Abstract
Mutations in LRIT3 lead to complete congenital stationary night blindness (cCSNB). The exact role of LRIT3 in ON-bipolar cell signaling cascade remains to be elucidated. Recently, we have characterized a novel mouse model lacking Lrit3 (no b-wave 6, (Lrit3nob6/nob6)), which displays similar abnormalities as patients with cCSNB with LRIT3 mutations. Here we compare the localization of components of the ON-bipolar cell signaling cascade in wild-type and Lrit3nob6/nob6 retinal sections by immunofluorescence confocal microscopy. An anti-LRIT3 antibody was generated. Immunofluorescent staining of LRIT3 in wild-type mice revealed a specific punctate labeling in the outer plexiform layer (OPL), which was absent in Lrit3nob6/nob6 mice. LRIT3 did not colocalize with ribeye or calbindin but colocalized with mGluR6. TRPM1 staining was severely decreased at the dendritic tips of all depolarizing bipolar cells in Lrit3nob6/nob6 mice. mGluR6, GPR179, RGS7, RGS11 and Gβ5 immunofluorescence was absent at the dendritic tips of cone ON-bipolar cells in Lrit3nob6/nob6 mice, while it was present at the dendritic tips of rod bipolar cells. Furthermore, PNA labeling was severely reduced in the OPL in Lrit3nob6/nob6 mice. This study confirmed the localization of LRIT3 at the dendritic tips of depolarizing bipolar cells in mouse retina and demonstrated the dependence of TRPM1 localization on the presence of LRIT3. Since tested components of the ON-bipolar cell signaling cascade and PNA revealed disrupted localization, an additional function of LRIT3 in cone synapse formation is suggested. These results point to a possibly different regulation of the mGluR6 signaling cascade between rod and cone ON-bipolar cells.
Keywords: retina, knock-out mice, complete CSNB, immunolocalization studies, mGluR6
INTRODUCTION
The visual ON-pathway is initiated at the first retinal synapse between photoreceptors and ON-bipolar cells. In darkness, glutamate released by the photoreceptors binds to the metabotropic glutamate receptor 6 (GRM6/mGluR6) on the ON-bipolar cell dendrites (Nakajima et al., 1993; Nomura et al., 1994). This binding leads to the activation of the heterotrimeric G-protein, Go (Nawy, 1999; Dhingra et al., 2000), which results in the closure of the transient receptor potential melastatin 1 (TRPM1) cation channel (Morgans et al., 2009; Shen et al., 2009; Koike et al., 2010). When the photoreceptors are stimulated by light, glutamate release is reduced, the cascade is deactivated, and TRPM1 channels open, resulting in cell membrane depolarization.
Mutations in several genes disrupt synaptic transmission between photoreceptors and ON-bipolar cells, leading to complete congenital stationary night blindness (cCSNB) (Zeitz, 2007; Zeitz et al., 2015). cCSNB has been associated with mutations in genes encoding proteins localized at the dendritic tips of ON-bipolar cells (Masu et al., 1995; Morgans et al., 2009; Orlandi et al., 2012; Peachey et al., 2012b; Orhan et al., 2013), including nyctalopin, a leucine-rich repeat protein (Morgans et al., 2006). Recently, we have identified mutations in LRIT3, a gene encoding the leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 protein, leading to cCSNB (Zeitz et al., 2013). The corresponding protein localizes at the dendrites of ON-bipolar cells in human (Zeitz et al., 2013).
The exact role of LRIT3 in the mGluR6 signaling cascade remains to be elucidated. It has been shown that nyctalopin binds to TRPM1 (Cao et al., 2011; Pearring et al., 2011) and is essential for its correct localization (Pearring et al., 2011). Membrane transport of proteins such as TRPM1 involves cytoskeletal scaffolding proteins, many of which contain PDZ domains (Feng & Zhang, 2009). Nyctalopin, being mainly an extracellular protein, does not contain an intracellular PDZ binding domain and is therefore probably not able to bring TRPM1 to the cell surface (Pearring et al., 2011). Interestingly, LRIT3 has a PDZ-binding motif and might fulfill this function (Zeitz et al., 2013).
Mouse models for cCSNB have been helpful in dissecting the visual ON-pathway in bipolar cells (Cao et al., 2011; Pearring et al., 2011; Orlandi et al., 2012; Orlandi et al., 2013). Seven mouse models with defects in four different genes have already been published (Masu et al., 1995; Pardue et al., 1998; Gregg et al., 2003; Pinto et al., 2007; Maddox et al., 2008; Morgans et al., 2009; Shen et al., 2009; Koike et al., 2010; Peachey et al., 2012a; Peachey et al., 2012b). Recently, we have characterized a novel mouse model lacking Lrit3 (no b-wave 6 (Lrit3nob6/nob6)), which displays similar abnormalities as patients with cCSNB due to LRIT3 mutations, with lacking or severely reduced b-wave amplitudes in the scotopic and photopic electroretinogram, respectively (Neuille et al., 2014).
Here we describe the localization of LRIT3 in mouse retina and compare the localization of known components of the mGluR6 signaling cascade in wild-type and Lrit3nob6/nob6 retinas to better understand the function of LRIT3.
MATERIALS AND METHODS
Ethics statements
All animal procedures were performed according with the Council Directive 2010/63EU of the European Parliament and the Council of 22 September 2010 on the protection of animals used for scientific purposes and were approved by the French Minister of Agriculture (authorization A-75–1863 delivered on 09th November 2011). All efforts were made to minimize suffering.
Animal Care
The generation and characterization of the Lrit3 knock-out mouse has been described elsewhere (Neuille et al., 2014) (http://www.taconic.com/knockout-mouse/lrit3/tf2034_-_model_lrit3). Ten wild-type (Lrit3+/+) and seven mutant (Lrit3nob6/nob6) six to seven-week-old male and female mice were used in this study. Mice were housed in a temperature-controlled room with a 12-h light/12-h dark cycle. Fresh water and rodent diet were available ad libitum.
LRIT3 antibody
To obtain an antibody to mouse LRIT3, the following procedure was performed by a company (Eurogentec, Seraing, Belgium). Two peptides corresponding to amino acids 180–195 and 613–628 of mouse LRIT3 (NP_001274153.1; peptide sequences: AVTPSRSPDFPPRRII and CTSKPFWEEDLSKETY, respectively) were synthesized and coupled to keyhole limpet haemocyanin (KLH). Two adult New Zealand White rabbits were immunized with both peptide-KLH conjugates. The rabbits received three more immunizations seven, ten and eighteen days later. Immune sera were collected ten days after the final immunization and stored at −20°C. We performed immunohistochemistry on Lrit3+/+ and Lrit3nob6/nob6 retinal sections with these two sera. One of them produced a specific punctate signal in the outer plexiform layer of the Lrit3+/+ retinal sections that was absent on Lrit3nob6/nob6 sections. However, strong non-specific signal and noise were also present on the whole Lrit3+/+ and Lrit3nob6/nob6 sections (data not shown). In order to determine which of the two peptides resulted in the specific staining, we performed immunohistochemistry on Lrit3+/+ retinal sections after preincubation of the serum with either the N- or the C-terminal peptide. The specific signal was absent when the serum was preincubated with the peptide localized at the N-terminus but noise remained (data not shown). Subsequently, affinity purification was performed with this peptide to decrease noise and to obtain a more specific antibody.
Preparation of retinal sections for immunohistochemistry
Mice were killed by CO2 administration and cervical dislocation. Eyes were removed and prepared following three methods. For method 1, we made two slits in a cross within the cornea and placed the eyeball in ice cold 4% (w/v) paraformaldehyde in 0.12 M phosphate buffer, pH 7.2 for 1 hour. After three 10-min-washes with ice cold phosphate buffer saline (PBS), we transferred the eyeball to cold 30% sucrose. Finally, the lens was removed and the eyecup was embedded in OCT (Sakura Finetek, AJ Alphen aan den Rijn, The Netherlands) and frozen in a dry ice-cooled bath of isopentane. For method 2, the anterior segment and lens were removed and the eyecup was fixed in ice cold 4% (w/v) paraformaldehyde in 0.12 M phosphate buffer, pH 7.2 for 20 min. The eyecup was washed three times in ice-cold PBS and cryoprotected with increasing concentrations of ice cold sucrose in 0.12 M phosphate buffer, pH 7.2 (10%, 20% for 1 h each and 30% overnight). Finally, the eyecup was embedded in 7.5% gelatin-10% sucrose and frozen in a dry ice-cooled bath of isopentane. For method 3, we made a hole just behind the ora serrata and placed the eyeball in 4% (w/v) paraformaldehyde in 0.12 M phosphate buffer, pH 7.2 for 5 min. We then removed the lens and the eyecup was again fixed for 20 min in paraformaldehyde at room temperature. The eyecup was washed three times in PBS and cryoprotected with increasing concentrations of ice cold sucrose in 0.12 M phosphate buffer, pH 7.2 (10% for 1 h and 30% overnight). Finally, the eyecup was embedded in 7.5% gelatin-10% sucrose and frozen in a dry ice-cooled isopentane bath. Sections were cut at a thickness of 18 µm on a cryostat and mounted onto glass slides (Super-Frost, Thermo Fisher Scientific, Waltham, MA, USA). The slides were air dried and stored at −80°C.
