Abstract
Evolve and resequence (E&R) experiments use experimental evolution to adapt populations to a novel environment, followed by next-generation sequencing. They enable molecular evolution to be monitored in real time at a genome-wide scale. We review the field of E&R experiments across diverse systems, ranging from simple non-living RNA to bacteria, yeast and complex multicellular Drosophila melanogaster. We explore how different evolutionary outcomes in these systems are largely consistent with common population genetics principles. Differences in outcomes across systems are largely explained by different: starting population sizes, levels of pre-existing genetic variation, recombination rates, and adaptive landscapes. We highlight emerging themes and inconsistencies that future experiments must address.
Introduction
The incredible diversity of life results from adaption in response to a changing environment. Our understanding of how adaptation occurs at the molecular level is surprisingly rudimentary and is derived mostly from comparisons within and between species. Identifying the causative beneficial mutations that give rise to species differences from such genomic comparisons remains a challenge, not to mention inferring their impact on selection and dynamics. As a result, the adaptive landscape upon which organisms evolve is still largely uncharacterized, and debates persist as to how rates, effects, and interactions among beneficial mutations and their environments determine allele frequency change1-4.
Over the last century, different fields of biology have converged towards the use of evolution in the laboratory to study the process of adaptation. Microbiologists5,6, geneticists7,8, biochemists9-11, and population geneticists12,13 found that experimental evolution, in which replicate populations of diverse model organisms are allowed to adapt to novel but controlled laboratory environments (Figure 1), could shed new light on the biological processes they studied. For a long time the different fields focused either on the dynamics of adaptation and/or on the phenotypic or physiological impact on organisms, rather than on the underlying genetic changes that were not easily accessible. Yet, with the rise of sequencing technologies, the “Evolve and Resequence”14 (E&R) approach can uncover the molecular determinants of adaptation in many different systems. The sequencing of evolved RNA molecules15 and viruses16 were followed by bacteria17-20, yeast21 and Drosophila melanogaster22, such that it is now possible to compare E&R experiments across systems. Indeed, E&R affords the possibility of watching evolution occurring in real time and at genome-scale, and fitting dynamic models to the resulting data23,24. Highly replicated E&R experiments, especially those using temporal sampling, now allow for several long-standing questions on the nature and dynamics of molecular evolution to be addressed across models. What is the role of standing genetic variation versus newly arising mutations in contributing to selection response? How reproducible is evolution at the molecular level25? Do the selection coefficients of beneficial alleles change during evolutionary time as a function of distance to a new fitness optimum1? What is the role of protein-coding versus regulatory variation in adaptation26? E&R experiments might begin to resolve these debates as the trajectories of loci contributing to adaptation can be observed in real time.
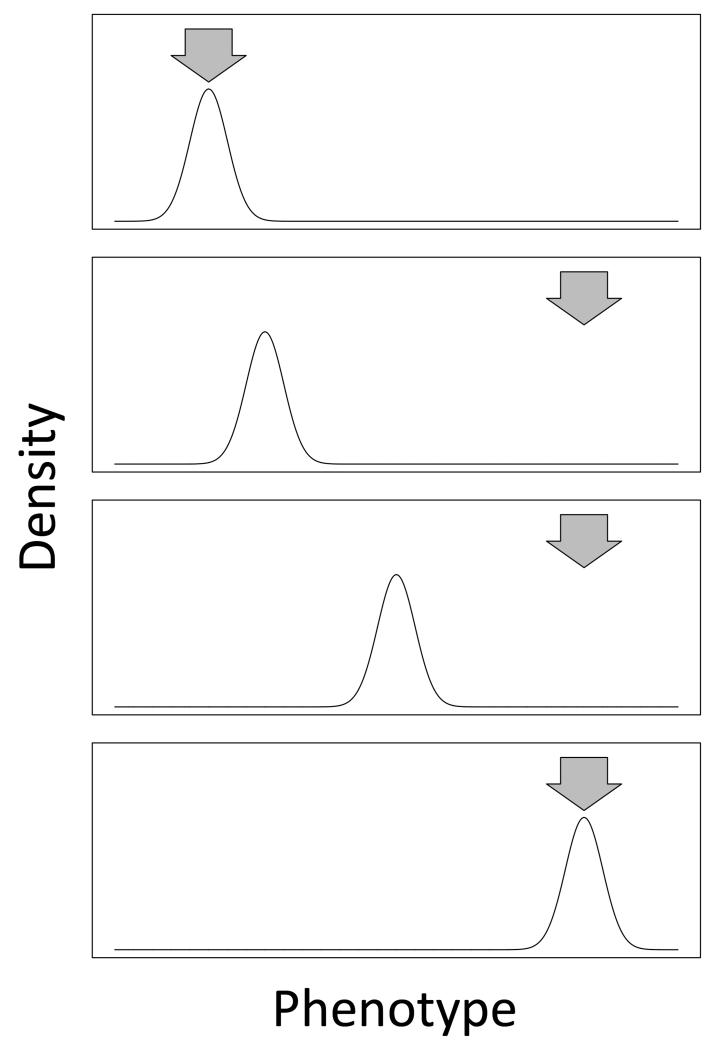
Figure 1.
A conceptual experimental evolution experiment. Starting with a population of organisms, cells, or in vitro molecules, initially the distribution of phenotypes will track the average fitness conditional on phenotype (arrow) in the ancestral environment (top panel). In an experimental evolution experiment the fitness optimum is manipulated through a shift in the environmental conditions under which the system is propagated (e.g., changing the temperature, adding a chemical to the media, forcing molecules to bind to a ligand; second panel). This shift redefines the phenotypic optimum relative to the population's average phenotype. The population will then attempt to track the new optima via natural selection, using standing variation and/or newly arising mutations (third and fourth panels). The speed and mode of adaptation will depend on the system.
E&R experiments are currently being performed in a variety of vastly different systems. The four main systems we will present are: in vitro evolution of libraries of oligonucleotides (RNA or DNA) selected under defined chemical conditions; asexually evolving bacteria or yeast with selection initiated from an initially isogenic founder strain; asexually or semi-sexually evolving yeast with selection initiated from a synthetic founder population derived via intercrossing a small number of diverse natural strains; and obligate sexual D. melanogaster with selection initiated from an outbred population of hundreds of individuals sampled from the wild and established in the laboratory (Table 1). Evolution of proteins27-29 or systematic evolution of ligands by exponential enrichment (SELEX)-type experiments used to study transcription factors binding sites30,31, are examples of other fields of in vitro evolution that have benefited from next-generation sequencing (NGS), but we will not focus on these systems here. The systems we review here are extremely diverse in various ways: the appearance on earth of the equivalent of these organisms spans billions of years (in the case of in vitro evolution the “organisms” are not even living); genome sizes span seven orders of magnitude (from ~50 nucleotides to ~108 bp); and reproduction and propagation range from fully asexual to fully sexual. Interestingly, the field of in vitro E&R has developed almost completely independently of in vivo work, despite the underlying evolutionary principles being identical. The outcome of E&R experiments across systems seems different. In vitro evolution experiments typically result in multiple evolved solutions, asexual microbial systems seem to exhibit fixation of a small number new mutations with divergence of solutions across replicates, and sexual systems initiated from population samples seem to exhibit a highly polygenic response with a high degree of parallel evolution across replicates. These observations create a theoretical dilemma: is the molecular basis of adaptation different in different systems, or are observations generally consistent with one another once different aspects of experimental design are taken into consideration? All these systems evolve according to the laws of natural selection (Box 1), thus the framework of population genetics should capture the details of their evolution. Nevertheless, population genetics is only predictive when applied to a given adaptive landscape (Box 2), which presumably differs across systems.
Table 1.
The four main E&R experimental paradigms discussed in this Review.
This table highlights salient information on type of model, sexual recombination, population size and initial level of genetic diversity in the different model systems.
| System | In vitro | Microbial Isogenic | Microbial Outbred | Obligate Sexual Higher Eukaryotes |
|---|---|---|---|---|
| Example Models | Synthetic DNA, RNA molecules |
Bacteria, haploid yeast, diploid yeast |
Diploid yeast | Drosophila |
| Sexual Recombination |
Can be mimicked | Mostly absent but some experiments with plasmid exchange in bacteria Optional in yeast |
Starting population obtained by crossing different strains. Optional during evolution |
Obligate |
| Population Size | Up to 1016 | Up to 1010 per ml | Up to 109 per ml | 102-103 |
| Initial Genetic Variation Present |
Extremely high, limited by oligonucleotide synthesis technology |
None | Unique haplotypes genome- wide obtained via recombination from 2-16 naturally occurring founders |
Typically initiated from ~100 naturally occurring strains obtained from the wild |
| Initial Variation in Fitness |
Extremely high with the vast majority of molecules have a fitness of zero |
None | High, only limited by natural variation in fitness |
High, only limited by natural variation in fitness |
| Selection Response Mostly Due to |
Variation present in the base population with some modifier mutations |
Newly arising mutations |
Standing genetic variation with newly arising mutations after several hundred generations |
Standing genetic variation |
| Role of Clonal Interference |
Strong | Strong | Strong when evolution is asexual, weak otherwise |
Weak |
Box 1. Population genetics of adaptation.
Population genetics can model the forces that contribute to rates of adaptation.
Emergence of beneficial mutations
Changes in the mean fitness of the population require variation in fitness in a population. In an initially clonal population, this diversity comes from newly arising mutations, although for adaptation to occur a subset of newly arising mutations must be beneficial. Population size then affects the rate of adaptation in two ways. First, the number of beneficial newly arising mutations is a linear function of population size. Second, population size determines the fluctuations in allele frequency from one generation to the next through a process known as random genetic drift. In a finite population a beneficial mutation with a selective advantage smaller than the reciprocal population size is effectively neutral (and its probability of ultimate fixation thus 1/N)145. Even if a beneficial allele has a selective advantage larger than 1/N, it can still be lost due to drift, with the probability of eventual fixation roughly proportional to its selective coefficient146. This being said, beneficial alleles that are lost tend to be lost early, as once an allele reaches a copy number greater than the reciprocal of its fitness effect, its dynamics are essentially deterministic. From these simple well-known population genetics results147 adaptation of an initially clonal population will be marked by a delay corresponding to the emergence of low-frequency beneficial mutations, their survival against drift, and their deterministic increase in frequency until they detectably affect the mean fitness of the population.
Fixation of beneficial mutations
The fate of mutations that survive drift is then highly affected by the existence of genetic exchange. In the absence of recombination, mutations stay coupled to the genetic background on which they arise. Consequently, the beneficial mutations that survived drift compete with one another in a process called clonal interference. Mutations with the highest fitness, or the combination of mutations with the highest fitness, will eventually reach fixation and other beneficial mutations will be lost. The number of beneficial mutations ultimately lost in clonal population is very large71. By contrast, in the presence of high levels of recombination mutations can switch backgrounds, and many mutations can simultaneously increase in frequency in a population. Intermediate levels of recombination can result in genetic hitchhiking, where neutral passenger mutations on the same background as (and closely linked to) a beneficial allele can be sweep to a high frequency due solely to linkage. Distinguishing a beneficial allele from a neutral hitchhiker may require functional assays.
