Abstract
Fibroids represent a major public healthcare problem as the most prevalent pelvic tumors in women of reproductive age and as the leading cause of gynecological surgeries in the U.S. Th recent advances in the genomic technologies including genome-wide association studies and high throughput sequencing provide insight into their pathogenesis and molecular classification. Understanding the molecular basis of fibroids may facilitate development of effective targeted treatment options of this very common disease.
Introduction
Fibroids (uterine leiomyomas or myomas) are benign smooth muscle neoplasm of the uterus and the most common pelvic tumors in women of reproductive age. They are the leading cause of hysterectomies worldwide and the most common indication for gynecological surgeries in the U.S. The lifetime prevalence of fibroids is over 80% among black women and nearly 70% among white women. The annual societal cost for fibroids is estimated up to 34 billion dollars, calculated through combined expenditures for medical management of symptomatic fibroids, lost work attributable to diagnosis of fibroids, and obstetrical complications of fibroids.1 Therefore, finding effective and improved therapeutical options is considered to be crucial for overcoming this major public health problem. An important step towards this goal is to explore the molecular basis of fibroids to understand and target the underlying specific pathophysiological pathways.
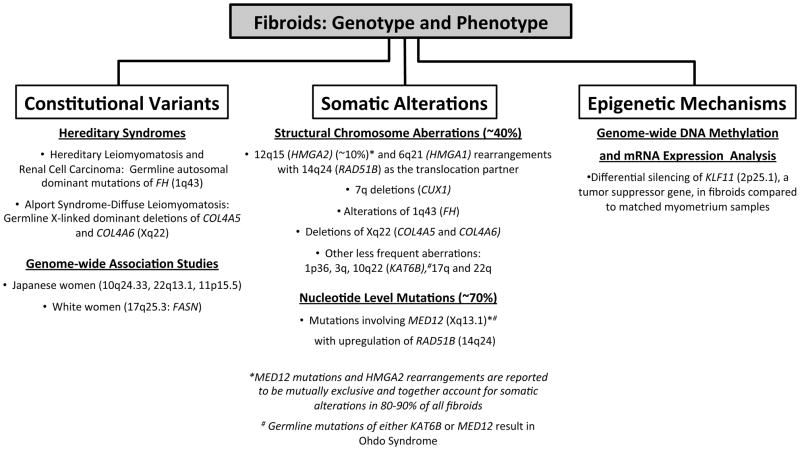
Genetic factors have been implicated to play an important role in the development of fibroids through twin and familial aggregation studies, as well as through the observations of ethnic disparities in the incidence and clinical presentation of fibroids as exemplified by black women having increased prevalence, more severe symptoms, and earlier age of onset in comparison to white women.2 Herein, the ever-increasing genomic evidence informing the phenotypic profile of this very common disease will be reviewed under the categories of constitutional genetic variants, somatic alterations, and epigenetic mechanisms (figure1).
Figure 1.
Summarized genomic evidence informing the phenotypic profile of fibroids.
Constitutional Genetic Variants
The constitutional genetic variants pertain to molecular aspects of the inherited genome and may be analyzed by evaluating the genetic basis of hereditary conditions or the association of a disease with genetic polymorphisms.
Several familial tumor susceptibility syndromes have been characterized by smooth muscle neoplasms in the uterus and other organ systems. In particular, hereditary leiomyomatosis and renal cell carcinoma (HLRCC), an autosomal dominant syndrome resulting from germline mutations of fumarate hydratase gene (FH), is associated with multiple early onset symptomatic uterine fibroids, in addition to cutaneous leiomyomas and renal cell cancer.3 Alport Syndrome and Diffuse Leimyomatosis (ATS-DL) is another hereditary condition combining the features of Alport Syndrome with diffuse leiomyomatosis of the esophageal, tracheobronchial, and genitourinary tract, with germline X-linked dominant deletions in COL4A5 and COL4A6 on Xq22.4
Genome-wide association studies (GWAS) is a powerful approach for mapping disease genes through a computational analysis of common variants in the constitutional human genome. A GWAS in Japanese women identified three chromosomal loci to be associated with susceptibility to fibroids: 10q24.33, 22q13.1, and 11p15.5.5 Another genome-wide linkage and association study of white women described fatty acid synthase gene (FASN) on 17q25.3 as a candidate gene involved in predisposition to fibroids.6 Lastly, a study analyzing the ancestry informative markers by an admixture-based genome-wide scan in black women showed that mean proportion of European ancestry markers is much lower in women with fibroid diagnosis in comparison to controls. The same study also analyzed a set of markers for the significant loci reported in the Japanese GWAS, but the associations were not replicated in this black women cohort.7 Future studies with larger cohorts may provide additional insights into specific genes predisposing to development of fibroids as well as the ethnic disparities in the disease presentation.
