Abstract
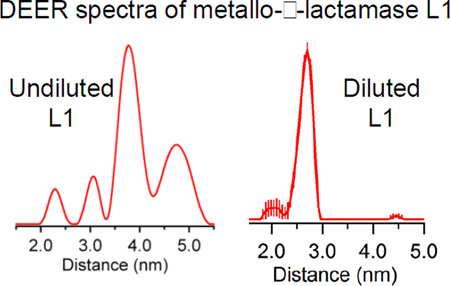
The metallo-β-lactamases (MβLs), which require one or two Zn(II) ions in their active sites for activity, hydrolyze the amide bond in β-lactam-containing antibiotics, and render the antibiotics inactive. All known MβLs contain a mobile element near their active sites, and these mobile elements have been implicated in the catalytic mechanisms of these enzymes. However little is known about the dynamics of these elements. In this study, we prepared a site-specific, double spin-labeled analog of homotetrameric MβL L1 with spin labels at position 163 and 286 analyzed the sample with DEER (double electron electron resonance) spectroscopy. Four unique distances were observed in the DEER distance distribution, and these distances were assigned to the desired intramolecular dipolar coupling (between spin labels at positions 163 and 286 in one subunit) and to intermolecular dipolar couplings. To rid the spin-labeled analog of L1 of the intermolecular couplings, spin-labeled L1 was “diluted” by unfolding/refolding the spin-labeled enzyme in the presence of excess wild-type L1. DEER spectra of the resulting, spin-diluted enzyme revealed a single distance corresponding to the desire intramolecular dipolar coupling.
Keywords: Metallo-β-lactamase (MβL), site-directed spin labeling (SDSL), double electron electron resonance (DEER), MTSL
Graphical abstract
A method is presented to prepare a tightly-associated, oligomeric metalloprotein with spin label(s) in only one subunit. The method involves unfolding metal-free, metallo-β-lactamase L1 and refolding L1 in the presence of the desired metal ion. DEER spectroscopy was used to demonstrate successful “dilution” of the spin-labeled protein.
1. Introduction
The antibiotics most commonly prescribed to treat infections are β-lactam-containing compounds, which inhibit peptidoglycan cross-linking in bacterial cell walls [1]. Since their clinical debut, resistance to these antibiotics has become increasingly prevalent. The most common mechanism for β-lactam resistance is the production of β-lactamases, which hydrolyze the β-lactam ring and render the drugs inactive [2]. A classification system has been developed that groups the 1,300 known β-lactamases into 4 classes: A, B, C, and D [3–5]. Class A, C, and D enzymes possess an active site serine that nucleophilically attacks the β-lactam carbonyl. Class B β-lactamases, which are known as metallo-β-lactamases (MβLs), are unique in that they require one or two Zn(II) ions in the active site for activity [6, 7]. MβLs are of increasing medical concern due to their potential for horizontal gene transfer on mobile plasmids and the lack of a clinical inhibitor against them [8–12].
Metal content and sequence homology data provide criteria for dividing the MβLs into the B1, B2, and B3 subclasses [3, 4]. B1 and B3 enzymes generally utilize two Zn(II) ions in their active sites and exhibit wide-spectrum β-lactamase activity. B2 enzymes carry only one active site Zn(II) and preferentially hydrolyze carbapenems. Despite structural and functional variance, certain characteristics are ubiquitous across the MβLs. Most notably is the αββα tertiary structure characteristic of the β-lactamase fold superfamily [6, 7]. Another important feature is a mobile motif near the active site of MβLs, which manifests itself as an unstructured loop in B1 and B3 enzymes and as an α-helix in B2 enzymes. Early crystallographic studies on B1 and B3 MβLs identified a position-conserved, highly-disordered loop near the active site of the enzymes; subsequent crystal structures of enzyme-inhibitor complexes have demonstrated migration of the loop towards the active site.[13–19] Dyson and coworkers conducted NMR studies and showed that Trp64 on the position-conserved loop of CcrA may play a role in inhibitor (and by analogy substrate) binding and further speculated that Trp64 and the loop play a role in promotion of catalysis.[20] Mutations and partial and complete deletions of the loop have resulted in marked changes in steady-state kinetics and formation of a reaction intermediate, as well as suggested a role for the loop in substrate binding [19–22]. Stopped-flow fluorescence experiments on L1 (a B3 enzyme) show a catalytically relevant rate of loop movement, further suggesting an important role for the loop in catalysis [23], and EPR studies showed movement of the position-conserved α-helix above the active site of ImiS, which belongs to the B2 class.[24]
We hypothesize that the mobile loop of L1 activates bound substrate for hydrolysis by “clamping down” and sterically-distorting the planar β-lactam ring and thereby raising the ground state energy of the substrate. One potential way to probe loop motion during catalysis is to use rapid-freeze quench double electron electron resonance (RFQ-DEER) spectroscopy. A necessary first step in the use of this technique is the generation of site-specifically, spin-labeled analogs of the enzyme. This work describes our efforts at spin-labeling a homotetrameric MβL in only one of the subunits.
2. Materials and Methods
2.1. Materials
Site-directed mutagenesis kits were purchased from Stratagene (Carlsbad, CA). E. coli strains DH5α and BL21(DE3) cells were purchased from Novagen (Madison, WI). Sequencing and mutagenesis primers were purchased from Integrated DNA Technologies. Isopropyl-β-D-galactoside (IPTG) was purchased from Anatrace (Muamee, OH). Q-Sepharose and Sephacryl S-200 chromatographic media were purchased from GE Healthcare. S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methylmethanesulfonothioate (MTSL) was purchased from Toronto Research Chemicals TRC (Canada). Substrate nitrocefin was purchased from Becton Dickinson. All buffer solutions and growth media were prepared by using Barnstead Nanopure water. The standard metallo-β-lactamase numbering scheme is used throughout this work.[25]
2.2 Methods
2.2.1. Preparation and characterization of L1 mutants
To generate single and double mutants of L1, site-directed mutagenesis was carried out using a QuikChange mutagenesis kit (Stratagene) according to manufacturer’s instructions. The T163C and K286C L1 mutants were generated using the L1 over-expression plasmid, pET26bL1 [26], as the template and the following primers: T163C (5'-cggcgatggcatctgctacccgcctgcc-3'), T163C_antisense (5'-ggcaggcgggtagcagatgccatcgccg-3'), K286C (5'-gccagggccggtgcctgcgcactgacctgcaag-3'), and K286C_antisense (5'-cttgcaggtcagtgcgcaggcaccggccctggc-3'). The T163C/K286C L1 double mutant was generated using pT163CL1 as the template and the K286C primers listed above. Mutated DNA was confirmed by DNA sequencing at CBFG facility Miami University, and plasmids containing the mutated DNA were transformed into E. coli BL21(DE3) cells. Wild-type L1 and the single mutants of L1 were over-expressed and purified as described previously[26] except that the induction temperature for the mutants was lowered from 37 °C to 28 °C.