Adaptation from standing genetic variation
When the population starts with a high genetic diversity, as opposed to a single clonal individual, much of the initial variation in fitness in the population is due to standing genetic variation. Current models assume that there are many loci at intermediate frequencies that are essentially neutral in the starting population, but once that population is placed in a novel environment, those alleles start to change in allele frequency due to natural selection. Initially change will be strongest for alleles at intermediate frequency (since they contribute the most to the variance in fitness), and as a result phenotypes or fitness can exhibit rapid change12. Furthermore, in synthetic populations resulting from crosses among a limited number of different clones, the initial allele frequency of each allele is markedly higher than the drift threshold and allele frequency change can be virtually deterministic (unless fitness is affected by de novo mutations or complex epistatic interactions between distant sites). For this reason experimental evolution experiments in D. melanogaster, whose starting populations are typically derived from around 100 individuals, may be very different than yeast populations derived from four isogenic founders. Additionally, experimental evolution populations of D. melanogaster will have effective population sizes several orders of magnitude smaller than yeast, thus requiring a higher initial frequency for each mutation to be above the drift threshold.
Long-term adaptation
Population genetics can be predictive for the early stage of adaptation, provided we know the distribution of fitness effects, the mutation rate and recombination rate. Once adaptation involves the combination of several adaptive mutations, we need to know further global properties of the adaptive landscape, in particular those characterizing microscopic and macroscopic epistasis (Box 2) that define the shape of the landscape.
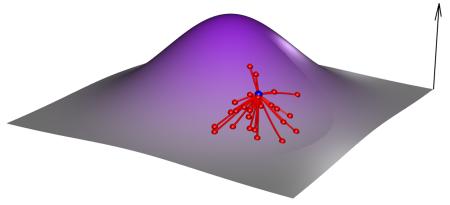
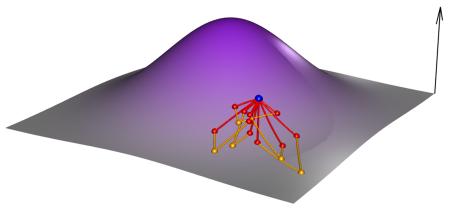
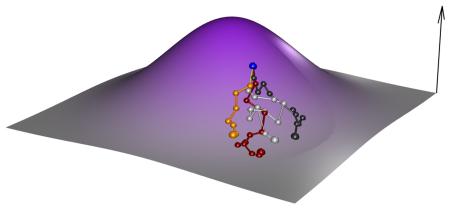
Box 2. Adaptive Landscape [Contains a figure].
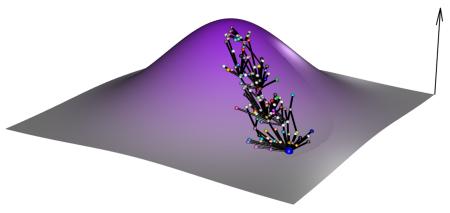
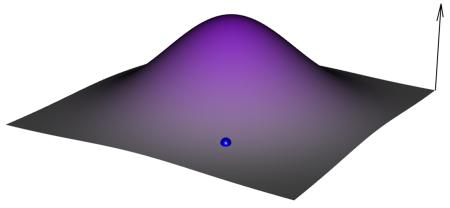
The evolutionary fate of a population depends on the particular mutations sampled during its evolution. The aim of the adaptive landscape metaphor is to find a visual way to illustrate such possibilities. The genetic space is virtually infinite. For a genome composed of n biallelic loci, it is a hypercube with 2n states, which cannot be visualized for more than five loci. Hence, a landscape metaphor is used in which the vertical axis is fitness and the horizontal axes represent a continuous vision of the genetic space in which proximity suggests genetic similarity (see the figure). Though metaphoric, analysis of the adaptive landscape can also be quantitative at a local and a global scale. In panels a-d of the figure we illustrate experimental approaches for exploring the adaptive landscape relative to a single genotype represented by the blue dot.
Local scale
A) The first characterization of the local scale is the fitness effects of neighboring mutations whose distribution is shown in red. It is explored via the fitness analysis of a collection of single mutants. B) A second layer of complexity to the adaptive landscape comes from pairwise epistasis, with yellow dots depicting the fitness of double mutants. It is uncovered by comparing the fitness effects of single mutants to the fitness of double mutants.
Global scale
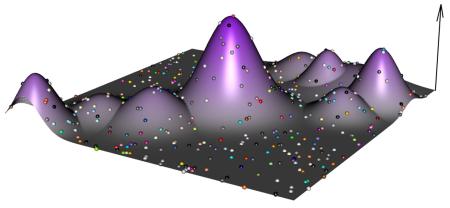
While exploring the whole landscape is out of reach, some global properties of the landscape can be explored via evolution. C) Mutation accumulation experiments — in which a lineage is regularly subjected to a population bottleneck of one or a few individuals — provide an estimate of the average effect of newly arising mutations; four such lineages are shown in different colours. D) By contrast, standard experimental evolution experiments — in which large populations are propagated in a given environment — estimate the cumulative effect of mutations favored by natural selection. Lines represent beneficial mutations sampled during the adaptive walk that survive drift. Mutations that survive ultimate extinction are plotted using new colours.
Starting points of different experimental systems
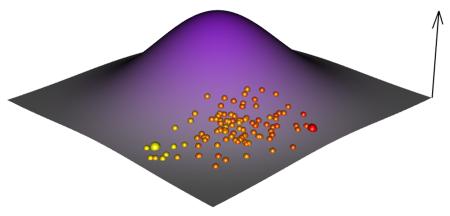
The four different experimental systems use different starting points to explore the adaptive landscape; these starting points are illustrated in parts e-h of the figure. E) in vitro selection samples random genotypes covering an extremely large part of the genotypic landscape. F) In stark contrast, asexual evolution initiated with a clone explores the landscape from a single initial genotype. G) Outbred microbial populations are initiated from a synthetic population obtained via several rounds of recombination from a small set of isogenic founders. For simplicity we depict two founder genotypes (large yellow and red dots) and many recombinants coloured according to founder proportions. H) Obligate sexual populations sample a large number of natural occurring genotypes. Since strategies G and H sample natural alleles (or recombinants between natural alleles) the initial variance in fitness is much less than strategy E.
In this Review, we aim to reconcile observations from E&R experiments across these disparate systems. We first describe the differences between systems in terms of experimental setting and population genetics parameters and explain how these may impact the dynamics of adaptation. We then review what we have learned using E&R across this diverse set of systems. We focus on the population dynamics of beneficial alleles, the molecular bases of adaptation, inferences that can be drawn from parallel adaptation in replicate evolving populations, and the role of epistasis in adaptation. Finally we attempt to identify experiments that can potentially clarify remaining discontinuities between the systems
E&R: the systems
In vitro systems
In vitro selection and evolution experiments32-34 start with highly diverse libraries of short DNA oligonucleotides (~1014–1016 different molecules) that typically consist of random regions of ~30–200 bp flanked by primer-binding sequences. In vitro evolution consists of a transcription step to generate a corresponding RNA pool, an enrichment step during which the RNA population is exposed to a defined chemical condition (e.g. binding to ATP-coated beads, or ligation to another nucleic acid), followed by an amplification step in which the enriched library is reverse transcribed back to DNA and PCR-amplified10. Enrichment and amplification steps are typically repeated for 10–20 cycles. The starting bulk population of molecules often does not show any detectable activity in the assay, but after three to five rounds there is appreciable activity, and after 8–12 rounds the population demonstrates strong activity in the assay and converges upon a few dominant motifs35. Similar experiments have been performed directly with single-stranded DNA10,36,37, ‘mosaic’ nucleic acids, in which the backbone was scrambled between ribose and deoxyribose38, nucleic acid analogues39,40, as well as proteins linked to their coding mRNAs or cDNAs41,42.
Due to the small ‘genome sizes” being considered and relatively low complexity of the population following in vitro selection, Sanger sequencing has been used for decades to identify the RNA motifs that ‘won’ the evolutionary competition. Furthermore, the total diversity of the population over rounds of selection could be followed using restriction enzymes43. NGS allows testing the selected population in earlier rounds and provides a much more complete picture of the final array of winning genotypes44. In fact, direct experimental measurement of the fitness landscapes of RNA ligase15, kinase45, Diels-Alderase46, and self-splicing ribozymes47 are obtained at much higher resolution than ever before. Combined with microfluidic analytical platforms, NGS can be directly coupled to activity measurement, revealing the fitness landscape of a functional RNA48,49.
Asexual microbes with an isogenic starting population
The dominant model consists of an initially isogenic population of 106 to 108 bacteria or yeast that is evolved asexually. The evolution protocol relies on the renewal of the media the microbes use to grow. This renewal can be made through serial transfers, in which a fraction of a saturated culture is regularly diluted into fresh media50, or continuously using a chemostat51. Based on the dilution factor or flow rate, the evolutionary rate can be computed (e.g., a daily 100-fold dilution implies log2(100) = 6.64 generations per day). Given estimated growth rates, hundreds to thousands of generations of evolution can occur in a few weeks or months in a microbial system. The keystone example of microbial experimental evolution is the Long-Term Evolution Experiment (LTEE) that has been running for more than 25 years and has now reached more than 60,000 generations of evolution52. To put this in perspective, in humans where a generation takes ~20 years, 60,000 generations corresponds to 1.2 million years ago, which predates the emergence of the species Homo sapiens.
Microbes with a synthetic outbred starting population
A less mature branch of microbial experimental evolution utilizes synthetic populations of yeast generated through the intercrossing of two or more isogenic highly characterized founder strains53,54. While these populations maintain all the desired features of the microbial model, including large population size and replicate populations, they also introduce the important innovations of sexual reproduction and standing genetic variation in the starting population. The evolution of these populations potentially mimics obligate sexual higher eukaryotes more closely than initially isogenic asexual systems. These synthetic outbred populations are evolved asexually via serial passage in liquid or in solid culture for several hundred generations at effective population sizes of 106–108, with entire pooled populations resequenced to high coverage. This “Pool-seq” protocol (which can be applied to diverse species and systems)20,55 results in an estimate of allele frequency in the population at every single nucleotide polymorphism (SNP) in the genome. Despite yeast synthetic populations being derived from a small number of founder genomes, multiple rounds of intercrossing results in a near infinite number of genome-wide haplotype combinations (Figure 2).