Somatic Alterations
The acquired changes in the genomic landscape of fibroids have been analyzed through the traditional cytogenetic methods for decades and have expanded further with the recent advances in the high throughput next-generation sequencing technologies. These somatic alterations detected in the tumor genome can be grouped into “structural chromosome aberrations” and “nucleotide level mutations”.
Based on conventional cytogenetic studies approximately 40% of uterine leiomyomas have recurrent structural chromosome aberrations including rearrangements of 12q15 and 6q21, as well as deletions of 7q, involving 20%, 5%, and 17% of the cases with a chromosomal abnormality, respectively. Other less frequent aberrations are rearrangements of 1p36, 1q43, 3q, 10q22, 17q24 and 22q.8 High-mobility group AT-hook genes are upregulated in tumors with 12q15 (HMGA2) and 6q21 (HMGA1) rearrangements, having RAD51B, a DNA repair protein encoding gene located on 14q24, as their most common translocation partner.9, 10 For the cases with deletions of 7q, CUX1 is found to be disrupted by inversions and located in the minimally deleted region of 7q22.1.11 The fibroids of HLRCC patients typically harbor biallelic loss of FH (1q43), which is also reported in 1.3% of sporadic fibroids.12 KAT6B is mapped to the 10q22 breakpoint of the recurrent t(10;17), which is present in 2% of fibroids.13
Whole exome sequencing provides nucleotide level precision for detecting mutations in the protein-coding regions. A striking finding of this advanced high throughput technology has been the discovery of the MED12 mutations (Xq13.1) in approximately 70% of fibroids.14 In addition, characterization of fibroids by whole genome sequencing revealed further insight into the previously reported structural chromosome aberrations and identified cryptic genomic rearrangements that were not apparent by conventional G-banded karyotyping and/or chromosomal microarrays. One such finding is the phenomenon described as “complex chromosome rearrangements” with multiple interconnected breakage and reunion events, as illustrated in cases with 12q15 (HMGA2) and 14q24 (RAD51B) rearrangements. In addition, aberrations involving 7q are discovered to be more complex events with inversions, translocations, and deletions at various loci. However, CUX1 remains to be the most commonly deleted gene associated with 7q rearrangements. Another significant finding of the whole genome sequencing analysis was that a subset of fibroid cases is discovered to harbor somatic deletions within the COL4A5-COL4A6 locus of Xq22.3, which corresponds to the germline deletion locus detected in ATS-DL syndrome.15
In light of these recent molecular discoveries through high throughput sequencing, fibroids can be classified into different molecular subtypes. Fibroids with HMGA2 rearrangements and MED12 mutations are mutually exclusive with distinct gene expression profiles, suggesting two separate molecular pathways and together they account for 80 to 90% of all fibroid cases. While RAD51B is disrupted by HMGA2 translocations, it is upregulated in cases with MED12 mutations. Fibroids with HMGA2 rearrangements are larger in size;16 whereas the MED12 mutations are the most common somatic alteration detected in fibroids. HMGA1 alterations co-occur with MED12 mutations, and 7q rearrangements can be detected in both groups. Complex chromosome rearrangements are observed mainly in cases without MED12 mutations. Lastly, cases with somatic rearrangements in the loci associated with germline alterations in hereditary syndromes (FH for HLRCC and COL4A5-COL4A6 for ATS-DL) present unique molecular profiles in comparison to the HMGA2 and MED12 groups despite their much lower frequency.14, 15 Following this molecular classification of fibroids with HMGA2, MED12, FH, and COL4A5-COL4A6 somatic alterations, only a small fraction of cases (<10%) remain to be without an identifiable driver mutation. Somatic alterations of 10q22 and 1p36 are two other less frequent structural rearrangements that may be within this small fraction. Interestingly, 10q22 aberrations occur frequently in leiomyosarcomas17 and fibroids with 1p36 deletions cluster together with leiomyosarcomas in a gene expression profiling study,18 suggesting that these groups might be involved in malignant progression. Of note, germline mutations of KAT6B (10q22) is associated with Ohdo Syndrome19, a heterogenous group of disorders with intellectual disability and craniofacial anomalies, which also characterized by germline mutations in MED12.20 Taken together with the mapping of KAT6B to 10q22 breakpoint of the recurrent t(10;17) aberrations in fibroids,13 this gene may also represent a target for candidate driver mutations in fibroids.
