | 131(68.9) | 88 | 43 | 32.8 | ||
| Histology subtype | ||||||||||
| SCC | 202(45.8) | 142 | 60 | 29.7 | <0.001 | 102(53.7) | 27 | 102 | 79.1 | <0.001 |
| ADC | 194(44.0) | 44 | 150 | 77.3 | 71(37.4) | 41 | 71 | 63.4 | ||
| others | 45(10.2) | 23 | 22 | 48.9 | 17(8.9) | 8 | 17 | 68 | ||
| TNM stage | ||||||||||
| I–II | 284(64.4) | 130 | 154 | 54.2 | 0.36 | 114(60.0) | 83 | 31 | 24.6 | 0.02 |
| III–IV | 157(35.6) | 79 | 78 | 49.7 | 76(40.0) | 31 | 33 | 51.6 | ||
| Tumor infiltration | ||||||||||
| T1T2 | 380(86.2) | 170 | 210 | 55.3 | 0.005 | 164(86.3) | 98 | 66 | 40.2 | 0.863 |
| T3T4 | 61(13.8) | 39 | 22 | 36.1 | 26(13.7) | 16 | 10 | 38.5 | ||
| Lymph node metastasis a | ||||||||||
| N0 | 241(55.5) | 113 | 128 | 53.1 | 0.871 | 107(56.6) | 71 | 36 | 33.6 | 0.053 |
| N1–N3 | 193(44.5) | 92 | 101 | 52.3 | 82(43.4) | 43 | 39 | 47.6 | ||
| Distant metastasis | ||||||||||
| M0 | 415(94.1) | 196 | 219 | 52.8 | 0.784 | 183(96.3) | 112 | 71 | 38.8 | 0.084 |
| M1 | 26(5.9) | 13 | 13 | 50 | 7(3.7) | 2 | 5 | 71.4 | ||
Number not equal to the total number due to missing data; SCC: Squamous cell carcinoma; ADC: Adenocarcinoma
2.2. Tissue microarray and immunohistochemistry (IHC)
Tissue microarrays were produced as described previously [17]. All NSCLC cases were histologically reviewed by HE staining, and representative tumor areas were premarked in the paraffin blocks, away from necrotic and hemorrhagic materials. The immunohistochemical staining of serial TMAs was carried out as described previously [17]. The details and quantification of the RYBP expression are provided in the Supplementary Methods.
2.3. Chemicals, plasmids, siRNA, reagents, and cell lines
All chemicals and solvents were of analytical grade. Antibodies, plasmids, and siRNAs were obtained commercially or were generated by our lab. Human NSCLC cell lines A549, H1299, H358 and H838 were obtained from American Type Culture Collection (Manassas, VA). Normal human lung epithelial cell line NL-20 was a gift from Dr. Yi Sun (University of Michigan Health System, Ann Arbor, MI). Further details can be found in the Supplementary Methods.
2.4. Adenovirus construction, generation, purification and viral titers determination
The pAd-Easy1 adenovirus system was kindly provided by Dr. T-C He (University of Chicago). RYBP was subcloned into the pAd-track vector and then recombined with BJ5183/pAd-Easy1 competent cells by using calcium transformation method [17, 19]. The dilution which have 10–20% GFP-positive cell was used for titer calculation [17]. More details can be found in the Supplementary Methods.
2.5. Cell viability, colony formation, and apoptosis assays
Cells were transfected plasmids or infected with AdRYBP or exposed to various concentrations of Paclitaxel, and cell viability, colony formation, and apoptosis assays were performed as described previously [17, 20–23].
2.6. Western blotting analysis and Real-time reverse transcription PCR
The protein and mRNA expression of RYBP and related molecules was analyzed by Western blotting and real-time quantitative PCR, respectively [16, 17, 20–23]. Further details can be found in the Supplementary Methods.
2.7. Mouse xenograft model of NSCLC and treatment
The animal protocol was approved by the Institutional Animal Use and Care Committee (IACUC) of the Texas Tech University Health Sciences Center. The A549 and H1299 xenograft models were established as described previously [17, 20–23]. The treatment of the animals with AdRYBP and conventional chemotherapy, the evaluation of their tumor growth and clinical status, and the tissue pathology studies are detailed in the Supplemental Methods.
2.8. Hematoxylin and Eosin (H&E) Staining and TUNEL assay
The HE staining was performed as described previously [17, 22, 23]. In vivo apoptosis was measured by TUNEL staining [17, 22, 23]. All sections were analyzed under a phase-contrast Olympus microscope (Olympus America Inc, Central Valley, PA). Further details can be found in the Supplementary Methods.