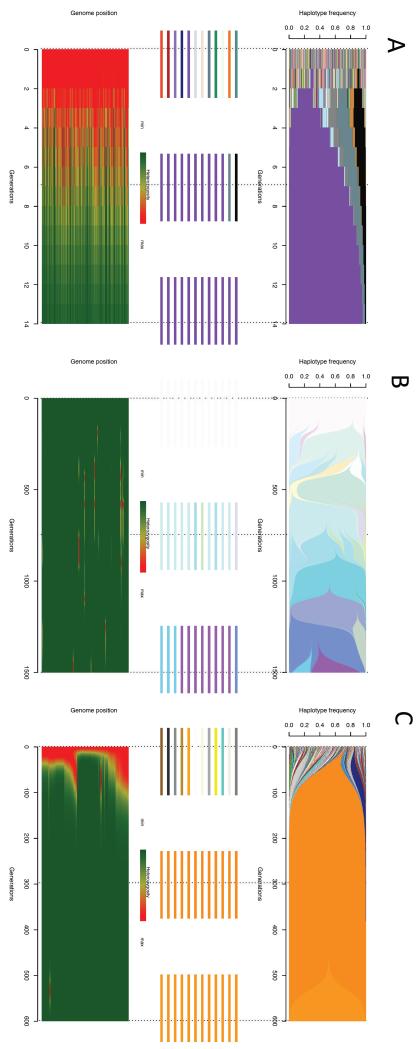
Figure 2.
E&R experiments reveal the dynamics of adaptation at a genome-wide scale. From sequencing datasets of individuals or pooled populations (Pool-seq), the evolution of haplotype diversity (upper panel of each figure part) and that of allele diversity at each site of the genome (bottom panel of each figure part) can be uncovered. Haplotypes are coloured according to the initial genome-wide haplotypes, and the middle panels of each figure part show example haplotypes from the start, middle and end of the E&R experiment. Population heterozygosity is shown at each site of the simulated genome, with red transitioning to green heat colours indicating decreasing levels of heterozygosity. A) For in vitro experiments, the great initial haplotypic diversity is lost within a few generations as the best haplotype(s) quickly increase in frequency, and the large majority of initial haplotypes have a fitness of near zero. Per-site heterozygosity is homogeneous and mostly decays through the adaptive process. B) In asexual microbial evolution from an isogenic starting population, the initial diversity is minimal, and can only build up through newly arising mutations. In the clonal interference regime depicted here, several haplotypes compete with one another to reach fixation (upper panel). Diversity in the genome is only maintained at the beneficial sites and is therefore highly heterogeneous across the different sites of the genome (lower panel). New mutations are eventually lost or fixed hence heterozygosity is transitory. C) In yeast asexual evolution from a synthetic outbred starting population, the initial diversity is high but organized in blocks resulting from recombination between the founding parents. Similar to in vitro studies adaptation is characterized by a genome-wide loss of haplotype diversity. However, the rate of heterozygosity loss is lower than in the in vitro case as there is much lower variation in initial fitness and perhaps selection is not as strong. The regional loss of diversity will depend on the variance in fitness of the different haplotype blocks present, with regions not affecting fitness losing diversity later. For details on the parameters used to generate the panels in this figure, see Supplementary information S1 (box).
Obligate sexual higher eukaryotes with an outbred starting population
Drosophila species are long established as a system for the study of experimental evolution in the laboratory56,57. In the past decade several groups have resequenced pooled DNA samples from experimentally evolved laboratory populations of D. melanogaster to identify regions of the genome responding to laboratory-imposed selection7,14,22,58-60. The details of E&R experiments in D. melanogaster are quite different from those previously discussed. The starting populations are founded from collections of flies sampled from the wild, which can harbor hundreds of natural haplotypes, hence individual genetic variants can be rare. Population sizes are modest by the standards of microbes, or RNA molecules, and range from 100 to 1,000 individuals. Evolution experiments in D. melanogaster are also modest in terms of the number of generations of selection carried out: a one to three year experimental evolution experiment in D. melanogaster is only 25–75 generations, since the egg-to-egg life cycle of a fly is almost 2 weeks. Nevertheless, there are a few much longer-term (200–300 generation) ongoing experiments that are being actively investigated22,61,62.
Population dynamics of loci during selection
Despite population genetics theory having a wealth of models dealing with the dynamics of alleles responding to selection there is a relative dearth of empirical datasets where allele trajectories are directly observed. E&R experiments coupled with temporal sampling of populations allow the direct study of how allele frequencies change over time. The speed and the magnitude of allele frequency changes tell us a great deal about the selective advantages associated with different variants. Other patterns of change, such as alleles plateauing at intermediate frequencies, suggest that simple population genetics models of adaption do not fully capture the dynamics of adaptation.
In vitro selection systems are characterized by dramatic selective advantages associated with beneficial alleles. A variant starting at a frequency of 10−16 can fix in 10 rounds of selection43, suggesting that the selective coefficient (s) associated with beneficial oligonucleotide haplotypes is greater than 50%. Initially there are extremely high levels of genetic diversity, but since oligonucleotides are randomly synthetized the initial fitness of the vast majority of molecules is essentially zero (Figure 2a). Starting from initially random sequences, and selecting for a simple biochemical function results in a rapid loss of diversity and dramatic fitness improvement.
The dynamics of allele frequency change in initially isogenic microbes (such as bacteria or yeast) evolving asexually is surprisingly complex. In the absence of genetic exchange, these mutations compete with one another through a process called clonal interference12,63 (Box 1). The total number of potential beneficial mutations genome-wide is large enough that it is virtually impossible for a beneficial mutation to arise and reach fixation without having to compete with some other beneficial mutation that arose in a competing lineage in the population. Hence, as shown elegantly in yeast64, in large asexual populations evolution is not characterized by single beneficial mutations reaching fixation, but instead by ‘lucky’ combinations of mutations34,65 that manage to out-compete other such combinations. Consequently, the allele frequency of a beneficial mutation may initially increase but then may decrease or even reverse as alternative combinations of mutations are being selected for 66-68 (Figure 2b). Although the selective advantage of the beneficial mutations observed depends drastically on the selective regime69 it is quite common to observe s ranging from 1 to 20% 70,71. These lower values, in comparison to in vitro evolution, may be due to pleiotropic constraints imposed on newly arising mutations. Those pleiotropic effects may increase with organismal complexity and favor alleles of more modest effect. On the other hand, since fitness effects are measured relative to average fitness of the population as a whole, fitness effects may seem larger in in vitro evolution purely as a result of most oligonucleotides in the initial population having a fitness of essentially zero.
The dynamics of allele frequency change is quite different if selection is initiated from an outbred starting population as in yeast (Figure 2c). As initial genetic diversity is high and every allele starts from a high initial frequency (e.g., ~25% in ‘4-way’ populations derived from four isogenic founders), the response to selection is rapid. The selection coefficients inferred from the rate of allele frequency change are generally ~1%, unless the selective pressure is intense, in which case de novo mutations of strong effect may also come into play. Genome-wide patterns of allele frequency change over time allow inferences about how variation present at the start of the experiment is sorted out during adaptation. Specifically, regions of a few kilobases in size harbouring beneficial or detrimental alleles can be precisely identified53,72. Furthermore, for the first several hundred generations of evolution, virtually all adaptation is from standing genetic variation present in the starting population53,54. Through manipulations to the culturing regime, yeast populations can be forced to participate in sexual reproduction at regular intervals, providing insights into long-term effect of sex in evolving populations73,74. Yeast synthetic populations adapting to novel laboratory environment with weekly episodes of sexual recombination show that adaptation patterns are very comparable to asexually evolving synthetic yeast populations, at least for the first several hundred generations75.
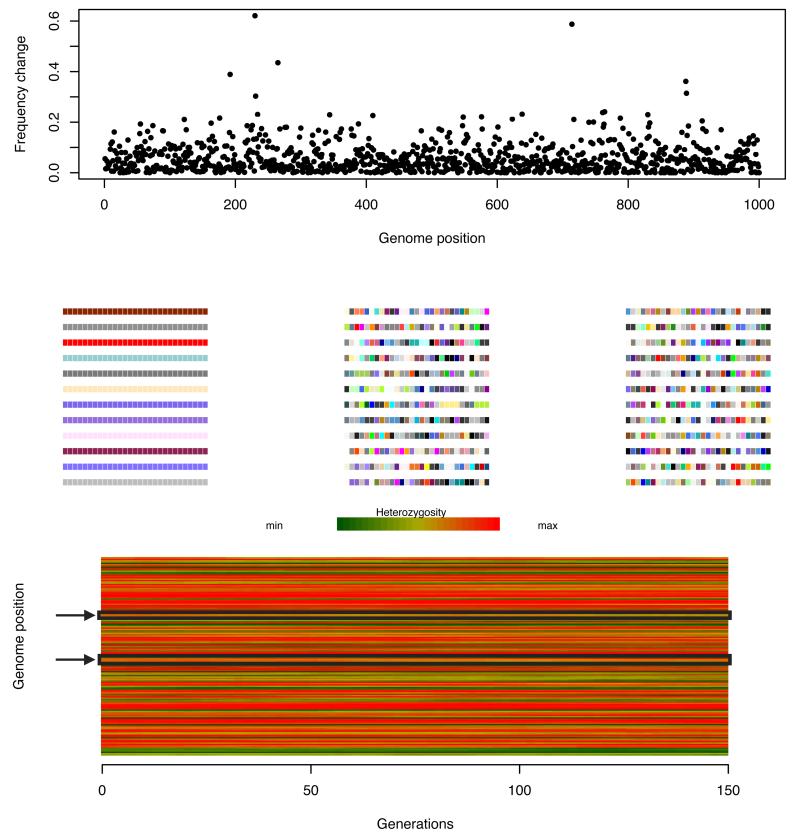
Experiments in D. melanogaster are initiated from outbred starting populations harbouring hundreds of natural haplotypes at each genetic locus. Thus they are similar to the outbred yeast discussed above, except many more natural haplotypes are used to found the starting population. Furthermore, because recombination is obligate in D. melanogaster, clonal interference is unlikely to impede evolution, and two regions of the genome can independently respond to natural selection. Finally, the populations of D. melanogaster used to found E&R experiments are typically derived from hundreds of wild haplotypes, thus alleles with minor allele frequencies of 1–5% can contribute to selection response, unlike the yeast outbred system. Consistent with these properties, sliding window plots of heterozygosity versus genome position tend to show several dozen regions of the genome experiencing dramatic reductions in heterozygosity punctuated by the vast majority of the genome maintaining substantial variation (Figure 3) with changes in absolute allele frequency showing similar local spikes22. Notably, regions showing reductions in heterozygosity never show a drop to zero, consistent with the ideas that adaptation is due to selection on standing genetic variation, or that selective sweeps are happening but have had insufficient time to reach fixation, or that advantageous alleles plateau in frequency before they reach fixation.
Figure 3.
E&R experiments in sexually reproducing species. Evolutionary patterns are different in obligate sexual E&R experiments initiated from an outbred population, compared to the asexually evolving examples of Figure 2 As a result of recombination, haplotypes at the beginning of the experiments are shuffled and therefore genome-wide haplotype evolution cannot be tracked by sequencing pools of individuals (Pool-seq). Instead, investigators tend to track sliding window haplotype change over the course of the entire experiment as a function of genome position 75, as presented in the top panel. Population sizes are also typically much smaller than the systems of Figure 2 as a result the variance in haplotype and allele frequency change is an important consideration. Recombination occurring in the course of the adaptation further shuffles the initial haplotypes as shown in the middle panels of example haplotypes. The pattern of heterozygosity presented in the lower panel contrasts with the ones of Figure 2. Heterozygosity remains globally high over the genome apart from the few regions harbouring the variants that are important for adaptation that show reduced diversity (two cases are indicated by arrows). For details on the parameters used to generate the lower panels in this figure, see Supplementary information S1 (box).
