Abstract
Among the prokaryotic genomic islands (GIs) involved in horizontal gene transfer (HGT) are the classical pathogenicity islands, including the integrative and conjugative elements (ICEs), the gene-transfer agents (GTAs), and the staphylococcal pathogenicity islands (SaPIs), the primary focus of this review. While the ICEs and GTAs mediate HGT autonomously, the SaPIs are dependent on specific phages. The ICEs transfer primarily their own DNA the GTAs exclusively unlinked host DNA and the SaPIs combine the capabilities of both. Thus the SaPIs derive their importance from the genes they carry (their genetic cargo) and the genes they move. They act not only as versatile high frequency mobilizers, but also as mediators of phage interference, and consequently are major benefactors of their host bacteria.
Keywords: Pathogenicity Island, mobile genetic element, phage interference, horizontal gene transfer, staphylococcus, transduction
GIs and their mobility
Genomic islands (GIs) in prokaryotes are discrete inserted DNA segments, many of which are mobile, carry genes that impact the pathobiology of their host organisms, and contribute significantly to host fitness [1] [2]. Three major classes are the staphylococcal pathogenicity islands (SaPIs), the gene transfer agents (GTAs) and the integrative and conjugative elements (ICEs). The first two were probably derived from ancestral prophages and have retained certain key elements of the prophage design. The third group was almost certainly derived primarily from conjugative plasmids, acquiring other features and becoming mosaics. Immobile GIs are presumed to be variants of mobile ones.
In this review we focus on the SaPIs, which form a widespread family in Staphlococcus aureus, and allude to families of similar elements in other Gram-positive cocci. We also contrast the SaPIs with the gene transfer agents (GTAs), both of which generate small infectious phage-like particles, and with the ICEs, which use the conjugative mode of transfer.
A key feature of Gram-positive cocci, especially the staphylococci, streptococci and lactococci, as revealed by inspection of their genomes, is that they contain large cohesive families of phage-related islands and very few other phage-related elements, aside from intact prophages. This is remarkable since genomic islands, being non-essential, are vulnerable to whatever the gods of DNA wish to sabotage them with. Consequently, the typical bacterial chromosome is littered with surviving remnants of prophages and other islands. For example, the genome of the epidemic Escherichia coli 0157:H7 [3] contains a nearly 1 Mb collection of inserted DNA elements, including 20 that are prophage-related [4].
Among the phage-related islands of the Gram-positive cocci, the SaPIs of S. aureus have been studied in the most detail. They are highly mobile, mediate several types of horizontal gene transfer (HGT) [5], interfere with helper phages [6, 7], and apparently affect the expression of certain chromosomal genes (unpublished data). Those of the streptococci have recently occasioned considerable interest, as they form at least two major subgroups: those presumably involved in HGT, and those involved in local gene regulation [8, 9]. Those of the lactococci are known thus far only at the genomic level.
The SaPI story
The story begins with the notorious outbreak of toxic shock syndrome in the early 1980s, related to high-absorbency menstrual tampons. The syndrome was soon attributed to a newly discovered staphylococcal toxin, TSST-1 [10]. As a cause of death among healthy young women and as one of the first known bacterial superantigens, TSST-1 occasioned considerable clinical and experimental interest [11], eventually leading to its cloning [12], sequence determination, and genetic analysis [13, 14]. It was found that the gene was embedded in a 15 kb chromosomal DNA segment that was absent from tst− strains, suggesting an inserted genetic element, which was designated staphylococcal pathogenicity island 1 (SaPI1) (see Fig. 1) [13] [14]. It is flanked by an 18 nt direct repeat and is organized, prophage-like, as two divergent transcription units. It contains homologs of phage integrase (int), excisionase (xis), primase (pri), replication initiator (rep), and terminase small subunit (terS), consistent with its relation to, and probable derivation from, a prophage. Although it is functionally related to phages, which enable it to be transferred at extremely high frequencies, it has developed a distinctive lineage and life style. This life style is characterized not only by the SaPI’s ability to parasitize the phage life cycle, but also by a remarkable ability to interfere with phage reproduction. This interference is determined by a set of genes that are not phage related. They form an operon that is regulated independently of the rest of the SaPI genome and has no known homologs [15] (see Fig. 1). In addition to tst, the gene for TSST-1, SaPI1 carries other superantigen genes plus a variety of genes that seem beneficial to the host organism.
Fig. 1. SaPI1 genome.
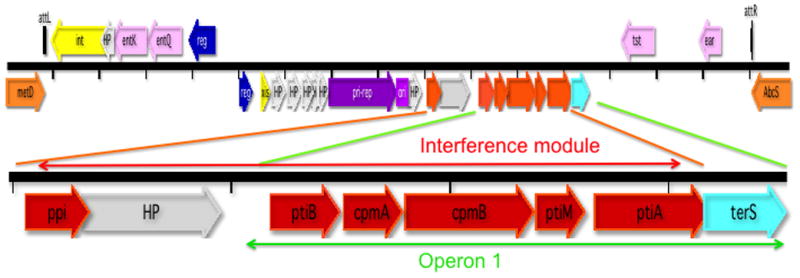
Scheme: orange – flanking chromosomal genes; yellow – int/xis; blue – regulatory genes; purple – replication module; red – interference module; aqua – terS; pink –accessory genes; gray – hypothetical proteins; black – att sites. Tick marks represent nucleotide scale with a spacing of 1 kb. Abbreviations: int – integrase; HP – hypothetical protein; ent –enterotoxin; reg – regulation; xis – excision function; - pri-rep – primase-replication initiation; ori –replication origin; ppi – phage packaging inhibition; pti – phage transcription inhibition; terS –terminase small subunit; tst – toxic shock syndrome toxin 1 (TSST-1); ear – extracellular β-lactamase (ampicillin resistance) homolog. The region including operon 1 and the interference module is amplified for clarity. Most of the gene assignments are based on experimental data [16] [6]; a few on GenBank annotations.
These initial findings suggested a novel and possibly very important mobile genetic element, which has been analyzed in some detail, in the authors’ lab and in the collaborating labs of José Penadés and Gail Christie. These studies, involving primarily four individual SaPIs (SaPIs 1, 2, bov1 and bov2, which differ in interesting and informative ways) have delineated the SaPI life style in depth.