Immunostaining of retinal cryosections
Primary antibodies used for immunostaining are listed in Table 1. The TRPM1 immunoreactive serum from a patient suffering from melanoma-associated retinopathy (MAR), the rat mGluR6 antiserum raised in sheep, the purified goat polyclonal antibody to mouse Gβ5 and the mouse RGS11 antibody raised in rabbit were used as previously described (Chen et al., 2003; Morgans et al., 2006; Morgans et al., 2007; Xiong et al., 2013). Immunohistochemistry on retinal sections was performed following two protocols depending on the section preparation. For eyecups embedded in OCT, we used a previously published protocol (Morgans et al., 2006). Briefly, eyecup sections were blocked by incubation with antibody incubation solution (AIS) (3% (v/v) normal horse serum, 0.5% (v/v) Triton X-100, 0.025% (w/v) NaN3 in PBS) at room temperature for 60 min. Subsequently, the sections were incubated with primary antibodies in AIS for 1 h at room temperature. After washing in PBS, the sections were incubated with secondary antibodies coupled to Alexa Fluor 488 or Alexa Fluor 594 (Life Technologies, Grand Island, NY, USA) at a dilution of 1:1000 in PBS for 1 h at room temperature. The slides were stained with 4’,6-diamidino-2-phenylindole (DAPI) and subsequently cover-slipped with mounting medium (Aqua-Mount mounting medium, Thermo Fisher Scientific, Waltham, MA, USA). For eyecups embedded in gelatin-sucrose, sections were blocked by incubation at room temperature for 60 min in 10% (v/v) donkey serum, 0.3% (v/v) Triton X-100 in PBS. Subsequently, the sections were incubated with primary antibodies in blocking solution overnight at room temperature. After washing in PBS, the sections were incubated with secondary antibodies coupled to Alexa Fluor 488 or Cy3 (Jackson ImmunoResearch, West Grove, PA, USA) at a dilution of 1:1000 in PBS for 1.5 h at room temperature. The slides were stained with DAPI and subsequently cover-slipped with mounting medium (Mowiol, Merck Millipore, Billerica, MA, USA). None of the secondary antibodies used gave recordable signal when used without primary antibodies (data not shown).
Table 1.
Primary antibodies used in immunohistochemistry in this study
| Antibody | Species | Dilution | Reference | Retinal section preparation |
|---|---|---|---|---|
| LRIT3 | rabbit | 1:500 | This paper | Method 2 |
| TRPM1 | human | 1:2000 | Xiong et al., 2013 | Method 1 |
| mGluR6 | sheep | 1:100 | Morgans et al., 2006 | Method 1 |
| mGluR6 | guinea pig | 1:200 | AP20134SU-N Acris | Methods 2 and 3 |
| GPR179 | mouse | 1:200 | Ab-887-YOM Primm | Method 1 |
| RGS11 | rabbit | 1:4000 | Chen et al., 2003 | Method 1 |
| Gβ5 | goat | 1:100 | Morgans et al., 2007 | Method 1 |
| RGS7 | mouse | 1:1000 | sc-271643 Santa-Cruz | Method 2 |
| Lectin PNA 488 conjugate | Arachis hypogaea | 1:1000 | L21409 Life Technologies | Method 2 |
| Lectin PNA 594 conjugate | Arachis hypogaea | 1:1000 | L32459 Life Technologies | Method 2 |
| Cone arrestin | rabbit | 1:2000 | ab15282 Abcam | Method 2 |
| PKCα | mouse | 1:1000 | P5704 Sigma-Aldrich | Method 2 |
| Goα | mouse | 1:200 | MAB3073 Merck Millipore | Method 3 |
| Calbindin D-28k | mouse | 1:5000 | 300 Swant | Method 2 |
| ribeye | mouse | 1:10000 | 612044 BD Biosciences | Method 2 |
Image acquisition and quantification
Fluorescent staining signals were captured with a confocal microscope (FV1000, Olympus, Hamburg, Germany) equipped with 405, 488, and 559 nm lasers.
For localization studies as well as for quantification of LRIT3, confocal images were acquired with a 40x objective compatible with oil (lens NA: 1.3) imaging pixels of 310 nm and 77 nm in width and height for zoom 1 and 4, respectively, and using a 0.52 µm step size. For localization, each image corresponds to the projection of three optical sections. For figures, brightness and contrast were optimized (ImageJ, version 1.49; National Institutes of Health, Bethesda, MD, USA). The method for quantification of LRIT3 on 4 independent Lrit3+/+ retinal sections was adapted from a previously published protocol (Ramakrishnan et al., 2014), using Imaris (Bitplane, Zurich, Switzerland) and is illustrated in Figure S1. Optical sections centered on the outer plexiform layer (OPL) were stacked to obtain a 3D reconstruction of the entire retinal section thickness at this level. An initial region of interest (ROI) was drawn around the OPL (Figure S1A and S1B). We used peanut agglutinin (PNA) staining as a marker to define the ROI of cone ON-bipolar cell dendritic tips (Figure S1C). The mean intensity per voxel of LRIT3 staining at the dendritic tips of cone ON-bipolar cells was obtained by quantifying LRIT3 staining enclosed within this region. The ROI corresponding to LRIT3 staining at the dendritic tips of rod bipolar cells was formed by drawing spheres of 2 µm radius around LRIT3-labeled puncta that are not associated with PNA (Figure S1D). The mean intensity per voxel of LRIT3 staining at the dendritic tips of rod bipolar cells was then measured. Finally, these two intensities were compared in the same retinal section.
For quantification of PNA, confocal images were acquired with a 20x objective compatible with oil (lens NA: 0.85) imaging pixels of 621 nm in width and height, and using a 1.55 µm step size. We reconstructed three Lrit3+/+ and three Lrit3nob6/nob6 retinas by assembling multiple confocal images of the same retina. For each Lrit3+/+ mouse, a Lrit3nob6/nob6 mouse of the same breeding pair was chosen and their retinas were processed in parallel and in the same way. Quantification of PNA in the OPL and in inner and outer segments (IS/OS) was adapted from a previously published protocol (Xu et al., 2012) and was performed with ImageJ (version 1.49). The background value was defined as the mean intensity per pixel in a ROI comprising the inner nuclear layer (INL) in which no PNA staining has been described. This value was then subtracted from the measurements of OPL and IS/OS intensities to give corrected measurements. For quantification of PNA in the OPL, the corrected mean intensity per pixel was measured for a segmented line of 30 pixels thickness drawn along the OPL. To obtain the number of PNA-labeled synaptic clefts between cones and corresponding cone ON-bipolar cells per mm of OPL (density of particles), the line was straightened and a threshold applied to isolate each labeled structure. The same threshold was applied for the wild-type and the mutant retinas of the same pair. We then counted the number of labeled structures with a size between 4 and 45 µm² and divided this value by the length of the selected line. The corrected mean intensity per pixel and the density of particles were compared between wild-type and mutant mice in each pair. To quantify the PNA staining in the IS/OS compartment, the corrected mean intensity per pixel was measured in a ROI surrounding these compartments. The values obtained were compared between Lrit3+/+ and Lrit3nob6/nob6 mice of each pair.
Preparation of retinal lysates and Western blotting
Retinas were prepared according to a previously published protocol (Cao et al., 2012). Mice were killed by CO2 administration and cervical dislocation. Whole retinas were removed from 3 Lrit3+/+ and 3 Lrit3nob6/nob6 mice and lysed by sonication in ice-cold PBS supplemented with 150 mM NaCl, 1% Triton X-100, and protease inhibitor (complete protease inhibitor tablets, Roche, Meylan, France). Supernatants were collected after a 15 min centrifugation step at 21,000 g at 4°C and total protein concentration was measured by using a bicinchoninic acid protein assay kit (BCA Protein Assay Kit, Thermo Fisher Scientific, Rockford, IL, USA). SDS sample buffer (pH 6.8) containing 8 M urea was added to the supernatants and 8.5 µg of protein were subjected to 4–15% SDS-PAGE (Bio-Rad Laboratories, Hercules, CA, USA). Protein bands were transferred onto nitrocellulose membranes (GE Healthcare, Little Chalfont, UK). Membranes were blocked for 1 hour at room temperature in 5% dry milk diluted in PBS, 0.1% Tween 20 and subjected to primary antibodies in 1% dry milk diluted in PBS, 0.1% Tween 20 overnight at 4°C. After washing in PBS, 0.1% Tween 20, the membranes were incubated with HRP-conjugated secondary antibodies (Jackson ImmunoResearch) in 1% dry milk diluted in PBS, 0.1% Tween 20, and subsequently bands were detected by using a chemiluminescent substrate (ECL2, Thermo Fisher Scientific). Primary antibodies were mouse anti TRPM1 antibody raised in sheep diluted at 1:1000 (Cao et al., 2011) and anti β-actin antibody raised in mouse diluted at 1:10000 (MAB1501, Merck Millipore, Darmstadt, Germany). Bands intensities were quantified by using ImageJ (version 1.49). For each lane, TRPM1 staining intensity was normalized to β-actin staining intensity and compared between 3 Lrit3+/+ and 3 Lrit3nob6/nob6 mice. The experiment was repeated 7 times.