Overall, the strength of the selective advantage of beneficial alleles seems to vary across systems, as well as within systems, and depends on the experimental system and on the nature of the selective pressure. Once these factors are taken into account, population genetics provides a good qualitative description of the dynamics of adaptation of these highly variable systems. An important observation that is currently being actively debated is the extent to which allele frequency change can plateau in longer-term experimental evolution experiments initiated from outbred sexual populations58,75,76. If the alleles that are important in adaptation plateau before reaching fixation, then average selection coefficients in longer-term experiments will not accurately reflect an initially very rapid change in allele frequency at selected sites.
Types of mutations recruited
By virtue of sequencing the entire genome of evolved clones or populations, E&R experiments can potentially unravel the molecular nature of beneficial alleles and determine some notion on their functions, frequency, and interactions.
The complexity of the DNA pools used for in vitro experimental evolution is limited by the capabilities of DNA synthesis platforms, which typically yield ~1016 different molecules. The theoretical diversity of a random DNA polymer is 4N, where N is the length of the random region. For DNA pools longer than 26 random nucleotides (nts), this number is larger than 1016, which means that the actual diversity is smaller, often vastly so, than the theoretical limit (e.g. for a 60 nt random pool, the theoretical limit is 460~1036 — twenty orders of magnitude above the actual complexity). Thus many in vitro experimental evolution experiments are initiated with pools that greatly under-sample the sequence space, leaving much room for fine-tuning during the subsequent selection rounds. In one example, selection for GTP aptamers directly yielded an optimal family of aptamer sequences, suggesting that these sequences were present in the initial pool; by contrast, other aptamer families improved their binding affinity by several orders of magnitude upon mutagenesis and re-selection, suggesting that the initial sequences of these aptamers were relatively far from optimal and required subsequent newly arising mutations for optimal activity35,77. To date, the evidence from NGS of in vitro selected pools suggest that individual families of functional RNAs exist in the starting pools, but greatly benefit from mutagenesis15,46,77 or synthetic shuffling45 to uncover the most active variants, which then do not tend to drift towards other peaks in the fitness landscape78.
In initially isogenic asexual systems, despite clonal interference increasing the time to fixation, genetic diversity within the populations remains low compared to the other systems we discuss in this Review. As these systems are initially isogenic, total variation in the population is limited to newly arising mutations. In a highly replicated E&R experiment in bacteria carried out for ~2,000 generations the average number of genetic events distinguishing evolved clones from the ancestral strain was 11, with several lines of evidence suggesting the majority of these events were adaptive 20,79 (see the parallel evolution section).
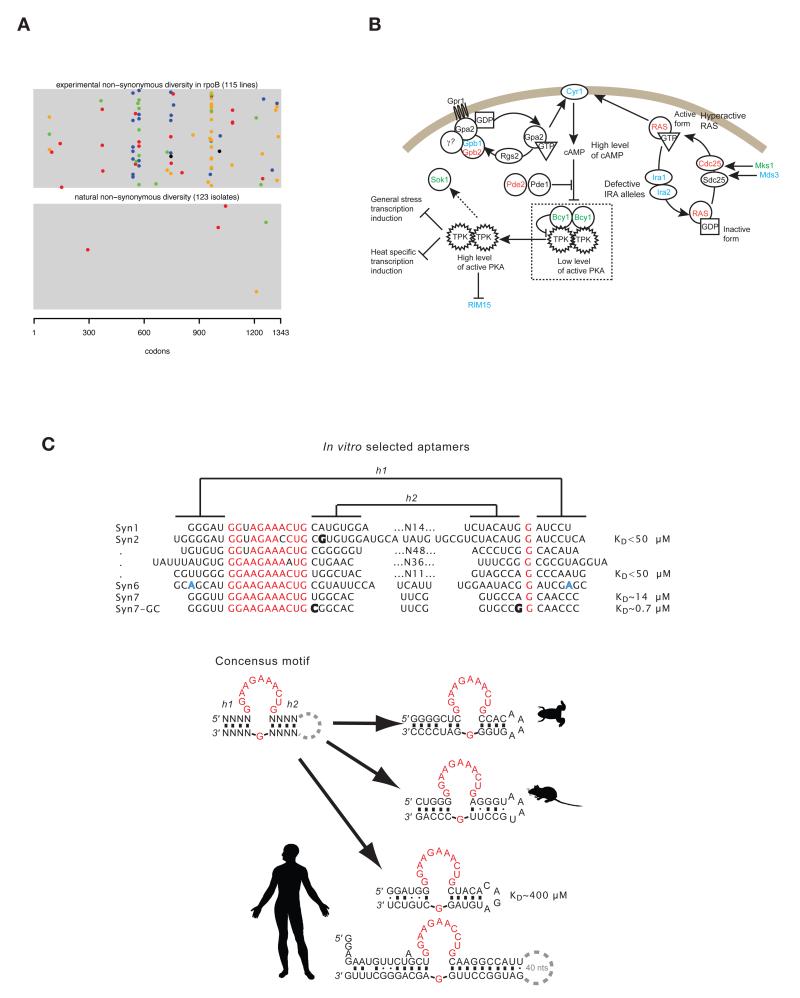
Strikingly these experiments suggest that for virtually every novel environment a myriad of beneficial mutations are accessible. Bacterial and yeast asexual E&R experiments starting from an isogenic population have recovered many different types of mutations in evolved lines. While point mutations dominate in number 79-83, small insertions or deletions (indels), large duplications or deletions, and transposition events also contribute to the selection response79,84. Most selected alleles appear to be clear loss-of-function mutations, such as premature stop codons, gene deletions, and transposable element insertions into genes79,83,85. Presumably the selective regime favours the loss of certain cellular functions. There are however also clear examples of non-synonymous nucleotide changes being positively selected64,79. An interesting observation in bacteria is that among the first large-effect adaptive steps, selection seems to recruit mutations in global regulators. For example, the RNA polymerase (rpo) is a major mutational target in many different adaptations ranging from high temperature to glycerol minimal media86. Several distinct novel rpoBC alleles are found in many different experimental evolution contexts, which is surprising given that this operon is generally highly conserved among bacterial species (Figure 4a). It is unclear why changes in this operon are so often favored in experimental evolution settings despite the function of these genes being highly conserved in nature.
Figure 4.
The molecular bases of adaptation. (A) rpoB sequence alignments for replicate populations (rows) evolved at high temperature79 (top panel) and naturally occurring isolates148 (bottom panel) showing that rpoB is targeted repeatedly in laboratory-based adaptation to high temperature, yet is largely invariant between naturally occurring E. coli strains. Colors correspond to different base changes (A, green; C, blue; G, orange; T red; deletions black) (B) The RAS–cAMP pathway is targeted by both de novo mutations and standing variants involved in yeast adaptation to multiple stress conditions. Colours indicate alleles detected in experiments initiated from a single clone (red), an outbred synthetic population (green), or both (blue). (C) Adenosine aptamer sequences recovered from an in vitro selection experiment and subsequently optimized for strong binding116. The pattern of sequence evolution is different for the ligand-binding loop (red) and the helical segments (marked as h1 and h2), which are defined by solely by sequence covariation. A structure-based search for the identified motif and genomic systematic evolution of ligands by exponential enrichment (SELEX) for adenosine/ATP novel aptamers revealed the adenosine aptamer sequences in bullfrog, mouse, and humans, suggesting molecular convergence between in vitro evolved molecules and genomic sequences120. Part B is adapted from Parts et al. 2011 53.
In asexual yeast evolving from an isogenic starting population, large-scale duplications are often observed in response to environmental stresses87-89, and similar genomic signatures can be observed in extant populations with association to phenotypes90-92. Amplification of gene regions is thought to be a general mechanism for compensation of deleterious mutations93 or adaptation to limiting substrate. For instance, laboratory evolved yeast populations and synthetic yeast libraries harbouring chromosomal amplifications showed that increased copies of whole chromosomes or chromosomal regions have beneficial effects for adapting to limiting substrate conditions94. A recent study evolved 180 strains with different single-gene deletions and observed compensatory evolution via duplicated regions in 22% of the initial strains81. One potential explanation for the ubiquity of this response is that the rate of mutation to aneuploidy or amplified gene regions is several orders of magnitude higher than that for point mutations95. Thus selection may be favoring available, as opposed to optimal, solutions.
The few reports of E&R on outbred microbial populations with standing genetic variation paints a different picture in which selection acts mainly on standing genetic variation53,54,75. Large-scale structural and copy-number variants are not observed apart from dramatic depletion of mitochondrial DNA copy number triggered by the accumulation of reactive oxygen species (ROS) during heat stress53. There are several potential explanations for this apparent discrepancy. These include differences in experimental design that could lead to different rates of formation of complex rearrangements, different selective regimes, dominance in haploid versus diploid yeast, and the extent to which meiosis tolerates such events. Alternatively, large-effect structural mutations with deleterious pleiotropic side-effects may be tolerated in experimental evolution experiments that opportunistically rely on beneficial newly arising mutations, whereas if standing genetic variation is available it may be preferred since those variants are less-likely to have strong deleterious pleiotropic side-effects (as they are segregating in natural populations). Another possibility is that structural variants are occurring but are poorly queried via Pool-seq experiments. Sliding window plots of sequence coverage versus genome location suggest large insertions and deletions are not present, but the Pool-seq data await more sophisticated analyses55.
To date, most E&R experiments on D. melanogaster have been unable to pin-point the causative regions responding to selection to single genes; instead, they typically identify several dozen genomic regions of hundreds of kilobases in size that show reductions in heterozygosity and/or frequency change of SNPs7,14,22,58,59. That being said when adaptation is due to causative loci of large effect relative to standing variation, single nucleotide resolution is achievable60. The failure to identify the genes harboring causative variants in E&R experiments on D. melanogaster should not come as a surprise, as simulation studies show that for modest-effect causative alleles, precise localization is only possible with higher levels of experimental replication, larger populations sizes, and/or more generations of evolution than is typically achieved96,97. More highly replicated (Number of experimental replicates >15) larger population size (Effective population size >1,000) E&R experiments in D. melanogaster are possible and should allow for finer localization of beneficial alleles, but their execution requires considerable fortitude.