The SaPI life style
SaPIs are maintained in the integrated state by a master repressor, Stl, which is analogous to the λc1 repressor but is not inducible by the SOS response to DNA damage [16]. This repressor is counteracted by helper phages, which encode specific, nonessential antirepressor proteins. These bind to the repressor and induce the repressor-controlled SaPI excision, replication, and packaging cycle. Repressor-antirepressor interactions are SaPI- and phage-specific. For example, one helper phage (80α) encodes at least 4 different antirepressor proteins, which act differentially on SaPIs 1,2, bov1 and bov2. Three of these have been identified thus far [17]. It has been demonstrated that deletion of stl results in the excision and autonomous replication of SaPI DNA [16] which can be packaged by non-helper phages, indicating that the primary role of the helper is to counter Stl-mediated repression. Autonomous SaPI replication indicates the existence of a SaPI replicon. This replicon consists of a specific replication origin (ori) and an initiator protein (Rep) that recognizes and binds to it. All known SaPI oris consist of two sets of short iterons flanking an ~80 bp AT-rich region [18]. The Rep protein, like typical phage initiators [19], has helicase activity, which is required for initiation [18]. The Rep-ori interaction is SaPI-specific and is determined by a matching interaction between the iterons and a specificity determinant in the Rep C-terminus [18]. The Rep-ori complex is variable and at least 7 different Rep-ori specificities have been identified [18]. Following initiation, replication is continued by host functions and results in a linear concatemer [18], which is packaged by the headful mechanism [20]. Packaging is initiated from the post-replicative concatemeric DNA by the terminase complex, which consists of the phage-coded terminase large subunit (TerL) and either the phage- or SaPI-coded terminase small subunit (TerS) [16, 21]. Phages and most SaPIs encode specific TerS proteins, which recognize the phage and SaPI pac sites, respectively. The phage pac site is usually embedded within the terS coding sequence [22], whereas the SaPI pac site is some distance upstream [23]. SaPI DNA is packaged in proheads formed from helper phage virion proteins [24] [25]; pre-assembled helper phage tails are then attached. Many SaPIs remodel the proheads to form smaller capsids (Fig. 2A) [14], commensurate with their smaller genome size, using SaPI genes cpmA and cpmB [25]. However, both SaPI and phage DNAs can be packaged in either full- or small-sized proheads [26]. Deletion of the phage-coded terS eliminates phage DNA packaging, but does not affect lysis so that lysates are produced with particles of both sizes containing only SaPI DNA [27].
Fig. 2. Electron micrographs of SaPI and GTA particles.
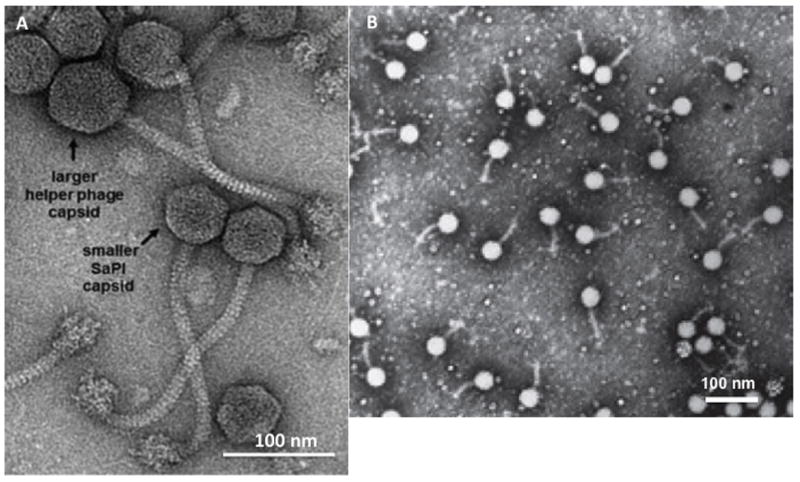
A. Electron micrograph of a mixed helper phage-SaPI lysate (Kindly provided by Dr. Terje Dokland, U. Alabama) B. Electron micrograph of infectious GTA particles, (reproduced from reference 31 with the kind permission of Nature Publishing Group)
A few unusual SaPIs lack an intact terS and a SaPI-specific pac site, and use the helper phage packaging system [28]. Among these, SaPIbov5 and its close relatives have a prophage cos site in addition to their phage-specific pac site, and can be transferred by helper phages of either type [28].
SaPIs are extremely common in S. aureus and occupy 5 different specific chromosomal attachment (attc) sites in S. aureus, and at least one other in non-aureus staphylococci, designated by roman numerals [29] (See online supplementary material, Fig. S1). These sites are at the 3′ ends of genes, which are not disrupted by the insertions. In S. aureus, none of these sites is occupied by a prophage, nor is any known prophage site occupied by a SaPI. However, in some Bacillus strains, the groEL site, corresponding to SaPI attc-V, can be occupied by prophages containing genes that are orthologous to SaPI genes (see online supplementary material, Table S1, genes SAV2028-2025). This suggests the possibility that such a prophage may represent the SaPI progenitor. It was predicted that SaPIs occupying any one att site would be more closely related to one another than to those occupying other sites. A survey of staphylococcal genomes supported this prediction for only a few SaPIs (See online supplementary material, Fig. S2). Perhaps it was once generally true but has been obscured by recombination, as illustrated by two sets of paired islands shown in the online supplementary material, Fig. S3. Following deletion of its primary chromosomal att site, incoming SaPIs integrate rather efficiently into secondary att sites, which are common in S. aureus [30](see online supplementary material, Fig. S4). These, again, do not represent prophage att sites, as all of 8 known staphylococcal prophage att sites [31] are completely unrelated to the primary and secondary SaPI1 att sites. However, the possibility cannot be ruled out that someday an exception to this rule will be found.
Accessory genes
Accessory genes, including genes for superantigens and other virulence and resistance factors, have evidently been inserted into the SaPIs by an unknown non-homology-based recombination mechanism. A remarkable case is that of tst (TSST-1) and seb (enterotoxin B), which are each inserted at precisely the same site in SaPIs 1 & 3 respectively, but in opposite orientations [32].
Our detailed understanding of the basic molecular biology of the SaPIs has confirmed their nature as remarkable molecular parasites, which are exquisitely designed to exploit their helper phages with utmost precision and efficiency to effect their own high-frequency transfer along with their accessory (virulence) genes Recently, we have initiated studies on the evolution of the SaPIs and their relation to the “outside” world (i.e., the bacteriophages and their host bacteria), and have discovered several additional, and remarkable, aspects of their biology: transgeneric SaPI transfer, interference with helper phage reproduction, and SaPI-mediated generalized transduction. These are described in the next three sections.
Transgeneric SaPI transfer
SaPIs as well as certain staphylococcal plasmids can be transferred at very high frequencies to Listeria monocytogenes, where they integrate into sites that resemble their secondary att sites in S. aureus [30]. A compendium of secondary att sites in S. aureus and in L. monocytogenes is shown in online supplementary material, Fig. S4. Additionally, staphylococcal plasmids are maintained autonomously in L. monocytogenes. Since the helper phage cannot replicate in L. monocytogenes [30], transgeneric transfer represents a “stealth” mechanism of phage-mediated HGT.