Statistical analyses
Statistical analyses were performed using SPSS Statistics (version 19.0, IBM, Armonk, NY, USA). A Student’s t-test was used to compare TRPM1 protein amounts between Lrit3+/+ and Lrit3nob6/nob6 mice, the intensity of LRIT3 staining at the dendritic tips of rod bipolar cells versus cone ON-bipolar cells in Lrit3+/+ mouse as well as PNA staining in the OPL and in the IS/OS complex between Lrit3+/+ and Lrit3nob6/nob6 mice. Tests were considered as significant when p<0.05.
RESULTS
Validation of the LRIT3 antibody
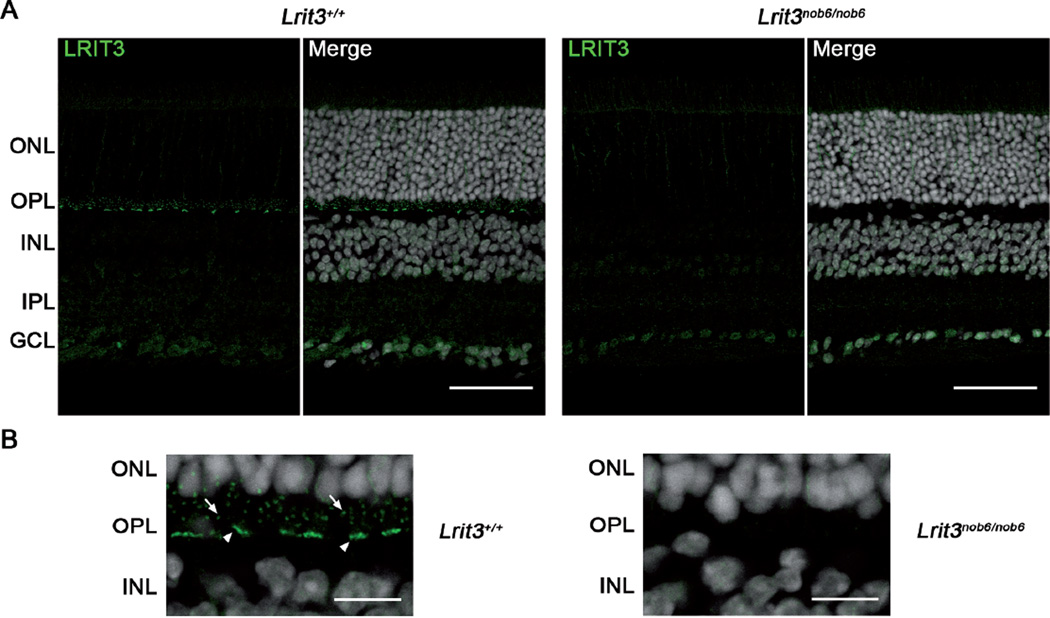
Immunofluorescent staining of LRIT3 on Lrit3+/+ mouse retinal sections revealed punctate labeling in the OPL (Figure 1A, two pictures on the left and Figure 1B left). On Lrit3nob6/nob6 mouse retinal sections, this punctate labeling was absent, demonstrating the specificity of the LRIT3 labeling (Figure 1A, two pictures on the right and Figure 1B, right). Some non-specific signal remained in the inner plexiform and the ganglion cell layers (IPL and GCL, respectively) in both Lrit3+/+ and Lrit3nob6/nob6 mice (Figure 1A, LRIT3 staining alone). The specific staining in the OPL most likely represents the dendritic tips of rod bipolar cells (arrows, Figure 1B, left) and cone ON-bipolar cells (arrowheads, Figure 1B, left).
Figure 1. Validation of mouse LRIT3 antibody.
(A) Representative confocal images of cross sections of Lrit3+/+ and Lrit3nob6/nob6 retina stained with mouse LRIT3 antibody (green) with or without DAPI (grey). Scale bar, 50 µm. (B) 4× zoom of (A) focused on outer plexiform layer (OPL). Arrows represent putative dendritic tips of rod bipolar cells, arrowheads represent putative dendritic tips of cone ON-bipolar cells. Scale bar, 10µm.
LRIT3 localizes at the dendritic tips of depolarizing bipolar cells in the mouse retina
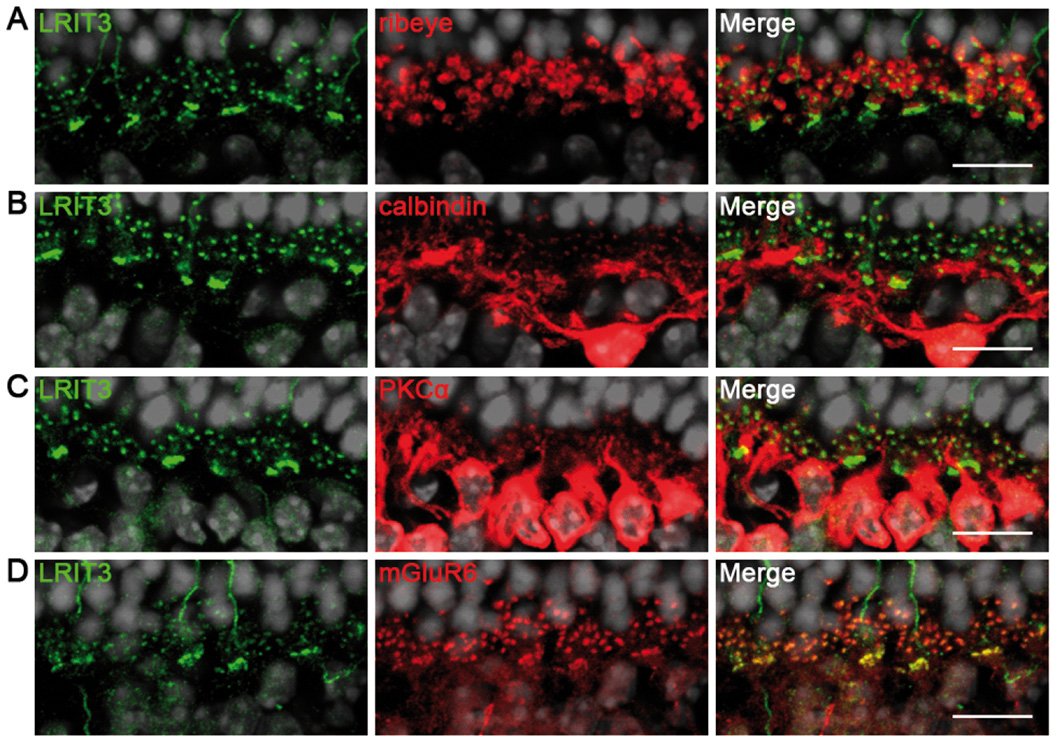
In order to precisely localize LRIT3 in mouse retina, and particularly in the OPL, we performed co-immunolocalization studies with the anti-LRIT3 antibody and antibodies against ribeye, a component of the photoreceptor synaptic ribbon (Schmitz et al., 2000; tom Dieck et al., 2005), calbindin, which labels the post-synaptic processes of horizontal cells (Haverkamp & Wassle, 2000), PKCα to label rod bipolar cells (Negishi et al., 1988; Greferath et al., 1990) and mGluR6 to label dendritic tips of ON-bipolar cells (Vardi et al., 2002; Morgans et al., 2007; Cao et al., 2008).
The LRIT3 puncta did not colocalize with ribeye but were nested within the arcs of the synaptic ribbons (Figure 2A). LRIT3 staining was also adjacent to calbindin staining but without colocalization (Figure 2B). Thus, LRIT3 was present in the OPL but did not localize presynaptically and was absent in the horizontal cells. PKCα was present in both the dendritic tips as well as in the cell bodies of rod bipolar cells, whereas LRIT3 staining was solely concentrated in the OPL (Figure 2C). Strong colocalization of LRIT3 and mGluR6 confirmed that LRIT3 is present at the dendritic tips of rod and cone ON-bipolar cells (Figure 2D).
Figure 2. Localization of LRIT3 in mouse retina.
Representative confocal images of cross sections centered on the OPL of Lrit3+/+ retina stained with antibodies against LRIT3 (green) and (A) ribeye (red) (B) calbindin (red) (C) PKCα (red) or (D) mGluR6 (red) and merge (yellow). Scale bar, 10 µm.