The T163C/K286C mutant was found to over-express as an insoluble protein. After protein over-expression, E. coli cells were centrifuged at 8000 ×g for 10 minutes. The supernatant was discarded, and the cell pellets were resuspended in 100 mL of 50 mM HEPES pH 7.0. Lysis was achieved by passage through a French press three times at 1000 psi. The resulting solution was centrifuged (25 minutes at 23,400 ×g), and the supernatant was discarded. The pale white inclusion bodies were resuspended in 80 mL of 7 M guanidinium hydrochloride containing 100 µM Zn(II). The solution was subjected to vortexing for 5 minutes and then centrifuged (25 minutes at 23,400 ×g) to remove insoluble debris. The supernatant was dialyzed vs. 4 × 1 L 50 mM HEPES, pH 7.0, containing 100 mM NaCl. The resulting sample was then centrifuged (25 minutes at 23,400 ×g) to remove insoluble species. The supernatant was concentrated to 4 mL using an Amicon apparatus equipped with a YM-10 membrane, and the concentrated protein was purified using a G-25 size exclusion column with 50 mM HEPES, pH 7.0, containing 100 mM NaCl as the chromatography buffer. Fractions containing the purified T163C/K286C mutant were identified by using SDS-PAGE.
Wild-type L1 and L1 mutants were characterized using metal analyses and steady state kinetic studies [26]. Metal content of L1 samples was determined with a Perkin-Elmer Optima 7300 DV inductively coupled plasma spectrometer with atomic emission detection as described previously [26]. Steady-state kinetic studies were conducted at 25 °C with an Agilent 8453A UV-visible (UV-Vis) Diode Array spectrophotometer, with nitrocefin as the substrate and 50 mM cacodylate, pH 7.0, containing 50 µM Zn(II) as the buffer.
2.2.2. Preparation of spin-labeled L1 mutants
L1 mutants were dialyzed versus 1 L of 50 mM HEPES, pH 7.0, containing 100 mM NaCl. One equivalent of dithiothreitol (DTT) was added to the samples 30 minutes prior to addition of (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate (MTSSL). A 15-molar excess of MTSSL was dissolved in 100 µL of neat dimethyl sulfoxide, and the entire solution of MTSSL was added into DTT-pretreated L1 samples. The spin-labeling reaction was carried out in the dark on a rocking platform overnight at 4 °C. Unbound MTSSL was removed by passing the sample through a G-25 (1.5 cm × 40 cm of bed volume 60 ml) chromatography column using 50 mM HEPES, pH 7.0, containing 100 mM NaCl as the buffer. The efficiency of the spin labeling was estimated by using cw-EPR at room temperature. Briefly, 35 mm quartz capillary tubes (1.1 mm inner diameter and 1.6 mm outer diameter) were filled with ca. 30 µL of 100 µM spin-labeled protein samples. Capillary tubes were placed in 3 mm inner diameter quartz EPR tubes and inserted in the microwave cavity. CW EPR spectra were collected at X-band on a Bruker EMX continuous wave (CW) EPR spectrometer using an ER041xG microwave bridge and ER4119-HS cavity coupled with a BVT 3000 nitrogen gas temperature controller at the Ohio Advanced EPR Laboratory. CW EPR spectra were collected by using parameters previously published by Feldmann et al. [27]. Spectra were collected by signal averaging 25 scans (consisting of 1024 points and 40 ms time constants). The instrument parameters were a center field of 3370 G and a sweep width of 100 G, microwave frequency of 9.5 GHz, modulation frequency of 100 kHz, modulation amplitude of 1 G, and microwave power of 1 mW at 298 K. Spin label concentrations of protein samples were quantified as previously described.[27]
2.2.3. Preparation of spin-diluted, spin-labeled L1 mutants
Wild-type metallo-β-lactamase L1 was over-expressed and purified according to the procedure by Crowder et al. [26]. Metal-free (apo) L1 samples, both wild-type and mutants, were generated as described by Hu and coworkers [28]. The metal-free, double mutant of L1 (1 mL, 200 µM) was mixed with 1 mL of 800 µM apo-wild-type L1, and the mixture was unfolded in 18 mL of 7 M Gdn-HCl containing 100 µM Zn(II). After incubation on ice for 30 minutes, the mixture was dialyzed versus 4 × 1 L of 50 mM HEPES, pH 7.0, containing 100 mM NaCl (six hours for each dialysis step). The refolded protein was centrifuged (23,400 ×g for 25 minutes) to remove any insoluble species. The refolded double mutant of L1 was labeled with MTSSL using the procedure described.
