Abstract
Previous crystallographic and mutagenesis studies have implicated the role of a position-conserved hairpin loop in the metallo-β-lactamases in substrate binding and catalysis. In an effort to probe the motion of that loop during catalysis, rapid-freeze-quench double electron electron resonance (RFQ-DEER) spectroscopy was used to interrogate metallo-β-lactamase CcrA, which had a spin label at position 49 on the loop and spin labels (at positions 82, 126, or 233) 20–35 Å away from residue 49, during catalysis. At 10 milliseconds after mixing, the DEER spectra show distance increases of 7, 10, and 13 Å between the spin label at position 49 and the spin labels at positions 82, 126, and 233, respectively. In contrast to previous hypotheses, these data suggest that the loop moves nearly 10 Å away from the metal center during catalysis and that the loop does not clamp down on the substrate during catalysis. This study demonstrates that loop motion during catalysis can be interrogated on the millisecond time scale.
Keywords: Double Electron-Electron Resonance (DEER), Metallo-β-lactamase (MβLs), site-directed spin labeling (SDSL), Rapid Freeze Quench (RFQ), MTSL
Introduction
The β-lactamase-fold superfamily of metalloenzymes binds 1 or 2 transition metal ions per protein and has an αββα tertiary structure [1]. The prototypical members, the metallo-β-lactamases (MβLs), hydrolyze amide bonds found in β-lactam containing antibiotics. The Zn(II)-containing MβLs constitute an ever-growing and troubling class of β-lactamases that have been found in clinical isolates of Bacillus anthracis (anthrax), Pseudomonas aeruginosa, Klebsiella pneumoniae, and a host of other pathogenic organisms [2–6]. The MβLs contain either 1 or 2 moles of Zn(II) per mole of enzyme, hydrolyze all known cephalosporins, carbapenems, and penicillins, and are not inhibited by clavulanic acid or any other clinically-useful inhibitor. Previous studies have shown that there is significant structural and mechanistic diversity among the MβLs, leading to the grouping of the enzymes into three distinct subclasses: B1, B2, and B3 [2, 5, 7, 8]. The B1 enzymes have one Zn(II) site (the Zn1 site) consisting of His116, His118, and His196, a second Zn(II) site (the Zn2 site) consisting of Asp120, Cys221, and His263, and are typified by MβL CcrA from Bacteroides fragilis [9]. The B2 enzymes are mono-zinc enzymes chiefly found only in species of Aeromonas [10, 11], with the same Zn2 binding site as the B1 enzymes (His116 is replaced by a conserved asparagine, which abolishes metal binding at the Zn1 site), and include MβL ImiS from Aeromonas sobria [12]. The B3 enzymes have the same metal binding sites as the B1 enzymes except that Cys221 is replaced with a conserved histidine, and include MβL L1 from Stenotrophomonas maltophilia [13]. The B1 and B3 enzymes most often require two bound Zn(II) ions for full catalytic activity [14–16]. The diversity of the MβLs is best exemplified by the enzymes’ vastly differing susceptibilities towards inhibitors [4, 5, 7, 8, 17–23], metal binding properties (cooperative versus sequential) [15], and reaction mechanisms (i.e., whether a ring-opened nitrogen anionic intermediate is formed when using nitrocefin or chromacef as substrate (Scheme 1)) [24].
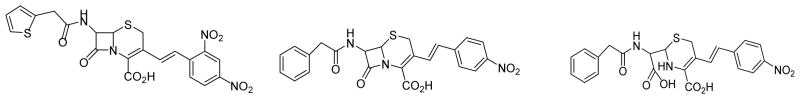
Scheme 1.
Structures of nitrocefin (left), chromacef (center), and hydrolyzed chromacef (right)
Crystal structures of several B1 and B3 MβLs identified a position-conserved loop that extends over the metal binding site [13, 25–28], and similar loops have been observed in other enzymes belonging to the β-lactamase fold superfamily, suggesting a common role for these loops [29–31]. Crystal structures of MβL-inhibitor complexes showed decreased flexibility and reorientation of the loop towards the metal center [5, 7, 8, 25–28, 32]. NMR studies indicated that Trp49 on the loop in CcrA may play a role in inhibitor (and by analogy substrate) binding and suggested that Trp49 and the loop plays a role in promotion of catalysis [33, 34]. These results are supported by mutagenesis studies in which mutations of Trp to other amino acids resulted in over 50-fold decreases in kcat/Km (depending on the identity of the residue that replaced Trp49) [34]. Deletion of residues 47–49 induced a >100-fold decrease in kcat/Km, for CcrA [34] and deletion of the entire loop led to a reduction of kcat/Km by factors of up to 5,000 [35]. It should be noted, however, that Trp49 is not conserved across the MβLs.
Studies of variants of IMP-1 and BcII containing deletions or substitutions in the loop region identified altered kinetic parameters and suggested that Trp64 plays a role in substrate binding by interacting with hydrophobic portions of the substrate [34, 36], thus extending the mechanistic importance of the loop beyond CcrA. It has been speculated, however, that enzyme molecules from which the loop was deleted may have altered folding, and caution should be exercised when interpreting the data in terms of structure-function relationship [36]. Vila has speculated that the differences in reaction mechanisms indicated for BcII relative to other B1 and B3 MβLs may be due to a comparatively smaller loop over the BcII active site [37].
The B2 enzymes have an α-helix in the same position as the loop in the B1 and B3 enzymes that appears to have the same function [38, 39]. The helix in the resting state of the B2 MβL CphA has been structurally characterized by X-ray diffraction, but mechanistic data are lacking, whereas an earlier EPR spectrokinetic study of the related enzyme ImiS identified rotation of the helix about its axis during the catalytic cycle [39]. The available data suggest that the loop in B1 and B3 MβLs and the position-conserved α-helix in B2 MβLs play a role in substrate binding and catalysis. In this study, further information was sought on the role of the hairpin loop in CcrA by the use of pulsed double electron-electron resonance (pELDOR or “DEER”) spectroscopy of trapped catalytic intermediates of doubly spin-labeled CcrA containing one spin label on the putative mobile loop and another in one of three presumed immobile sites in α-helices.