Epigenetic Mechanisms
DNA methylation and histone modification are epigenetic mechanisms regulating the gene expression independent from the DNA sequence of the genome. A genome wide study analyzing the DNA methylation and mRNA expression in fibroids with matched myometrium tissue from black women revealed 55 genes that are different in between the two tissue types. The majority of these genes are silenced in fibroids (62%), including KLF11 (2p25.1), a tumor suppressor gene and also a target of progesterone or antiprogestins in the fibroid tissue.21 Therefore, KLF11 might have a significant role in the fibroid pathogenesis.
Conclusion
The constitutional, somatic, and epigenetic alterations observed in fibroids elucidate distinct molecular pathways involved in the pathogenesis of fibroids, that may inform the future targeted treatment options for fibroids.
References
- 1.Cardozo ER, Clark AD, Banks NK, Henne MB, Stegmann BJ, Segars JH. The estimated annual cost of uterine leiomyomata in the United States. Am J Obstet Gynecol. 2012;206:211e211–219. doi: 10.1016/j.ajog.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okolo S. Incidence, aetiology and epidemiology of uterine fibroids. Best Pract Res Clin Obstet Gynaecol. 2008;22:571–588. doi: 10.1016/j.bpobgyn.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci U S A. 2001;98:3387–3392. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Torres R, Cruz D, Orozco L, Heidet L, Gubler MC. Alport syndrome and diffuse leiomyomatosis. Clinical aspects, pathology, molecular biology and extracellular matrix studies A synthesis. Nephrologie. 2000;21:9–12. [PubMed] [Google Scholar]
- 5.Cha PC, Takahashi A, Hosono N, Low SK, Kamatani N, Kubo M, Nakamura Y. A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nat Genet. 2011;43:447–450. doi: 10.1038/ng.805. [DOI] [PubMed] [Google Scholar]
- 6.Eggert SL, Huyck KL, Somasundaram P, Kavalla R, Stewart EA, Lu AT, Painter JN, Montgomery GW, Medland SE, Nyholt DR, Treloar SA, Zondervan KT, Heath AC, Madden PA, Rose L, Buring JE, Ridker PM, Chasman DI, Martin NG, Cantor RM, Morton CC. Genome-wide linkage and association analyses implicate FASN in predisposition to Uterine Leiomyomata. Am J Hum Genet. 2012;91:621–628. doi: 10.1016/j.ajhg.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wise LA, Ruiz-Narvaez EA, Palmer JR, Cozier YC, Tandon A, Patterson N, Radin RG, Rosenberg L, Reich D. African ancestry and genetic risk for uterine leiomyomata. Am J Epidemiol. 2012;176:1159–1168. doi: 10.1093/aje/kws276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandberg AA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: leiomyoma. Cancer Genet Cytogenet. 2005;158:1–26. doi: 10.1016/j.cancergencyto.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Van de Ven WJ. Genetic basis of uterine leiomyoma: involvement of high mobility group protein genes. Eur J Obstet Gynecol Reprod Biol. 1998;81:289–293. doi: 10.1016/s0301-2115(98)00204-8. [DOI] [PubMed] [Google Scholar]
- 10.Quade BJ, Weremowicz S, Neskey DM, Vanni R, Ladd C, Dal Cin P, Morton CC. Fusion transcripts involving HMGA2 are not a common molecular mechanism in uterine leiomyomata with rearrangements in 12q15. Cancer Res. 2003;63:1351–1358. [PubMed] [Google Scholar]