The relative importance of newly arising mutations versus standing genetic variation as contributors to adaptation is currently debated. Patterns of within species variation and between species divergence in humans suggest that standing variation is more important in adaptation than newly arising mutations98, whereas in flies the opposite appears to be the case99, and more generally the role of newly arising mutations appears larger in species with large populations sizes100. In E&R experiments initiated from an isogenic base only newly arising mutations matter, so these systems cannot address this question. E&R experiments in D. melanogaster have concluded that most adaptation is due to standing variation7,14,22,58-60, but populations sizes are extremely small, a situation that favours standing variants2. Experiments in outbred yeast could begin to address this question, but if the base populations are derived from a small number of founders (as they have been) rare alleles are not effectively captured. Furthermore, if the evolution itself is carried out without recombination, then the experiment is effectively sorting lineages as opposed to modeling evolution in sexuals. Despite these shortcomings, outbred yeast population can be created with more founders, and evolution can be carried out at large populations sizes with sex75. There is an opportunity to directly experimentally address the importance of standing variation versus newly arising mutations in contributing to selection response.
Parallel evolution
Parallel evolution is said to have occurred if two lineages independently evolving from the same starting population converge on the same solution at some level of organization. Unlike evolution in the wild, the ability of E&R experiments to study multiple evolving lineages under identical selection conditions provides a powerful opportunity to directly detect parallel evolution. The relative prevalence of this event will help to address a long-standing question regarding the replicability of evolution at the molecular level. Parallel evolution in independent lineages can also be used as a tool to identify functionally relevant changes16,79,101,102 and distinguish them from non-selected passenger mutations that may be found in the evolved genomes.
When evolution is initiated from an isogenic starting population the observation of similar changes recovered in independent lineages suggests either a large mutation rate or the filtering action of natural selection. For experimental evolution of asexually evolving isogenic yeast or bacteria, in most cases and at most loci, the large mutation rate hypothesis can be rejected as replicate populations have different mutations affecting the same target. Indeed, in most targeted genes, numerous adaptive mutations appear to exist 79,103,104 even if the gene is essential (for example the rpoB example shown in Figure 4a). Consequently, parallelism is rarely observed at the level of individual mutations79,105, but instead at the gene level and often at higher levels of functional integration such as the operon or sets of functionally related genes (e.g., genes involved in cell shape)79,103,104. The genomic precision of targeting seems to depend on the selective regime employed including the media used for selection 89 as well as the mode of culture (chemostats versus batch culture)106. For example, antibiotic treatments select almost exclusively for mutations in the active site of very specific target genes107, whereas for less-specific environmental stressors in highly replicated E&R experiments many genes and alternative mutations among genes are recovered64,79. With less-specific environmental stressors, most genes are hit in less than half of the evolved replicates. These observations suggest there are perhaps thousands of mutations available that improve fitness in some novel environment and the particular set utilized in any given evolutionary realization depends on the particular mutations that occur early on in the experiment, the genetic backgrounds they occur on, and the action of clonal interference.
Despite the stochastic nature of evolutionary change in asexual systems initiated from an isogenic starting population, there is enough parallelism that many functional targets have been identified. The functions identified to date as targets of selection are very diverse, ranging from genes important in metabolism18,89 to genes involved in cell shape20,79, stress response104,108,109, as well as genes of unknown function79. Interestingly, mutations in global regulators are often observed to be of large effect110, beneficial, occur early in the experimental evolution experiment, and may even be recovered across different laboratory-induced selective regimes111, with different alleles being discovered in different settings112. For example, in yeast, the RAS–cAMP signaling pathway64,66,85,113 is a target of selection both in experimental evolution initiated from an isogenic population as well as experiments initiated from synthetic outbred population, where the pathway appears to harbor multiple deleterious SNPs53 (Figure 4b).
The observation of convergence can be equally as illuminating when selection is initiated from an outbred population potentially harboring millions of polymorphic sites, as only a very small fraction are likely to be the target of selection and hence show convergent change across replicate populations. Indeed, modeling experiments of evolution initiated from an outbred population suggest that replication is perhaps the easiest way to increase the power to detect causative sites96,97. In contrast to microbial systems where parallelism at the base-pair level is almost non-existent and gene-level parallelism is detectable but modest, in systems with standing genetic variation the degree of parallelism is high. In published replicated experiments in yeast and D. melanogaster, convergence is almost always observed. The likely explanation for this result is perhaps obvious: outbred sexual systems start with the same standing variation available for natural selection to act upon in independent replicates, the total number of pre-existing beneficial alleles of modest-to-large effect is somewhat limited, and these same alleles are targeted in replicate populations. Sexual recombination results in an absence of clonal interference, which means that unlinked beneficial alleles can independently and simultaneously increase in frequency.
A caveat in the sexual systems is that even small amounts of accidental gene flow between replicate populations will virtually guarantee parallel allele frequency change across replicate populations, with allele frequency drift in the metapopulation masquerading as parallel adaptive change22. Population genetics theory predicts that exchanging one migrant per generation between two populations is sufficient enough to homogenize neutral allele frequencies114. Because replicate populations are generally not marked in any way, it is difficult to know if low levels of gene flow are an issue, and future experiments would benefit from barcoding of replicate populations. In outbred yeast this could be accomplished using barcodes that are identical for every individual within a population, but different between populations. A simple PCR multiplexed NGS sequencing reaction could detect contamination events with some sensitivity (such a barcoding system is described in ref71). However in a system such as D. melanogaster it is technically more challenging to easily barcode an outbred population.
Although parallel evolution dominates the landscape in the outbred systems, this is a novel and recent observation, and one that would not necessarily have been predicted a priori. If selection tends to operate on rare alleles present in the starting population, the adaptive response may not be so replicable, as the probability of initially rare alleles being stochastically lost in a given replicate evolving population is ~1-2s (see Box 1). By contrast, if an allele starts out at a frequency of >5% the probability of fixation is almost certain provided s>>1/N (where N is the population size). Taking these two theoretical considerations into account, the observation of highly parallel evolution in yeast synthetic outbred E&R experiments should not be surprising at all. Since the synthetic populations are derived from four isogenic founders all alleles start out at a frequency of roughly ¼, ½, or ¾, and populations sizes are >106, the loss of beneficial alleles is highly unlikely. Convergence is more surprising in D. melanogaster, where minor allele frequencies in the starting population can be less than 1%, and population sizes are less (or much less) than 1,000 individuals. If gene flow is absent in E&R experiments in D. melanogaster, the observation of strong parallel evolution suggests that the targeted beneficial alleles start at minor allele frequencies of >5% and selection is fairly strong. However, we note that studies in outbred systems (particularly those in flies) often have few replicates and are thus typically underpowered to confidently distinguish beneficial from neutral alleles based on parallelism across replicates. Thus, apparent evidence of parallelism should be interpreted cautiously, and E&R studies in outbred yeast and flies should ultimately aim for highly replicated formats analogous to those in bacteria80.
Beyond parallel evolution at the primary sequence or higher functional level, parallelism is also apparent when considering molecular structures. In vitro evolution experiments suggest that chemically functional RNAs (or DNAs and their analogues) fold into specific structures, often dominated by secondary structures, and it is usually these structures, rather than a specific sequence, that is selected for. Hence, independent populations may converge toward a secondary structure that may be revealed by sequence co-variation (e.g. G-C vs C-G base-pairs) but not primary sequence conservation, whereas key tertiary interactions and active/binding sites tend to be conserved on a primary sequence level (Figure 4c). Thus genotypes that evolve are dominated by strong sequence conservation in key loops, surrounded by strongly co-varying (but generally sequence-independent) helical structures. The information content of co-varying segments is therefore higher than the primary sequence would suggest77; as a result, the degree to which the evolution of functional RNAs shows convergence may not be fully appreciated. Yet, strong functional convergence is observed across experiments. For example, in vitro selection starting from a short, 24-nt pool that extensively sampled the theoretical sequence space yielded the same GTP aptamers in two independent experiments115. Functional convergence can even be extended across biological systems: the same adenosine aptamer motif has for instance been independently selected in vitro from random sequences at least four times116-119 and it also occurs in genomes spanning bacteria to humans120. Similarly, the hammerhead ribozyme, a specific type of self-cleaving ribozyme with a conserved catalytic core and secondary structure, has been identified in vitro several times121 and is widespread in nature122-124.
Epistasis
Epistasis measures the extent to which allelic effects depend on the genetic background in which they appear. It is a property of the adaptive landscape that conditions the dynamics of adaptation. Epistasis can be defined at a microscopic level as a functional interaction between alleles such that the fitness of a double mutant differs from what is expected based on the combined fitness effects of the single mutants, or at a macroscopic level where the fitness of a mutant depends on some higher-level property of the overall genetic background it occurs on. An emerging question is whether robust statistical properties may be apparent at a higher macroscopic level24 once all microscopic epistatic interactions are accounted for. If rules exist at the macroscopic epistasis level, long-term predictions on the adaptive process may be possible despite a poor knowledge of the details of epistasis at the microscopic level24. What have E&R experiments told us about microscopic and macroscopic epistasis?
Microscopic epistasis
As discussed above, in vitro evolution experiments select RNA molecules with sequence-conserved loops flanked by strongly co-varying but generally sequence-independent helical structures (Figure 4c). The epistatic interactions among the partners of the helical structures can be identified using NGS reads: covariation is extracted from the entire population-wide haplotype of the adapting 30–100–mers. It is worth noting that these epistasic interactions are easily detected in this system because the distances between interacting sites in the linear nucleotide chain are typically shorter than sequencing read lengths, hence interaction sites can be collectively analyzed in single sequencing reads. It is important to appreciate that all evolutionary change occurring in the stem part of a ribozyme or an aptamer is epistatic in nature, thus epistasis is pervasive in this system77,78,116.
E&R experiments also reveal microscopic epistasis among beneficial mutations in asexual isogenic systems. For instance, once a particular gene, pathway or function is targeted by some mutation, especially loss-of-function mutations, little advantage is gained from additional mutations in the same gene, pathway or function. The old adage that “there is little use in beating a dead horse” comes to mind. Moreover, populations evolving more than a few hundred generations will eventually fix multiple beneficial mutations contributing to adaptation. Provided enough E&R replicates have been carried out, order-of-fixation epistasis can be detected as non-random orders of fixation events in replicate evolved populations. Tenaillon et al79 observed many significant epistatic interactions between independent functional targets of selection. The strongest such example was that early substitution events in rpoB tended to preclude later events in rho (and vice versa). Other studies coupling E&R with experiments have revealed many cases of mutations with sign epistasis, that is beneficial in the ancestral background but deleterious when coupled to another one125-128.
Despite the evidence for pervasive epistasis in in vitro systems and in microbial systems initiated from an isogenic population, there is little evidence for epistasis in systems when experimental evolution is initiated from an outbred population. This might be telling us that epistasis is less important in sexual systems since any favorable allele increasing in frequency with time will find itself in numerous different genetic backgrounds, whereas in asexual systems a newly arising mutation is always associated with the genome-wide haplotype on which it arises. Thus a beneficial allele in an asexual system only needs to be advantageous in the background on which it arises, whereas in a sexual population the advantage of an allele is its average effect over all backgrounds it is likely to encounter. It follows that in sexual systems natural selection will favor only those alleles that combine beneficially across all backgrounds, whereas in asexual systems natural selection will favor alleles that are beneficial only in the background in which they occur. Contrarily, epistasis may be commonplace yet rarely observed in experiments initiated from outbred populations due to it being difficult to detect given the way experiments are carried out in these systems. The easiest way to detect fitness epistasis in an outbred population is to identify unlinked loci in linkage disequilibrium (LD)129. E&R experiments from an outbred starting population prepare genomic DNA from an entire population, sequence it as a pool, and then estimate the frequency of every SNP in the genome as a function of treatment and/or time. Given that these data report population-wide allele frequencies, but not which alleles co-occur in individuals, LD between unlinked loci is not estimated, and hence it is very difficult to show that particular combinations of alleles are more favored than others. That being said, at least in the yeast system, once causal sites are identified, gene-replacement experiments could be used to shed light on the problem.