Materials and Methods
Materials
Site-directed mutagenesis kits were purchased from Strategene (Carlsbad, CA). E. coli strains DH5α and BL21(DE3) cells were purchased from Novagen (Madison, WI). Sequencing and mutagenesis primers were purchased from Integrated DNA Technologies. Isopropyl-β-D-galactoside (IPTG) was purchased from Anatrace (Muamee, OH). Q-Sepharose and Sephacryl S-200 chromatographic media were purchased from GE Healthcare. S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methylmethanesulfonothioate (MTSL) was purchased from Toronto Research Chemicals (Toronto, Canada). The substrates nitrocefin and chromacef were purchased from Becton Dickinson and Sopharmia, respectively. All buffer solutions and growth media were prepared with Barnstead Nanopure water.
Design and generation of site-directed variants of CcrA
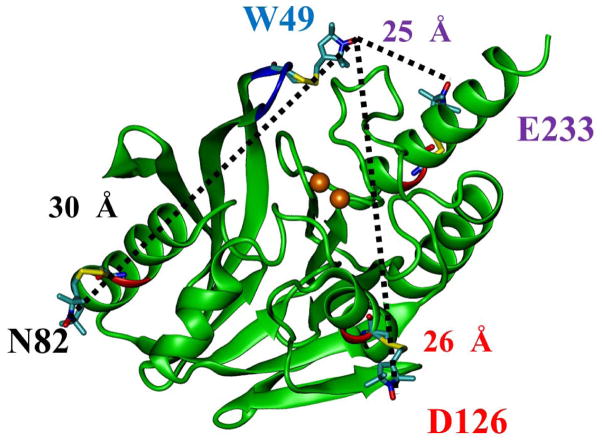
The site-directed variants CcrA:C155S/W49C/N82C, CcrA:C155S/W49C/D126C, and CcrA:C155SW49C/E233C (referred to as CcrA*(49/82), CcrA*(49/126) and CcrA*(49/233), respectively, hereafter) were generated using the primers in Table S1 and the Stratagene Site-Directed Mutagenesis kit, using previously reported procedures and manufacturer’s instructions. Plasmids were sequenced at the Center for Bioinformatics and Functional Genomics (CBFG) facility at Miami University.[40] W49 is a residue on the mobile loop and N82, D126 and E233 are immobile residues that form the base of a pyramid in the resting state, with W49 at the apex. Any displacement of the spin label at the 49 position during the reaction can therefore be calculated by triangulation of the W49 to N82, D126, and E233 distances measured by DEER, respectively (Figure 1).
Figure 1.
Positions of site-directed spin labels in CcrA (PDB id: 2BMI) used in this study. Figure generated using previously described procedures [51]. Positions of the 49, 82, 126, and 233 residues in the CcrA and estimated distances from the position 49 to the 82, 126, and 233 are shown.
Over-expression and purification of CcrA
Recombinant CcrA was over-expressed and purified as previously reported [41]. The homogeneities of the preparations were estimated by visual inspection of SDS-PAGE, and protein was quantitated spectrophotometrically using an extinction coefficient (ε280nm) of 39,000 M−1cm−1 as previously reported [41]. The procedure yielded > 100 mg of > 95 % CcrA from each 4 L growth culture.
Metal analyses
Metal content was estimated by inductively-coupled plasma optical emission spectrophotometry (Perkin-Elmer Optima 7300 DV), using reference calibration curves (R2 > 0.999) generated with standard solutions of Zn(II), Co(II), Cu(II), Fe, Mn(II), and Ni(II) [24].
Steady-state kinetic studies
All steady state kinetic studies were conducted on an Agilent 8453 diode array spectrophotometer at 25 °C. Michaelis constants (Km) and turnover numbers (kcat) were determined by monitoring product formation at 442 nm using chromacef as substrate in 50 mM cacodylate, pH 7. Rate data were converted to concentration data using the extinction coefficient of hydrolyzed chromacef (ε442nm = 18,600 M−1cm−1). Concentrations versus time data were then fitted to the Michaelis-Menton equation, as previously reported [24].
Spin labeling of CcrA and quantification by CW-EPR spectroscopy
Recombinant CcrA (100 μM in 10 mL volume) was incubated with 1 eq. DTT per mol CcrA for 30 mins in 50 mM Tris, pH 7, followed by 10 – 20 molar equivalents of MTSL in 50 – 100 μL DMSO at 4 °C for 3 to 4 days in the dark with stirring. Unbound spin label was subsequently removed by size exclusion chromatography on a Sephacryl S-200 column (1.5 cm X 40 cm of bed volume 60 ml). The efficiency of the spin labeling was estimated from the intensity of the cw-EPR signal at 25 °C, as described elsewhere [42].
Samples for DEER spectroscopy
Concentrations of 0.06 to 0.10 mM CcrA were employed in stable samples (i.e. the resting state, and the product complex that was prepared by incubating resting enzyme and substrate on the ice for 1 hour), which were concentrated by ultrafiltration prior to substrate addition and frozen in liquid nitrogen. All initial enzyme and substrate concentrations were 0.4 and 2 mM, respectively, and the samples were prepared in 50 mM Tris, pH 7.0. A model 715 Update Instruments ram controller was used to drive a PMI-Kollmorgen stepping motor (model 00D12F-02001-1) connected to a ram that in turn drove two Update Instrument syringes of the same inner diameter. The syringes, mixer, and tubing were all contained in a water bath that was maintained at 2 °C [39, 43, 44]. 10 ms intermediate samples were collected in isopentane at −100 °C contained in a glass funnel attached to 4 mm O.D. EPR sample tube (Wilmad 706-SQ-250M, 7 cm length). The resulting concentration of CcrA in the frozen aqueous phase was 0.2 mM (the effective spin concentration was further diluted by a factor of about two due to the ≈ 50 % immiscible isopentane matrix).