2.2.4. Double Electron Electron Resonance (DEER) studies
Spin-labeled mutants of L1 were concentrated to 60–100 µM by using an Amicon ultrafiltration concentrator equipped with an YM-10 membrane. The samples were analyzed by using either Q-band DEER spectroscopy at the Ohio Advanced EPR Laboratory or X-band DEER at the National Biomedical EPR Center, Medical College of Wisconsin. DEER was performed at 80 K using Bruker EleXsys E-580 Pulse EPR spectrometers equipped with nitrogen cooling and either a Bruker SuperQFTu bridge, 10 W AmpQ microwave amplifier and Q-band EN 5107D2 dielectric resonator (34.2 GHz), or a Bruker SuperXFT bridge, Applied Systems Engineering 2 kW traveling wave tube amplifier and X-band EN4118X-MD4 resonator (9.7 GHz). The MD4 resonator is designed for 3.8 mm O.D. tubes but was used here with 4 mm O.D. tubes (Wilmad 706-SQ-250M) that were cut to 7 cm length and loaded through the bottom of the resonator.[29] A four-pulse π/2O − τ1 − πO − τE − πP − (τ1 + τ2 − τE) − πO DEER sequence was employed, where the superscripts "O" and "P" denote pulses at the observe and pump frequencies, respectively, τE is the time between the first inversion pulse and the pump pulse, and τ2 is the dipolar evolution time. At X-band, π/2O and πO were 16 and 32 ns, respectively, with, τ2 = 1200, and πP = 32 ns; at Q-band, π/2O and πO were 20 and 40 ns, respectively, with, τ2 = 2200, and πP = 48 ns. Spectra were pumped at the ml = 0 center line and observed at the low-field ml = 1 line, with a Δν = 73 MHz at X-band and 61 MHz at Q-band. Shot repetition times of 1200 µs (X-band), and 500 µs (Q-band) were used. Distances were obtained using DEERAnalysis v.2009 and v.2011.[30] The use of two frequencies deserves comment. In principle, exactly the same information is available at both frequencies provided that the relaxation times allow for reasonable dipolar evolution times. In the present study, we were able to capture at least one full oscillation of the dominant DEER modulation at each frequency. Q-band DEER is a much more efficient technique in terms of time and material and is the preferred method. However, it is important to demonstrate that high quality interpretable data could be also obtained at X-band, as one of the goals of this work is to develop a magnetically-diluted labeled tetramer, free of inter-subunit dipolar interactions, that can be probed using rapid-freeze-quench (RFQ) to trap conformational intermediates in the catalytic cycle. RFQ sample preparation, and maintenance of samples at cryogenic temperatures during loading and transfer, is significantly simpler and more reliable with the larger and thermally-massive X-band samples, and X-band DEER is being used in ongoing RFQ-DEER studies.[31] In the present study, useful X-band DEER data were observed out to 1.1 µs; data collected for longer times actually resulted in quantitatively poorer fits (higher uncertainty) that were qualitatively identical to the fits to 1.1 µs data. With these data, we would expect to be able to confidently assign distances of up to about 4.3 nm, with decreasing confidence in longer distances.
2.2.5. Molecular Dynamics Simulations
The atomic coordinates for the L1 crystal structure (PDB ID: 1SML) from Fischerella were downloaded from the Protein Data Bank and used to generate the structures of various spin-labeled L1 mutants with the Nanoscale Molecular Dynamics (NAMD) program.[32] The T163C and K286C mutations were created using the molecular graphics software VMD.[33] The nitroxide spin-probe MTSL was attached using CHARMM force field topology files incorporated into NAMD. The modified protein assembly was solvated into a spherical water environment and further equilibrated and minimized by running NAMD simulations at room temperature using CHARMM force field parameters.[27] The distance distribution between the T163C and K286C residues was predicted with rotamer library modeling of MTSL conformations using Multiscale modeling of macromolecular systems (MMM version 2010).[32]
2.3. Results
2.3.1. Preparation and characterization of L1 mutants
Our overall goal in this project was to prepare analogs of metallo-β-lactamase L1 that contained site-specific spin labels that could be used to probe motions of the protein during catalysis. One label was designed to be positioned on the catalytic loop that extends over the active site, and the other label was designed to be positioned on a more static part of the enzyme, such as on an α-helix, 20–35 Å away from the loop. Using the crystal structure of L1 as a guide [18], we initially identified Asp160 on the loop as the optimum position to introduce a cysteine residue for the site-specific spin label (Figure S3). The side chain of Asp160 is in the center of the flexible loop in L1 and appeared to protrude away from the active site, and we reasoned that this position might tolerate a mutation. The D160C mutant was over-expressed and purified. While the D160C mutant bound 2 equivalents of Zn(II), steady-state kinetic studies with nitrocefin as the substrate demonstrated that the mutant exhibited a kcat of 5.5 s−1 and a Km of 7 µM, as compared to the kcat and Km values of 40 s−1 and 4 µM, respectively, for wild-type L1 (Table 1). In an effort to identify a mutant with activity more similar to that of the wild-type enzyme, we prepared and characterized the S153C mutant. Given our results with the D160C mutant, we reasoned that the introduction of a mutation on a part of the loop that was not as flexible (closer to an α-helix) would result in a mutant that exhibited steady-state kinetic behavior closer to that of the wild-type enzyme. We chose position 153 because there is a serine in this position in the wild-type enzyme, and a Ser to Cys mutation would not be a huge structural change. Unfortunately, this mutant exhibited a low kcat value (Table 1). We chose a T163C mutant for similar reasons as we used for the S153C mutant. The T163C mutant was over-expressed and purified, and the protocol described in Materials and Methods yielded approximately 30 mg of >95% pure, soluble T163C mutant per 4 liters of growth culture, compared with the 80 mg typically obtained from preparations of wild-type L1. The T163C mutant bound 1.7 ± 0.2 equivalents of Zn(II) and exhibited a kcat of 40 ± 1 s−1 and a Km of 11 ± 2 µM; these values are similar to those of wild-type L1 (Table 1). Since Ser153, Asp160, and Thr163 are found on the “mobile loop” in L1, it is clear that the mobile loop on L1 is very sensitive to point mutations. Nonetheless, the T163C mutant exhibited sufficient catalytic properties to be used in the proposed double labeling studies.
Table 1.