TRPM1 localization is disrupted at the dendritic tips of ON-bipolar cells in Lrit3nob6/nob6 retina
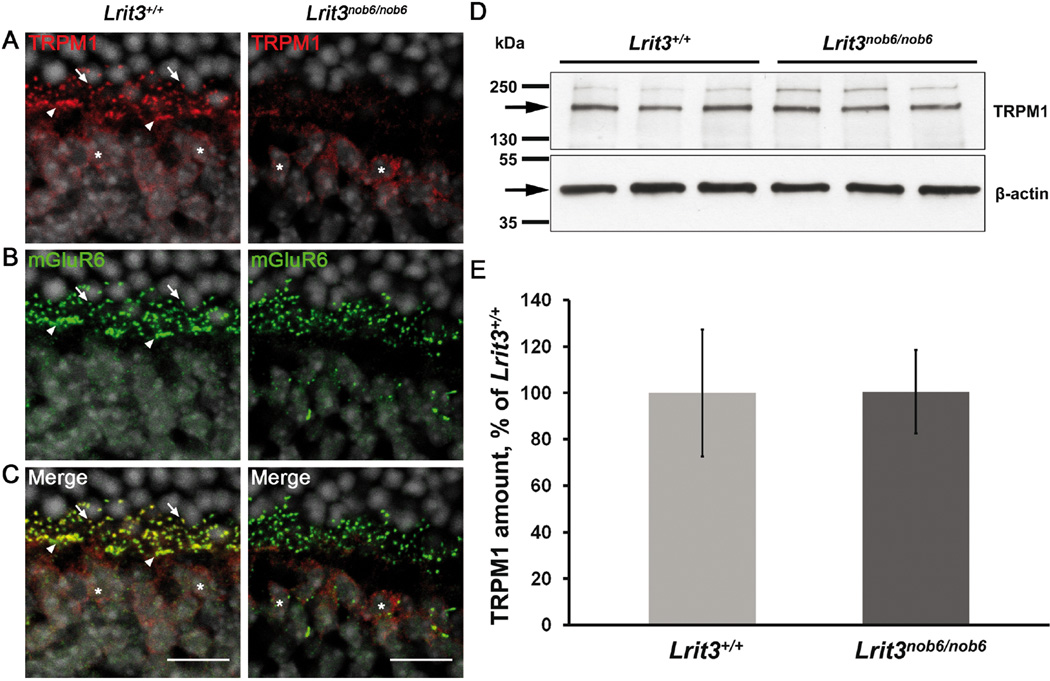
In order to test our hypothesis that LRIT3 is required for the correct localization of TRPM1, we performed immunohistochemistry on Lrit3+/+ and Lrit3nob6/nob6 mouse retinal sections with serum from a patient suffering from melanoma-associated retinopathy (MAR) that was previously demonstrated to show specific TRPM1 immunoreactivity on mouse retinal sections (Xiong et al., 2013). In Lrit3+/+ retinal sections, the serum revealed punctate labeling in the OPL (Figure 3A and 3C, left) that colocalized with mGluR6 staining (Figure 3B and 3C), confirming the localization of TRPM1 to the dendritic tips of rod bipolar cells (arrows, Figure 3B and 3C, left) and cone ON-bipolar cells (arrowheads, Figure 3B and 3C, left). We also observed immunofluorescent labeling of bipolar cell bodies (asterisks) in the INL (Figure 3A and 3C, left). In Lrit3nob6/nob6 mice, the intensity of the TRPM1 puncta co-localizing with mGluR6 was dramatically decreased, but somatic staining (asterisks) seemed unchanged (Figure 3A and 3C, right), suggesting that TRPM1 localization at the dendritic tips of ON-bipolar cells is dependent on LRIT3.
Figure 3. Localization of TRPM1 and mGluR6 as well as TRPM1 protein quantification in Lrit3nob6/nob6.
Representative confocal images of cross sections centered on OPL of Lrit3+/+ and Lrit3nob6/nob6 retina stained with antibodies against (A) TRPM1 (red) and (B) mGluR6 (green) and (C) merge (yellow). Arrows represent putative dendritic tips of rod bipolar cells, arrowheads represent putative dendritic tips of cone ON-bipolar cells, asterisks represent putative somas of ON-bipolar cells. Scale bar, 10µm. (D) Western blots of 3 Lrit3+/+ and 3 Lrit3nob6/nob6 total retinal lysates probed with antibodies to TRPM1 (top) and β-actin (bottom). Bars on the left side of blots represent molecular weight in kDa. Arrows represent bands of interest. (E) Quantification of TRPM1 Western blotting data in panel (D). For each lane, TRPM1 staining intensity was normalized to β-actin staining intensity. Values were plotted as the percentage of the level in Lrit3+/+. Error bars represent standard deviation values.
To investigate whether the decrease in TRPM1 localization at the dendritic tips of depolarizing bipolar cells is also reflected by reduction of total amount of TRPM1 protein, we compared TRPM1 protein levels in Lrit3+/+ and Lrit3nob6/nob6 retinas by Western blot analysis. TRPM1 protein was still present in the retina of Lrit3nob6/nob6 mice and the protein amount was not significantly altered (Figure 3D and 3E). These findings suggest that elimination of LRIT3 results in a severe reduction of TRPM1 at the dendritic tips of all ON-bipolar cells but does not significantly affect the total TRPM1 protein amount.
Other components of the ON-bipolar signaling cascade are mislocalized in Lrit3nob6/nob6 retina
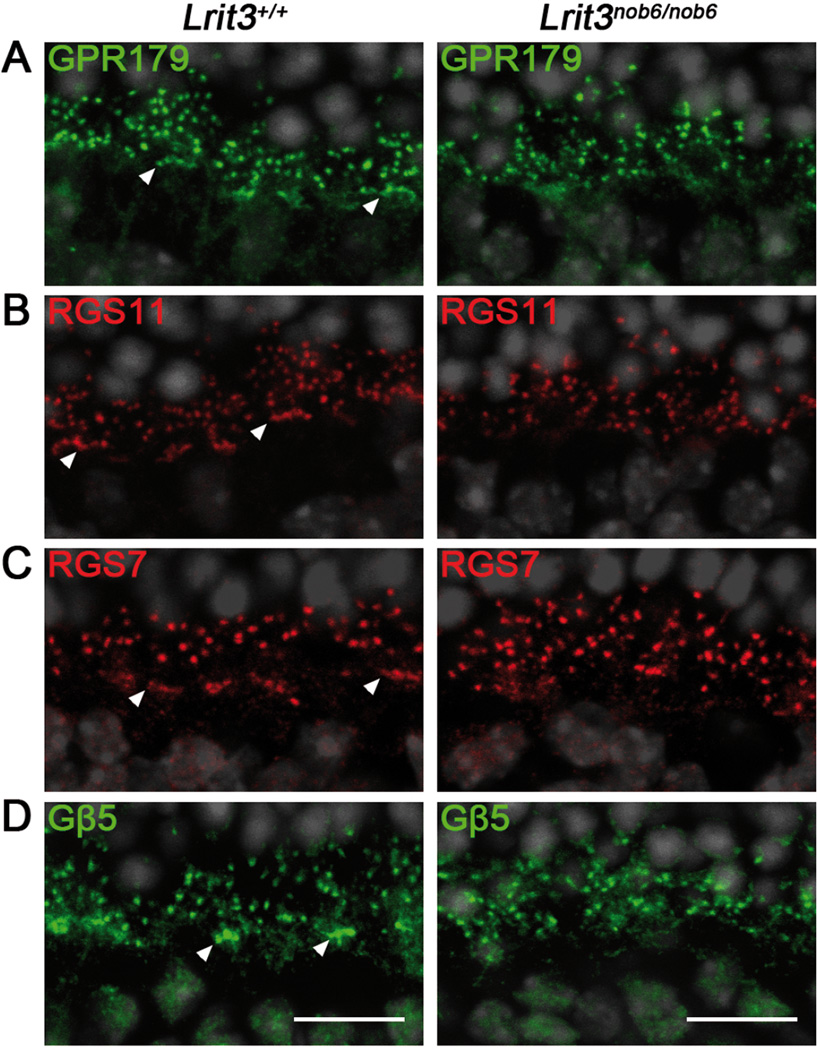
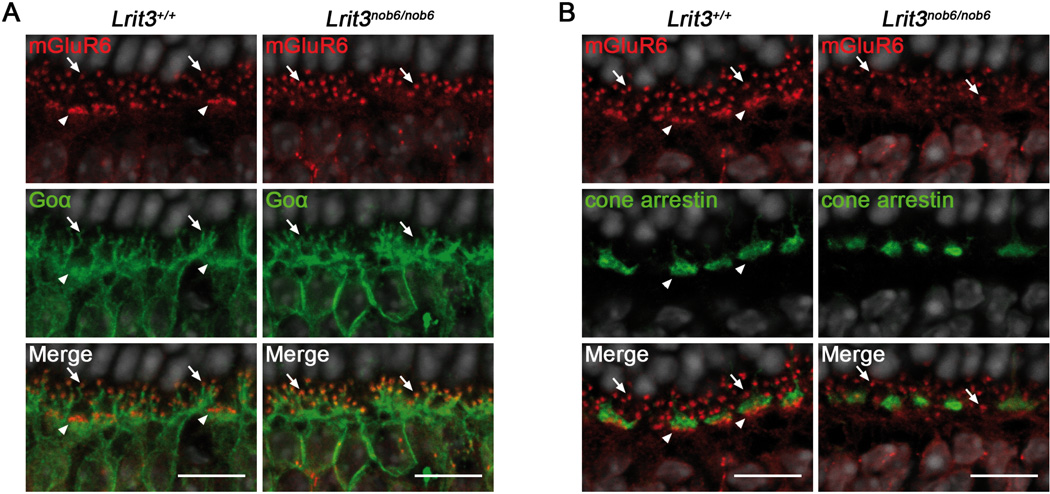
Interestingly, mGluR6 staining in Lrit3nob6/nob6 retina showed an apparently normal punctate labeling of the dendritic tips of rod bipolar cells (arrows, Figure 3B and 3C), but immunofluorescence seemed nearly absent at the dendritic tips of cone ON-bipolar cells (arrowheads, Figure 3B and 3C, left versus right) with some mGluR6 immunofluorescence appearing instead over bipolar cell bodies in the INL. This dramatic decrease of staining at the dendritic tips of Lrit3nob6/nob6 cone ON-bipolar cells was also observed for GPR179, RGS7, RGS11 and Gβ5 (arrowheads, Figure 4 A-D), which are also components of the mGluR6 signaling cascade and normally localize at the dendritic tips of both rod and cone ON-bipolar cells (Morgans et al., 2007; Rao et al., 2007; Orlandi et al., 2012; Peachey et al., 2012b; Orhan et al., 2013). Co-immunostaining of mGluR6 with Goα (Figure 5A) and with cone arrestin (Figure 5B) confirmed these findings. While Goα is a marker of the somas and dendrites of both rod and cone ON-bipolar cells (Vardi, 1998), cone arrestin labels cone photoreceptors including the cone pedicles (Sakuma et al., 1996), which are contacted by the dendritic tips of cone ON-bipolar cells. On Lrit3+/+ retinal sections co-stained with Goα, mGluR6 was present as small puncta at the dendritic tips of rod bipolar cells (arrows, Figure 5A, left) and as larger patches, where each patch represents the dendritic tips of multiple cone ON-bipolar cells contacting a single cone pedicle (arrowheads, Figure 5A, left). On Lrit3+/+ retinal sections co-stained with cone arrestin, mGluR6 was again normally present as small puncta at rod bipolar cell dendrites (arrows, Figure 5B, left) and as larger patches at the dendritic tips of cone ON-bipolar cells (arrowheads, Figure 5B, left). Interestingly, on Lrit3nob6/nob6 sections, in the presence of unaltered Goα staining, cone-associated mGluR6 staining was absent whereas rod-associated mGluR6 puncta were still present (arrows, Figure 5A, right). The cone arrestin immunofluorescence showed that cone pedicles are present in the Lrit3nob6/nob6 retina, but that mGluR6 in close vicinity of cone arrestin was absent (Figure 5B, right).