SaPI-helper phage interactions
Phage interference and its mechanisms
All SaPIs thus far analyzed interfere with the reproduction of their helper phages. Although bacteria and other mobile elements block phage reproduction as a defense against phage predation, the SaPIs interfere in order to gain an advantage over their helpers. Their own reproduction demands that they do not disable the production of virion proteins and lysins. Thus far, three different interference mechanisms have been identified and characterized, and are used in different combinations by different SaPIs
i) Many SaPIs divert up to 90% of the phage’s virion proteins to form small procapsids [14] [7]. This reduces the availability of full-sized procapsids so that phage DNA is packaged in the small ones, which is a dead-end for the phage since only 1/3 of its genome can fit. This mechanism is used to a greater or lesser extent by all SaPIs that produce small capsids. ii) All SaPIs encode a protein, Ppi (phage packaging interference), that binds to and directly blocks phage TerS but not SaPI TerS function [6], reducing phage particle production by ~10-fold [6]. Ppi homologs fall into two families with different phage inhibition specificities [6]. SaPIs 2, bov1 and bov2 all use this mechanism, with SaPIbov2 using it to greatest effect. SaPI1 does not use it against any of its known helper phages [6]. iii) Many SaPIs encode a protein, PtiA (phage transcription interference), that binds to and inhibits LtrC (an analog of λQ) [7]. LtrC is essential for late phage gene transcription and therefore essential for SaPI as well as phage particle production. To prevent complete blockage, a protein encoded by the adjacent gene, ptiM, binds to and partially blocks the activity of PtiA, ensuring the production of virion proteins and lysins [7]. There is also a third pti gene, ptiB, that interferes with LtrC by an unknown mechanism [7]. The pti system of SaPI2 is extremely effective; ptiA and ptiM of SaPIs 1 and bov1 are inactive owing to nucleotide substitutions. SaPIbov2 does not have the system. Remarkably, all of these interference mechanisms are encoded in the region between pri/rep and terS - operon 1 or the immediate upstream region (ppi) (red genes in Fig. 1). Note that mechanisms i) and ii) do not require modulation; since they inhibit only the formation of viable phage particles but not the expression of phage genes, their inhibitory activities would not impact the SaPI. These interactions are summarized diagrammatically in Fig. 3.
Fig. 3. SaPI-helper phage interactions.
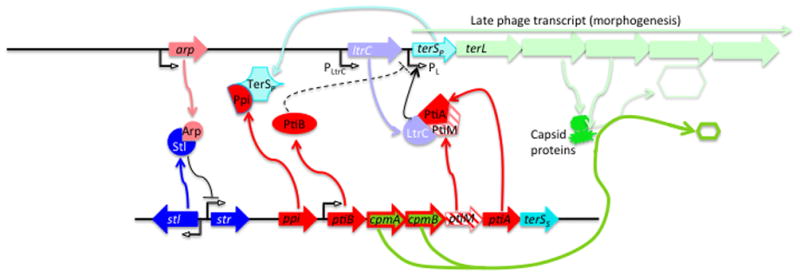
Key interacting genes and proteins for typical prophages (top) and SaPIs (bottom), with lighter colors for phage elements, darker for SaPI. See Table 2 for explanation of the interactions involving the interference genes.
The SaPIs represent an interesting contrast to the xenophobic CRISPRs, which totally block the reproduction of phages and other mobile elements and thus prevent HGT [33]. It is remarkable that CRISPRs are extremely rare in S. aureus, having been found in only a very few strains, whereas SaPIs are everywhere; one might argue that in the gladiatorial evolutionary contest, the SaPIs have won this round. Similarly, with the exception of a single plasmid-carried element [34], CRISPRs are not found in Lactococcus lactis, nor are functional CRISPRS found in Streptococcus pneumoniae [35], though, as described later, both of these species are replete with SaPI-like elements, supporting this concept.
Paucity of helper phages
Although most S. aureus strains are multiply lysogenic and most carry at least one SaPI, very few strains have a resident prophage that acts as a helper for a co-resident SaPI (unpublished data). A possible explanation for this is that, because of interference, SaPI induction is counterselective for the phage. Consequently, it is likely that phages which may once have served as helpers have undergone mutations of their anti-repressors enabling them to avoid SaPI induction. In a recent in vitro “evolution” study, a standard helper phage (80α) was cycled on a SaPI-carrying and a SaPI-negative host strain. After several successive lytic cycles, the phage grown on the SaPI-containing strain were over 1000-fold more likely to contain inactivating mutations in their antirepressor gene (in this case dut, the gene for dUTPase) [36, 37, 38] than those grown on the SaPI-negative host [39]. There was no difference in mutation frequency for any other phage gene.
Host cell survival
Not only do the SaPIs interfere with phage reproduction, they also benefit the host by greatly enhancing host cell survival following phage infection [7, 14]. Here, a significant fraction of phage-infected cells do not lyse; nor, however, do they become lysogenic [7, 14]. The effect is seen for all of the three interference mechanisms and is eliminated by deletion of the responsible interference gene(s), suggesting a common mechanism. This may indicate something fundamental but as yet unknown about phage biology, and it must have significant evolutionary potency; its elucidation represents a fascinating challenge.
SaPIs as couriers of HGT
SaPI-mediated generalized transduction
Generalized transduction, which is mediated by pac phages, involves the recognition of pac-site homologs (pseudo-pac sites), resulting in mispackaging of host DNA. Typically about 1% of pac phage particles carry host DNA, resulting in a transduction frequency of about 10−7 for the average host gene. Not surprisingly, pseudo-pac sites recognized by the SaPI TerS also occur with considerable frequency, also resulting in mispackaging of host DNA [40]. Pseudo-pac sites vary greatly in their resemblance to the primary pac site, resulting in dramatic variations in transduction frequency for different host genes [41]. Remarkably, SaPI pseudo-pac sites are significantly associated with chromosomal genes involved in adaptation to the animal tissue environment (Fig. 4) [40]. The biological basis of this association is unknown.
Fig. 4. S. aureus strain NCTC8325 chromosome showing SaPI pseudo-pac (ppac) sites.