Steady state kinetic constants and metal content of wild-type L1 and L1 mutants
| Enzyme | kcat (s−1) | Km (µM) | Metal content (eq) |
|---|---|---|---|
| Wild-type L1 | 40 ± 1 | 4 ± 1 | 1.9 ± 0.2 |
| D160C | 5.5 ± 0.5 | 7 ± 1 | 2.0 ± 0.1 |
| S153C | 13 ± 1 | 2.3 ± 0.2 | 2.0 ± 0.1 |
| T163C | 40 ± 1 | 11 ± 2 | 1.7 ± 0.2 |
| Spin-labeled T163C | 51 ± 4 | 4 ± 1 | 2.0 ± 0.2 |
| K286C | 13 ± 1 | 0.5 ± 0.1 | 1.7 ± 0.1 |
| Spin-labeled K286C | 9 ± 1 | 0.9 ± 0.3 | 1.8 ± 0.1 |
| T163C/K286C | 21 ± 1 | 3.7 ± 0.5 | 1.7 ± 0.1 |
| Spin-labeled T163C/K286C | 9.2 ± 2.2 | 1.4 ± 0.1 | 2.1 ± 0.1 |
| Spin-diluted, unlabeled T163C/K286C | 20 ± 1 | 2.5 ± 0.5 | 1.7 ± 0.1 |
| Spin-diluted, spin-labeled T163C/K286C | 23 ± 1 | 2.1 ± 0.7 | 1.8 ± 0.1 |
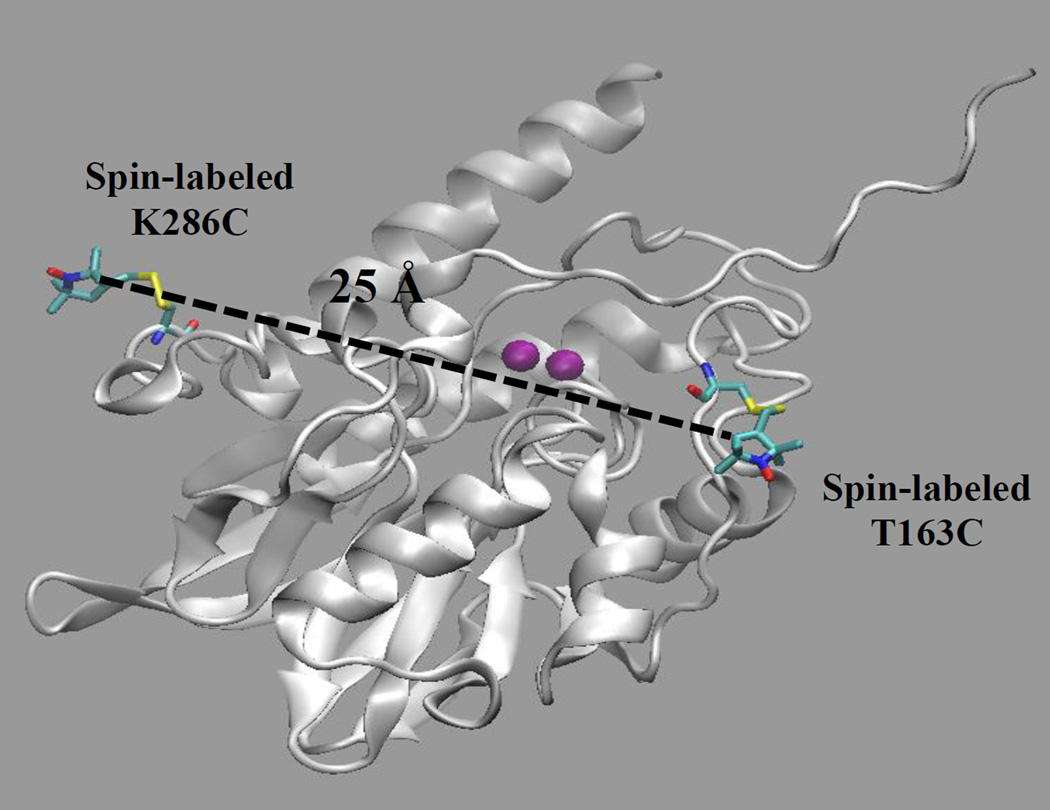
We successfully generated the T163C/K286C double mutant. The spin label on Thr163 is estimated to be 25 Å away from the spin label on Lys286 (Figure 1), and this distance is amenable to interrogation by DEER spectroscopy. Lys286 is found on one of the 310 α-helices in L1[34], and we reasoned that the motion of residues on α-helices would be less than those on unordered loops. In other words, any changes in distances between the introduced spin labels could be attributed to motions of the spin label on the loop (Thr163). The K286C mutant exhibited a kcat of 13 ± 1 s−1 and a Km of 0.5 ± 0.1 µM and bound 1.7 ± 0.1 equivalents of Zn(II) (Table 2). The T163C/K286C double mutant was shown to bind 1.7 ± 0.1 equivalents of Zn(II) and exhibit a kcat of 21 ± 1 s−1 and a Km of 3.7 ± 0.5 µM.
Figure 1.
L1 monomer labeled with MTSL at positions 163 and 286. The Zn(II) ions are purple. The figure was generated using molecular graphing software VMD and molecular dynamics simulation starting from the L1 monomer crystal structure (PDB ID: 1SML) as described in the Methods section.[33].
2.3.2. Spin-labeling of the L1 mutants
Wild-type L1 and the T163C, K286C, and T163C/K286C mutants of L1 were spin-labeled as described in the Methods section. Unbound spin label was removed by gel filtration. The efficiency of spin labeling was evaluated by using cw-EPR spectroscopy, as described previously [27], and the spectra demonstrate that spin-labeled T163C, K286C, and T163C/K286C mutants were labeled with 95%, 91%, and 92% efficiency, respectively. Since wild-type L1 has two cysteines (Cys252 and Cys280) that form a disulfide bond, cw-EPR spectroscopy was used to show that wild-type L1 was not spin-labeled with MTSSL and that the T163C/K286C double mutant binds 2 MTSL groups (Figure S1). Spin-labeled T163C L1 bound 2.0 ± 0.2 eq of Zn(II) and exhibited a kcat of 51 ± 4 s−1 and a Km of 4 ± 1 µM, which are values similar to those of wild-type L1 (Table 1). Spin-labeled K286C bound 1.8 ± 0.1 eq of Zn(II) and exhibited a kcat of 9 ± 1 s−1 and a Km of 0.9 ± 0.3 µM, which are values similar to those of wild-type L1 (Table 1). The spin-labeled T163C/K286C mutant bound 2.1 ± 0.1 eq. of Zn(II) and exhibited a kcat of 9.2 ± 2.2 s−1 and a Km of 1.4 ± 0.1 µM.