DEER spectroscopy
DEER was performed at 80 K using Bruker EleXsys E-580 pulsed EPR spectrometers equipped with nitrogen cooling and either a Bruker SuperQFTu bridge, 10 W AmpQ microwave amplifier and Q-band EN5107D2 dielectric resonator (34.2 GHz) or a Bruker SuperXFT bridge, Applied Systems Engineering 2 kW traveling wave tube amplifier and X-band EN4118X-MD4 resonator (9.7 GHz). The MD4 resonator is designed for 3.8 mm O.D. tubes but was used here with 4 mm O.D. tubes (Wilmad 706-SQ-250M) that were cut to 7 cm length and loaded through the bottom of the resonator [45]. A four-pulse π/2O − τ1 − πO − τE − πP − (τ1 + τ2 −τE) − πO DEER sequence was employed, where the superscripts “O” and “P” denote pulses at the observe and pump frequencies, respectively, τE is the time between the first inversion pulse and the pump pulse, and τ2 is the dipolar evolution time. At X-band, π/2O and πO were 16 and 32 ns, respectively, with, τ2 = 1200 ns, and πP = 32 ns; at Q-band, π/2O and πO were 24 and 48 ns, respectively, with, τ2 = 1600 to 1800 ns, and πP = 48 ns. Spectra were pumped at the ml = 0 center line and observed at the low-field ml = 1 line, with a Δν = 73 MHz at X-band and 61 MHz at Q-band. Shot repetition times of 1200 μs (X-band), and 500 μs (Q-band) were used. The use of two frequencies deserves comment. As pointed out earlier [42], the same information is available at both frequencies provided that the relaxation rates allow for reasonable dipolar evolution times. Q-band DEER is a much more efficient technique in terms of time and material [46, 47], and was used whenever possible. However, the RFQ sample tubes were of too large a diameter for Bruker Q-band resonators and necessitated the use of X-band. Signal averaging was carried out for 8 to 12 h at Q-band data and 20 h at X-band. Fits presented were obtained using DEERAnalysis v.2009 and v.2011 [48], and additional verification was carried out using LongDistance (Dr. Christian Altenbach, UCLA) [49]. Auto-phasing was used for consistency, with the flatness of the highest-t 85 % of the imaginary data as the phasing criterion, though phasing resulted in very little change in the appearance of the data or the distance distributions. A homogeneous three-dimensional model was used for background correction, where the background contribution reduces to a simple exponential, e−kt, and where k is the only fit parameter. Default suggestions were adopted for low-t data cut-off: data thus treated represent the time-domain traces presented herein. The distance distributions P(r) were obtained by Tikhonov regularization in the distance domain, incorporating the constraint P(r) > 0. The regularization parameter in the L curve was optimized by examining the fit of the time domain. The dipolar evolution times, t, were used to calculate limits for the distances, d, that provided (i) reliable distance distributions, that allow deconvolution of overlapping distances; (ii) reliable distribution widths, that describe the overall heterogeneity of distances around the mean; (iii) reliable mean distances, the most important limit for the present study; and (iv) reliable distance recognitions, that describe the maximum distance that can be observed but not necessarily accurately measured. Calculations were based on the relationship d ∝ t1/3, and constants of proportionality for each of the four limits were calculated from Jeschke’s empirical calibrations, described in the user manual for DEERAnalysis (http://www.epr.ethz.ch/software/DeerAnalysis2013_manual.pdf) and based on fitting simulated data with known distances and distributions.
Molecular dynamics simulations
The atomic coordinates for the CcrA crystal structure (PDB id: 2BMI) from Bacteroides fragilis were downloaded from the Protein Data Bank and used to generate the structures of various spin-labeled CcrA mutants with the Nanoscale Molecular Dynamics (NAMD) program [50]. All mutated CcrA structures were created using the molecular graphics software VMD [51]. The nitroxide spin-probe MTSL was attached using CHARMM force field topology files incorporated into NAMD. The modified protein assembly was solvated into a spherical water environment and further equilibrated and minimized by running NAMD simulations at room temperature using CHARMM force field parameters [52]. The distance distribution from the W49C to N82C, D126C, or E233C residues were predicted with rotamer library modeling of MTSL conformations using Multiscale Modeling of Macromolecular systems (MMM version 2010) [50].
Results
Properties of spin-labeled recombinant CcrA
Native CcrA has cysteine residues at positions 104, 181, and 155 [53]. An examination of several crystal structures of CcrA suggested that Cys104 and 181 would likely not be accessible to the MTSL label, whereas Cys155 did appear to be solvent-accessible; therefore, Cys155 was substituted with serine (referred to as CcrA* hereafter). From examination of the CcrA structure with VMD [51], three distinct doubly-labeled CcrA species were identified (Figure 1) as being likely to provide useful structure-function information on the loop: CcrA:C155S/W49C/N82C, CcrA:C155S/W49C/D126C, and CcrA:C155SW49C/E233C (referred to as CcrA*(49/82), CcrA*(49/126) and CcrA*(49/233), respectively, hereafter). The label at residue 49 provides the dynamic probe of the loop, while residues 82, 126, and 233 reside on more rigid α-helices at 4.0, 3.6, and 2.3 nm (40, 36, and 23 Å) distances, respectively, from residue 49. Residues 82, 126, and 233 were chosen to “triangulate” the position of the residue 49 on the loop (Figure 1).
CcrA* was shown to bind 2.0 ± 0.1 equivalents of Zn(II) and < 0.1 eq. MTSL. It exhibited a kcat value of 65 ± 3 s−1 and a Km value of 6 ± 3 μM, when using chromacef as the substrate (Table 1). CcrA*(49/82), CcrA*(49/126) and CcrA*(49/233) were shown to bind 1.7 ± 0.1, 1.8 ± 0.1 and 2.0 ± 0.1 equivalents of Zn(II), respectively (Table 1). Observed Km values were 2–3 fold higher than for wild-type CcrA, but similar to those for CcrA*(49) (Table 1). Observed kcat values for CcrA*(49/82), CcrA*(49/126) and CcrA*(49/233) were similar to those of wild-type CcrA and CcrA*(49) (Table 1). CcrA*(49/82), CcrA*(49/126) and CcrA*(49/233) were found to bind 1.6, 1.7, and 2.0 eq. of MTSL, respectively, under the conditions employed; poorer labeling efficiency was observed with shorter incubation times. Spin-labeling did not significantly affect the metal content or kinetic parameters for the CcrA species (Table 1). Ambient-temperature EPR (Figure S1) indicated that the spin labels on residues 126 and 233 experienced somewhat, though not severely, restricted local motion whereas the local motion of the label on residue 82 was barely restricted at all. Molecular modeling of the possible rotamers of the spin labels suggested likely interspin distances of 3.0, 2.6 and 2.5 nm (30, 26, and 25 Å) for CcrA*(49/82), CcrA*(49/126), and CcrA*(49/233), respectively (Figure 1).
Table 1.