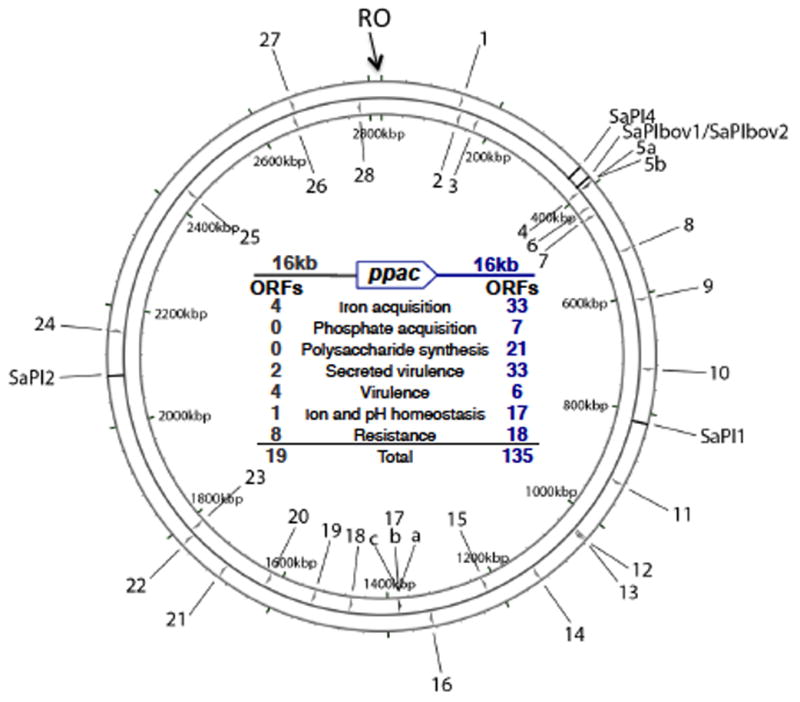
Chromosomal replication origin is at 0 kbp (RO). The 3 prophage genomes are omitted and several of the well-characterized SaPIs are indicated. Numbers represent ppac sites, with those on the outside oriented clockwise, on the inside, counterclockwise. Tabulation in the center lists the types of genes within 16 kb of each ppac site, on either side. Note that those downstream of any ppac site will be transduced by the SaPI at much higher frequency than those upstream (reprinted from [40] with the kind permission of Elsevier).
Generalized transduction by uninduced SaPI
The SaPI terS gene is located in SaPI operon 1, which is, inexplicably, activated by the SOS response [15], although the SaPI itself is not SOS induced. SOS induction of operon 1 causes the expression of terSS, plus two of the three interference systems, which are thereby expressed in the absence of SaPI induction. If there is concurrent phage growth, host DNA will be recognized by TerSS at SaPI pseudo-pac sites and cleaved and packaged by the phage TerL. This will generate transducing particles even if the phage is not a helper phage, provided that its TerL can function with the SaPI TerS. Interestingly, SOS induction of a non-helper prophage would be expected to activate the interference genes in operon 1, whereas infection by the same phage would not be expected to. This could result in severe interference of an SOS-induced prophage but no interference of the same phage when infecting. We suggest that SOS induction of operon1 is of benefit to the host bacterium, as the SOS-induced prophage, while suffering interference, would unwittingly package and transduce the chromosomal fragments produced by SaPI-specific cleavage [5], rather like the cuckoo, which parasitizes other birds by secretly laying eggs in their nests, for the unsuspecting mother bird to hatch and nurture.
SaPI occurrence and evolution
As described in the first part of this review, the SaPIs have developed a set of shared properties that sharply differentiates them from their putative prophage progenitor. A genome-based analysis suggests that they have spread widely and diversified while retaining their distinctive biotype.
Occurrence
The SaPIs have an easily recognizable genome organization [as typified by SaPI1 (Fig. 1)], share gene functions, and are extraordinarily common. Examination of the staphylococcal genomes in GenBank (of which there are by now several hundred) has revealed that SaPIs are by far the most common phage-related elements, aside from the prophages themselves. This finding has been reinforced by Sato’o, et al. [42], who have identified many SaPIs occupying the known SaPI att sites but no other type of inserted element in these sites. Why are SaPIs and other similar elements so well conserved in comparison to other GIs despite their non-essentiality? One reason could be the low recombination frequency in S. aureus and other genera with cohesive families of such elements. Further, as noted earlier, the separation between the terS coding sequence and the SaPI pac site would serve as a crossover suppressor, interfering with recombination in this region. Additionally, SaPIs are under strong positive selection owing to their multiple roles in host biology, especially the enhancement of survival following helper phage infection (as noted later) and the regulation of key host genes (unpublished data).
SaPIs as a large cohesive family: an evolutionary paradigm
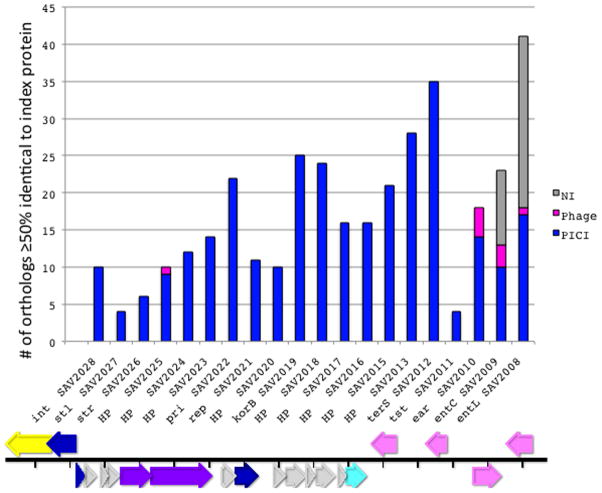
There is an average of one SaPI per natural S. aureus strain and many strains contain two or more. Although they have undergone considerable sequence divergence, they remain easily recognizable as a cohesive family on the basis of the orthology patterns of open reading frames, especially of those annotated as “hypothetical proteins” (HPs). Fig. 5 (Key figure) is a summary of the KEGG-based [43] ortholog lists for all of the 22 ORFs of SaPI-Mu50(V)*. Orthologs in other SaPIs are shown in blue, those matching prophage genes in red, and those matching genes that do not belong to any identifiable inserted element in gray. The actual lists, extended as far as 50% identity to the corresponding SaPI-Mu50(V) ORF, are shown in online supplementary material, Table S1. The elements to which the orthologs belong were identified by examining the corresponding KEGG genome pattern. See online supplementary Fig. S5 A & B for examples of SaPI and prophage genomic patterns. The observation that most HPs do not have orthologs outside of the SaPIs suggests that these were acquired or developed de novo by the SaPIs after the establishment of their unique lineage.
Fig. 5. Ortholog analysis of SaPI ORFs.
An ortholog list [43] was prepared for each of the 22 ORFs of SaPI-Mu50(V) listed along the abscissa. Each vertical bar represents all orthologs with ≥50% nucleotide sequence identity to the index ORF. The type of insert corresponding to each ortholog was determined by inspecting the KEGG genome pattern in the regions flanking the ORF. The data on which this chart is based are listed in online supplementary material, Table S1. Typical KEGG genome patterns for SaPIs and prophages are shown in online supplementary material Figs. 5A and B, respectively. Gray – NI - no insert; pink – prophage; blue – SaPI. Below the graph is a diagram of the genome of SaPI-Mu50(V) showing the genes corresponding to those in the graph.