Macroscopic epistasis
In all the different systems, there seems to be a change in the dynamics of adaptation through time.
For in vitro E&R experiments, after the early stages of fast adaptation, further selective cycles result in diminishing returns with respect to biochemical activity in the assay130. Similarly, in isogenic asexual populations, rates of adaptation slow over the course of the experiment52. The explanation for diminishing returns over the course of an experimental evolution experiment has often been based on Fisher’s geometric model of adaptation131. Under this model, as an evolving population approaches a new optimum in phenotypic space the beneficial alleles favored by natural selection are both less common and of smaller selective effect131,132. According to the model, in the initial phases of evolution large-effect mutations, which may also have deleterious pleiotropic side-effects, are favored, but as the optimum is approached fine-tuning mutations are instead favored133. In yeast, allele-replacement techniques are straightforward and they have revealed a form of macroscopic epistasis where the effect of a beneficial mutation is a function of the fitness of the clone into which it was inserted, as opposed to the specific combination of other mutations present in that clone82. The higher the initial fitness of the recipient background, the lower the incremental effect of the introduced beneficial mutation on fitness134. This overall diminishing rate of adaptation as fitness increases seems to be conserved from viral systems through yeast81,82,135-139, and is consistent with the analysis of epistasis among combinations of beneficial mutations70,140-142. An interesting question is the extent to which this observation applies to evolution in outbred sexual populations.
Conclusions and future perspectives
Despite the vastly different organismal complexity, ranging from 30 bp oligonucleotides transcribed into RNA to complex multicellular higher eukaryotes, the principles governing evolution are constant. The large differences we see in how quickly these systems respond to environmental challenges and the molecular bases of adaptation are largely outcomes of the nature of the starting populations employed and the intensity of natural selection experienced in each system. Thus each population is exploring its adaptive landscape in a slightly different manner, and that exploratory process governs evolutionary outcomes. In asexual systems clonal interference results in genome-wide haplotypes competing with one another for eventual fixation, whereas in sexual systems different genomic regions can respond to evolution somewhat independently. In systems starting from standing variation adaptation can be much more deterministic, since the same palette of alleles is available to all replicate populations. By contrast, in systems starting from a single isogenic population, all evolution must proceed from newly arising mutations, thus it is somewhat predicated on the stochastic nature and timing of these events. Even when evolution is initiated from populations with standing variation, it will proceed differently if that starting variation is randomly synthesized oligonucleotides (where the vast majority of alleles have an initial fitness of near-zero) compared to the situation where adaptation is initiated from naturally occurring standing variation and the variance in fitness among starting alleles is probably much more subtle. But the role of very strong and/or molecularly targeted selection cannot be discounted. In bacteria the nature of evolutionary response can be very different when the selection pressure is relatively specific and strong (e.g., antibiotic resistance) versus more weak yet pervasive (e.g., high temperature). In in vitro systems the selective agent directly queries a specify biochemical activity, so it is difficult to imagine a system that consistently experiences a more specific selective pressure.
There is still a rather large disconnect between the communities who believe the principles of evolutionary change can be completely elucidated via experimental evolution in the laboratory and those who believe that the rules in nature are somehow different. The often observed phenomena of mutations in global regulators (such as rpoB) being important in laboratory microbial evolution, despite these proteins being highly conserved over evolutionary time, supports the idea that evolution in the laboratory does not fully recapitulate what is occurring in the wild. That being said, the controlled and replicated nature of laboratory experiments are extremely appealing features of E&R. In a few cases replicated evolution has certainly occurred in nature and these systems can yield incredible insights143, but it is difficult to routinely identify such ‘natural experiments’. Another avenue might be to introduce isogenic bacteria to replicate sets of more natural environments144 than a chemostat or Erlenmeyer flask, or to introduce outbred laboratory populations back into natural replicated environments.
The overall comparability between experimental evolution systems, once the underlying population genetics and nature of the adaptive landscape are taken into account, belies some nagging differences. Both microscopic and macroscopic epistasis seems pervasive in all the lower complexity systems, yet epistasis seems much less common in the outbred sexual higher complexity systems. It is an important future question to determine if this difference is a feature of the systems or imposed by different experimental designs. Individual-based sequencing seems cost prohibitive in these systems, but epistasis could be studied via gene-replacement experiments. A second somewhat striking finding is that larger-scale structural variants often play a role in adaptation in asexual evolution initiated from an outbred population, but these same events are not generally observed in sexual systems initiated from an outbred population. Future experiments should address if these observations are replicable in systems where recombination is occurring during evolution (currently it is more difficult to evolve these systems for as many generations). Finally, it is fascinating that the field of in vitro E&R has ‘evolved’ largely independently of the in vivo systems. Given the increasing desire to understand E&R experiments within a systems biology framework the distinction between in vivo and in vitro systems seems increasingly arbitrary. The time is ripe for practitioners in the different systems to learn from one another.
Supplementary Material
Supplementary information S1 (box): Simulation parameters used to generate data in figures 2 and 3.
At-a-glance summary.
Evolve and Resequence (E&R) is a powerful paradigm for understanding the molecular basis of adaptation
Several systems exist, ranging from in vitro RNA and DNA molecules, to microbes evolving from an isogenic clone, or sexual eukaryotes harbouring standing variation. E&R experiments are producing different results in the different systems. Can observed differences be reconciled with evolutionary theoretical models.
The systems differ in: population size, levels of standing variation, initial variance in fitness and levels of genetic exchange. We argue that when these differences between systems are taken into account many of the apparent differences can be explained.
There remain enigmas. Why do ploidy changes and/or large duplications and deletions seem more important in asexual microbes and sexual eukaryotes? At what point do sexually reproducing organisms need newly arising mutations? In sexually reproducing organisms, does allele frequency change often plateau before fixation? How much can macroscopic epistasis help us understand evolution in microbes, and what is the role of epistasis in sexually reproducing organisms?
Acknowledgements
This work was supported by: the Borchard Scholar-in-Residence Program; ATIP-Avenir (CNRS/INSERM), FP7- PEOPLE-2012-CIG (322035), ANR (ANR-13-BSV6-0006-01; ANR-11-LABX-0028-01), ARC (SFI20111203947) and La Ligue contre le cancer; NSF MCB1330606; European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant 310944.
Glossary definitions
- Clonal Interference:
A phenomenon observed in asexually evolving systems. Due to a lack of recombination, clones harboring different combinations of mutations compete against one another to reach fixation.
- Selection coefficient:
Represented by s, the proportional change in the fitness of a genotype owing to a mutation. 1-s is the fitness of that genotype.
- Average Fitness:
The average fitness of a population is defined as the weighted sum of the fitness values associated with each genotype, where the weights are the frequencies of those genotypes. In an in vitro evolution experiment initially there could be several million genotypes, the vast majority having fitness values close to zero.
- Linkage disequilibrium (LD):
The condition in which the frequency of a particular haplotype for two loci is significantly different from that expected if the loci were assorting independently.
- Ribozyme:
An RNA that is capable of catalyzing a chemical reaction. Natural ribozymes include ribosomal RNAs, spliceosomal RNAs, RNase P RNA, self-splicing introns and self-cleaving ribozymes.
- Fixation:
When an allele of an initially polymorphic locus or haplotype increases in frequency to reach 100% frequency in the population.
- Standing genetic variation:
Genetic diversity that preexists in a population of interest.
- Aneuploid:
Abnormal chromosome number due to a gain or loss of entire chromosomes.
- Pleiotropic:
A genetic change affecting more than one phenotype.
- Selective sweep:
When selection drives a genetic polymorphism to fixation closely linked regions of the genome will follow along to fixation with the adaptive allele. The size of the swept region depends on the starting allele frequency of the beneficial allele, the strength of selection, and local recombination rate.
- Tertiary interactions:
Molecular interactions stabilizing the overall (tertiary) structure of a functional RNA.
- Haplotype:
The ordered collection of alleles along a single chromosome.
- Mutation accumulation experiment:
An experiment in which an initially isogenic strain is propagated for many generations with severe population size bottlenecking (often a single cell or individual) without voluntary selection. The mutations that distinguish the accumulation strain from its ancestor can be used to estimate mutation rates.
Biographies
Author biographies
Anthony Long is a Professor at the University of California Irvine, USA. His research addresses questions in population, quantitative and statistical genetics using a variety of model systems. Recent work has explored the utility of Evolve and Resequence Experiments for unraveling the genetic basis of complex trait variation as well as elucidating the dynamics of adaptation over ecological timescales in outbred populations.
Gianni Liti studied biology and obtained his PhD from the University of Perugia, Italy. He holds a CNRS researcher position at the Institute for Research on Cancer and Ageing of Nice (IRCAN), France, where he leads the team “Population genomics and complex traits analysis”. His research group uses the budding yeast, S. cerevisiae to dissect the genetic architecture of multiple traits related to ageing and cancer. His team is involved in several yeast population-level sequencing projects aimed at understanding how genetic variation in a population influences phenotypic variation and evolution.
Andrej Luptak is an Associate Professor at the University of California, Irvine, USA. His research group uses in vitro selection experiments to discover novel functional RNAs, including genomic aptamers and ribozymes, and diagnostic and potentially therapeutic DNA aptamers. His work extends to the development of new in vitro evolution and molecular display techniques, as well as structure-based bioinformatics and functional analyses of genomic aptamers and ribozymes.
Olivier Tenaillon is a researcher at INSERM, University Denis Diderot Paris 7, and associate professor at Ecole Polytechnique. His team, “Quantitative Evolutionary Microbiology” is focusing on E. coli evolution both in the laboratory and in the wild and studying how models such as Fisher’s geometric model of adaptation can be used to characterize bacterial adaptation. Experimental evolution, epidemiology, animal models, genomics, high-throughput mutagenesis and phenotyping are used to challenge some of the theoretical predictions of population genetics and uncover the molecular bases of the E. coli adaptive landscape.
Footnotes
Competing Financial Interests: The authors declare no competing financial interests.
Papers to add comments to
Sherlock 2015: Adding molecular markers to 500,000 yeast and following their frequency through time allowed the authors uncover the diversity of beneficial mutations simultaneously present in a yeast population.