Metal content and steady state kinetic constants of CcrA mutants.
| Enzyme(Abbreviation) | Metal content (eq) | Km (μM) | kcat (s−1) | Metal content after spin-labeling | Km (μM) after spin-labeling | kcat (s−1) after spin labeling |
|---|---|---|---|---|---|---|
| Wild-type(CcrA) | 1.9 ± 0.1 | 7 ± 1 | 65 ± 3 | 1.8 ± 0.1 | 8 ± 3 | 57 ± 2 |
| C155S(CcrA*) | 2.0 ± 0.1 | 8 ± 1 | 62 ± 2 | 2.0 ± 0.1 | 6 ± 1 | 65 ± 1 |
| C155S/W49C(CcrA*49) | 2.1 ± 0.1 | 15 ± 3 | 31 ± 3 | 2.0 ± 0.1 | 17 ± 2 | 25 ± 1 |
| C155S/W49C/N82C(CcrA*49/82) | 1.7 ± 0.1 | 17 ± 5 | 41 ± 5 | 1.8 ± 0.1 | 22 ± 6 | 48 ± 1 |
| C155S/W49C/D126C(CcrA*49/126) | 1.8 ± 0.1 | 18 ± 4 | 63 ± 1 | 1.8 ± 0.1 | 17 ± 4 | 74 ± 4 |
| C155S/W49C/E233C(CcrA*49/233) | 2.0 ± 0.1 | 15 ± 4 | 42 ± 2 | 2.1 ± 0.1 | 16 ± 4 | 34 ± 2 |
DEER Spectroscopy
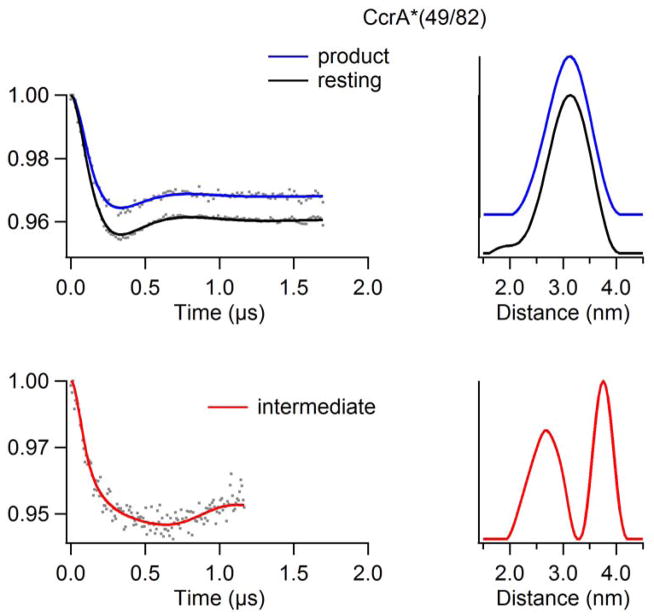
Time-domain DEER data of resting CcrA*(49/82) and CcrA*(49/82) after completed reaction with chromacef were collected to 1.7 μs, which allowed reliable measurement of a mean distance of up to 4.73 nm and a distribution overall width for distances up to 3.79 nm, but no further information could be reliably obtained from the distribution shape for distances beyond 2.84 nm [48]. The distance-domain DEER spectrum of resting CcrA*(49/82) indicates a broad interspin distance distribution centered at 3.1 nm (31 Å; Figure 2); this distance is consistent with the modeling studies that were used to identify the sites at which spin labels were introduced. The width of this peak [σ(r) ≈ 1.1 nm] indicates significant heterogeneity of the inter-spin distance. This is hardly surprising given that the X-ray diffraction indicated that one of the labeled residues, Trp49, did not occupy a defined volume of space within the resolution of the diffraction; residues 48 and 49 comprised the only disordered region of the structure [9]. Indeed, the hypothesis that this region represents a dynamic element in catalysis, and hence the reason for labeling this residue, may be considered to presuppose flexibility in solution and, therefore, heterogeneity in the frozen state [27]. The ambient temperature continuous-wave EPR signal of CcrA*(49/82) confirmed a high degree of mobility of at least one of the labels (Figure S1).
Figure 2.
DEER spectra of resting, 10 ms intermediate, and product analogs of doubly spin-labeled CcrA*(49/82) using a chromacef as a substrate. (Top) CcrA*(49/82) vs chromacef resting and product time domain spectra overlay (left) and corresponding distance domain DEER spectra overlay (right) (Q-Band). (Bot) CcrA*(49/82) vs chromacef 10 ms intermediate time domain spectrum (left) and corresponding distance domain (right) DEER spectrum (X-Band).
Time-domain DEER data of resting CcrA*(49/82) reacted with chromacef for 10 ms were collected to 1.2 μs, which allowed reliable measurement of a mean distance of up to 4.22 nm and a distribution overall width for distances up to 3.37 nm, but no further information could be reliably obtained from the distribution shape for distances beyond 2.53 nm. Upon reaction of CcrA*(49/82) with chromacef for 10 ms, a clear change in the time-domain DEER spectrum was observed and yielded two interspin distance distributions. The shorter mean distance accounted for about two-thirds of the sample and was 2.6 nm (26 Å), with a distribution σ(r) ≈ 0.8 nm. In addition to the 2.6 nm interspin distance, a longer distance of 3.8 nm (38 Å) was also observed that accounts for about one-third of the sample. After the reaction with chromacef was allowed to run to completion, the DEER spectrum indicated a reliable single mean interspin distance of 3.1 nm (31 Å), as in the resting enzyme, though with a narrower distribution, σ(r) ≈ 0.9 nm.