SaPI-like elements in other bacteria
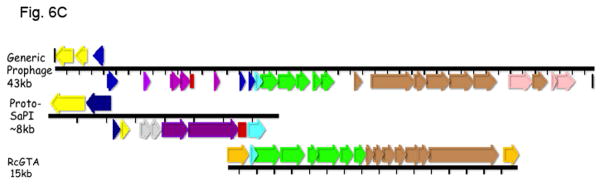
This evolutionary paradigm is not exclusive to the staphylococci: Not surprisingly, SaPI-like elements occur widely throughout the bacterial world. These were first identified in other Gram-positive cocci [44] and more recently it has been observed that streptococci and lactococci possess large families of phage-related elements genomically highly similar to the SaPIs. They have orthology patterns similar to those of the SaPIs in which elements in their own families appear first and those in other families or in other locations or species appear only very far down the ortholog lists, or not at all. A diagrammatic representation of the orthology patterns for one of the lactococcal elements, LlCI-CV56(0.05), is shown in online supplementary material Fig. S6. This analysis, especially of the hypothetical genes, reveals a cohesive family of elements similar to that in S. aureus. These families probably evolved from prophages independently within their own genera. A few genomic diagrams are presented in Fig. 6A. Note that these elements are architecturally similar to the SaPIs in the following respects: All have an int gene at the left end, and all show the SaPI-like transcriptional divergence, in that the rightward transcription unit contains the typical pri-rep pair (though in some cases, these two genes are fused, as with a few of the known SaPIs). In addition, many have a terS homolog; as in S. aureus, these elements occupy specific att sites that are never (thus far) occupied by prophages, and vice-versa. The full compilations will be published elsewhere. As with the staphylococci, there are very few other phage-related elements in the constituent species. The description of these, however, is thus far based almost entirely on genomic analysis; the only information on their functionality is based upon recent findings with a similar element in Enterococcus faecalis, EfCI-V583 ([45]). This element has the same genome organization as the SaPIs, is induced by a co-resident prophage to excise and replicate, is packaged in small phage-like particles composed of helper phage proteins, and also interferes, SaPI-like, with its helper phage. It does not, however, belong to any widespread cohesive family and E. faecalis harbors other types of phage-related elements such as those shown in Fig. 6B.
Fig. 6. A. Genomes of lactococcal PICIs.
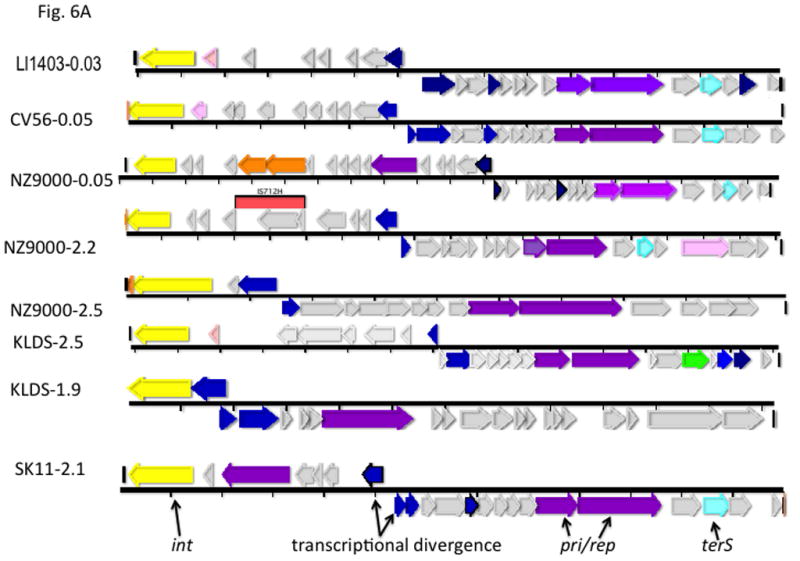
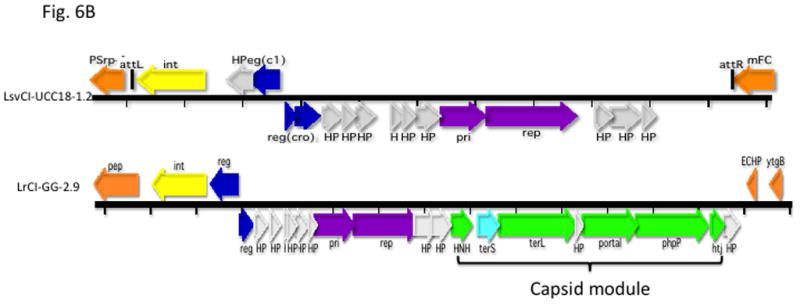
Gene coloring is the same as in Fig. 1 with the addition of green for procapsid and light red-orange for the transposon in the NZ9000(0.03) element. Numbers after the strain names represent approximate genome locations. B. Genomes of phage-related elements from lactobacilli. Color coding is as in Fig. 1, with the addition of green for capsid morphogenesis. Annotations are from GenBank. Abbreviations in addition to those in Fig. 1: LsvCI-UC1118 – L. salivarius UCC118; PSrp – ribosome-associated protein; reg (c1) –λc1-like repressor; reg (cro) - λcro-like regulator; mFC – comF protein 3; LrCI-GG – L. rhamnosus GG; pep – phosphomonomutase-like protein; reg – regulation; HNH – HNH nuclease; terL – terminase large subunit; portal – portal protein; php – prohead protease; htj – head-tail joining protein. C. Hypothetical origins of proto-SaPI and GTA. At top is shown the genome of a generic prophage that could be from either a Gram+ or a Gram- organism. Below is shown a possible “proto-SaPI” consisting of the 5′ region of the prophage from int to terS. Below the “proto-SaPI” is shown the RcGTA, consisting of most of the 3′ region of the prophage from terS to the tape measure gene, but lacking the extreme 3′ end. Gene annotations are from the KEGG orthology lists [43] or from GenBank. Colors: yellow – int/xis; blue – regulatory; purple – replication; aqua – terS; green – capsid morphogenesis; - brown – tail morphogenesis; pink – lysis; orange –flanking genes; red box – replication origin.
Based on their genomic similarity to the SaPIs, we have chosen to designate these elements overall as phage-inducible chromosomal islands (PICIs), of which SaPIs are therefore a subset.
Other families?
Many bacterial genomes are replete with phage-related elements that encode putative components of the phage prohead in addition to the usual features of the SaPIs and PICIs. These are common in bacilli and enterococci and also in the Enterobacteriaciae but have not thus far been seen in staphylococci, lactococci, or streptococci. A typical example is shown in Fig. 6B along with a SaPI-like element from a lactobacillus. There are as yet no experimental data on the functionality of these elements and they are included here solely to illustrate another common type of phage-related element.