2.9. Statistical analyses
All clinical statistical analyses were performed with the SPSS 18.0 software program for Windows (IBM). The value for mRNA expression level were calculated based on the formula of log102−ΔΔCt. Paired-Samples T-Test or Wilcoxon rank test was used to examine the differences in RYBP mRNA or protein levels. Kaplan-Meier method and log-rank test were used for survival analysis. Cox’s Proportional Hazard Model was used to estimate the relationship between mRNA or protein expression value and patients survival in univariate and multivariate analyses. All experimental data were analyzed using Prism software version 6 (Graph Pad software Inc., San Diego, CA, USA). The Student t test was used for comparisons between two groups. The quantitative data are reported as means ± SEM from at least three independent experiments. Differences were considered statistically significant at P < 0.05. All statistical tests were two-sided.
3. Results
3.1. RYBP is downregulated in NSCLC tissues
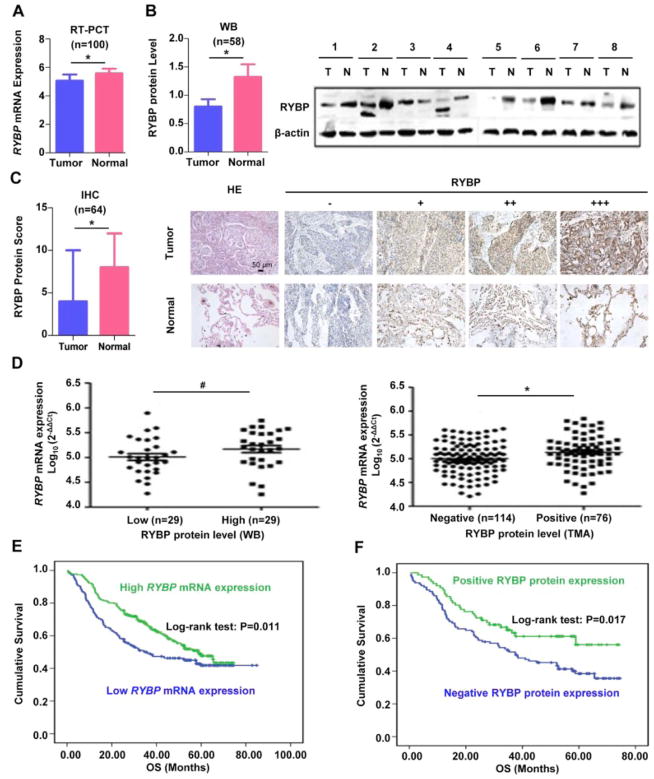
To determine the association of RYBP expression in NSCLC with disease outcome, we examined the relative RYBP mRNA levels in 100 pairs of human NSCLC tissue samples by quantitative real-time PCR. Our results showed that RYBP mRNA expression was significantly down-regulated (5.06 ± 0.44) in NSCLC specimens, as compared to that of corresponding adjacent normal lung tissues (5.58 ± 0.31) (P < 0.001; Figure 1A). To further investigate if the protein level of RYBP is associated with NSCLC, 58 pairs of human NSCLC tissue samples were examined by Western blot. We found that 38 of 58 (65.5 %) tumor samples had lower level than that in adjacent normal lung tissues (p < 0.05, Figure 1B). In addition, TMAs from 64 patients with NSCLC were also examined by IHC. As shown in Figure 1C, RYBP protein was positive in 41 cases (64 %) of tumor tissues, and 60 cases (94 %) of corresponding adjacent normal lung tissues. The RYBP protein level was significantly decreased in NSCLC compared with their corresponding adjacent normal lung tissues (P < 0.001). Moreover, Spearman’s rank correlation analysis showed that the RYBP mRNA expression was positively associated with RYBP protein level detected by Western blotting (R = 0.274, P = 0.038, Figure 1D left) and IHC (R = 0.191, P = 0.008, Figure 1D right), samples with down-regulated RYBP mRNA expression was presented with low RYBP protein expression.
Figure 1. Down-regulation of RYBP expression is common in primary NSCLC tissues.
(A) The quantitative real-time analyses of the RYBP mRNA level in training cohort of NSCLC patients and adjacent nonmalignant lung tissues (n = 100); (B) Western blot (WB) analyses were performed to determine the RYBP protein level in NSCLC patients and corresponding non-malignant adjacent tissue samples (n = 58). The graphs on the left showed the quantification of the Western blotting data; the graphs on the right showed the representative images of RYBP expression in malignant and adjacent normal lung tissues; (C) The immunohistochemical (IHC) analysis of the RYBP expression in tumor and normal adjacent tissue samples (n = 64). The graphs on the left showed the quantification of the IHC analysis; the graphs showed the representative images of RYBP expression with H&E and IHC staining on the right panel, in malignant and adjacent normal lung tissues ((−) no signal, (+) weak, (++) moderate, (+++) marked) (scale bar, 50 μm); (D) The correlation analyses of RYBP mRNA expression and RYBP protein expression levels detected by WB and IHC; (E) The overall survival (OS) rates of 441 patients with NSLCL were compared between the low or high RYBP mRNA level groups using the Kaplan–Meier method; (F) The correlation between RYBP protein expression level and survival in validation cohort of NSCLC patients (n=190). (# P < 0.05; *P < 0.01)
3.2. Low level of RYBP predicts poor prognosis in NSCLC patients
To confirm the results, the expression of RYBP mRNA was further evaluated in 441 human NSCLC tissue samples. Of these cases, 53 % (232/441) of which showed high RYBP expression, whereas 47 % (209/441) showed low RYBP expression. In addition, downregulation of RYBP was significantly associated with gender (P < 0.001), smoking history (P < 0.001), histological subtype (P < 0.001), and tumor infiltration (P = 0.005) (Supplementary Table 1). The value of RYBP protein level as a clinical predictor of NSCLC prognosis was also subsequently evaluated in 190 human NSCLC tissue samples. We found that 114 (60.0%) tumor samples were shown negative RYBP expression, while 76 (40.0%) tumor samples were shown positive. Chi-square test showed that the RYBP protein expression were associated with gender (P = 0.002), smoking history (P = 0.003), histology subtype (P < 0.001), and TNM stage (P = 0.020) (Supplementary Table 2). Log-rank test showed that NSCLC patients with low level of RYBP (both mRNA and protein) had shorter overall survival (OS) than those with high RYBP expression (Figures 1E and 1F). In Univariate and multivariate analysis, the tumor RYBP (both mRNA and protein) status was defined as an independent prognostic factor for OS (Supplementary Tables 1 and 2).