References
- 1.Chevin LM, Hospital F. Selective Sweep at a Quantitative Trait Locus in the Presence of Background Genetic Variation. Genetics. 2008;180:1645–1660. doi: 10.1534/genetics.108.093351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplan NL, Hudson RR, Langley CH. The ‘hitchhiking effect’ revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sellis D, Callahan BJ, Petrov DA, Messer PW. Heterozygote advantage as a natural consequence of adaptation in diploids. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20666–20671. doi: 10.1073/pnas.1114573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams J, Rosenzweig F. Experimental microbial evolution: history and conceptual underpinnings. Genomics. 2014;104:393–398. doi: 10.1016/j.ygeno.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 7.Zhou D, et al. Experimental selection of hypoxia-tolerant Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2349–2354. doi: 10.1073/pnas.1010643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassidy JJ, et al. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 2013;155:1556–1567. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nature Publishing Group. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 10.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 11.Joyce GF. Forty years of in vitro evolution. Angew. Chem. Int. Ed. Engl. 2007;46:6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- 12.Rose MR, Lauder GV. Adaptation. Academic Pr; 1996. [Google Scholar]
- 13.Kawecki TJ, et al. Experimental evolution. Trends in Ecology & Evolution. 2012;27:547–560. doi: 10.1016/j.tree.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Turner TL, Stewart AD, Fields AT, Rice WR, Tarone AM. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet. 2011;7:e1001336. doi: 10.1371/journal.pgen.1001336. [An evolve and resequence experiment in Drosophila melanogaster that introduces the term “evolve and resequence”.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitt JN, Ferré-D'Amaré AR. Rapid construction of empirical RNA fitness landscapes. Science. 2010;330:376–379. doi: 10.1126/science.1192001. [This is the first paper using NGS to measure the shape of the fitness landscape of an in vitro selected ribozyme and explain the mutations within the context of the ribozyme structure.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 17.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 18.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 19.Velicer GJ, et al. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8107–8112. doi: 10.1073/pnas.0510740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [This paper, sequencing E. coli lineages throughout 30,000 generations of evolution, is the first paper to link molecular evolution to fitness improvement.] [DOI] [PubMed] [Google Scholar]
- 21.Araya CL, Payen C, Dunham MJ, Fields S. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics. 2010;11:88. doi: 10.1186/1471-2164-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burke MK, et al. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature. 2010;467:587–590. doi: 10.1038/nature09352. [The first E&R experiment in an obligate sexual higher eukaryote. Despite several hundred generations of evolution fixation events are not observed.] [DOI] [PubMed] [Google Scholar]
- 23.Illingworth CJR, Parts L, Schiffels S, Liti G, Mustonen V. Quantifying selection acting on a complex trait using allele frequency time series data. Mol. Biol. Evol. 2012;29:1187–1197. doi: 10.1093/molbev/msr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Good BH, Desai MM. The impact of macroscopic epistasis on long-term evolutionary dynamics. Genetics. 2015;199:177–190. doi: 10.1534/genetics.114.172460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin A, Orgogozo V. The Loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- 26.Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 27.Hietpas RT, Jensen JD, Bolon DNA. Experimental illumination of a fitness landscape. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Firnberg E, Ostermeier MP. Funkel: efficient, expansive, user-defined mutagenesis. PLoS ONE. 2012;7:e52031. doi: 10.1371/journal.pone.0052031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 β-lactamase. 2015;160:882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 30.Slattery M, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu G, Wang T, Yang Y, Xu X, Wang J. An improved SELEX-Seq strategy for characterizing DNA-binding specificity of transcription factor: NF-κB as an example. PLoS ONE. 2013;8:e76109. doi: 10.1371/journal.pone.0076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 33.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 34.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 35.Davis JH, Szostak JW. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11616–11621. doi: 10.1073/pnas.182095699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 37.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 38.Trevino SG, Zhang N, Elenko MP, Lupták A, Szostak JW. Evolution of functional nucleic acids in the presence of nonheritable backbone heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13492–13497. doi: 10.1073/pnas.1107113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu H, Zhang S, Chaput JC. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem. 2012;4:183–187. doi: 10.1038/nchem.1241. [DOI] [PubMed] [Google Scholar]
- 40.Taylor AI, et al. Catalysts from synthetic genetic polymers. Nature. 2015;518:427–430. doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seelig B, Szostak JW. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature. 2007;448:828–831. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 44.Kobori S, Nomura Y, Miu A, Yokobayashi Y. High-throughput assay and engineering of self-cleaving ribozymes by sequencing. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtis EA, Bartel DP. Synthetic shuffling and in vitro selection reveal the rugged adaptive fitness landscape of a kinase ribozyme. RNA. 2013;19:1116–1128. doi: 10.1261/rna.037572.112. [NGS measurement of the fitness landscape of a kinase ribozyme, with explicit comparison of recombination versus point mutations in an RNA population.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ameta S, Winz M-L, Previti C, Jäschke A. Next-generation sequencing reveals how RNA catalysts evolve from random space. Nucleic Acids Research. 2014;42:1303–1310. doi: 10.1093/nar/gkt949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayden EJ, Bratulic S, Koenig I, Ferrada E, Wagner A. The effects of stabilizing and directional selection on phenotypic and genotypic variation in a population of RNA enzymes. J. Mol. Evol. 2014;78:101–108. doi: 10.1007/s00239-013-9604-x. [DOI] [PubMed] [Google Scholar]
- 48.Cho M, et al. Quantitative selection and parallel characterization of aptamers. Proc. Natl. Acad. Sci. U.S.A. 2013;110:18460–18465. doi: 10.1073/pnas.1315866110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buenrostro JD, et al. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat Biotechnol. 2014;32:562–568. doi: 10.1038/nbt.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atwood KC, Schneider LK, Ryan FJ. Periodic selection in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1951;37:146–155. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novick A, Szilard L. Experiments with the Chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. U.S.A. 1950;36:708–719. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiser MJ, Ribeck N, Lenski RE. Long-term dynamics of adaptation in asexual populations. Science. 2013;342:1364–1367. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]
- 53.Parts L, et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 2011;21:1131–1138. doi: 10.1101/gr.116731.110. [This paper introduces the yeast outbred population system to the E&R field.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cubillos FA, et al. High-resolution mapping of complex traits with a four-parent advanced intercross yeast population. Genetics. 2013;195:1141–1155. doi: 10.1534/genetics.113.155515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlötterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 2014;15:749–763. doi: 10.1038/nrg3803. [DOI] [PubMed] [Google Scholar]
- 56.L'Heritier PL, Neefs Y, Teissier G. Apterisme de insects et selection naturelle. Compt. Rend. Acad. Sci. 1937;204:907–909. [Google Scholar]
- 57.Powell JR. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford University Press; 1997. [Google Scholar]
- 58.Orozco-terWengel P, et al. Adaptation of Drosophila to a novel laboratory environment reveals temporally heterogeneous trajectories of selected alleles. Mol. Ecol. 2012;21:4931–4941. doi: 10.1111/j.1365-294X.2012.05673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teotónio H, Chelo IM, Bradić M, Rose MR, Long AD. Experimental evolution reveals natural selection on standing genetic variation. Nat. Genet. 2009;41:251–257. doi: 10.1038/ng.289. [DOI] [PubMed] [Google Scholar]
- 60.Martins NE, et al. Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5938–5943. doi: 10.1073/pnas.1400378111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teotónio H, Rose MR. Variation in the reversibility of evolution. Nature. 2000;408:463–466. doi: 10.1038/35044070. [DOI] [PubMed] [Google Scholar]
- 62.Passananti HB, Matos M, Rose MR. Methuselah Flies: A Case Study in the Evolution of Aging. World Scientific Publishing Company, Incorporated; 2004. [Google Scholar]
- 63.Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102-103:127–144. [PubMed] [Google Scholar]
- 64.Lang GI, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–574. doi: 10.1038/nature12344. [The sequencing of tens of yeast populations through time reveals the dynamics of clonal interference in which group of mutations compete with one another until one combination, possibly including neutral hitchhiker mutations, reaches fixation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Desai MM, Fisher DS. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics. 2007;176:1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lang GI, Botstein D, Desai MM. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics. 2011;188:647–661. doi: 10.1534/genetics.111.128942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodríguez-Verdugo A, Carrillo-Cisneros D, González-González A, Gaut BS, Bennett AF. Different tradeoffs result from alternate genetic adaptations to a common environment. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12121–12126. doi: 10.1073/pnas.1406886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan AIA, Dinh DMD, Schneider DD, Lenski RER, Cooper TFT. Negative epistasis between beneficial mutations in an evolving bacterial population. Science. 2011;332:1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 71.Levy SF, et al. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature. 2015;519:181–186. doi: 10.1038/nature14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ehrenreich IM, et al. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2011;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeyl C, Bell G. The advantage of sex in evolving yeast populations. Nature. 1997;388:465–468. doi: 10.1038/41312. [DOI] [PubMed] [Google Scholar]
- 74.Goddard MR, Godfray HCJ, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434:636–640. doi: 10.1038/nature03405. [DOI] [PubMed] [Google Scholar]
- 75.Burke MK, Liti G, Long AD. Standing genetic variation drives repeatable experimental evolution in outcrossing populations of Saccharomyces cerevisiae. Mol. Biol. Evol. 2014;31:3228–3239. doi: 10.1093/molbev/msu256. [A 4-way yeast population experimentally evolved with sexual recombination roughly once every 30 cell divisions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burke MK, Long AD. What paths do advantageous alleles take during short-term evolutionary change? Mol. Ecol. 2012;21:4913–4916. doi: 10.1111/j.1365-294x.2012.05745.x. [DOI] [PubMed] [Google Scholar]
- 77.Carothers JM, Oestreich SC, Davis JH, Szostak JW. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 2004;126:5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petrie KL, Joyce GF. Limits of neutral drift: lessons from the in vitro evolution of two ribozymes. J. Mol. Evol. 2014;79:75–90. doi: 10.1007/s00239-014-9642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tenaillon O, et al. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [The parallel adaptation of 115 lineages of E. coli reveals that convergence among replicates occurs mostly at a functional level and that many competing mutations may affect each of the functional targets of adaptation.] [DOI] [PubMed] [Google Scholar]