2.3.3. DEER studies on the spin-labeled T163C/K286C mutant of L1
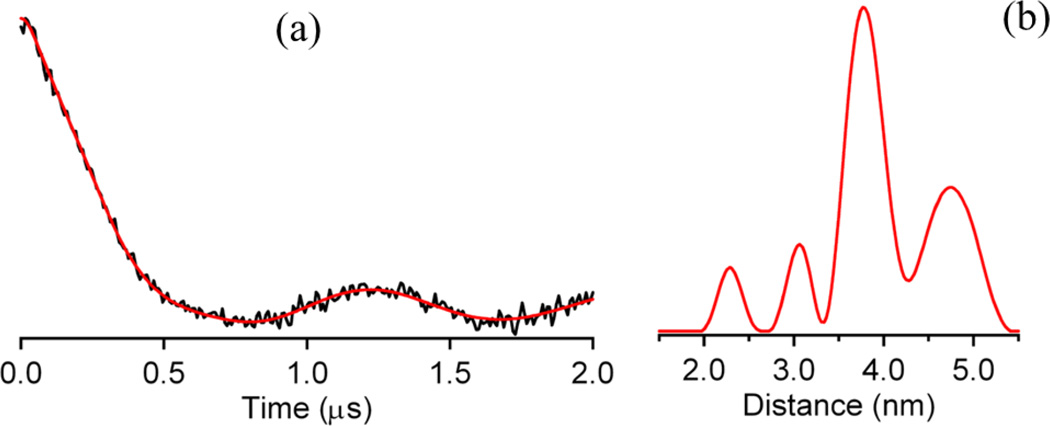
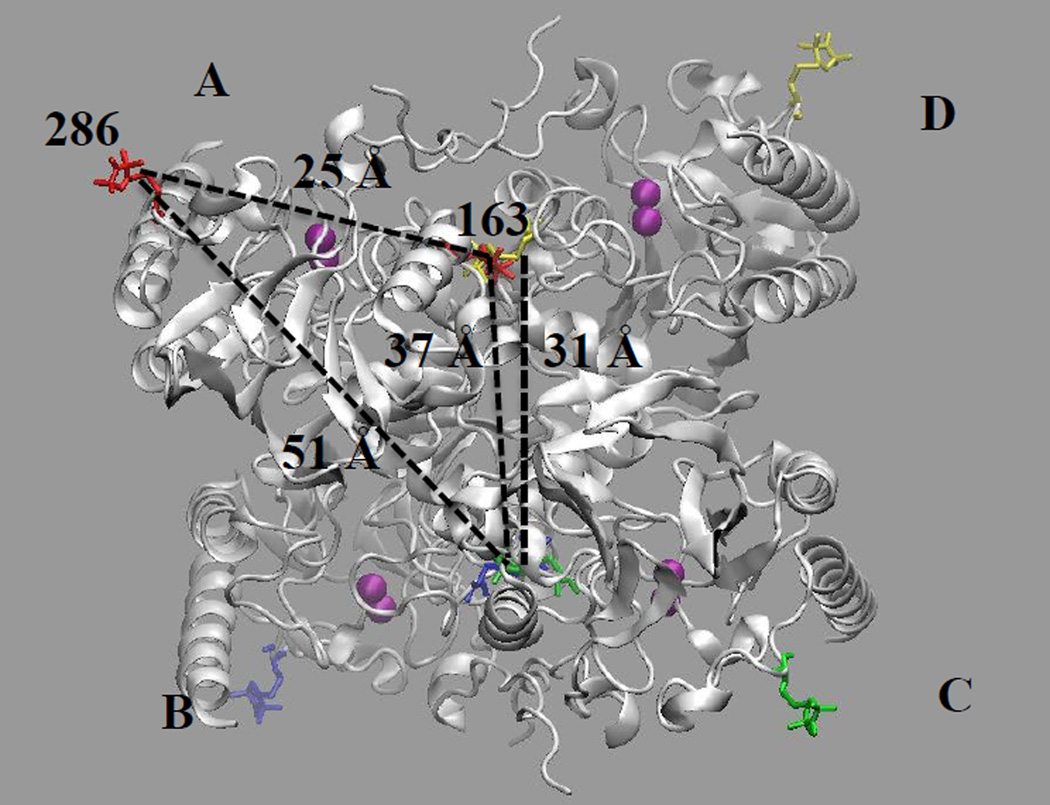
The time- and distance domain Q-band DEER spectra of the spin-labeled T163C/K286C mutant of L1 are shown in Figure 2. The spin-labeled T163C/K286C L1 mutant yielded four distinct distances at 23, 31, 37, and 48 Å (± 10%). An examination of the crystal structure of tetrameric L1 shows one intramolecular dipolar coupling between a spin label at position 163 with a spin label at position 286 and a number of potential intermolecular dipolar couplings (Figure 3). Judging by distances alone, we can tentatively assign the distance-domain DEER peaks at 31, 37, and 51 Å to Thr163 on subunit D coupled to Thr163 on subunit C, Thr163 on subunit A coupled to Thr163 on subunit C, and Lys286 on subunit A coupled to Thr163 on subunit C, respectively (Figure 3). We were unable to detect any dipolar couplings between spin-labeled Thr163 on A and D (or B and C) subunits, most likely because the distances were < 10 Å [30]. While we were able to detect the desired intramolecular dipolar couplings between spin-labeled Thr163 and Lys286, the other undesired intermolecular dipolar couplings render this form of the labeled enzyme unsuitable for ongoing mechanistic DEER studies.
Figure 2.
(a) Time-domain Q-band DEER spectrum of spin-labeled T163C/K286C mutant of homotetrameric L1. (b) Distance domain DEER spectrum of the spin-labeled T163C/K286C mutant of homotetrameric L1.
Figure 3.
Structure of spin-labeled homotetrameric L1. Subunits are labeled with A, B, C, or D. Spin labels are at position 163 (in center of tetramer) and position 286 (on outer corners of tetramer) in all four subunits. Distances between spin labels are included: intramolecular distance of 25 Å is shown in subunit A (spin labels at positions 163 and 286) and intermolecular distances (see text for description). The figure was generated using by using molecular graphing software VMD and molecular dynamics simulations starting with the L1 tetramer crystal structure (PDB ID: 1SML) as described in the Methods section.
2.3.4. Preparation and characterization of a “spin-diluted”, spin-labeled T163C/K286C mutant of L1
To prepare a spin-diluted, spin-labeled analog, wild-type L1 and the T163C/K286C mutant were over-expressed and purified. The enzymes were made metal-free according to previously reported procedures [28]. The apo-enzymes were unfolded with Gdn-HCl, and the spin-diluted sample was made by refolding 4 molar equivalents of unfolded wild-type L1 with 1 molar equivalent of the unfolded T163C/K286C in the presence of Zn(II). The spin-diluted, unlabeled T163C/K286C mutant bound 1.7 ± 0.1 eq of Zn(II) and exhibited a kcat of 20 ± 1 s−1 and a Km of 2.5 ± 0.5 µM (Table 1).