3.3. RYBP affects NSCLC cell growth and apoptosis in vitro
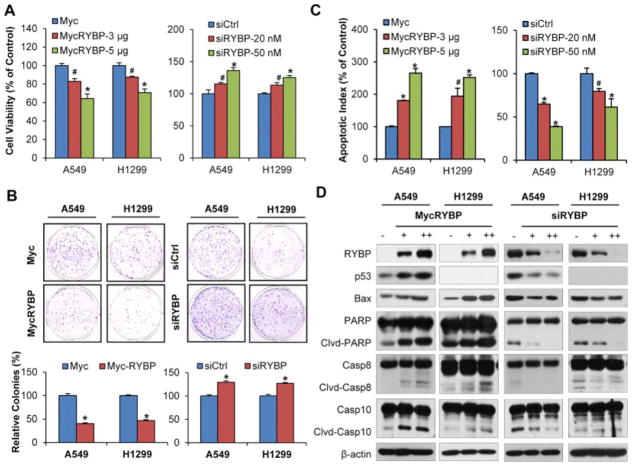
We next evaluated the expression of RYBP in normal lung epithelial cells (NL20 and BEAS-2B) and NSCLC cells (A549, H838, H358, H1299). The results showed that both RYBP mRNA expression and protein level were lower in most NSCLC cells than that in the normal lung epithelial cells (Supplemental Figures S1A and B). To investigate the potential role of RYBP in NSCLC cells, A549 (p53 wild-type) and H1299 (p53 null) cells were transfected with the Myc-RYBP or RYBP siRNA, and the cell viability was performed by using MTT and colony formation assays. As shown in Figure 2A and 2B, the enforced expression of RYBP resulted in reduced cell viability and inhibited cell colony formation in both A549 and H1299 cells, regardless of the p53 status. We then examined the apoptotic rate of control and RYBP-transfected, the results showed that RYBP overexpression induced high apoptosis in a dose-dependent manner (Figure 2C). Compared with the vehicle control, transfection of 5 μg RYBP increased the apoptotic index by 2.6-fold (P < 0.01) and 2.5-fold (P < 0.01) in the A549 and H1299 cells, respectively. In addition, RYBP overexpression also led to increased Bax, PARP cleavage, caspase-8 and -10 cleavages. The p53 levels were upregulated in the treated A549 cells (Figure 2D). In contrast, downregulation of RYBP markedly improved the cell viability and colony formation, inhibited apoptosis, and decreased the expression of proteins involved in apoptosis (Figures 2A–D).
Figure 2. RYBP affects the cell growth and apoptosis in NSCLC cells.
A549 and H1299 cells were transiently transfected with Myc-RYBP plasmid or corresponding empty vector for 24 h or with RYBP siRNA or a nontargeting negative control siRNA for 48 h. (A) The cell viability was performed by the MTT assay; (B) The cell survival was determined by the colony formation assay, graphs (lower panel) showed the quantification of the colonies; (C) The cell apoptosis was measured by the Annexin V-FITC method; and (D) RYBP and apoptosis-related proteins expression was detected by Western blot analysis. All assays were performed in triplicate, and all the experiments were repeated at least three times. (# P < 0.05; *P < 0.01).