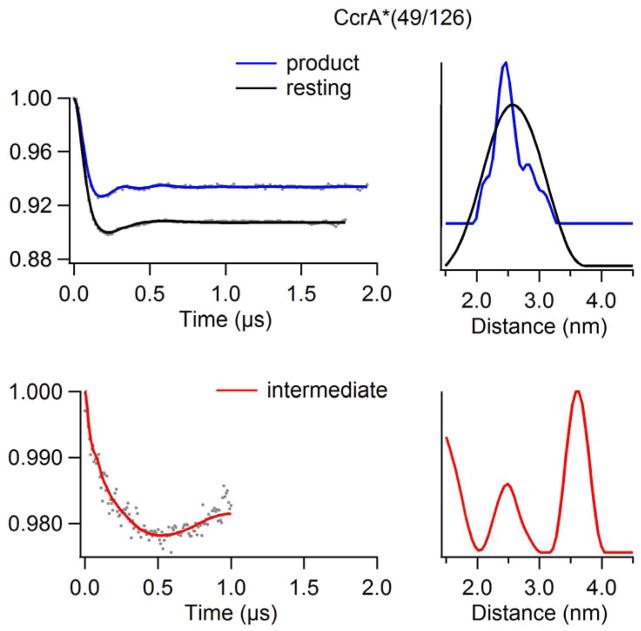
The DEER data for CcrA*(49/126) qualitatively reinforced the data from CcrA*(49/82) in that the resting species exhibited a broad interspin distance distribution centered around the predicted value [r = 2.5 nm (25 Å); σ(r) ≈ 1.2 nm], the catalytic intermediate exhibited two distances, and the product complex exhibited the same mean interspin distance as the resting enzyme but with a significantly smaller distribution (Figure 3). The spectra of the CcrA*(49/126) catalytic intermediate and product complex did differ quantitatively, however, from those of their CcrA*(49/82) analogues. For the CcrA*(49/126) catalytic intermediate, data were collected to 1.0 ≈ s, implying a maximum reliable mean distance measurement of 3.97 nm, and a reliable distribution width measurement of 3.16 nm. The shorter of the distances corresponded to the resting state mean interspin distance of 2.5 nm (25 Å), though with a much narrower distribution, σ(r) ≈ 0.6 nm, that is comparable to the corresponding CcrA*(49/82) data. Interestingly, the difference between the higher mean interspin distance of 3.7 nm (37 Å; within the reliable mean distance limit) in the CcrA*(49/126) catalytic intermediate and the resting distance is 3.7 − 2.5 = 1.2 nm (12 Å), significantly larger than the corresponding difference for CcrA*(49/82) (3.8 − 3.1 = 0.7 nm). The time domain DEER spectrum of the CcrA*(49/126) product complex shows resolved DEER modulations that reflect the narrow distribution, σ(r) ≈ 0.5 nm, of the dominant 2.5 nm peak in the distance domain spectrum. Additionally resolved features appeared at about 2.2, 2.9, and 3.3 nm appear to account for about one-third of the sample, although the latter two are close to the reliable distribution limit of 2.95 nm (for a 1.9 μs dipolar evolution time), and the phenomenon should be treated with caution.
Figure 3.
DEER spectra of resting, 10 ms intermediate, and product analogs of doubly spin-labeled CcrA*(49/126) using a chromacef as a substrate. (Top) CcrA*(49/126) vs chromacef resting and product time domain spectra overlay (left) and corresponding distance domain DEER spectra overlay (right) (Q-Band). (Bot) CcrA*(49/126) vs chromacef 10 ms intermediate time domain spectrum (left) and corresponding distance domain (right) DEER spectrum (X-Band).
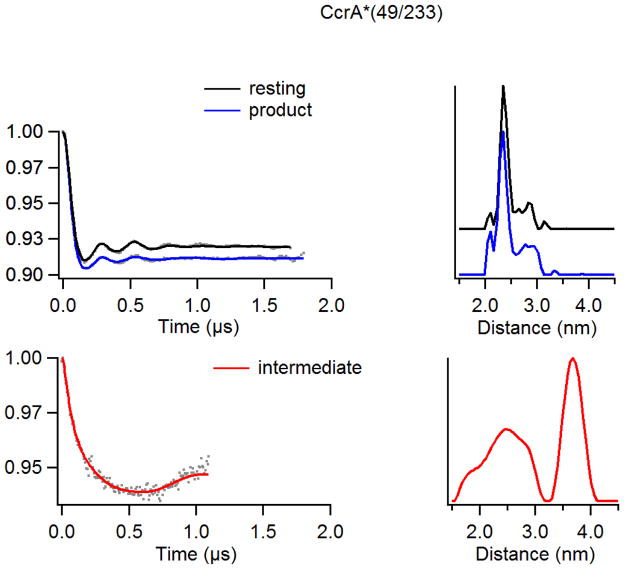
The DEER data for CcrA*(49/233) parallel those for CcrA*(49/126) very closely (Figure 4). The mean values of the interspin distance distributions are indistinguishable, although the widths of the dominant distributions for the resting and product-bound states are very narrow [σ(r) ≈0.3 nm]. Uniquely among the spin-labeled CcrA variants, the distance distribution for the resting state is as narrow as for the product complex. The observed distribution widths for the catalytic intermediate distances are much larger than for the other species, though the width of the longer distance cannot be taken as reliable [σ(r) ≈ 1.0 nm for r = 2.5 nm; σ(r) ≈ 0.6 nm for r = 3.7 nm]. There is some resolution of the 2.5 nm peak in the distance domain spectrum of the CcrA*(49/233) catalytic intermediate, suggesting distinct distances at 1.9, 2.5, and 2.9 nm; the resolution of the 1.9 and 2.5 nm distances is within the resolution limit whereas the resolution of the 2.5 and 2.9 nm distances is not. Minor populations with distances at 2.1 and 2.9 nm also appear in the spectra of the other CcrA*(49/233) species and are well within the resolution reliability limits of these spectra.
Figure 4.
DEER spectra of resting, 10 ms intermediate, and product analogs of doubly spin-labeled CcrA*(49/233) using a chromacef as a substrate. (Top) CcrA*(49/233) vs chromacef product and resting time domain spectra overlay (left) and corresponding distance domain DEER spectra overlay (right) (Q-Band). (Bot) CcrA*(49/233) vs chromacef 10 ms intermediate time domain spectrum (left) and corresponding distance domain (right) DEER spectrum (X-Band).
Discussion
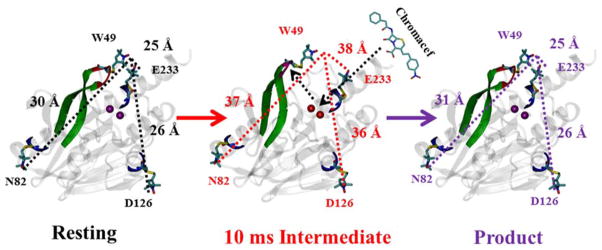
Molecular modeling suggested that the interspin distances in the resting-state spin-labeled recombinant CcrA variants CcrA*(49/82), CcrA*(49/126), and CcrA*(49/233) are 3.0, 2.6, and 2.5 nm, respectively. The corresponding dominant distances obtained experimentally by DEER spectroscopy were 3.1, 2.5, and 2.5 nm, i.e., within 0.1 nm (1 Å) of those predicted. The distance distribution in resting CcrA*(49/82) was broad, consistent with the flexibility of the label at residue 82 that was identified by EPR (Figure S1). The dominant distance distribution in resting CcrA*(49/233) was narrow but subpopulations with distinct distances were also observed. These data are consistent with the EPR spectrum of CcrA*(49/233) showing partially restricted motion and, interestingly, suggest that the label at residue 49 on the loop is not the main determinant of distance distribution width and must, therefore, be under motional constraint. The distance distribution in CcrA*(49/126) is far greater than in CcrA*(49/233). One possible explanation that reconciles these observations is that rotation of the label on residue 49 translates the spin density along the 49–126 connecting vector but perpendicular to the 29–233 connecting vector; this seems entirely reasonable in the light of the modeled structure (Figure 5) and the fact that the two interspin vectors are essentially orthogonal.