Other phage-related mediators of HGT: the gene transfer agent (GTA)
The SaPI’s evolutionary paradigm appears to be mirrored by the GTA. The GTA is a cluster of phage-derived genes that generates tiny phage-like particles (Fig. 2B) containing random 4–5 kb fragments of genomic DNA [46]. The classical GTA system is in Rhodobacter capsulatus (RcGTA) [46] and is, in a sense, the alter-ego of the SaPIs and PICIs: instead of encoding SaPI-like infectious particles, the GTA contains nearly an entire phage morphogenesis module with prophage-like organization [47] (see Fig. 6C). Presumably, the information for making tiny capsids is contained in this module. The GTA lacks the integration, excision, regulation, and replication genes carried by the SaPIs and PICIs, its TerS is non-sequence-specific, and transcription of the GTA gene cluster is unidirectional and is up-regulated postexponentially by unlinked chromosomal genes that also regulate other functions such as motility [48]. Only a small percentage of the population produces GTA particles and lyses to release them. Several of the genes involved in GTA particle production and release, including certain tail genes and a holin-lysin pair, are located elsewhere on the chromosome. GTA systems are widely distributed, especially among the alphaproteobacteria and spirochetes [47], and have also been identified in Archaea [49]. Rhodobacteriales genera and many other Gram-negative bacteria (GNB) all contain a gene cluster very similar to the RcGTA cluster in R. capsulatus and always at the same site (the serine o-acetyl transferase (cysE) site) (see online supplementary material, Table S2). All of the genes in the cluster are under strong purifying selectivity [47] and the GTAs represent a cohesive family. It is especially interesting that GTA particle-carried DNA requires genes of the competence (com) pathway for its uptake [50] as well as recA-mediated recombination for incorporation into cognate regions of the recipient chromosome [50].
Conjugative mobility of pathogenicity islands in other genera
Pathogenicity islands (PAIs) were first described in the late 1980s [51] as inserted DNA segments lacking essential genes (i.e., genes belonging to the basic core genome) and containing genes involved in disease causation, resistance, and other types of clinically relevant adaptations. Many were flanked by short direct repeats (10–20 nt) and contained integrases, transposases and/or other genes associated with mobility. But despite great interest in their potential mobility, and a vast descriptive literature, including a demonstration of a conjugation module as well as int/xis genes in an E. coli element [52] [53] [54] actual mobility was not demonstrated until 2006 when Lory and coworkers observed conjugative transfer of a Pseudomonas aeruginosa island, PAPI-1 [55]. Since that time, a wide variety of conjugative elements carrying virulence or virulence-related genes have been identified and characterized in many species, many more in GNB than in Gram-positive bacteria (GPB). A key example is the “high pathogenicity island” of Yersinia pestis [56] [52, 54], of which homologs are widely distributed among GNB and which is thought to be the progenitor of many ICE elements [52]. Actually, many elements that were originally recognized as pathogenicity islands are now classed as ICEs [57], including conjugative transposons (CTs) [58]. Many of these correspond to the elements in the extensive Schmidt-Hensel catalog [2], suggesting that the vast majority of transmissible islands in GNB are transferred by conjugation [59]. In contrast to the SaPIs, which are inert unless activated by a helper phage, their mobilization is entirely autonomous and they differ from plasmids in that they start their travels as chromosomal integrants rather than as extrachromosomal replicons and require activation signals to initiate transfer cycles. Some are activated by SOS induction, others by the antibiotic to which they carry resistance (especially tetracycline), and still others are activated by circularization-dependent transcriptional up-regulation [60]. Additionally there are hints that some of them may actually replicate autonomously [37]. Thus, reinforcing the hypothesis that their basic genome was derived primarily from conjugative plasmids, though they have acquired a wide variety of genes from other sources and are considered as mosaics. They can also mobilize adjacent chromosomal DNA, as well as non-conjugative plasmids [60] [61] [62].
Since the most important mode of HGT in Gram-negative bacteria (GNB) is conjugation, it is not surprising that most of their mobile elements utilize this mode. These elements are presumably derived from integrated conjugative plasmids, of which there are plenty – ‘granddaddy F’ celebrates his 63rd anniversary as our mascot this year – and Hemophilus and other species harbor reversibly integrating conjugative plasmids [63] that are modular and are closely related to ICE elements [64]. It is all but certain that they have common ancestry; most likely the ICEs have evolved from the plasmids; it seems rather unlikely that conjugative plasmids have evolved from ICEs. This is in contrast to the Gram-positive bacteria (GPB), for which the commonest means of HGT is via transducing phages and it is not surprising that the mobile islands that we have described are evolutionarily related to prophages. This differentiation is far from complete - GPB also have plenty of conjugative transposons and ICE elements [57, 65] and GNB (especially Vibrio cholerae) have a few phage-mediated island transfer systems [53] – but the latter are very much in the minority.
Concluding Remarks: Significance of GIs in HGT
In this review, we have touched on three major types of genomic islands that have all developed from pre-existing and widespread mobile elements, namely prophages and conjugative plasmids. Though each represents a successful evolutionary adaptation, they have radically different roles in HGT and in the economy of their host bacteria. Thus the conjugative GIs, which initiate and effect their self transfer autonomously, may be quite large and carry a wide variety of virulence and adaptivity genes. These genes enable formerly commensal bacteria to invade the eukaryotic tissue environment, causing some very serious diseases. In sharp contrast, the GTAs are immobile but generate tiny phage-like particles that carry random small genomic segments without any obvious role in the biology of their host bacteria, except for the obvious possibility of repairing mutations in the target chromosome by recombination. Nevertheless, they are widespread, well-conserved and therefore probably advantageous. The SaPIs are somewhere in between. Though highly mobile, they are not autonomous but depend on helper phages for their mobilization. They have a complex role in the economy of their host bacteria, largely based on their intimate relation with bacteriophages, which they not only parasitize for critical steps of their life cycle, but also interfere with in a carefully regulated manner. Phage interference enables the SaPI to enhance the transfer of adaptive chromosomal genes, many of which impact host virulence. It also enhances their own high frequency transfer of self-carried toxin genes, while moderately diminishing the ability of the phage to carry out its own HGT. Moreover, the SaPIs seem especially beneficial to their bacterial hosts by virtue of increasing survival of phage infection and of regulating the expression of key host genes. Overall, the SaPIs have hit upon a highly successful evolutionary strategy, since they have diversified widely, while retaining their basic design, and have spread to virtually the entire world of S. aureus. Preliminary observations suggest that the SaPI type of element is very widespread and has a major role in bacterial and archaeal evolutionary economy. The overall picture presented here raises many interesting and important questions, which are listed in an “outstanding questions” box.
Box 1. Outstanding questions.
What was the evolutionary pathway leading to SaPI development?
Do the PICIs of lactococci and streptococci function similarly to the SaPIs?
What was the origin of SaPI operon 1 and how is it regulated?
What is the mechanism of SaPI-induced survival of phage infection?
What is the mechanism of small capsid formation by the GTAs?
What was the evolutionary pathway leading to GTA development
What was the evolutionary pathway from conjugative plasmid to ICE element?
What is the function of the phage-related elements in GNB and GPB that contain modules for procapsid formation and packaging?
How do the GIs acquire accessory genes?
What are the dynamics of in vivo transfer of mobile GIs?
What is the biological basis for the presence in staphylococci, streptococci and lactococci of large families of PICIs and SaPIs, but very few other phage-related elements?
Supplementary Material
Trends Box.