3.4. RYBP affects the chemosensitivity of NSCLC cells to Paclitaxel in vitro
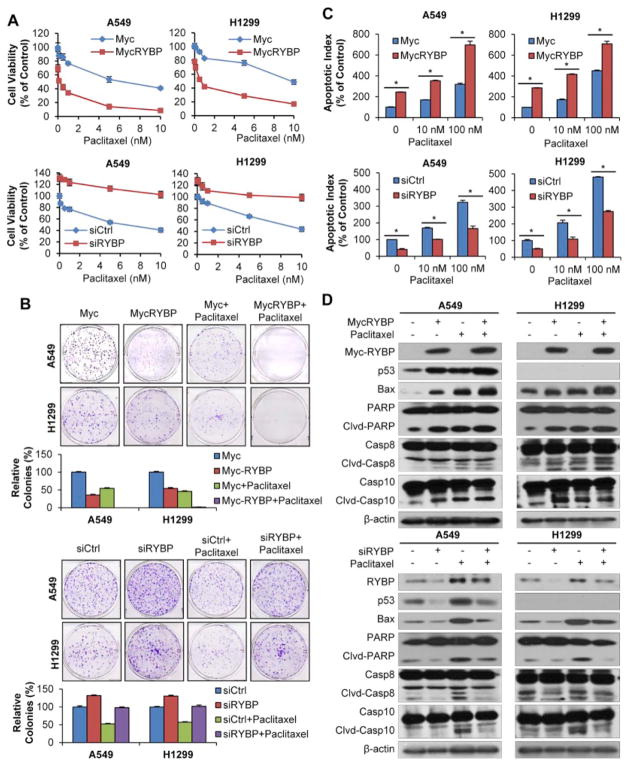
One of the strategies to enhance the cytotoxic effects of gene therapy is combinatory use of chemotherapeutic agents [24]. We tested the combined effects of RYBP and chemotherapy (Paclitaxel, the first-line anti-NSCLC agent used in clinic) in the treatment of NSCLC cells. As shown in Figure 3A, Paclitaxel inhibited the cell growth with an IC50 value 6.4 nM, enforced RYBP cooperated with Paclitaxel reduced the IC50 value to 0.1 nM. H1299 cells showed a similar sensitivity to combination treatment compared with Paclitaxel treated alone (from 13.1 nM to 0.6 nM). Contrastingly, reduced RYBP expression was associated with decreased sensitivity to Paclitaxel, the IC50 being 17.4- and 8.7-fold lower than that in A549- and H1299-RYBP-knockdown cells, respectively. Similar results were observed in colony formation assay (Figure 3B). To further explore whether the enhanced chemosensitivity by RYBP is linked to its role in regulating apoptosis, we determined the apoptosis in NSCLC cells in the presence or absence of chemotherapy. As shown in Figure 3C, 100 nM Paclitaxel alone increased the apoptotic index by about 3.2- and 4.1-fold, and combined with RYBP, enhanced the apoptotic index by about 7.0- and 7.1-fold in A549 and H1299 cells, respectively, compared to vehicle-treated cells. In contrast, RYBP knockdown resulted in less apoptotic cells, suggesting RYBP is critically involved in chemotherapy-induced apoptosis in NSCLC cells. We next wanted to know by which mechanism that RYBP mediates chemotherapy-induced cell apoptosis. As shown in Figure 3D, in both A549 and H1299 cells, Paclitaxel treatment induced RYBP expression. Paclitaxel or RYBP treatment activated Bax, Cleaved-PARP, Cleaved-Caspase 8 and -Caspase 10. In addition, RYBP overexpression enhanced, while RYBP knockdown inhibited the Paclitaxel-induced protein levels, indicating that RYBP may be involved in Paclitaxel-induced cell death through the regulation of pro-apoptotic protein expression.
Figure 3. RYBP sensitizes NSCLC cells to Paclitaxel.
A549 and H1299 cells were treated with Paclitaxel (0–100 nM for MTT assay, and 5 nM for other assays) and Myc-RYBP plasmid (5 μg, 24 h) or RYBP siRNA (50 nM, 48 h), then the cell viability was determined by the MTT assay (A); the cell survival was determined by the colony formation assay, graphs (lower panel) showed the quantification of the colonies (B); the cell apoptosis was determined by the Annexin V-FITC method (C); RYBP and apoptosis-related proteins expression was detected by Western blot analysis (D). All assays were performed in triplicate, and all the experiments were repeated at least three times. (# P < 0.05; *P < 0.01).