The spin-diluted T163C/K286C mutant was spin-labeled by reacting the double mutant with a 15-fold excess of MTSSL. Unbound spin label was removed by gel filtration chromatography. Cw-EPR spectroscopy showed that the spin-diluted, double mutant was spin-labeled with 92% efficiency. The spin-labeled, spin-diluted T163C/K286C mutant bound 1.8 ± 0.1 eq of Zn(II) and exhibited a kcat of 23 ± 1 s−1 and a Km of 2.1 ± 0.7 µM.
2.3.5. DEER studies on the “spin-diluted”, spin-labeled T163C/K286C mutant of L1
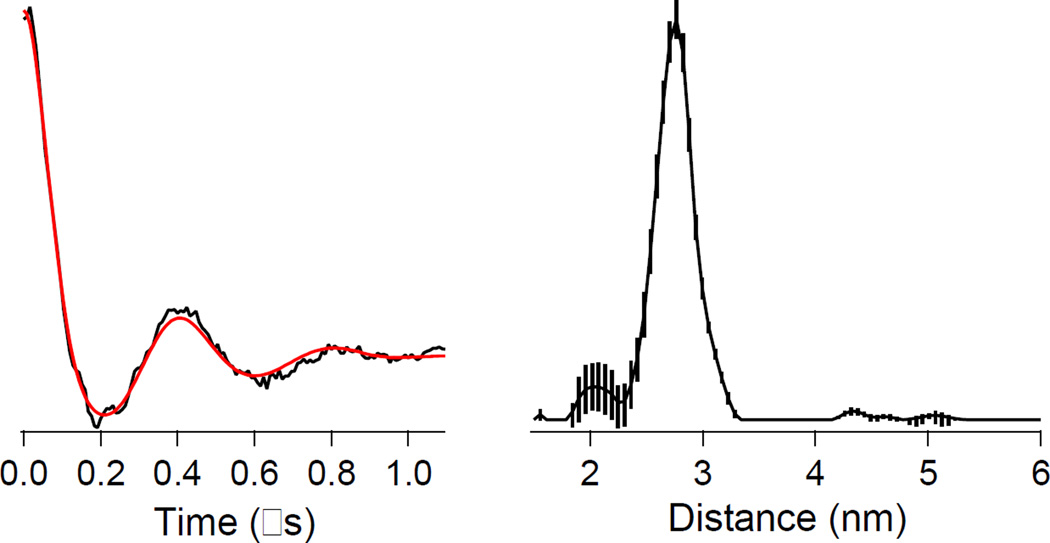
In marked contrast to the fully spin-labeled homotetramer, X-band DEER of the spin-diluted, spin-labeled T163C/K286C L1 mutant indicated only a single interspin distance at 26 ± 3 Å (Figure 4), which corresponds to the expected intramolecular dipolar coupling between a spin label at position 163 and a spin label at position 286. Other weak features in the distance domain trace were observed at 20, 43, 47 and 51 Å, though the significance of these is debatable due to the limited range of useful DEER data in the time domain, corresponding to uncertainty beyond 43 Å, and the calculated uncertainties in the magnitudes indicates dependence on the details of the data processing parameters. Low levels of intermolecular interactions at 31, 37, and 51 Å were expected. There is a shoulder on the 26 Å peak corresponding to 31 Å. Nothing, however, was observed at 37 Å, and although a peak at 51 Å was in fact detected, its significance is in doubt. A small but apparently significant peak was observed at 43 Å and may correspond to the peak observed at 37 Å in the undiluted sample if the limits of the errors are considered.
Figure 4.
(Left) Time-domain X-band DEER spectrum of spin-diluted, spin-labeled T163C/K286C mutant of homotetrameric L1. (Right) Distance domain DEER spectrum of spin-diluted, spin-labeled T163C/K286C mutant of homotetrameric L1. The vertical lines on the distance domain trace are the calculated uncertainties in the data due to the dependence on the data processing parameters.
2.4. Discussion
An understanding of enzyme dynamics is central to the characterization of protein function. Information about changes in enzyme structure during catalysis has been approached using several techniques, each with their advantages and shortcomings. One strategy is to determine the crystal structure of various enzyme-substrate complexes along the catalytic pathway, a technique which has been shown to provide a wealth of information about reaction mechanism.[35, 36] However, this strategy has had only limited application due the rarity of systems involving intermediate complexes sufficiently stable for the generation of crystals. A more common technique is fluorescence spectroscopy, which measures changes in fluorescence due to tryptophan residues.[37] While fluorescence spectroscopy provides information about the rate of a catalytic reaction, it fails to provide specific structural data. Fluorescence resonance energy transfer (FRET) can be used to probe the interaction between site-specific donor and acceptor fluorophores in a molecule and give distance information ranging from 10 to 75 Å.[38, 39] When combined with stopped-flow fluorescence, FRET can be used to measure changes in inter- or intramolecular distances over time.
We initially attempted to use stopped-flow FRET studies to probe loop motion in L1 during catalysis. However, our attempts to generate a doubly-labeled mutant were unsuccessful; we were unable to label any of the loop residues with the large fluorophores. As a result we turned to DEER spectroscopy, which utilizes relatively small spin-labels and provides distance distributions from approximately 20 to 80 Å.[30, 40] Results from CW EPR spectroscopic experiments demonstrated successful spin-labeling of L1 mutants. Steady-state kinetics and ICP-AES studies showed that spin-labeled L1 mutants exhibited similar steady state kinetic constant and metal content as wild-type L1. In addition, gel filtration chromatography and CD spectroscopy were used to verify that the refolded enzyme was tetrameric (Supplementary Figure S2).