Three major types of Horizontal Gene Transfer systems use radically different strategies for gene transfer at widely varying frequencies. ICE elements initiate their own transfer autonomously; SaPIs depend on helper phages; and GTAs generate and release tiny DNA-containing particles.
GTAs and SaPIs were probably derived from prophages by complementary deletions. ICE elements were probably derived from integrated conjugative plasmids by ‘trading’ replication for integration/excision.
SaPIs interfere with their helper phages by three carefully regulated strategies to ensure high frequency transfer.
SaPIs mediate generalized transduction while in situ, independently of their self-transfer.
SaPIs and their relatives in Gram-positive cocci exist as cohesive and widespread families as shown by orthology analysis of their hypothetical proteins, a method newly described for this Review.
Glossary
- Competence (com)
the genetically determined natural ability to take up and incorporate free DNA (i.e., transformation)
- Conjugation
Mode of intercellular bacterial DNA transfer involving cellular attachment followed by transfer of a single-stranded DNA molecule generated by rolling circle replication.
- Cos site
Specific sequence cleaved by the terminase of a cos phage
- CRISPRs
clustered regularly interspaced short palindromic repeats responsible for bacterial immunity against invading genetic units such as bacteriophages
- Genomic island (GI)
a discrete DNA segment from another organism, with defined boundaries, that has been inserted into a bacterial genome
- GTA
Phage-related chromosomal segment that encodes the production of tiny infectious phage-like particles
- Headful packaging
the pac phage DNA packaging mechanism in which concatemeric post-replicative DNA is cleaved at the pac site and fed into a pre-existing prohead. When the prohead is full, a non-specific cleavage completes the packaging.
- Iterons
short repeated nucleotide sequences required for replication of plasmids and related elements
- ICE element
a mobile GI that excises, circularizes, and is transferred by conjugation to a recipient cell, in which it integrates into a specific att site.
- Mobilization
Preparation for and intercellular transmission of DNA
- Orthologs
homologous genes in different organisms
- Pac site
specific sequence cleaved by the terminase of a pac phage
- Paralogs
homologous genes in the same organism
- Pathogenicity island
A GI containing one or more genes that could enhance pathogenicity
- Phage-inducible chromosomal island (PICI)
A GI that possesses phage-related genes analogous to those of the SaPIs and is likely to be inducible by a helper phage
- Procapsid
Hollow polyhedral protein shell into which phage DNA is inserted
- Prophage
the repressed and stable form of a temperate bacteriophage; most but not all are integrated into the host chromosomne
- Staphylococcal pathogenicity island (SaPI)
A GI that possesses phage-related genes and is known or likely to be inducible by a helper phage. The SaPIs are a subset of the PICIs
- SOS
the global bacterial stress response to DNA damage
- Superantigen (SAG)
an antigen that causes non-specific polyclonal T cell activation and massive cytokine release.
- Terminase
a two subunit bacteriophage enzyme that catalyzes the initiation of phage DNA packaging
- Transduction
Transfer of host DNA as a result of mis-packaging by a phage
- Virion
an infective virus particle
Footnotes
SaPI/PICI designations. Each designation starts with 2 or 3 letters denoting the species of origin, followed by PI or CI (for pathogenicity island or chromosomal island), then a hyphenated indication of the strain of origin and the genomic location of the island in parentheses. For SaPIs, the latter is a roman numeral denoting the att site; for elements from species other than staphylococci, it is a numerical indication of location in Mb in the respective genome. For example: SaPI-N315(V) refers to the SaPI in S. aureus strain N315 located in att site V; SpyCI-NZ131(0.37) refers to the PICI in S. pyogenes strain NZ131 located at 0.37 Mb. For the non-staphylococcal species, we do not have a complete list of att sites; hence, their locations are given as Mb coordinates in the available genomes.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blum G, et al. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seth-Smith HM, et al. Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae. J Bacteriol. 2012;194:1494–1504. doi: 10.1128/JB.06403-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 4.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, et al. Pathogenicity island-directed transfer of unlinked chromosomal virulence genes. Mol Cell. 2015;57:138–149. doi: 10.1016/j.molcel.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ram G, et al. Staphylococcal pathogenicity island interference with helper phage reproduction is a paradigm of molecular parasitism. Proc Natl Acad Sci U S A. 2012;109:16300–16305. doi: 10.1073/pnas.1204615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ram G, Ross HF, Novick RP. Precisely modulated SaPI interference with late phage gene transcription. Proc Natl Acad Sci, USA. 2014 doi: 10.1073/pnas.1406749111. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott J, et al. Phage-associated mutator phenotype in group A streptococcus. J Bacteriol. 2008;190:6290–6301. doi: 10.1128/JB.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott J, et al. Phage-Like Streptococcus pyogenes Chromosomal Islands (SpyCI) and Mutator Phenotypes: Control by Growth State and Rescue by a SpyCI-Encoded Promoter. Frontiers in microbiology. 2012;3:317. doi: 10.3389/fmicb.2012.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergdoll MS, Schlievert PM. Toxic shock syndrome toxin [letter] Lancet. 1984;2:691. [Google Scholar]
- 11.Proft T, Fraser JD. Bacterial superantigens. Clin Exp Immunol. 2003;133:299–306. doi: 10.1046/j.1365-2249.2003.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreiswirth B, et al. Genetic characterization and cloning of the toxic shock syndrome exotoxin. Surv Synth Path Res. 1984;3:73–82. [PubMed] [Google Scholar]
- 13.Lindsay JA, et al. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 14.Ruzin A, et al. Molecular genetics of SaPI1 - a mobile pathogenicity island in Staphylococcus aureus. Mol Microbiol. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 15.Ubeda C, et al. SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol Microbiol. 2007;65:41–50. doi: 10.1111/j.1365-2958.2007.05758.x. [DOI] [PubMed] [Google Scholar]
- 16.Ubeda C, et al. Characterization of mutations defining SaPI functions and enabling autonomous replication in the absence of helper phage. Mol Microbiol. 2008;67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 17.Tormo-Mas MA, Shrestha A, Mir I, Campoy S, Lasa I, Barbé J, Novick RP, Christie GE, Penadés JR. Moonlighting phage proteins de-repress staphylococcal pathogenicity islands. Nature. 2010;465:779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ubeda C, et al. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc Nat Acad Sci, USA. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghisotti D, et al. Multiple regulatory mechanisms controlling phage-plasmid P4 propagation. FEMS Microbiol Rev. 1995;17:127–134. doi: 10.1111/j.1574-6976.1995.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 20.Novick RPR, Ruzin A. Molecular Genetics of SaPI1-a Mobile Pathogenicity Island in Staphylococcus aureus. 2000 doi: 10.1046/j.1365-2958.2001.02488.x. submitted. [DOI] [PubMed] [Google Scholar]