3.5. AdRYBP suppresses the malignant properties of NSCLC cells and sensitizes NSCLC cells to chemotherapy in vitro
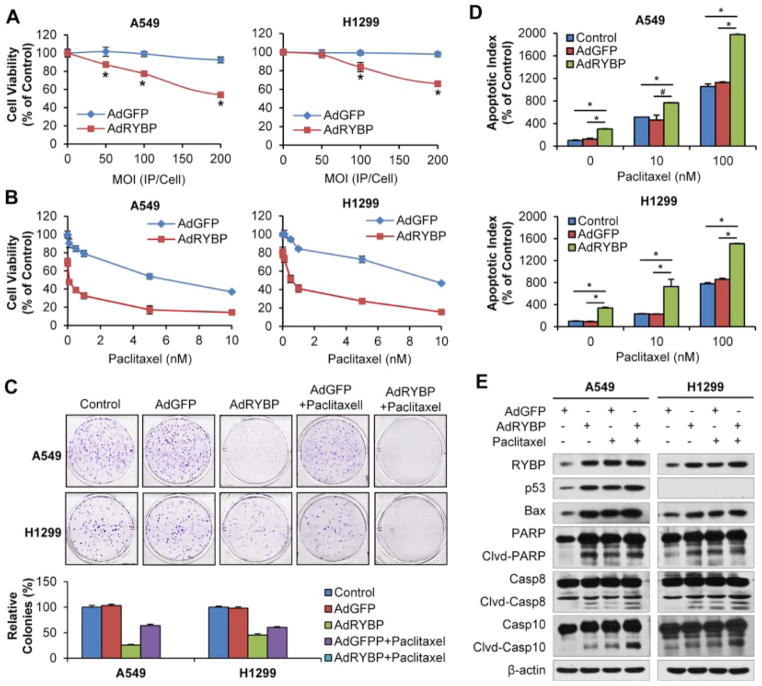
Gene therapy represents a novel treatment approach for cancer and adenoviral vectors have been used extensively in cancer gene therapy [25]. We generated a replication-deficient adenovirus driving the expression of RYBP (AdRYBP) to determine whether the virus will suppress or eradicate the tumor. Consistent with the results obtained by plasmid transfection method, AdRYBP infection, but not AdGFP, decreased the cell viability, inhibited cell colony formation, induced apoptosis and modulated protein expression in both cell lines, regardless of the p53 status (Figure 4A–E). Similarly, combining AdRYBP with conventional therapies significantly decreased cell viability and dramatically increased cell apoptosis and related protein levels compared to Paclitaxel treatment alone (all P < 0.01; Figure 4A–E). The results indicated that the combination of AdRYBP and Paclitaxel was more efficacious than AdRYBP or Paclitaxel alone and RYBP could sensitize the NSCLC cells to the chemotherapeutic agent.
Figure 4. AdRYBP infection induces NSCLC cell growth arrest and apoptosis, and increases the chemosensitivity of NSCLC cells to Paclitaxel.
(A) A549 and H1299 cells were infected with AdRYBP (0, 50, 100 and 200MOI) or AdGFP (0, 50, 100 and 200MOI) for 24 h, then the cell viability was determined by the MTT assay; (B) Cells were treated with Paclitaxel (0–10 nM) and infected AdRYBP (200 MOI, 24 h), then the cell viability were determined by the MTT assay; (C) Cells were treated with Paclitaxel (5 nM) or/and infected AdRYBP (200 MOI, 24 h), then maintained for additional 7–10 days. The cell survival was performed by the colony formation assay, graphs (lower panel) showed the quantification of the colonies; (D) Cells were treated with Paclitaxel (0, 10 and 100 nM) or/and infected AdRYBP (200 MOI, 24 h), the cell apoptosis was measured by the Annexin V-FITC method; (E) Cells were treated with Paclitaxel (5 nM) or/and infected AdRYBP (200 MOI, 24 h), RYBP and apoptosis-related proteins expression was detected by Western blot analysis. All assays were performed in triplicate, and all the experiments were repeated at least three times. (# P < 0.05; *P < 0.01).
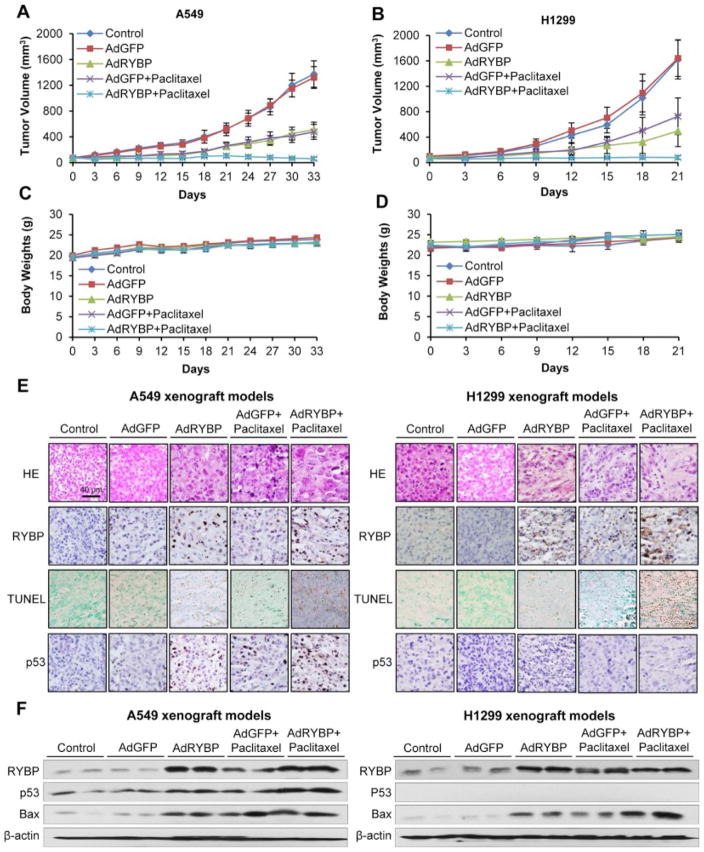
3.6. AdRYBP suppresses the NSCLC tumor growth and sensitizes NSCLC cells to chemotherapy in vivo
The anticancer effects of RYBP were then evaluated in vivo. As shown in Figure 5, AdRYBP treatment, not AdGFP, decreased the A549 and H1299 xenograft tumor growth by about 62% (P < 0.01) on Day 33 (Figure 5A), and 70% (P < 0.01) on Day 21 (Figure 5B), respectively. Tumor growth was inhibited by 63% and 66% in Paclitaxel treatment group (Figure 5A and B), respectively. When AdRYBP combined with Paclitaxel, the tumor growths were almost regressed in both models (Figure 5A and B). There were no remarkable differences in the body weight (as a surrogate marker of toxicity) (Figure 5C and D), and gross necropsy revealed no internal abnormalities in different tissues (Liver, lung, kidney, spleen, heart and brain) in all treatment groups (Supplementary Figure 2), indicating that there were no significant side effects of the treatments. We then compared the protein levels from different treatment groups to investigate the mechanism(s) by which RYBP inhibited tumor growth and sensitized chemotherapy. As shown in Figure 5E, in situ TUNEL assays showed substantially increased apoptosis in the tumors of AdRYBP- and Paclitaxel-treated mice, whereas the combination of AdRYBP and Paclitaxel dramatically enhanced the apoptotic cells, supporting the notion that RYBP performs anti-proliferative role through the induction of apoptosis in NSCLC cells. These observations were further confirmed by western blotting analyses, and similar results were obtained in both xenograft models. Paclitaxel and AdRYBP treatment induced RYBP and Bax protein levels, and AdRYBP enhanced the Paclitaxel-induced expression of p53 and Bax (Figure 5F).