Figure 5.
Proposed model with results of this study (Top). (PDB id: 2BMI) (Left) DEER distances from the spin label at position 49 to the spin labels at positions 82, 126 and 233 spin labels in resting CcrA (Center) DEER distances from the spin label at position 49 to the spin labels at positions 82, 126 and 233 in the CcrA + chromacef samples quenched at 10 ms. (Right) DEER distances from the spin label at position 49 to the spin labels at positions 82, 126, and 233 in the CcrA-chromacef product complexes. Distances between the spin labels are shown with dotted lines, and proposed loop movement in the 10 ms intermediate shown with the dotted line with arrow. (Bot) Surface structure of CcrA (PDB id: 2BMI). Metal center shown in the red color and hairpin loop shown in the blue color. Figure generated using VMD [51] software package in built surface representation option.
Incubation of CcrA with substrate that allows the reaction to run to completion may be expected to generate a product complex under the conditions employed, where the concentration of chromacef was about 100 × Km. While we have not measured the KD for binding of hydrolyzed chromacef to CcrA, the KD for hydrolyzed nitrocefin binding to MβL L1 is >300 μM [54]. Simulations of stopped-flow kinetic progress curves suggest weak binding of hydrolyzed nitrocefin or chromacef to all MβLs [24, 54–56]. Consistent with weak product binding, there was little evidence from DEER for product complex formation with either CcrA*(49/82) or CcrA*(49/233) other than the exhibition of narrower distance distributions in the product species. With CcrA*(49/126), the narrowing effect was more dramatic. Rotation of the spin label at residue 49 is expected to have a small effect on the 49–82 and 49–233 distances whereas it will result in a large translation of the spin density along the 49–126 connecting vector. It is likely, therefore, that product interacts with the spin label on residue 49 to restrict rotation of the nitroxide, while the position of residue 49 corresponds to that in the resting enzyme. The larger width of the distance distribution in the product complex of CcrA*(49/82) can be tentatively assigned to motion of the label at residue 82.
Of greatest interest are the data from the RFQ-trapped catalytic intermediates. Perhaps the simplest to interpret are the data from CcrA*(49/126), where the distance domain spectrum is entirely consistent with a dominant (65%) contribution from a species with an interspin distance of ≥ 3.7 nm and a smaller contribution exhibiting a distance that is consistent with the resting enzyme and/or the fully reacted enzyme. CcrA*(49/233) behaves in an analogous manner to CcrA*(49/126). Previous stopped-flow kinetic studies with CcrA (and other MβLs) showed that no detectable product is formed at 10 ms [16, 24, 54, 55, 57–59], and it is, therefore, tempting to assign the shorter of the two distances in CcrA*(49/126) and (49/233) to unreacted enzyme, though it is entirely possible that a second intermediate is present, albeit one in which the distances between residues 49, and 126 and 233, respectively, remain unchanged. Such an intermediate may correspond to substrate forming an initial complex prior to loop movement and binding at the active site. With CcrA*(49/82), a population with r = 3.8 nm is observed, which corresponds to the loop-opened intermediate. An additional population is observed with r = 2.6 nm, which represents a change in the distance between residues 49 and 82 but does not correspond to loop opening. The origin of this shorter distance is unclear but may be tentatively assigned to an additional pre-Michaelis intermediate that is associated with allosteric substrate recognition prior to loop opening and formation of the Michaelis complex at the active site, with concomitant movement of either or both the loop and the residue 82-bearing helix. Further studies with additional CcrA variants CcrA*(82/233) and CcrA*(82/126) are needed to identify any movement of residue 82, and additional work is necessary to confirm (or refute) and characterize the putative allosteric substrate recognition site. Control experiments have been carried out with the related L1 enzyme from S. maltophilia to determine the influence of the method of sample generation by RFQ on the DEER signal, using spin labeled variants in which distance changes would not be expected due to the reaction (Figure S2). The data indeed indicated the expected distances; although, the data quality were poorer with RFQ samples due to dilution of the spin-containing frozen aqueous suspension in the immiscible isopentane matrix [60].
Conclusion
One can consider the labeled residues 82, 233, and 126 in CcrA as forming the base of a (distorted) tetrahedron with reside 49 at the apex and the metal center at the centroid (see Figure 1). The DEER data clearly identify a catalytically-competent species, which is formed upon incubation with substrate for a time that is short compared to the turnover time, in which the distances between residue 49 and residues 82, 126, and 233 have increased by 0.7, 1.0, and 1.3 nm, respectively (Figure 5). Since previous NMR studies identified the loop, which contains residue 49, as being very flexible [33, 34], the increases in distances are most likely due to movements of the loop, and residue 49, away from the other residues. This corresponds to a movement of the spin label on residue 49, at the extremity of the hairpin loop, of 0.94 nm (9.4 Å) away from the plane described by residues 82, 216, and 233, and a further translation of 0.21 nm (2.1 Å) roughly along the direction from residue 233 towards residue 82. Thus the residue 49 moves a net 0.92 nm (9.2 Å) away from the metal center during the catalytic reaction. It is important to note that we cannot unambiguously rule out contributions from movements of domains relative to each other to explain the distance increases. The additional distances observed in DEER are due to a small amount of the product-bound species in the variant with the highest kcat, CcrA*(49/126), and are consistent with either or both unreacted enzyme and product complex in CcrA*(49/82) and CcrA*(49/233).
Examination of the surface plot of CcrA (Figure 5) illustrates how well the mechanistic model provided by DEER complements the three-dimensional structure information. In the resting state, the active site is effectively guarded by the hairpin loop. This may be to prevent promiscuous reaction of metal-bound nucleophile with non-substrate molecules that could otherwise diffuse into the active site. Substrate recognition appears to somehow trigger the DEER-observed retreat of the hairpin loop from the active site, allowing substrate access. Following reaction, the loop is reinstated, even in the product-bound complex.
Supplementary Material
Highlights.