- 21.Maiques E, et al. Role of Staphylococcal Phage and SaPI Integrase in Intra- and Interspecies SaPI Transfer. J Bacteriol. 2007;189:5608–5616. doi: 10.1128/JB.00619-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, et al. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol Microbiol. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- 23.Bento JC, et al. Sequence determinants for DNA packaging specificity in the S. aureus pathogenicity island SaPI1. Plasmid. 2014;71:8–15. doi: 10.1016/j.plasmid.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poliakov A, et al. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. J Mol Biol. 2008;380:465–475. doi: 10.1016/j.jmb.2008.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damle PK, et al. The roles of SaPI1 proteins gp7 (CpmA) and gp6 (CpmB) in capsid size determination and helper phage interference. Virology. 2012;432:277–282. doi: 10.1016/j.virol.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ubeda C, et al. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol Microbiol. 2009;72:98–108. doi: 10.1111/j.1365-2958.2009.06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christie GE, Dokland T. Pirates of the Caudovirales. Virology. 2012;434:210–221. doi: 10.1016/j.virol.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quiles-Puchalta N, Carpenaa N, Alonsoc JC, Novick RP, Marina A, Penadés JR. A novel cos-site-based SaPI DNA packaging system involves phage-encoded HNH endonucleases. Proc Natl Acad Sci, USA. 2014 doi: 10.1073/pnas.1320538111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novick RP, Subedi A. The SaPIs: mobile pathogenicity islands of Staphylococcus. Chem Immunol Allergy. 2007;93:42–57. doi: 10.1159/000100857. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323:139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy AJ, et al. Staphylococcus aureus temperate bacteriophage: carriage and horizontal gene transfer is lineage associated. Frontiers in cellular and infection microbiology. 2012;2:6. doi: 10.3389/fcimb.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novick RP. Mobile genetic elements of staphylococci. In: Schmidt H, Hensel M, editors. Horizontal gene transfer and the evolution of pathogenesis. Cambridge University Press; 2007. [Google Scholar]
- 33.Marraffini LA. CRISPR-Cas immunity against phages: its effects on the evolution and survival of bacterial pathogens. PLoS Pathog. 2013;9:e1003765. doi: 10.1371/journal.ppat.1003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millen AM, et al. Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis. PLoS One. 2012;7:e51663. doi: 10.1371/journal.pone.0051663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore JE, et al. Comparison of clustered, regularly interspaced short palindrome repeats (CRISPRs) in viridans streptococci (Streptococcus gordonii, S. mutans, S. sanguinis, S. thermophilus) and in S. pneumoniae. British journal of biomedical science. 2008;65:104–108. doi: 10.1080/09674845.2008.11978109. [DOI] [PubMed] [Google Scholar]
- 36.Tormo-Mas MA, et al. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465:779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quiles-Puchalt N, et al. A super-family of transcriptional activators regulates bacteriophage packaging and lysis in Gram-positive bacteria. Nucleic Acids Res. 2013;41:7260–7275. doi: 10.1093/nar/gkt508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szabo JE, et al. Highly potent dUTPase inhibition by a bacterial repressor protein reveals a novel mechanism for gene expression control. Nucleic Acids Res. 2014;42:11912–11920. doi: 10.1093/nar/gku882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frígols B, Quiles-Puchalt N, IMir-Sanchis I, Donderis j, Santiago FE, Buckling A, Novick RP, Marina A, Penadés JR. Virus satellites drive viral evolution and ecology. PLoS genetics. 2015 doi: 10.1371/journal.pgen.1005609. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, et al. Pathogenicity Island-Directed Transfer of Unlinked Chromosomal Virulence Genes. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmieger H, Backhaus H. The origin of DNA in transducing particles in P22-mutants with increased transduction-frequencies (HT-mutants) Mol Gen Genet. 1973;120:181–190. doi: 10.1007/BF00267246. [DOI] [PubMed] [Google Scholar]
- 42.Sato’o Y, et al. A novel comprehensive analysis method for Staphylococcus aureus pathogenicity islands. Microbiol Immunol. 2013;57:91–99. doi: 10.1111/1348-0421.12007. [DOI] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novick RP, et al. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matos RC, et al. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS genetics. 2013;9:e1003539. doi: 10.1371/journal.pgen.1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solioz M, Marrs B. The gene transfer agent of Rhodopseudomonas capsulata. Purification and characterization of its nucleic acid. Archives of biochemistry and biophysics. 1977;181:300–307. doi: 10.1016/0003-9861(77)90508-2. [DOI] [PubMed] [Google Scholar]
- 47.Lang AS, et al. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol. 2012;10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frígols B, Quiles-Puchalt N, IMir-Sanchis I, Donderis j, Santiago FE, Buckling A, Novick RP, Marina A, Penadés JR. Virus satellites drive viral evolution and ecology. PLoS genetics. 2015 doi: 10.1371/journal.pgen.1005609. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertani G. Transduction-like gene transfer in the methanogen Methanococcus voltae. J Bacteriol. 1999;181:2992–3002. doi: 10.1128/jb.181.10.2992-3002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brimacombe CA, et al. Homologues of Genetic Transformation DNA Import Genes Are Required for Rhodobacter capsulatus Gene Transfer Agent Recipient Capability Regulated by the Response Regulator CtrA. J Bacteriol. 2015;197:2653–2663. doi: 10.1128/JB.00332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hacker J, et al. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog. 1990;8:213–225. doi: 10.1016/0882-4010(90)90048-u. [DOI] [PubMed] [Google Scholar]
- 52.Schubert S, et al. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol Microbiol. 2004;51:837–848. doi: 10.1046/j.1365-2958.2003.03870.x. [DOI] [PubMed] [Google Scholar]
- 53.Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schubert S, et al. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int J Med Microbiol. 2004;294:83–94. doi: 10.1016/j.ijmm.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 55.Qiu X, et al. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103:19830–19835. doi: 10.1073/pnas.0606810104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rakin A, et al. Integrative module of the high-pathogenicity island of Yersinia. Mol Microbiol. 2001;39:407–415. doi: 10.1046/j.1365-2958.2001.02227.x. [DOI] [PubMed] [Google Scholar]
- 57.Burrus V, et al. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 58.Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salyers AA, et al. Conjugative Transposons: an unusual and diverse set of integrated gene transfer elements. Microbiological Reviews. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 61.Wozniak RA, et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS genetics. 2009;5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carraro N, et al. Differential regulation of two closely related integrative and conjugative elements from Streptococcus thermophilus. BMC Microbiol. 2011;11:238. doi: 10.1186/1471-2180-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stuy JH. Chromosomally integrated conjugative plasmids are common in antibiotic-resistant Haemophilus influenzae. J Bacteriol. 1980;142:925–930. doi: 10.1128/jb.142.3.925-930.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohd-Zain Z, et al. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J Bacteriol. 2004;186:8114–8122. doi: 10.1128/JB.186.23.8114-8122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burrus V, et al. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48:77–97. doi: 10.1016/s0147-619x(02)00102-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.