Figure 5. RYBP suppresses NSCLC tumor growth and sensitizes NSCLC cells to chemotherapy in vivo.
AdGFP or AdRYBP (2×109 infectious particles (IP)) was administered by intratumoral injection every three days, and Paclitaxel (15 mg/kg) was administered by intraperitoneal injection every two weeks to nude mice bearing A549 (A) or H1299 (B) tumors. The animals were monitored for changes in body weight as a surrogate marker for toxicity in both the A549 (C) and H1299 (D) xenograft models. At the end of the experiment, xenograft tumors were exercised, fixed or homogenized and analyzed for protein expression by H&E, immunohistochemistry and TUNEL straining (scale bar, 40 μm) (E) and Western blot (F).
4. Discussion
In this study, we used an extensive collection of NSCLC tumors and in vitro and in vivo experiments, and described a novel role of RYBP in the progression of NSCLC and the correlation between RYBP expression and survival in NSCLC patients. Herein, we have demonstrated at least four novel points: First, both RYBP mRNA expression and protein level were downregulated in NSCLC tissues, the low level of RYBP predicted poor patient survival in NSCLC; Second, RYBP overexpression decreased cell viability, inhibited colony formation and induced apoptosis in NSCLC cells, and promotion of apoptosis was one of the main mechanism of RYBP-mediated cell growth inhibition in NSCLC; Third, successful AdRYBP infection led to a decreased NSCLC tumor xenograft growth; Forth, chemotherapeutic agent induced RYBP expression, and RYBP sensitized NSCLC cells to conventional chemotherapeutic agent by apoptosis induction both in vitro and in vivo. These results demonstrated that RYBP may be a novel prognostic factor in patients with NSCLC and be a target for cancer therapy.
Although the previous studies from the other researchers and our lab have implicated RYBP as a potential tumor suppressor gene [16, 17, 26–28], the physiological and pathological role(s) of RYBP are still poorly understood. Integrative genomic profiling is shown the loss of RYBP gene in prostate and cervical cancers and the decrease of RYBP transcript has been linked to poor prognosis [27, 28]. We demonstrate that the low RYBP expression is an independent predictor of a poor prognosis in patients with HCC [17]. In contrary, RYBP is suggested to be overexpressed in primary classic Hodgkin’s lymphoma and adult T-cell leukemia/lymphoma cases [29, 30]. Although the discrepancy between solid tumors and lymphoma has not been addressed, it seems that RYBP participates in carcinogenesis and cancer progression via distinct mechanisms. In the present study, to our best knowledge, we presented the first large-scale study to examine the prognostic value of RYBP expression in NSCLC patients. The results highlighted RYBP as a signature molecule correlated with the prognosis of NSCLC patients. We found that NSCLC patients with low levels of RYBP (both mRNA and protein) were at significantly high risk of overall survival and RYBP was an independent prognostic factor.