Invariant hairpin loop of the MβLs moves during catalysis
RFQ-DEER can be used to probe conformational dynamics of a loop during catalysis
Hairpin loop moves away from the active site during catalysis, in contrast to previous predictions
Acknowledgments
Funding and support from Miami University, National Science Foundation (CHE1151658 to MWC), and the National Institutes of Health (GM108026 to GAL) are gratefully acknowledged. Zahilyn Roche was an NSF-REU student in 2012. The National Biomedical EPR Center (James S. Hyde, Medical College of Wisconsin) is supported by a NIH P41 EB001980 grant. The pulsed EPR spectrometer at Miami was purchased through NSF MRI-0722403 and the Ohio Board of Regents grants. The authors thank Christian Altenbach, UCLA, for providing LongDistance and Candice S. Klug, Medical College of Wisconsin, for assistance with LongDistance and discussions.
Abbreviations
- CcrA*
refers to the site-directed variant CcrA:C155S
- CcrA*(49)
refers to the site-directed variant CcrA:C155S/W49C
- CcrA*(49/82)
refers to the site-directed variant; CcrA:C155S/W49C/N82C
- CcrA*(49/126)
refers to the site-directed variant; CcrA:C155S/W49C/D126C
- CcrA*(49/233)
refers to the site-directed variant; CcrA:C155SW49C/E233C
- DEER
Double electron-electron resonance/pulsed electron-electron double resonance
- EPR
Electron paramagnetic (spin) resonance
- MTSL
(S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate)
- MβL
metallo-β-lactamase
- RFQ
Rapid freeze quench
Footnotes
The online version of this contains supplementary material, including raw DEER data (Figure S3), which is available to authorized users.
References
- 1.Daiyasu H, Osaka K, Ishino Y, Toh H. FEBS Lett. 2001;503:1–6. doi: 10.1016/s0014-5793(01)02686-2. [DOI] [PubMed] [Google Scholar]
- 2.Bush K, Jacoby GA. Antimicro Agents Chemo. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornaglia G, Giamarellou H, Rossolini GM. Lancet Infect Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 4.Fast W, Sutton LD. Biochim Biophys Acta. 2013;1834:1648–1659. doi: 10.1016/j.bbapap.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 5.Palzkill T. Ann N Y Acad Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palzkill T. In: Metallo-β-lactamase structure and function. Bush K, editor. New York Academy of Sciences; New York: 2013. pp. 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King DT, Strynadka NC. Future Med Chem. 2013;5:1243–1263. doi: 10.4155/fmc.13.55. [DOI] [PubMed] [Google Scholar]
- 8.Wang JF, Chou KC. Curr Topics Med Chem. 2013;13:1242–1253. doi: 10.2174/15680266113139990011. [DOI] [PubMed] [Google Scholar]
- 9.Concha NO, Rasmussen BA, Bush K, Herzberg O. Structure. 1996;4:823–836. doi: 10.1016/s0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- 10.Villadares MH, Galleni M, Frere JM, Felici A, Perilli M, Franceschini N, Rossolini GM, Oratore A, Amicosante G. Micro Drug Resist. 1996;2:253–256. doi: 10.1089/mdr.1996.2.253. [DOI] [PubMed] [Google Scholar]
- 11.Crawford PA, Sharma N, Chandrasekar S, Sigdel T, Walsh TR, Spencer J, Crowder MW. Prot Express Purif. 2004;36:272–279. doi: 10.1016/j.pep.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Walsh TR, Gamblin S, Emery DC, MacGowan AP, Bennett PM. J Antimicro Chemo. 1996;37:423–431. doi: 10.1093/jac/37.3.423. [DOI] [PubMed] [Google Scholar]
- 13.Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS, Gamblin SJ, Spencer J. J Mol Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 14.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. Antimicro Agents Chemo. 1998;42:921–926. doi: 10.1128/aac.42.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Aitha M, Marts AR, Hetrick A, Bennett B, Crowder MW, Tierney DL. J Am Chem Soc. 2014;136:7273–7285. doi: 10.1021/ja410376s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aitha M, Marts A, Bergstrom A, Moller A, Moritz L, Turner L, Nix J, Bonomo R, Page R, Tierney DL, Crowder MW. Biochemistry. 2014;53:7321–7331. doi: 10.1021/bi500916y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush K, Macielag MJ. Exp Opin Thera Patents. 2010;20:1277–1293. doi: 10.1517/13543776.2010.515588. [DOI] [PubMed] [Google Scholar]
- 18.Drawz SM, Bonomo RA. Clin Microbiol Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lassaux P, Hamel M, Gulea M, Delbruck H, Mercuri PS, Horsfall L, Dehareng D, Kupper M, Frere JM, Hoffmann K, Galleni M, Bebrone C. J Med Chem. 2010;53:4862–4876. doi: 10.1021/jm100213c. [DOI] [PubMed] [Google Scholar]
- 20.Rolain JM, Parola P, Cornaglia G. Clin Microbiol Infect. 2010;16:1699–1701. doi: 10.1111/j.1469-0691.2010.03385.x. [DOI] [PubMed] [Google Scholar]
- 21.Vella P, Hussein WM, Leung EW, Clayton D, Ollis DL, Mitic N, Schenk G, McGeary RP. Bioorg Med Chem Lett. 2011;21:3282–3285. doi: 10.1016/j.bmcl.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Prescott JF. In: Other β-lactam antibiotics: Beta-lactamase inhibitors, carbapenems, and monobactams. Giguere S, Prescott JF, Dowling PM, editors. John Wiley & Sons; New York: 2013. pp. 175–188. [Google Scholar]
- 23.Yang KW, Feng L, Yang SK, Aitha M, LaCuran AE, Oelschlaeger P, Crowder MW. Bioorg Med Chem Lett. 2013;23:5855–5859. doi: 10.1016/j.bmcl.2013.08.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, Aitha M, Hetrick AM, Richmond TK, Tierney DL, Crowder MW. Biochemistry. 2012;51:3839–3847. doi: 10.1021/bi300056y. [DOI] [PubMed] [Google Scholar]