Figure 4. Localization of GPR179, RGS11, RGS7 and Gβ5 in Lrit3nob6/nob6 retina.
Representative confocal images of cross sections centered on OPL of Lrit3+/+ and Lrit3nob6/nob6 retina stained with antibodies against (A) GPR179 (green) (B) RGS11 (red) (C) RGS7 (red) or (D) Gβ5 (green). Arrowheads represent putative dendritic tips of cone ON-bipolar cells. Scale bar, 10 µm.
Figure 5. Localization of mGluR6, Goα and cone arrestin in Lrit3nob6/nob6 retina.
Representative confocal images of cross sections centered on OPL of Lrit3+/+ and Lrit3nob6/nob6 retina stained with antibodies against mGluR6 (red) and (A) Goα (red) and merge (yellow) or (B) cone arrestin (green) and merge (yellow). Arrows represent putative dendritic tips of rod bipolar cells, arrowheads represent putative dendritic tips of cone ON-bipolar cells. Scale bar, 10 µm.
Peanut agglutinin staining is disrupted in the OPL in Lrit3nob6/nob6 retina
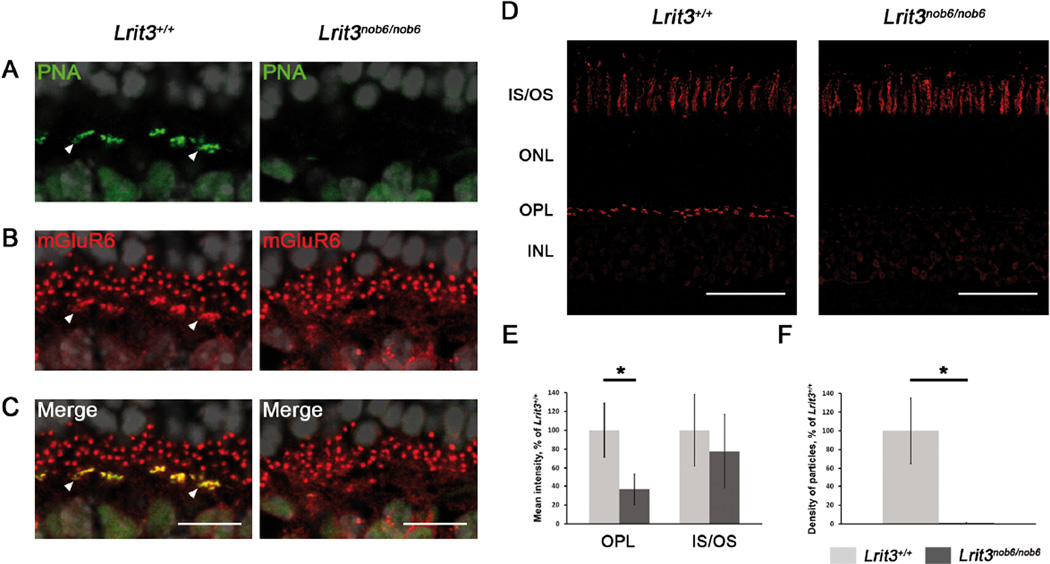
We performed immunostaining on wild-type and mutant retinas with peanut agglutinin (PNA), which binds specific glycosylated residues localized extracellularly to cone inner and outer segments (IS and OS, respectively), and at the synaptic cleft between cone pedicles and cone ON-bipolar cells located in the OPL (Blanks & Johnson, 1983; 1984; Wu, 1984) and was shown to overlap with GPR179 (Ray et al., 2014). In wild-type mice, PNA staining overlapped with mGluR6 staining at the dendritic tips of cone ON-bipolar cells in the OPL (arrowheads, Figure 6A–C, left). In contrast, PNA staining was dramatically decreased in the OPL on Lrit3nob6/nob6 retinal sections (Figure 6A–C, right). On low-power views of retinal sections, this decrease is obvious in the OPL with no apparent difference of staining intensity in the IS and OS of the cones (Figure 6D). Quantitatively, PNA fluorescence intensity was significantly decreased (64%, p=0.041) in the whole OPL of Lrit3nob6/nob6 retinas compared to wild-type mice (Figure 6E). More strikingly, the number of synaptic clefts stained with PNA between cone pedicles and corresponding cone ON-bipolar cells, per mm of OPL, was dramatically decreased with only 1% remaining (p=0.008) in Lrit3nob6/nob6 retinas compared to wild-type (Figure 6F). In contrast, a measured 23% decrease of the PNA staining at the IS/OS complex in Lrit3nob6/nob6 retinas compared to wild-type was not statistically significant (p=0.515) (Figure 6E).
Figure 6. Localization and quantification of PNA in Lrit3nob6/nob6 retina.
Representative confocal images of cross sections centered on OPL of Lrit3+/+ and Lrit3nob6/nob6 retina stained with antibodies against (A) PNA (green) and (B) mGluR6 (red) and (C) merge (yellow). Arrowheads represent putative dendritic tips of cone ON-bipolar cells. Scale bar, 10 µm. (D) Representative confocal images of cross sections of Lrit3+/+ and Lrit3nob6/nob6 retina stained with PNA. Scale bar, 50 µm. (E) Quantification of the intensity of PNA staining in the OPL and in the IS/OS complex in 3 independent Lrit3+/+/Lrit3nob6/nob6 pairs of retina. Error bars represent standard deviation values. Asterisks represent results that are significantly different (p<0.05). (F) Quantification of the number of PNA stained synaptic clefts between cone pedicles and corresponding cone ON-bipolar cells per mm of OPL in 3 independent Lrit3+/+/Lrit3nob6/nob6 pairs of retina. Error bars represent standard deviation values. Asterisks represent results that are significantly different (p<0.05).
LRIT3 staining is similar at the dendritic tips of rod and cone ON-bipolar cells
As localization of several components of the cone to cone ON-bipolar cell synapse seemed to be disrupted in Lrit3nob6/nob6 retina whereas their localization at the rod to rod bipolar cell synapse seemed to be unaffected, and because LRIT3 staining at the dendritic tips of cone ON-bipolar cells seemed to be more intense than that at the dendritic tips of rod bipolar cells, we performed LRIT3 staining quantification at the dendritic tips of rod and cone ON-bipolar cells in 4 Lrit3+/+ mice. LRIT3 staining is 7% less intense at the dendritic tips of rod bipolar cells compared to the dendritic tips of cone ON-bipolar cells but this difference was not significant (p=0.504).
DISCUSSION
In this work, we have produced a specific antibody against mouse LRIT3, localized the corresponding protein in the mouse retina and studied how the localization of most of the known components of the ON-bipolar signaling cascade are affected in a mouse model of cCSNB lacking Lrit3 (Lrit3nob6/nob6). Our results show that LRIT3 localizes at the dendritic tips of both rod and cone ON-bipolar cells, that the correct localization of TRPM1 is dependent on LRIT3 and that the localization of other partners is disrupted in Lrit3nob6/nob6 retina particularly at the cone to cone ON-bipolar cell synapse.