In a recent study, we have observed that RYBP positively regulates the levels and function of p53 and involves in cell cycle progression and cellular response to DNA damage [16]. These findings suggest that RYBP may have an essential role in regulating the p53 pathway. In the present study, we demonstrated that enforced RYBP significantly reduced the cell viability and induced cell apoptosis in NSCLC cells regardless of p53 status. Intriguingly, we also observed the effect of RYBP on cell cycle progression, consistent with our previous study, our results showed that enforced RYBP arrested cells in G1 phase in A549 cells, while no significant effects were observed in H1299 cells (data not shown). Although our previous studies show that RYBP inhibits MDM2-mediated ubiquintation of p53, we cannot exclude other possibilities. Since MDM2 has E3 ligase activity and RYBP can be monoubiquitinated [31], it is possible that MDM2 promotes RYBP ubiquitination, thus modulating the ability of RYBP to induce apoptosis or regulate transcription. Actually, MDM2 is capable of repressing transcription when tethered to a reporter gene promoter in the absence of p53 [32]. Therefore, it is possible that RYBP and MDM2 (with or without other PcG members) may form a protein complex in which RYBP recruits MDM2 to its target gene promoter(s). Alternatively, MDM2 may ubiquitinate RYBP or other members of the PcG family to directly regulate the trans-repressional activities of the PcG complex. In p53 non-functional cells, it is also possible that RYBP directly binds to Bax or other targets. As examples of the potential p53-independent effects of RYBP, RYBP physically binds to several apoptotic mediators, such as caspases-8 and -10, and Fas-associated protein with death domain (FADD) through their cytoplasmic death effector domains to form the death inducing signaling complex (DISC), resulting in the proteolytic cleavage and activation of these caspases, and subsequent induction of apoptosis [11]. Based on our observation, it is possible that RYBP regulates cell cycle arrest and apoptosis induction by different mechanisms. It is also possible that the apoptosis induction by RYBP is a process independent of p53 and needs other partners. In the future, the possible p53-independent functions of RYBP need to be explored, which will provide a complete picture for RYBP’s role in carcinogenesis and cancer development.
The multifaceted function of RYBP in inducing apoptosis and regulating the p53 tumor suppressor provides an ideal target for cancer therapy, and may be useful either alone or in combination with conventional chemotherapeutics. In this study, we also demonstrated that chemotherapeutic agent can induce RYBP expression and gene delivery of RYBP combined with chemotherapy may provide a better efficacy for NSCLC therapy. However, the mechanisms underlying RYBP-mediated chemosensitization are still not fully understood. Further studies are needed to explore the molecular mechanisms by which RYBP inhibits cancer progression and increases chemosensitization, and whether other members are capable of regulating RYBP.
In conclusion, we demonstrate the downregulation of RYBP and an inverse correlation between RYBP expression levels and prognosis in NSCLC patients. Based on the observed specific effects of RYBP on NSCLC cells, reactivating RYBP in cancer cells that are RYBP-deficient or express low levels of RYBP may provide an effective and safe therapeutic approach to NSCLC therapy.
Supplementary Material
Highlights.
Both RYBP mRNA expression and protein level were downregulated in NSCLC tissues, the low level of RYBP predicted poor patient survival in NSCLC.
Enforced RYBP inhibits cell viability and induces apoptosis in NSCLC cell.
The one of the main mechanism of RYBP-mediated cell growth inhibition is promotion of apoptosis.
Successful AdRYBP infection inhibits NSCLC tumor xenograft growth.
Chemotherapeutic agent induces the expression of RYBP.
RYBP sensitizes NSCLC cells to conventional chemotherapy both in vitro and in vivo.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grant R01 CA186662 (to R.Z.). The content is solely the responsibility of the authors, and do not necessarily represent the official views of the National Institutes of Health. This work was also supported by the National Natural Science Foundation of China (NSFC) grants No. 81372229 (to B.Q.). This work was also supported by the American Cancer Society (ACS) grant RSG-15-009-01-CDD (to W.W.).