- 11.Schoenmakers EF, Bunt J, Hermers L, Schepens M, Merkx G, Janssen B, Kersten M, Huys E, Pauwels P, Debiec-Rychter M, van Kessel AG. Identification of CUX1 as the recurrent chromosomal band 7q22 target gene in human uterine leiomyoma. Genes Chromosomes Cancer. 2013;52:11–23. doi: 10.1002/gcc.22001. [DOI] [PubMed] [Google Scholar]
- 12.Lehtonen R, Kiuru M, Vanharanta S, Sjoberg J, Aaltonen LM, Aittomaki K, Arola J, Butzow R, Eng C, Husgafvel-Pursiainen K, Isola J, Jarvinen H, Koivisto P, Mecklin JP, Peltomaki P, Salovaara R, Wasenius VM, Karhu A, Launonen V, Nupponen NN, Aaltonen LA. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am J Pathol. 2004;164:17–22. doi: 10.1016/S0002-9440(10)63091-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore SD, Herrick SR, Ince TA, Kleinman MS, Dal Cin P, Morton CC, Quade BJ. Uterine leiomyomata with t(10;17) disrupt the histone acetyltransferase MORF. Cancer Res. 2004;64:5570–5577. doi: 10.1158/0008-5472.CAN-04-0050. [DOI] [PubMed] [Google Scholar]
- 14.Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M, Aavikko M, Katainen R, Virolainen E, Bohling T, Koski TA, Launonen V, Sjoberg J, Taipale J, Vahteristo P, Aaltonen LA. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 15.Mehine M, Kaasinen E, Makinen N, Katainen R, Kampjarvi K, Pitkanen E, Heinonen HR, Butzow R, Kilpivaara O, Kuosmanen A, Ristolainen H, Gentile M, Sjoberg J, Vahteristo P, Aaltonen LA. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med. 2013;369:43–53. doi: 10.1056/NEJMoa1302736. [DOI] [PubMed] [Google Scholar]
- 16.Rein MS, Powell WL, Walters FC, Weremowicz S, Cantor RM, Barbieri RL, Morton CC. Cytogenetic abnormalities in uterine myomas are associated with myoma size. Mol Hum Reprod. 1998;4:83–86. doi: 10.1093/molehr/4.1.83. [DOI] [PubMed] [Google Scholar]
- 17.Quade BJ, Pinto AP, Howard DR, Peters WA, 3rd, Crum CP. Frequent loss of heterozygosity for chromosome 10 in uterine leiomyosarcoma in contrast to leiomyoma. Am J Pathol. 1999;154:945–950. doi: 10.1016/S0002-9440(10)65342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christacos NC, Quade BJ, Dal Cin P, Morton CC. Uterine leiomyomata with deletions of Ip represent a distinct cytogenetic subgroup associated with unusual histologic features. Genes Chromosomes Cancer. 2006;45:304–312. doi: 10.1002/gcc.20291. [DOI] [PubMed] [Google Scholar]
- 19.Clayton-Smith J, O’Sullivan J, Daly S, Bhaskar S, Day R, Anderson B, Voss AK, Thomas T, Biesecker LG, Smith P, Fryer A, Chandler KE, Kerr B, Tassabehji M, Lynch SA, Krajewska-Walasek M, McKee S, Smith J, Sweeney E, Mansour S, Mohammed S, Donnai D, Black G. Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am J Hum Genet. 2011;89:675–681. doi: 10.1016/j.ajhg.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vulto-van Silfhout AT, de Vries BB, van Bon BW, Hoischen A, Ruiterkamp-Versteeg M, Gilissen C, Gao F, van Zwam M, Harteveld CL, van Essen AJ, Hamel BC, Kleefstra T, Willemsen MA, Yntema HG, van Bokhoven H, Brunner HG, Boyer TG, de Brouwer AP. Mutations in MED12 cause X-linked Ohdo syndrome. Am J Hum Genet. 2013;92:401–406. doi: 10.1016/j.ajhg.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Chiang TC, Richard-Davis G, Barrett JC, McLachlan JA. DNA hypomethylation and imbalanced expression of DNA methyltransferases (DNMT1, 3A, and 3B) in human uterine leiomyoma. Gynecol Oncol. 2003;90:123–130. doi: 10.1016/s0090-8258(03)00194-x. [DOI] [PubMed] [Google Scholar]