LRIT3 localizes at the dendritic tips of ON-bipolar cells in mouse retina
The punctate labeling observed in the OPL of wild-type retina with the mouse LRIT3 antibody was absent on Lrit3nob6/nob6 retina, demonstrating the specificity of this antibody and validating the knock-out mouse model. This labeling resembles those obtained on human retina with an LRIT3 antibody (Zeitz et al., 2013) as well as those obtained with antibodies directed against various components of the ON-bipolar signaling cascade on mouse retina such as mGluR6, GPR179, NYX, TRPM1, RGS7, RGS11, Gβ5, and R9AP (Masu et al., 1995; Bahadori et al., 2006; Morgans et al., 2006; Morgans et al., 2007; Rao et al., 2007; Cao et al., 2008; Cao et al., 2009; Morgans et al., 2009; Jeffrey et al., 2010; Orlandi et al., 2012; Peachey et al., 2012b; Orhan et al., 2013). Colocalization studies with either ribeye or calbindin (Haverkamp & Wassle, 2000; Schmitz et al., 2000; tom Dieck et al., 2005) confirmed the absence of LRIT3 presynaptically in photoreceptors and in horizontal cells. In contrast, LRIT3 localizes at the dentritic tips of both rod and cone ON-bipolar cells where it colocalizes with mGluR6.
TRPM1 localization at the dendritic tips of ON-bipolar cell is dependent on LRIT3
The cation channel TRPM1 is the endpoint of the ON-bipolar cell signaling cascade at the dendritic tips of ON-bipolar cells. TRPM1 opening during light stimulation results in the depolarization of ON-bipolar cells and the formation of the ERG b-wave (Morgans et al., 2009; Shen et al., 2009; Koike et al., 2010). In some mouse models of cCSNB, TRPM1 staining is severely reduced or absent at the dendritic tips of depolarizing bipolar cells. This is the case in the Trpm1 knock-out mouse itself (Morgans et al., 2009; Koike et al., 2010), but also in three different Grm6 mutants: the Grm6 knock-out model (Masu et al., 1995; Xu et al., 2012), the nob3 mouse (Grm6 splice mutation (Maddox et al., 2008; Cao et al., 2011)) and the nob4 mouse (Grm6 missense mutation (Pinto et al., 2007; Cao et al., 2011)). Similarly, the TRPM1 staining at the dendritic tips was also absent in the nob mouse (Nyx deletion (Pardue et al., 1998; Gregg et al., 2003; Pearring et al., 2011)). These results suggest that mGluR6 and nyctalopin, encoded by Nyx, are required for the correct localization of TRPM1. In contrast, other mouse models deficient for proteins participating in the same signaling cascade (Rgs7−/−/Rgs11−/−, nob5 (deficient for GPR179)) do not show such a mislocalization of TRPM1 (Orlandi et al., 2012; Ray et al., 2014). Therefore these proteins likely play a role in regulating G-protein signaling, rather than a role in trafficking of TRPM1. Although it has been previously shown that nyctalopin is essential for the correct localization of TRPM1 at the dendritic tips of ON-bipolar cells, it was suggested that nyctalopin alone is probably not able to bring the cation channel to the cell surface as it is mainly an extracellular protein (Pearring et al., 2011). We hypothesized that LRIT3 with its intracellular PDZ-binding domain might interact with scaffolding proteins to traffic TRPM1 to its correct localization and, together with nyctalopin, LRIT3 maintains TRPM1 at this localization (Zeitz et al., 2013; Neuille et al., 2014). Indeed, here we show that TRPM1 localization at the dendritic tips of both rod and cone ON-bipolar cells is dramatically decreased in Lrit3nob6/nob6 mice, while the total quantity of TRPM1 was not significantly altered, indicating that localization but not the protein amount of TRPM1 is dependent on LRIT3. Previous studies have also investigated the localization and protein amount of TRPM1 in mouse models for cCSNB. Quantitative analysis of total intensity of TRPM1 in all retina layers in the knock-out model for mGluR6 was slightly lower compared to wild-type mouse (15% reduction by immunostaining and 23% reduction by Western blot) (Xu et al., 2012). However the authors concluded that the main effect of eliminating mGluR6 is the severe reduction of TRPM1 localization at the dendritic tips of ON-bipolar cells (intensity 30% reduced and number of puncta 60% decreased in the OPL) (Xu et al., 2012). However, this decrease of the total protein amount was not found in another study using the same mouse model (Ray et al., 2014). In the two other mouse models with mGluR6 mutations, nob3 and nob4, in addition to the lack of TRPM1 staining at the dendritic tips of ON-bipolar cells, a moderate 30% reduction of TRPM1 levels in both mouse lines was described by Western blot analysis (Cao et al., 2011). In contrast, the absence of TRPM1 at the dendritic tips of ON-bipolar cells in nyctalopin mutant (nob) mice was accompanied with a severe reduction in total TRPM1 protein quantified by Western blot analysis (Pearring et al., 2011). The observation that the total amount of TRPM1 protein seems unchanged in Lrit3nob6/nob6 mouse compared to the nob mouse suggests that LRIT3 is necessary for correct trafficking of TRPM1, but that nyctalopin in necessary for both trafficking and either synthesis or stability of TRPM1. It has been shown that nyctalopin, mGluR6, GPR179 and TRPM1 form a macromolecular complex (Cao et al., 2011; Pearring et al., 2011; Orlandi et al., 2013; Ray et al., 2014). It is not clear if LRIT3 directly interacts with TRPM1 or via another component of the complex. Motif analysis suggests that the predicted intracellular PDZ-binding domain present in LRIT3 interacts with an as yet unknown scaffolding protein to bring TRPM1 to the surface (Zeitz et al., 2013).
LRIT3 may play a role in cone synapse formation
Surprisingly, we found that most of the known components of the ON-bipolar cell signaling cascade including mGluR6, GPR179, RGS7, RGS11 and Gβ5 are nearly absent at the dendritic tips of cone ON-bipolar cells in Lrit3nob6/nob6 mouse, whereas their staining at the dendritic tips of rod bipolar cells appears normal. In the Lrit3nob6/nob6 mouse, some punctate mGluR6 labeling was present in the INL, possibly indicating a defect in trafficking mGluR6 in cone ON-bipolar cells. We also revealed a dramatic decrease of PNA staining at the synaptic clefts between cones pedicles and corresponding cone ON-bipolar cells in Lrit3nob6/nob6 mouse, whereas cone pedicles are still present. This decrease was qualitatively evident and was well reflected in the quantification of the number of PNA stained synaptic clefts per mm of OPL, while PNA staining was unchanged in IS and OS of cones. Together, these results suggest that LRIT3 may have an additional function in cone synapse formation. To our knowledge, this is the first report of absence of PNA in the OPL in a mouse model of cCSNB. Indeed, PNA staining in the OPL on Grm6 knock-out, nob (Nyx), Trpm1 knock-out and nob5 (Gpr179) retinal sections is not different from wild-type (Ray et al., 2014). However, alterations in ribbon synapse formation or bipolar dendrite invagination have been described previously in Grm6 mutants and Gβ5 knock-out mice (Rao et al., 2007; Cao et al., 2009; Ishii et al., 2009; Tsukamoto & Omi, 2014). Electron microscopy studies are needed to confirm if there is indeed an ultrastructural defect in Lrit3nob6/nob6 retina, which might also explain the slight but significant thinning of inner retinal layers in Lrit3nob6/nob6 mouse that we previously highlighted (Neuille et al., 2014). We quantified LRIT3 staining at the dendritic tips of rod versus cone ON-bipolar cells and showed that the staining is similar at the dendritic tips of both rod and cone ON-bipolar cells. These findings are in accordance with our previous LRIT3 staining on human retina, which revealed equal intensities at the dendritic tips of both rod and cone ON-bipolar cells (Figure 2 in (Zeitz et al., 2013). Thus, the additional function of LRIT3 in cone ON-bipolar cells does not seem to be correlated with a difference in the amount of LRIT3 protein in these cells compared to rod bipolar cells. Rather, the mGluR6 signaling cascade may be differently regulated in rod and cone ON-bipolar cells.
To conclude, this study reveals that several of the known components of the ON-bipolar cell signaling cascade display a disrupted localization in Lrit3nob6/nob6 mouse, a mouse model of cCSNB lacking Lrit3. In particular, TRPM1 is reduced at the dendritic tips of ON-bipolar cells, explaining the no b-wave phenotype and supporting our hypothesis that LRIT3 might bring TRPM1 to the cell surface. Moreover, LRIT3 may have an additional function in synapse formation as most of the known components of the cascade and PNA are absent at the cone to cone ON-bipolar cell synapse. Putative interactions between LRIT3 and partners of the cascade highlighted here have now to be confirmed to clarify the exact role of LRIT3 in the mGluR6 signaling cascade and to elucidate the pathogenic mechanism(s) of cCSNB.
Supplementary Material
Representative 3D reconstructions of cross sections centered on OPL of Lrit3+/+ retina stained with antibodies against LRIT3 (green), PNA (red) and DAPI (white). (A) The yellow box represents the ROI drawn to isolate the OPL in which quantification was made. (B) Zoom on a region of the selected OPL. The dendritic tips of cone ON-bipolar cells appear yellow because of the colocalization of PNA and LRIT3 staining at this localization, whereas the dendritic tips of rod bipolar cells appear only green. (C) The red surface represents the ROI drawn to surround PNA staining and to quantify LRIT3 staining at the dendritic tips of cone ON-bipolar cells. (D) The green spots represent the ROI drawn to isolate each dendritic tip of rod bipolar cells and to quantify LRIT3 staining at this localization. Red and green surfaces do not overlay.