- 25.Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP, Lewis C, Galleni M, Frere JM, Payne DJ, Bateson JH, Abdel-Meguid SS. Biochemistry. 2000;39:4288–4298. doi: 10.1021/bi992569m. [DOI] [PubMed] [Google Scholar]
- 26.Mollard C, Moali C, Papamicael C, Damblon C, Vessilier S, Amicosante G, Schofield CJ, Galleni M, Frere JM, Roberts GCK. J Biol Chem. 2001;276:45015–45023. doi: 10.1074/jbc.M107054200. [DOI] [PubMed] [Google Scholar]
- 27.Scrofani SD, Chung J, Huntley JJ, Benkovic SJ, Wright PE, Dyson HJ. Biochemistry. 1999;38:14507–14514. doi: 10.1021/bi990986t. [DOI] [PubMed] [Google Scholar]
- 28.Toney JH, Fitzgerald PM, Grover-Sharma N, Olson SH, May WJ, Sundelof JG, Vanderwall DE, Cleary KA, Grant SK, Wu JK, Kozarich JW, Pompliano DL, Hammond GG. Chem Biol. 1998;5:185–196. doi: 10.1016/s1074-5521(98)90632-9. [DOI] [PubMed] [Google Scholar]
- 29.Cameron AD, Ridderstrom M, Olin B, Mannervik B. Structure. 1999;7:1067–1078. doi: 10.1016/s0969-2126(99)80174-9. [DOI] [PubMed] [Google Scholar]
- 30.de la Sierra-Gallay IL, Pellegrini O, Condon C. Nature. 2005;433:657–661. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- 31.Marasinghe GPK, Sander IM, Bennett B, Periyannan G, Yang KW, Makaroff CA, Crowder MW. J Biol Chem. 2005;280:40668–40675. doi: 10.1074/jbc.M509748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitzgerald PMD, Wu JK, Toney JH. Biochemistry. 1998;37:6791–6800. doi: 10.1021/bi9730339. [DOI] [PubMed] [Google Scholar]
- 33.Huntley JJA, Scrofani SDB, Osborne MJ, Wright PE, Dyson HJ. Biochemistry. 2000;39:13356–13364. doi: 10.1021/bi001210r. [DOI] [PubMed] [Google Scholar]
- 34.Huntley JJA, Fast W, Benkovic SJ, Wright PE, Dyson HJ. Protein Sci. 2003;12:1368–1375. doi: 10.1110/ps.0305303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Keeney D, Tang XJ, Canfield N, Rasmussen BA. J Biol Chem. 1999;274:15706–15711. doi: 10.1074/jbc.274.22.15706. [DOI] [PubMed] [Google Scholar]
- 36.Moali C, Anne C, Lamotte-Brasseur J, Groslambert S, Devreese B, Van Beeumen J, Galleni M, Frere JM. Chem Biology. 2003;10:319–329. doi: 10.1016/s1074-5521(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 37.Dal Peraro M, Vila AJ, Carloni P. J Biol Inorg Chem. 2002;7:704–712. doi: 10.1007/s00775-002-0346-2. [DOI] [PubMed] [Google Scholar]
- 38.Garau G, Bebrone C, Anne C, Galleni M, Frere JM, Dideberg O. J Mol Biol. 2005;345:785–795. doi: 10.1016/j.jmb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 39.Sharma N, Hu Z, Crowder MW, Bennett B. J Am Chem Soc. 2008;130:8215–8222. doi: 10.1021/ja0774562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanchak MP, Taylor RA, Crowder MW. Biochemistry. 2000;39:11330–11339. doi: 10.1021/bi0010524. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Benkovic SJ. J Biol Chem. 1998;273:22402–22408. doi: 10.1074/jbc.273.35.22402. [DOI] [PubMed] [Google Scholar]
- 42.Aitha M, Richmond TK, Hu Z, Hetrick A, Reese R, Gunther A, McCarrick R, Bennett B, Crowder MW. J Inorg Biochem. 2014;136:40–46. doi: 10.1016/j.jinorgbio.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrity JD, Bennett B, Crowder MW. Biochemistry. 2005;44:1078–1087. doi: 10.1021/bi048385b. [DOI] [PubMed] [Google Scholar]
- 44.Sharma NP, Hajdin C, Chandrasekar S, Bennett B, Yang KW, Crowder MW. Biochemistry. 2006;45:10729–10738. doi: 10.1021/bi060893t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett B. Sample Preparation for service at the National Biomedical EPR Center. International EPR (ESR) Society EPR Newsletter. 2012:10–12. [Google Scholar]
- 46.Bohme S, Steinhoff HJ, Klare JP. Spectrosc Int J. 2010;24:283–288. [Google Scholar]
- 47.Zou P, Mchaourab HS. Biophys J. 2010;98:L18–20. doi: 10.1016/j.bpj.2009.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeschke G. Annu Rev Phys Chem. 2012;63:19.11–19.28. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 49.Toledo Warshaviak D, Khramtsov VV, Cascio D, Altenbach C, Hubbell WL. J Mag Res. 2013;232:53–61. doi: 10.1016/j.jmr.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 52.Feldmann EA, Ni S, Sahu ID, Mishler CH, Risser DD, Murakami JL, Tom SK, McCarrick RM, Lorigan GA, Tolbert BS, Callahan SM, Kennedy MA. Biochemistry. 2011;50:9212–9224. doi: 10.1021/bi201226e. [DOI] [PubMed] [Google Scholar]
- 53.Rasmussen BA, Gluzman Y, Tally FP. Antimicro Agents Chemo. 1990;34:1590–1592. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McManus-Munoz S, Crowder MW. Biochemistry. 1999;38:1547–1553. doi: 10.1021/bi9826512. [DOI] [PubMed] [Google Scholar]
- 55.Wang Z, Benkovic SJ. J Biol Chem. 1998;273:22402–22408. doi: 10.1074/jbc.273.35.22402. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Fast W, Benkovic SJ. Biochemistry. 1999;38:10013–10023. doi: 10.1021/bi990356r. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Fast W, Benkovic SJ. Biochemistry. 1999;38:10013–10023. doi: 10.1021/bi990356r. [DOI] [PubMed] [Google Scholar]
- 58.Garrity JD, Pauff JM, Crowder MW. J Biol Chem. 2004;279:39663–39670. doi: 10.1074/jbc.M406826200. [DOI] [PubMed] [Google Scholar]
- 59.Griffin DH, Richmond TK, Sanchez C, Moller AJ, Breece RM, Tierney DL, Bennett B, Crowder MW. Biochemistry. 2011;50:9125–9134. doi: 10.1021/bi200839h. [DOI] [PubMed] [Google Scholar]
- 60.Georgieva ER, Roy AS, Grigoryants VM, Borbat PP, Earle KA, Scholes CP, Freed JH. J Mag Res. 2012;216:69–77. doi: 10.1016/j.jmr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.