Abstract
Background:
The vitamin D receptor (VDR) gene has been identified as a candidate gene for susceptibility to Parkinson's disease (PD), but results from genetic association studies to date are inconsistent. Here, we conducted a meta-analysis of published case-control studies to evaluate the association of the extensively studied VDR ApaI (G/T), BsmI (G/A), FokI (C/T), and TaqI (T/C) gene polymorphisms with risk of PD.
Methods:
Electronic search at PubMed, EMBASE, EBSCO, China National Knowledge Infrastructure, Weipu database, and Wanfang database was conducted to identify all relevant studies. Odds ratio (OR) with 95% confidence interval (CI) values was applied to evaluate the strength of the association.
Results:
A total of seven studies with 2034 PD cases and 2432 controls were included in the meta-analysis following the inclusion and exclusion criteria. Overall, no significant association between ApaI, BsmI, and TaqI gene polymorphisms and PD susceptibility in all four genetic models was found (T vs. G: OR = 1.00, 95% CI: 0.89–1.12, P = 0.97; A vs. G: OR = 0.94, 95% CI: 0.77–1.15, P = 0.53; C vs. T: OR = 1.03, 95% CI: 0.85–1.25, P = 0.77) while a significant association between FokI (C/T) and PD risk was observed (C vs. T: OR = 1.41, 95% CI: 1.14–1.75, P = 0.001; CC vs. TT: OR = 2.45, 95% CI: 1.52–3.93, P = 0.0002; CT vs. TT: OR = 2.21, 95% CI: 1.38–3.52, P = 0.0009, CC vs. CT+TT: OR = 2.32, 95% CI: 1.49–3.61, P = 0.0002).
Conclusions:
Polymorphisms of ApaI, BsmI, and TaqI may not be associated with the susceptibility to PD while the FokI (C/T) polymorphism is possibly associated with increased PD risk. However, conclusions should be cautiously interpreted due to the relatively small number of studies included.
Keywords: Meta-analysis, Parkinson's Disease, Polymorphism, Vitamin D Receptor
INTRODUCTION
Parkinson's disease (PD) is the second most common neurodegenerative disorder after Alzheimer's disease in the elderly.[1] Clinically, PD is characterized by tremor, rigidity, slowness of movement, and postural imbalance. Although its etiology is not fully understood, PD is suggested to involve interactions between a susceptible genetic background and environmental factors.[2]
Vitamin D, which is now considered a hormone rather than a vitamin, is suggested to be an environmentally modifiable factor in the pathogenesis of PD recently.[3] A long-term cohort study from Finland suggested that low serum vitamin D is a predictive factor for high risk of PD.[4] The vitamin D receptor (VDR) is the key mediator of vitamin D's functions. Upon binding to 1, 25-dihydroxy vitamin D3, the biologically active form of vitamin D, VDR is activated and interacts with vitamin D responsive elements in the promoters of vitamin D target genes to regulate their expression.[5] Interestingly, VDR is most highly expressed in dopaminergic neurons of the substantia nigra[6] and an earlier animal study with knockout of VDR gene resulted in rats with muscular and motor impairments.[7] All these suggested a role of VDR in the pathogenesis of PD. Previous results showed that variations in the VDR gene influence receptor activation by altering the affinity of the receptor for vitamin D.[8,9] Recently, Butler et al.[10] suggested that the VDR gene is a candidate gene for promoting PD and then a number of restriction fragment length polymorphisms (RFLP) in this gene have been studied.[11] Among the accumulating studies on the association between VDR gene and PD in recent years, polymorphisms of ApaI (rs7975232), BsmI (rs1544410), FokI (rs10735810), and TaqI (rs731236) have been most extensively investigated, but results to date are inconsistent. The discrepancies in findings may be attributed to small sample sizes, clinical heterogeneity, statistical power or a combination of these factors. To overcome the limitations of individual studies, we performed a meta-analysis to investigate the effects of these polymorphisms on PD.
METHODS
Literature search
Relevant publications were identified by conducting a literature search in PubMed, EMBASE, EBSCO, China National Knowledge Infrastructure, Weipu database, and Wanfang database up to January 2015, with the following terms and combinations: VDR, calcitriol receptors, polymorphism, single nucleotide polymorphism, variant, PD. The references of the identified studies and reviews were additionally screened to determine other potential eligible studies.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (1) Case–control studies; (2) Studies analyzing the association of ApaI, BsmI, FokI, and TaqI gene polymorphisms and PD risk; (3) Those reporting sufficient genotype data to calculate the odds ratio (OR) and corresponding 95% confidence interval (CI) and; (4) Cases in the control group are publication-based or hospital-based which are matched for gender and age. Studies were excluded for the following reasons: (1) Case reports or reviews; (2) Family-based studies; (3) Insufficient genotype data for calculation and; (4) Genotype distribution of controls not in Hardy-Weinberg equilibrium (HWE), indicating the chance of bias during control selection or genotyping errors. In addition, we checked the polymerase chain reaction primer sequence in GeneBank (http://www.ncbi.nlm.nih.Gov/genebank/index.html) to confirm whether the selected genes were the target genes.
Quality assessment
The quality of studies was independently assessed by the two reviewers using the Newcastle-Ottawa Scale (NOS)[12] based on three aspects: Selection, comparability, and exposure of cases and controls. NOS scores ranged from 0 to 9, and studies with a score equal to or higher than six were regarded as high quality.