Acknowledgments
The authors are grateful to the platform of animal housing at the Institut de la Vision, to Stéphane Fouquet and David Godefroy for their support on acquisition and analysis of confocal images (Institut de la Vision platform), to Marie-Laure Niepon for technical help on histology (Institut de la Vision platform), to Yvrick Zagar for technical help on Western blotting (Insitut de la Vision platform), and to Alain Chédotal, Olivier Goureau and Deniz Dalkara for providing some commercial antibodies (Institut de la Vision). The project was supported by Agence Nationale de la Recherche [ANR-12-BSVS1-0012-01_GPR179] (CZ), Fondation Voir et Entendre (CZ), Prix Dalloz for “la recherche en ophtalmologie” (CZ), The Fondation pour la Recherche Médicale (FRM) in partnership with the Fondation Roland Bailly (CZ), Ville de Paris and Région Ile de France, LABEX LIFESENSES [reference ANR-10-LABX-65] supported by French state funds managed by the Agence Nationale de la Recherche within the Investissements d’Avenir programme [ANR-11-IDEX-0004-0], Foundation Fighting Blindness center grant [C-CMM-0907-0428-INSERM04], grants from the National Eye Institute (EY019907, EY009534 to RMD, EY018625 to CWM and EY018139 to KAM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- AIS
antibody incubation solution
- BCA
bicinchoninic acid
- cCSNB
complete form of congenital stationary night blindness
- CO2
carbon dioxide
- DAPI
4’,6-diamidino-2-phenylindole
- Go
heterotrimeric G-protein
- GCL
ganglion cells layer
- Gpr179/GPR179
G-protein receptor 179
- Grm6/GRM6/mGluR6
metabotropic glutamate receptor 6
- Gβ5/Gβ5
G-protein beta 5
- HRP
horseradish peroxidase
- INL
inner nuclear layer
- IPL
inner plexiform layer
- IS
inner segments
- KLH
keyhole limpet haemocyanin
- Lrit3/LRIT3/LRIT3
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 protein
- MAR
melanoma-associated retinopathy
- NA
numerical aperture
- NaCl
sodium chloride
- NaN3
sodium azide
- nob
no b-wave
- Nyx
nyctalopin
- ONL
outer nuclear layer
- OPL
outer plexiform layer
- OS
outer segments
- PAGE
polyacrylamide gel electrophoresis
- PBS
phosphate buffer saline
- PDZ
PSD95, DLG1 and ZO1
- PKCα
protein kinase C alpha
- PNA
peanut agglutinin
- R9AP
RGS9 anchor protein
- Rgs11/RGS11
regulator of G-protein signaling 11
- Rgs7/RGS7
regulator of G-protein signaling 7
- ROI
region of interst
- SDS
sodium dodecylsulfate
- Trmp1/TRPM1
transient receptor potential melastatin 1 cation channel
Footnotes
The authors declare no competing financial interests.
References
- Bahadori R, Biehlmaier O, Zeitz C, Labhart T, Makhankov YV, Forster U, Gesemann M, Berger W, Neuhauss SC. Nyctalopin is essential for synaptic transmission in the cone dominated zebrafish retina. Eur J Neurosci. 2006;24:1664–1674. doi: 10.1111/j.1460-9568.2006.05053.x. [DOI] [PubMed] [Google Scholar]
- Blanks JC, Johnson LV. Selective lectin binding of the developing mouse retina. J Comp Neurol. 1983;221:31–41. doi: 10.1002/cne.902210103. [DOI] [PubMed] [Google Scholar]
- Blanks JC, Johnson LV. Specific binding of peanut lectin to a class of retinal photoreceptor cells. A species comparison. Invest Ophthalmol Vis Sci. 1984;25:546–557. [PubMed] [Google Scholar]
- Cao Y, Masuho I, Okawa H, Xie K, Asami J, Kammermeier PJ, Maddox DM, Furukawa T, Inoue T, Sampath AP, Martemyanov KA. Retina-specific GTPase accelerator RGS11/G beta 5S/R9AP is a constitutive heterotrimer selectively targeted to mGluR6 in ON-bipolar neurons. J Neurosci. 2009;29:9301–9313. doi: 10.1523/JNEUROSCI.1367-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Pahlberg J, Sarria I, Kamasawa N, Sampath AP, Martemyanov KA. Regulators of G protein signaling RGS7 and RGS11 determine the onset of the light response in ON bipolar neurons. Proc Natl Acad Sci U S A. 2012;109:7905–7910. doi: 10.1073/pnas.1202332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Posokhova E, Martemyanov KA. TRPM1 forms complexes with nyctalopin in vivo and accumulates in postsynaptic compartment of ON-bipolar neurons in mGluR6-dependent manner. J Neurosci. 2011;31:11521–11526. doi: 10.1523/JNEUROSCI.1682-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Song H, Okawa H, Sampath AP, Sokolov M, Martemyanov KA. Targeting of RGS7/Gbeta5 to the dendritic tips of ON-bipolar cells is independent of its association with membrane anchor R7BP. J Neurosci. 2008;28:10443–10449. doi: 10.1523/JNEUROSCI.3282-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra A, Lyubarsky A, Jiang M, Pugh EN, Jr, Birnbaumer L, Sterling P, Vardi N. The light response of ON bipolar neurons requires G[alpha]o. J Neurosci. 2000;20:9053–9058. doi: 10.1523/JNEUROSCI.20-24-09053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat Rev Neurosci. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- Greferath U, Grunert U, Wassle H. Rod bipolar cells in the mammalian retina show protein kinase C-like immunoreactivity. J Comp Neurol. 1990;301:433–442. doi: 10.1002/cne.903010308. [DOI] [PubMed] [Google Scholar]
- Gregg RG, Mukhopadhyay S, Candille SI, Ball SL, Pardue MT, McCall MA, Peachey NS. Identification of the gene and the mutation responsible for the mouse nob phenotype. Invest Ophthalmol Vis Sci. 2003;44:378–384. doi: 10.1167/iovs.02-0501. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- Ishii M, Morigiwa K, Takao M, Nakanishi S, Fukuda Y, Mimura O, Tsukamoto Y. Ectopic synaptic ribbons in dendrites of mouse retinal ON- and OFF-bipolar cells. Cell Tissue Res. 2009;338:355–375. doi: 10.1007/s00441-009-0880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BG, Morgans CW, Puthussery T, Wensel TG, Burke NS, Brown RL, Duvoisin RM. R9AP stabilizes RGS11-G beta5 and accelerates the early light response of ON-bipolar cells. Vis Neurosci. 2010;27:9–17. doi: 10.1017/S0952523809990319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C, Obara T, Uriu Y, Numata T, Sanuki R, Miyata K, Koyasu T, Ueno S, Funabiki K, Tani A, Ueda H, Kondo M, Mori Y, Tachibana M, Furukawa T. TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc Natl Acad Sci U S A. 2010;107:332–337. doi: 10.1073/pnas.0912730107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox DM, Vessey KA, Yarbrough GL, Invergo BM, Cantrell DR, Inayat S, Balannik V, Hicks WL, Hawes NL, Byers S, Smith RS, Hurd R, Howell D, Gregg RG, Chang B, Naggert JK, Troy JB, Pinto LH, Nishina PM, McCall MA. Allelic variance between GRM6 mutants, Grm6nob3 and Grm6nob4 results in differences in retinal ganglion cell visual responses. J Physiol. 2008;586:4409–4424. doi: 10.1113/jphysiol.2008.157289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masu M, Iwakabe H, Tagawa Y, Miyoshi T, Yamashita M, Fukuda Y, Sasaki H, Hiroi K, Nakamura Y, Shigemoto R, et al. Specific deficit of the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell. 1995;80:757–765. doi: 10.1016/0092-8674(95)90354-2. [DOI] [PubMed] [Google Scholar]
- Morgans CW, Ren G, Akileswaran L. Localization of nyctalopin in the mammalian retina. Eur J Neurosci. 2006;23:1163–1171. doi: 10.1111/j.1460-9568.2006.04647.x. [DOI] [PubMed] [Google Scholar]
- Morgans CW, Wensel TG, Brown RL, Perez-Leon JA, Bearnot B, Duvoisin RM. Gbeta5-RGS complexes co-localize with mGluR6 in retinal ON-bipolar cells. Eur J Neurosci. 2007;26:2899–2905. doi: 10.1111/j.1460-9568.2007.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW, Zhang J, Jeffrey BG, Nelson SM, Burke NS, Duvoisin RM, Brown RL. TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proc Natl Acad Sci U S A. 2009;106:19174–19178. doi: 10.1073/pnas.0908711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Iwakabe H, Akazawa C, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel retinal metabotropic glutamate receptor mGluR6 with a high agonist selectivity for L-2-amino-4-phosphonobutyrate. J Biol Chem. 1993;268:11868–11873. [PubMed] [Google Scholar]
- Nawy S. The metabotropic receptor mGluR6 may signal through G(o), but not phosphodiesterase, in retinal bipolar cells. J Neurosci. 1999;19:2938–2944. doi: 10.1523/JNEUROSCI.19-08-02938.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi K, Kato S, Teranishi T. Dopamine cells and rod bipolar cells contain protein kinase C-like immunoreactivity in some vertebrate retinas. Neurosci Lett. 1988;94:247–252. doi: 10.1016/0304-3940(88)90025-0. [DOI] [PubMed] [Google Scholar]