DEER of the spin-labeled T163C/K286C mutant revealed four distinct interspin distances at 23, 31, 37 and 48 Å) (Figure 2). Based on the crystal structure of L1, the expected intramolecular distance between spin labels at position 163 and 286 was 25 Å (Figure 1). The other observed distances were assigned to dipolar coupling between spin labels on different subunits of tetrameric L1. The use of the fully spin-labeled, tetrameric L1 in future spectrokinetic studies to probe conformational change during reaction would be significantly complicated by the presence of these additional DEER modulations due to inter-subunit dipolar couplings. Given the position of the spin-labels on the loop in L1, it would also be impossible to probe for motion using dipolar couplings between Co(II) in a Co(II)-substituted L1 and a spin label on the loop, as we reported for ImiS.[23] The resulting broadening of the EPR signals of the spin labels would require that all four subunits were synchronized to obtain meaningful spectrokinetic information. A monomeric form of L1 would circumvent these problems, and it is indeed possible to generate monomeric L1 by introducing an M175→D substitution.[41] Unfortunately, the M175D variant exhibits markedly different kinetic behavior (e.g., Km = 900 µM for nitrocefin) that raises the possibility of an altered catalytic mechanism. Previously, several groups have reported adding unlabeled protein to spin-labeled oligomeric proteins to remove intermolecular dipolar couplings.[42, 43] For example, Kim et al. added fully-folded, cysteine-free arrestin-1 to fully-folded, spin-labeled arrestin-1 tetramers to remove the dipolar couplings of spin labels on adjacent subunits.[44] Xu et al. used a similar method to “dilute” BtuB [45]. We attempted a similar strategy with L1; however, we were unable with L1 to “dilute” the intermolecular dipolar couplings using this method.
Here, we have described a novel method of unfolding and monomerization of a spin-labeled homomultimeric metalloprotein, followed by dilution with unlabeled protein, and reassembly of the spin-diluted multimer. The use of a metalloprotein complicated this approach. In our hands, we could not unfold Zn(II)-containing L1 with Gdn-HCl. Therefore, the metal-free analog of L1 was used in the unfolding step. This method has the potential advantage of being able to generate metal-substituted analogs of the spin-label, spin-diluted protein since metal ion is added back to the protein during the refolding step. This approach, therefore, could be used to prepare single spin-labeled proteins containing paramagnetic metal ions, and the resulting proteins could be interrogated with cw-EPR.[23]
Here, with a dilution of a labeled tetramer, we can predict the composition of the reassembled protein. Taking the symbols “X” and “O” to represent labeled (doubly) and unlabeled monomers, we expect 42% of the desired XOOO species, along with 32% of the unlabeled OOOO species, 21% of the XXOO species, 5% of XXXO and 0.4% of XXXX. The unlabeled OOOO species can be ignored, and the very small amounts of XXXO and XXXX may also be neglected. For purposes of magnetic resonance, we can consider our population as one of singly- and doubly-labeled tetramers in a ratio of 2:1. For a generalized fully-labeled homotetramer with equivalent subunits A, B, C and D, one could expect up to six sets of intersubunit interactions, AB, BC, CD, AD, BD, and AC, compared to the four sets of intrasubunit interactions, giving a signal-to-interference ratio (SIR) of 4:6, or 1:1.5. Applying our dilution method to this general population, we now have only one intrasubunit interaction in two-thirds of the population, and have two intrasubunit and one intersubunit interaction in one-third of the population. This technique yields an SIR of , or 4:1, a six-fold improvement over the fully-labeled system. In the present case, the improvement is better yet, because the intersubunit interactions are specific for either AB (or AD), or else AC; that is to say that XXOO is not equivalent to XOXO. The expected relative intensities of the desired ~ 25 Å signal and the undesirable 31 Å signal are 6:1, and those for the 25 Å signal versus the 37 and 51 Å signals are 12:1. Thus we have improved the SIRs by factors of 9 and 18 by the use of the dilution method for L1. In practice, our DEER data show that these improvements are sufficient to render the unwanted couplings, at 8 and 17% prevalence, undetectable among the much stronger ~ 25 Å modulations.
In general, we anticipate that this method will have great utility in the design of specifically-labeled multimers where inter-subunit interactions contaminate spectroscopic data. This technique can be used with multimers that are tightly-associated, such as hemocyanin[46], xanthine oxidase[47], nitrogenase[48], and aspartate transcarbamoylase[49], and cannot be diluted with dialysis. Specific to L1, we anticipate that this method will provide much higher confidence in the interpretation of changes in the DEER spectra observed as a function of reaction time in ongoing RFQ spectrokinetic studies.
Supplementary Material
Highlights.
Most of the MβLs contain a dynamic loop that is involved in catalysis
Development of a method to double spin label a metalloenzyme
Development of a method to spin-dilute double spin labeled, tightly-bound oligomeric metalloprotein
Acknowledgments
Funding and support from Miami University, Arnold and Mabel Beckman Foundation (TKR), and the National Science Foundation (CHE1151658 to MWC) are gratefully acknowledged. Althea Gunther was an NSF-REU student in 2011.
Abbreviations
- MβL
Metallo-β-lactamase
- SDSL
site-directed spin labeling
- DEER
double electron electron resonance
- MTSL
(1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid
- RFQ
Rapid freeze quench
- ITPG
Isopropyl β-D-1-thiogalactopyranoside
- DTT
dithiothreitol
- FRET
Fluorescence resonance energy transfer
- SIR
signal-to-interference ratio
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The online version of this contains supplementary material, which is available to authorized users.