Footnotes
Conflict of interest
We do not have any conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Miller KD, Jemal A. Cancer statistics, 2015. C A Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–19. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 3.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 4.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 5.Wang W, Qin JJ, Voruganti S, Nag S, Zhou J, Zhang R. Polycomb group (PcG) proteins and human cancers: multifaceted functions and therapeutic implications. Med Res Rev. 2015 Jul 30; doi: 10.1002/med.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int J Dev Biol. 2009;53:355–70. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- 7.Wang R, Taylor AB, Leal BZ, Chadwell LV, Ilangovan U, Robinson AK, et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure. 2010;18:966–75. doi: 10.1016/j.str.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalenik JL, Chen D, Bradley ME, Chen SJ, Lee TC. Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Res. 1997;25:843–9. doi: 10.1093/nar/25.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia E, Marcos-Gutierrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999;18:3404–18. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bejarano F, Gonzalez I, Vidal M, Busturia A. The Drosophila RYBP gene functions as a Polycomb-dependent transcriptional repressor. Mech Dev. 2005;122:1118–29. doi: 10.1016/j.mod.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Zheng L, Schickling O, Peter ME, Lenardo MJ. The death effector domain-associated factor plays distinct regulatory roles in the nucleus and cytoplasm. J Biol Chem. 2001;276:31945–52. doi: 10.1074/jbc.M102799200. [DOI] [PubMed] [Google Scholar]
- 12.Danen-van Oorschot AA, Voskamp P, Seelen MC, van Miltenburg MH, Bolk MW, Tait SW, et al. Human death effector domain-associated factor interacts with the viral apoptosis agonist Apoptin and exerts tumor-preferential cell killing. Cell Death Differ. 2004;11:564–73. doi: 10.1038/sj.cdd.4401391. [DOI] [PubMed] [Google Scholar]
- 13.Stanton SE, Blanck JK, Locker J, Schreiber-Agus N. Rybp interacts with Hippi and enhances Hippi-mediated apoptosis. Apoptosis. 2007;12:2197–206. doi: 10.1007/s10495-007-0131-3. [DOI] [PubMed] [Google Scholar]
- 14.Novak RL, Phillips AC. Adenoviral-mediated Rybp expression promotes tumor cell-specific apoptosis. Cancer Gene Ther. 2008;15:713–22. doi: 10.1038/cgt.2008.25. [DOI] [PubMed] [Google Scholar]
- 15.McGarry LC, Winnie JN, Ozanne BW. Invasion of v-Fos(FBR)-transformed cells is dependent upon histone deacetylase activity and suppression of histone deacetylase regulated genes. Oncogene. 2004;23:5284–92. doi: 10.1038/sj.onc.1207687. [DOI] [PubMed] [Google Scholar]
- 16.Chen D, Zhang J, Li M, Rayburn ER, Wang H, Zhang R. RYBP stabilizes p53 by modulating MDM2. EMBO Rep. 2009;10:166–72. doi: 10.1038/embor.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Cheng J, Qin JJ, Voruganti S, Nag S, Fan J, et al. RYBP expression is associated with better survival of patients with hepatocellular carcinoma (HCC) and responsiveness to chemotherapy of HCC cells in vitro and in vivo. Oncotarget. 2014;5:11604–19. doi: 10.18632/oncotarget.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 19.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–47. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Rayburn ER, Hang J, Zhao Y, Wang H, Zhang R. Anti-lung cancer effects of novel ginsenoside 25-OCH(3)-PPD. Lung Cancer. 2009;65:306–11. doi: 10.1016/j.lungcan.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Rayburn ER, Velu SE, Nadkarni DH, Murugesan S, Zhang R. In vitro and in vivo anticancer activity of novel synthetic makaluvamine analogues. Clin Cancer Res. 2009;15:3511–18. doi: 10.1158/1078-0432.CCR-08-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Qin JJ, Voruganti S, Wang MH, Sharma H, Patil S, et al. Identification of a new class of mdm2 inhibitor that inhibits growth of orthotopic pancreatic tumors in mice. Gastroenterology. 2014;147:893–902. doi: 10.1053/j.gastro.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Qin JJ, Voruganti S, Srivenugopal K, Nag S, Patil S, et al. The pyrido[b]indole MDM2 inhibitor SP-141 exerts potent therapeutic effects in breast cancer models. Nat Commun. 2014;5:5086. doi: 10.1038/ncomms6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang B, Roth JA. The role of gene therapy in combined modality treatment strategies for cancer. Curr Opin Mol Ther. 2003;5:475–82. [PubMed] [Google Scholar]
- 25.Kaplan JM. Adenovirus-based cancer gene therapy. Curr Gene Ther. 2005;5:595–605. doi: 10.2174/156652305774964677. [DOI] [PubMed] [Google Scholar]
- 26.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lando M, Holden M, Bergersen LC, Svendsrud DH, Stokke T, Sundfor K, et al. Gene dosage, expression, and ontology analysis identifies driver genes in the carcinogenesis and chemoradioresistance of cervical cancer. PLoS Genet. 2009;5:e1000719. doi: 10.1371/journal.pgen.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lando M, Wilting SM, Snipstad K, Clancy T, Bierkens M, Aarnes EK, et al. Identification of eight candidate target genes of the recurrent 3p12-p14 loss in cervical cancer by integrative genomic profiling. J Pathol. 2013;230:59–69. doi: 10.1002/path.4168. [DOI] [PubMed] [Google Scholar]
- 29.Seitz V, Thomas PE, Zimmermann K, Paul U, Ehlers A, Joosten M, et al. Classical Hodgkin’s lymphoma shows epigenetic features of abortive plasma cell differentiation. Haematologica. 2011;96:863–70. doi: 10.3324/haematol.2010.031138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki D, Imaizumi Y, Hasegawa H, Osaka A, Tsukasaki K, Choi YL, et al. Overexpression of Enhancer of zeste homolog 2 with trimethylation of lysine 27 on histone H3 in adult T-cell leukemia/lymphoma as a target for epigenetic therapy. Haematologica. 2011;96:712–9. doi: 10.3324/haematol.2010.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006;580:6233–41. doi: 10.1016/j.febslet.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 32.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–9. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.