- Neuille M, El Shamieh S, Orhan E, Michiels C, Antonio A, Lancelot ME, Condroyer C, Bujakowska K, Poch O, Sahel JA, Audo I, Zeitz C. Lrit3 deficient mouse (nob6): a novel model of complete congenital stationary night blindness (cCSNB) PLoS One. 2014;9:e90342. doi: 10.1371/journal.pone.0090342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura A, Shigemoto R, Nakamura Y, Okamoto N, Mizuno N, Nakanishi S. Developmentally regulated postsynaptic localization of a metabotropic glutamate receptor in rat rod bipolar cells. Cell. 1994;77:361–369. doi: 10.1016/0092-8674(94)90151-1. [DOI] [PubMed] [Google Scholar]
- Orhan E, Prezeau L, El Shamieh S, Bujakowska KM, Michiels C, Zagar Y, Vol C, Bhattacharya SS, Sahel JA, Sennlaub F, Audo I, Zeitz C. Further insights into GPR179: expression, localization, and associated pathogenic mechanisms leading to complete congenital stationary night blindness. Invest Ophthalmol Vis Sci. 2013;54:8041–8050. doi: 10.1167/iovs.13-12610. [DOI] [PubMed] [Google Scholar]
- Orlandi C, Cao Y, Martemyanov K. Orphan receptor GPR179 forms macromolecular complexes with components of metabotropic signaling cascade in retina ON-bipolar neurons. Invest Ophthalmol Vis Sci. 2013 doi: 10.1167/iovs.13-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, Posokhova E, Masuho I, Ray TA, Hasan N, Gregg RG, Martemyanov KA. GPR158/179 regulate G protein signaling by controlling localization and activity of the RGS7 complexes. J Cell Biol. 2012;197:711–719. doi: 10.1083/jcb.201202123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue MT, McCall MA, LaVail MM, Gregg RG, Peachey NS. A naturally occurring mouse model of X-linked congenital stationary night blindness. Invest Ophthalmol Vis Sci. 1998;39:2443–2449. [PubMed] [Google Scholar]
- Peachey NS, Pearring JN, Bojang P, Jr, Hirschtritt ME, Sturgill-Short G, Ray TA, Furukawa T, Koike C, Goldberg AF, Shen Y, McCall MA, Nawy S, Nishina PM, Gregg RG. Depolarizing bipolar cell dysfunction due to a Trpm1 point mutation. J Neurophysiol. 2012a;108:2442–2451. doi: 10.1152/jn.00137.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peachey NS, Ray TA, Florijn R, Rowe LB, Sjoerdsma T, Contreras-Alcantara S, Baba K, Tosini G, Pozdeyev N, Iuvone PM, Bojang P, Jr, Pearring JN, Simonsz HJ, van Genderen M, Birch DG, Traboulsi EI, Dorfman A, Lopez I, Ren H, Goldberg AF, Nishina PM, Lachapelle P, McCall MA, Koenekoop RK, Bergen AA, Kamermans M, Gregg RG. GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2012b;90:331–339. doi: 10.1016/j.ajhg.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring JN, Bojang P, Jr, Shen Y, Koike C, Furukawa T, Nawy S, Gregg RG. A role for nyctalopin, a small leucine-rich repeat protein, in localizing the TRP melastatin 1 channel to retinal depolarizing bipolar cell dendrites. J Neurosci. 2011;31:10060–10066. doi: 10.1523/JNEUROSCI.1014-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Vitaterna MH, Shimomura K, Siepka SM, Balannik V, McDearmon EL, Omura C, Lumayag S, Invergo BM, Glawe B, Cantrell DR, Inayat S, Olvera MA, Vessey KA, McCall MA, Maddox D, Morgans CW, Young B, Pletcher MT, Mullins RF, Troy JB, Takahashi JS. Generation, identification and functional characterization of the nob4 mutation of Grm6 in the mouse. Vis Neurosci. 2007;24:111–123. doi: 10.1017/S0952523807070149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan H, Dhingra A, Tummala SR, Fina ME, Li JJ, Lyubarsky A, Vardi N. Differential Function of Ggamma13 in Rod Bipolar and ON Cone Bipolar Cells. J Physiol. 2014 doi: 10.1113/jphysiol.2014.281196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Dallman R, Henderson S, Chen CK. Gbeta5 is required for normal light responses and morphology of retinal ON-bipolar cells. J Neurosci. 2007;27:14199–14204. doi: 10.1523/JNEUROSCI.4934-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray TA, Heath KM, Hasan N, Noel JM, Samuels IS, Martemyanov KA, Peachey NS, McCall MA, Gregg RG. GPR179 is required for high sensitivity of the mGluR6 signaling cascade in depolarizing bipolar cells. J Neurosci. 2014;34:6334–6343. doi: 10.1523/JNEUROSCI.4044-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma H, Inana G, Murakami A, Higashide T, McLaren MJ. Immunolocalization of X-arrestin in human cone photoreceptors. FEBS Lett. 1996;382:105–110. doi: 10.1016/0014-5793(96)00163-9. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Konigstorfer A, Sudhof TC. RIBEYE, a component of synaptic ribbons: a protein’s journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857–872. doi: 10.1016/s0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- Shen Y, Heimel JA, Kamermans M, Peachey NS, Gregg RG, Nawy S. A transient receptor potential-like channel mediates synaptic transmission in rod bipolar cells. J Neurosci. 2009;29:6088–6093. doi: 10.1523/JNEUROSCI.0132-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tom Dieck S, Altrock WD, Kessels MM, Qualmann B, Regus H, Brauner D, Fejtova A, Bracko O, Gundelfinger ED, Brandstatter JH. Molecular dissection of the photoreceptor ribbon synapse: physical interaction of Bassoon and RIBEYE is essential for the assembly of the ribbon complex. J Cell Biol. 2005;168:825–836. doi: 10.1083/jcb.200408157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Omi N. Effects of mGluR6-deficiency on photoreceptor ribbon synapse formation: comparison of electron microscopic analysis of serial sections with random sections. Vis Neurosci. 2014;31:39–46. doi: 10.1017/S0952523813000473. [DOI] [PubMed] [Google Scholar]
- Vardi N. Alpha subunit of Go localizes in the dendritic tips of ON bipolar cells. J Comp Neurol. 1998;395:43–52. [PubMed] [Google Scholar]
- Vardi N, Dhingra A, Zhang L, Lyubarsky A, Wang TL, Morigiwa K. Neurochemical organization of the first visual synapse. Keio J Med. 2002;51:154–164. doi: 10.2302/kjm.51.154. [DOI] [PubMed] [Google Scholar]
- Wu AM. Differential binding characteristics and applications of DGal beta 1----3DGalNAc specific lectins. Molecular and cellular biochemistry. 1984;61:131–141. doi: 10.1007/BF00222491. [DOI] [PubMed] [Google Scholar]
- Xiong WH, Duvoisin RM, Adamus G, Jeffrey BG, Gellman C, Morgans CW. Serum TRPM1 autoantibodies from melanoma associated retinopathy patients enter retinal on-bipolar cells and attenuate the electroretinogram in mice. PLoS One. 2013;8:e69506. doi: 10.1371/journal.pone.0069506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Dhingra A, Fina ME, Koike C, Furukawa T, Vardi N. mGluR6 deletion renders the TRPM1 channel in retina inactive. J Neurophysiol. 2012;107:948–957. doi: 10.1152/jn.00933.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz C. Molecular genetics and protein function involved in nocturnal vision. Expert Rev Ophtalmol. 2007;2:467–485. [Google Scholar]
- Zeitz C, Jacobson SG, Hamel CP, Bujakowska K, Neuille M, Orhan E, Zanlonghi X, Lancelot ME, Michiels C, Schwartz SB, Bocquet B, Congenital Stationary Night Blindness C, Antonio A, Audier C, Letexier M, Saraiva JP, Luu TD, Sennlaub F, Nguyen H, Poch O, Dollfus H, Lecompte O, Kohl S, Sahel JA, Bhattacharya SS, Audo I. Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2013;92:67–75. doi: 10.1016/j.ajhg.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz C, Robson AG, Audo I. Congenital stationary night blindness: An analysis and update of genotype-phenotype correlations and pathogenic mechanisms. Prog Retin Eye Res. 2015;45C:58–110. doi: 10.1016/j.preteyeres.2014.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative 3D reconstructions of cross sections centered on OPL of Lrit3+/+ retina stained with antibodies against LRIT3 (green), PNA (red) and DAPI (white). (A) The yellow box represents the ROI drawn to isolate the OPL in which quantification was made. (B) Zoom on a region of the selected OPL. The dendritic tips of cone ON-bipolar cells appear yellow because of the colocalization of PNA and LRIT3 staining at this localization, whereas the dendritic tips of rod bipolar cells appear only green. (C) The red surface represents the ROI drawn to surround PNA staining and to quantify LRIT3 staining at the dendritic tips of cone ON-bipolar cells. (D) The green spots represent the ROI drawn to isolate each dendritic tip of rod bipolar cells and to quantify LRIT3 staining at this localization. Red and green surfaces do not overlay.