References
- 1.Fisher JF, Meroueh SO, Mobashery S. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 2.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Antimicro. Agents Chemo. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA. Antimicro. Agents Chemo. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K. J. Infect. Chemother. 2013;19:549–559. doi: 10.1007/s10156-013-0640-7. [DOI] [PubMed] [Google Scholar]
- 5.Ambler RP. Phil. Trans. R. Soc. Lond. 1980;B 289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 6.Crowder MW, Spencer J, Vila AJ. Acc. Chem. Res. 2006;39:721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 7.Heinz U, Adolph HW. CMLS, Cell. Mol. Life Sci. 2004;61:2827–2839. doi: 10.1007/s00018-004-4214-9. [DOI] [PubMed] [Google Scholar]
- 8.Drawz SM, Bonomo RA. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw RW, Kim SK. 7,772,388 B2. U.S. Patent. 2010
- 10.Sutton L, Yu S, Fountain KR. 2010/0261700 A1. U.S. Patent Application. 2010
- 11.Cornaglia G, Giamarellou H, Rossolini GM. Lancet Infect. Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 12.Dmitrienko GI, Johnson JW, Ramadhar TR, Viswanatha T, Viswanatha S. 2011/0046101 A1. U.S. Patent Application. 2011
- 13.Scrofani SDB, Chung J, Huntley JJA, Benkovic SJ, Wright PE, Dyson HJ. Biochemistry. 1999;38:14507–14514. doi: 10.1021/bi990986t. [DOI] [PubMed] [Google Scholar]
- 14.Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP, Lewis C, Galleni M, Frere JM, Payne DJ, Bateson JH, Abdel-Meguid SS. Biochemistry. 2000;39:4288–4298. doi: 10.1021/bi992569m. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald PMD, Wu JK, Toney JH. Biochemistry. 1998;37:6791–6800. doi: 10.1021/bi9730339. [DOI] [PubMed] [Google Scholar]
- 16.Mollard C, Moali C, Papamicael C, Damblon C, Vessilier S, Amicosante G, Schofield CJ, Galleni M, Frere JM, Roberts GCK. J. Biol. Chem. 2001;276:45015–45023. doi: 10.1074/jbc.M107054200. [DOI] [PubMed] [Google Scholar]
- 17.Toney JH, Fitzgerald PMD, Grover-Sharma N, Olson SH, May WJ, Sundelof JG, Vanderwall DE, Cleary KA, Grant SK, Wu JK, Kozarich JW, Pompliano DL, Hammond GG. Chem. Biol. 1998;5:185–196. doi: 10.1016/s1074-5521(98)90632-9. [DOI] [PubMed] [Google Scholar]
- 18.Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS, Gamblin SJ, Spencer J. J. Mol. Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 19.Huntley JJ, Fast W, Benkovic SJ, Wright PE, Dyson HJ. Protein Sci. 2003;12:1368–1375. doi: 10.1110/ps.0305303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huntley JJA, Scrofani SDB, Osborne MJ, Wright PE, Dyson HJ. Biochemistry. 2000;39:13356–13364. doi: 10.1021/bi001210r. [DOI] [PubMed] [Google Scholar]
- 21.Moali C, Anne C, Lamotte-Brasseur J, Groslambert S, Devreese B, Van Beeumen J, Galleni M, Frere JM. Chem. Biol. 2003;10:319–329. doi: 10.1016/s1074-5521(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Keeney D, Tang XJ, Canfield N, Rasmussen BA. J. Biol. Chem. 1999;274:15706–15711. doi: 10.1074/jbc.274.22.15706. [DOI] [PubMed] [Google Scholar]
- 23.Garrity JD, Pauff JM, Crowder MW. J. Biol. Chem. 2004;279:39663–39670. doi: 10.1074/jbc.M406826200. [DOI] [PubMed] [Google Scholar]
- 24.Sharma N, Hu Z, Crowder MW, Bennett B. J. Am. Chem. Soc. 2008;130:8215–8222. doi: 10.1021/ja0774562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garau G, Garcia-Saez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frere JM, Dideberg O. Antimicrob. Agents Chemother. 2004;48:2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. Antimicrob Agents Chemother. 1998;42:921–926. doi: 10.1128/aac.42.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feldmann EA, Ni S, Sahu ID, Mishler CH, Risser DD, Murakami JL, Tom SK, McCarrick RM, Lorigan GA, Tolbert BS, Callahan SM, Kennedy MA. Biochemistry. 2011;50:9212–9224. doi: 10.1021/bi201226e. [DOI] [PubMed] [Google Scholar]
- 28.Hu Z, Periyannan GR, Crowder MW. Anal. Biochem. 2008;378:177–183. doi: 10.1016/j.ab.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett B. International EPR (ESR) Society, EPR Newsletter. 2012;22:10–12. [Google Scholar]
- 30.Jeschke G. Annu. Rev. Phys. Chem. 2012;63:19.11–19.28. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 31.Georgieva ER, Roy AS, Grigoryants VM, Borbat PP, Earle KA, Scholes CP, Freed JH. J. Mag. Res. 2012;216:69–77. doi: 10.1016/j.jmr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey W, Dalke A, Schulten K. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 34.Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS, Galleni M, Spencer J. J. Mol. Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 35.He Y, Macauley MS, Stubbs KA, Vocadlo DJ, Davies GJ. J. Am. Chem. Soc. 2010;132:1807–1809. doi: 10.1021/ja9086769. [DOI] [PubMed] [Google Scholar]
- 36.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 37.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd. New York: Springer; 2006. [Google Scholar]
- 38.Allen WJ, Rothwell PJ, Waksman G. Protein Sci. 2008;17:401–408. doi: 10.1110/ps.073309208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy R, Hohng S, Ha T. Nature Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fajer PG, Brown L, Song L. In: Biological Magnetic Resonance ESR Spectroscopy in Membrane Biophysics. Hemminga MA, Berliner LJ, editors. New York: Springer; 2007. pp. 95–128. [Google Scholar]
- 41.Simm AM, Higgins CS, Carenbauer AL, Crowder MW, Bateson JH, Bennett PM, Clarke AR, Halford SE, Walsh TR. J. Biol. Chem. 2002;277:24744–24752. doi: 10.1074/jbc.M201524200. [DOI] [PubMed] [Google Scholar]
- 42.Langen R, Isas JM, Hubbell WL, Haigler HT. Proc. Natl. Acad. Sci. USA. 1998;95:14060–14065. doi: 10.1073/pnas.95.24.14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuello LG, Jogini V, Cortes DM, Perozo E. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim M, Vishnivetskiy SA, Van Eps N, Alexander NS, Cleghorn WM, Zhan X, Hanson SM, Morizumi T, Ernst OP, Meiler J, Gurevich VV, Hubbell WL. Proc. Natl. Acad. Sci. USA. 2012;109:18407–18412. doi: 10.1073/pnas.1216304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Q, Ellena JF, Kim M, Cafiso DS. Biochemistry. 2006;45:10847–10854. doi: 10.1021/bi061051x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hazes B, Magnus KA, Bonaventura C, Bonaventura J, Dauter Z, Kalk KH, Hol WGJ. Protein Science. 1993;2:597–619. doi: 10.1002/pro.5560020411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF. Proc. Natl. Acad. Sci. USA. 2000;97:10723–10728. doi: 10.1073/pnas.97.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J, Woo D, Rees DC. Biochemistry. 1993;32:7104–7115. doi: 10.1021/bi00079a006. [DOI] [PubMed] [Google Scholar]
- 49.Gouaux JE, Stevens RC, Lipscomb WN. Biochemistry. 1990;29:7702–7715. doi: 10.1021/bi00485a020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.