- 80.Blank D, Wolf L, Ackermann M, Silander OK. The predictability of molecular evolution during functional innovation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3044–3049. doi: 10.1073/pnas.1318797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szamecz B, et al. The genomic landscape of compensatory evolution. PLoS Biol. 2014;12:e1001935. doi: 10.1371/journal.pbio.1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. Microbial evolution. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science. 2014;344:1519–1522. doi: 10.1126/science.1250939. [The existence of macroscopic epistasis leading to a global diminishing return rate of adaptation as a function of fitness is shown using yeast] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Payen C, et al. Empirical determinants of adaptive mutations in yeast experimental evolution. biorxiv.org. 2015. p. 014068. [DOI]
- 84.Raeside C, et al. Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. MBio. 2014;5:e01377–14. doi: 10.1128/mBio.01377-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kvitek DJ, Sherlock G. Whole genome, whole population sequencing reveals that loss of signaling networks is the major adaptive strategy in a constant environment. PLoS Genet. 2013;9:e1003972. doi: 10.1371/journal.pgen.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Conrad TM, et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Di Rienzi SC, et al. The dynamics of diverse segmental amplifications in populations of Saccharomyces cerevisiae adapting to strong selection. Genes/Genomes/Genetics. 2014;4:399–409. doi: 10.1534/g3.113.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang S-L, Lai H-Y, Tung S-Y, Leu J-Y. Dynamic large-scale chromosomal rearrangements fuel rapid adaptation in yeast populations. PLoS Genet. 2013;9:e1003232. doi: 10.1371/journal.pgen.1003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bergström A, et al. A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 2014;31:872–888. doi: 10.1093/molbev/msu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strope PK, et al. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 2015;25:762–774. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Warringer J, et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koszul R, Caburet S, Dujon B, Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sunshine AB, et al. The fitness consequences of aneuploidy are driven by condition-dependent gene effects. PLoS Biol. 2015;13:e1002155. doi: 10.1371/journal.pbio.1002155. [This paper introduces a new methodology to dissect the underlying molecular basis of aneuploidy advantages] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dujon B. Yeast evolutionary genomics. Nat. Rev. Genet. 2010;11:512–524. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 96.Baldwin-Brown JG, Long AD, Thornton KR. The power to detect quantitative trait loci using resequenced, experimentally evolved populations of diploid, sexual organisms. Mol. Biol. Evol. 2014;31:1040–1055. doi: 10.1093/molbev/msu048. [A simulation study that examined the power of E&R experiments in sexual outbreds, a conclusion of the paper is that current experiments are under-powered to routinely identify regions responding to selection.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kofler R, Schlötterer C. A guide for the design of evolve and resequencing studies. Mol. Biol. Evol. 2014;31:474–483. doi: 10.1093/molbev/mst221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hernandez RD, et al. Classic selective sweeps were rare in recent human evolution. Science. 2011;331:920–924. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sattath S, Elyashiv E, Kolodny O, Rinott Y, Sella G. Pervasive adaptive protein evolution apparent in diversity patterns around amino acid substitutions in Drosophila simulans. PLoS Genet. 2011;7:e1001302. doi: 10.1371/journal.pgen.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Corbett-Detig RB, Hartl DL, Sackton TB. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 2015;13:e1002112. doi: 10.1371/journal.pbio.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cooper TF, Rozen DE, Lenski RE. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pelosi L, et al. Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics. 2006;173:1851–1869. doi: 10.1534/genetics.105.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee M-C, Marx CJ. Synchronous waves of failed soft sweeps in the laboratory: remarkably rampant clonal interference of alleles at a single locus. Genetics. 2013;193:943–952. doi: 10.1534/genetics.112.148502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maharjan RP, Liu B, Feng L, Ferenci T, Wang L. Simple phenotypic sweeps hide complex genetic changes in populations. Genome Biol Evol. 2015;7:531–544. doi: 10.1093/gbe/evv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ostrowski EA, Woods RJ, Lenski RE. The genetic basis of parallel and divergent phenotypic responses in evolving populations of Escherichia coli. Proc. Biol. Sci. 2008;275:277–284. doi: 10.1098/rspb.2007.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gresham D, Dunham MJ. The enduring utility of continuous culturing in experimental evolution. 2014;104:399–405. doi: 10.1016/j.ygeno.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lindsey HA, Gallie J, Taylor S, Kerr B. Evolutionary rescue from extinction is contingent on a lower rate of environmental change. Nature. 2013;494:463–467. doi: 10.1038/nature11879. [DOI] [PubMed] [Google Scholar]
- 108.Riehle MM, Bennett AF, Long AD. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2001;98:525–530. doi: 10.1073/pnas.021448998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Charusanti P, et al. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet. 2010;6:e1001186. doi: 10.1371/journal.pgen.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hindré T, Knibbe C, Beslon G, Schneider D. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat. Rev. Microbiol. 2012;10:352–365. doi: 10.1038/nrmicro2750. [DOI] [PubMed] [Google Scholar]
- 111.Conrad TM, Lewis NE, Palsson BØ. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 2011;7:509–509. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Achaz G, Rodríguez-Verdugo A, Gaut BS, Tenaillon O. The reproducibility of adaptation in the light of experimental evolution with whole genome sequencing. Adv. Exp. Med. Biol. 2014;781:211–231. doi: 10.1007/978-94-007-7347-9_11. [DOI] [PubMed] [Google Scholar]
- 113.Wenger JW, et al. Hunger Artists: Yeast Adapted to Carbon Limitation Show Trade-Offs under Carbon Sufficiency. PLoS Genet. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hartl DL, Clark AG. Principles of Population Genetics. Sinauer Associates Incorporated; 2007. [A population genetics textbook that should be read as often as it is cited.] [Google Scholar]
- 115.Jiménez JI, Xulvi-Brunet R, Campbell GW, Turk-MacLeod R, Chen IA. Comprehensive experimental fitness landscape and evolutionary network for small RNA. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14984–14989. doi: 10.1073/pnas.1307604110. [NGS analysis of two parallel aptamer selections starting from high-coverage pools and presenting isolation versus connectivity between peaks in the fitness landscape.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 117.Burke DH, Gold L. RNA aptamers to the adenosine moiety of S-adenosyl methionine: structural inferences from variations on a theme and the reproducibility of SELEX. Nucleic Acids Research. 1997;25:2020–2024. doi: 10.1093/nar/25.10.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gebhardt K, Shokraei A, Babaie E, Lindqvist BH. RNA aptamers to S-adenosylhomocysteine: kinetic properties, divalent cation dependency, and comparison with anti-S-adenosylhomocysteine antibody. Biochemistry. 2000;39:7255–7265. doi: 10.1021/bi000295t. [DOI] [PubMed] [Google Scholar]
- 119.Burgstaller P, Famulok M. Isolation of RNA Aptamers for Biological Cofactors by In Vitro Selection. Angew. Chem. Int. Ed. Engl. 1994;33:1084–1087. [Google Scholar]
- 120.Vu MMK, et al. Convergent evolution of adenosine aptamers spanning bacterial, human, and random sequences revealed by structure-based bioinformatics and genomic SELEX. Chem. Biol. 2012;19:1247–1254. doi: 10.1016/j.chembiol.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Salehi-Ashtiani K, Szostak JW. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature. 2001;414:82–84. doi: 10.1038/35102081. [DOI] [PubMed] [Google Scholar]
- 122.Seehafer C, Kalweit A, Steger G, Gräf S, Hammann C. From alpaca to zebrafish: hammerhead ribozymes wherever you look. RNA. 2011;17:21–26. doi: 10.1261/rna.2429911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jimenez RM, Delwart E, Lupták A. Structure-based search reveals hammerhead ribozymes in the human microbiome. J. Biol. Chem. 2011;286:7737–7743. doi: 10.1074/jbc.C110.209288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Perreault J, et al. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011;7:e1002031. doi: 10.1371/journal.pcbi.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maharjan RP, Ferenci T. Epistatic interactions determine the mutational pathways and coexistence of lineages in clonal Escherichia coli populations. Evolution. 2013;67:2762–2768. doi: 10.1111/evo.12137. [DOI] [PubMed] [Google Scholar]
- 126.Chou H-H, Delaney NF, Draghi JA, Marx CJ. Mapping the fitness landscape of gene expression uncovers the cause of antagonism and sign epistasis between adaptive mutations. PLoS Genet. 2014;10:e1004149. doi: 10.1371/journal.pgen.1004149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chiotti KE, et al. The Valley-of-Death: reciprocal sign epistasis constrains adaptive trajectories in a constant, nutrient limiting environment. 2014;104:431–437. doi: 10.1016/j.ygeno.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kvitek DJ, Sherlock G. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 2011;7:e1002056. doi: 10.1371/journal.pgen.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Corbett-Detig RB, Zhou J, Clark AG, Hartl DL, Ayroles JF. Genetic incompatibilities are widespread within species. Nature. 2013;504:135–137. doi: 10.1038/nature12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- 131.Fisher R. The Genetical Theory of Natural Selection. Oxford University Press; 1930. [Google Scholar]
- 132.Tenaillon O. The Utility of Fisher's Geometric Model in Evolutionary Genetics. Annu. Rev. Ecol. Evol. Syst. 2014;45:179–201. doi: 10.1146/annurev-ecolsys-120213-091846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Orr HA. Theories of adaptation: what they do and don't say. Genetica. 2005;123:3–13. doi: 10.1007/s10709-004-2702-3. [DOI] [PubMed] [Google Scholar]
- 134.Couce A, Tenaillon OA. The rule of declining adaptability in microbial evolution experiments. Front. Genet. 2015;6:1–6. doi: 10.3389/fgene.2015.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sanjuán R, Cuevas JM, Moya A, Elena SF. Epistasis and the adaptability of an RNA virus. Genetics. 2005;170:1001–1008. doi: 10.1534/genetics.105.040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rokyta DR, Abdo Z, Wichman HA. The genetics of adaptation for eight microvirid bacteriophages. J. Mol. Evol. 2009;69:229–239. doi: 10.1007/s00239-009-9267-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Perfeito L, Sousa A, Bataillon T, Gordo I. Rates of fitness decline and rebound suggest pervasive epistasis. Evolution. 2014;68:150–162. doi: 10.1111/evo.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moore FB, Rozen DE, Lenski RE. Pervasive compensatory adaptation in Escherichia coli. Proc. Biol. Sci. 2000;267:515–522. doi: 10.1098/rspb.2000.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.MacLean RC, Perron GG, Gardner A. Diminishing returns from beneficial mutations and pervasive epistasis shape the fitness landscape for rifampicin resistance in Pseudomonas aeruginosa. Genetics. 2010;186:1345–1354. doi: 10.1534/genetics.110.123083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bull JJ, Badgett MR, Wichman HA. Big-benefit mutations in a bacteriophage inhibited with heat. Mol. Biol. Evol. 2000;17:942–950. doi: 10.1093/oxfordjournals.molbev.a026375. [DOI] [PubMed] [Google Scholar]
- 141.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 142.Chou H-H, Chiu H-C, Delaney NF, Segrè D, Marx CJ. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science. 2011;332:1190–1192. doi: 10.1126/science.1203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Colosimo PF, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 144.Barroso-Batista J, et al. The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 2014;10:e1004182. doi: 10.1371/journal.pgen.1004182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 146.Haldane JB. A Mathematical Theory of Natural and Artificial Selection Part X. Some Theorems on Artificial Selection. Genetics. 1934;19:412–429. doi: 10.1093/genetics/19.5.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Crow JF, Kimura M. An Introduction to Population Genetics Theory. Harper and Row; 1970. [Google Scholar]
- 148.Lescat M, et al. The conserved nhaAR operon is drastically divergent between B2 and non-B2 Escherichia coli and is involved in extra-intestinal virulence. PLoS ONE. 2014;9:e108738. doi: 10.1371/journal.pone.0108738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information S1 (box): Simulation parameters used to generate data in figures 2 and 3.