Data extraction
Two investigators reviewed the publications independently and extracted the relevant information from each eligible study based on criteria in a standard data form, including name of first author, year of publication, ethnicity of the study population, number of cases and controls, matching criteria, source of controls, allele and genotype frequency, genotyping method, and evidence of HWE in controls. Disagreements between reviewers were resolved by reaching a consensus or consulting a third reviewer.
Statistical analysis
Cochrane Review Manager Version 5.3 (Cochrane Library, Oxford, UK) was used to calculate available data from each study. The pooled OR and 95% CI were calculated to measure the genetic association between VDR polymorphisms and risk of PD in four genetic models, including additive genetic model (M vs. W), codominant model (MM vs. WW and MW vs. WW), dominant model (MM + MW vs. WW) and recessive model (MM vs. MW + WW). Here, “W” and “M” represent “wild-type” and “mutant type,” respectively. The significance of OR was determined with the Z-test, in which P < 0.05 was considered statistically significant. The degree of heterogeneity between studies was detected using the Q-test and I2-statistics.[13] I2 values of 25%, 50%, and 75% were assigned as low, moderate, and high estimates, respectively. The fixed-effects model was adopted at PQ > 0.05 or I2 < 50%. Alternatively, the random-effects model was used. Sensitivity analysis was conducted by omitting each study to assess the stability of the results. Publication bias was assessed using Begger's funnel plots and Egger's linear regression test[14] by Stata 12.0 software (Stata Coporation, TX, USA). P < 0.05 was considered as statistically significant publication bias in Egger's test.
RESULTS
Characteristics of eligible studies
In total, 60 studies relevant to search keywords were identified, from which 30 repeated publications, 10 reviews and 8 irrelevant polymorphism studies were excluded, leaving 12 studies. After a full-text review, three of these studies were excluded, as they were not case-control analyses, and two were excluded owing to insufficient genotype data [Figure 1]. Ultimately, seven case-control studies involving 2034 PD patients and 2432 healthy individuals met our inclusion criteria.[15,16,17,18,19,20,21] Detailed characteristics of the eligible studies are presented in Table 1. Among these studies, four focused on ApaI,[18,19,20,21] four on BsmI,[15,17,20,21] two on FokI,[17,20] and four on TaqI gene polymorphisms.[16,18,20,21] Overall, five studies were conducted on Asian and two on Caucasian population. All genotype distributions in the controls in this meta-analysis did not deviate from HWE [Table 1].
Figure 1.
Flow chart of study selection.
Table 1.
Characteristics of the eligible studies
| Author, year | Ethnicity | Sample size | SNP | Case | Control | Genotyping method | HWE (P) | Control from | NOS | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case/control | MM | MW | WW | MM | MW | WW | |||||||
| Kim et al., 2005 | Asian | 85/231 | BsmI (G > A) | 2 | 11 | 72 | 3 | 60 | 168 | PCR-RFLP | 0.357 | HB | 7 |
| Lv et al., 2013 | Asian | 483/498 | TaqI (T > C) | 0 | 46 | 437 | 0 | 52 | 446 | PCR-RFLP | 0.219 | PB | 8 |
| Han et al., 2012 | Asian | 260/282 | BsmI (G > A) | 4 | 34 | 222 | 2 | 36 | 244 | PCR-RFLP | 0.599 | HB | 7 |
| FokI (T > C) | 114 | 124 | 22 | 109 | 126 | 47 | 0.306 | ||||||
| Liu et al., 2013 | Asian | 285/285 | ApaI (G > T) | 20 | 135 | 130 | 24 | 112 | 149 | PCR-RFLP | 0.651 | PB | 7 |
| TaqI (T > C) | 0 | 33 | 252 | 0 | 30 | 255 | 0.348 | ||||||
| Lin et al., 2014 | Asian | 700/792 | ApaI (G > T) | 105 | 361 | 234 | 141 | 401 | 250 | TaqMan | 0.365 | HB, PB | 6 |
| Török et al., 2013 | Caucasian | 100/109 | ApaI (G > T) | 42 | 43 | 15 | 42 | 46 | 21 | PCR-RFLP | 0.198 | HB | 7 |
| BsmI (G > A) | 24 | 49 | 27 | 25 | 57 | 27 | 0.629 | ||||||
| FokI (T > C) | 42 | 48 | 10 | 35 | 49 | 25 | 0.330 | ||||||
| TaqI (T > C) | 17 | 48 | 35 | 16 | 46 | 47 | 0.394 | ||||||
| Petersen et al., 2014 | Caucasian | 121/235 | ApaI (G > T) | 34 | 62 | 25 | 58 | 120 | 56 | TaqMan | 0.694 | HB | 6 |
| BsmI (G > A) | 20 | 53 | 48 | 34 | 117 | 84 | 0.510 | ||||||
| TaqI (T > C) | 20 | 54 | 47 | 34 | 119 | 81 | 0.360 | ||||||
SNP: Single nucleotide polymorphism; PB: Population-based; HB: Hospital-based; HWE: Hardy-Weinberg equilibrium; NOS: Newcastle-Ottawa Scale; PCR-RFLP: Polymerase chain reaction-restriction fragment length polymorphism.
Quantitative synthesis
The results of the assessment of correlations between VDR polymorphisms and PD are shown in Tables 2 and 3. Meta-analysis was performed for a combined Asian and Caucasian population and by each individual ethnic category. In overall population, no evidence of significant association between ApaI (G/T), BsmI (G/A) or TaqI (T/C) polymorphisms and PD was observed under all genetic models [Table 2]. For FokI (C/T) polymorphism, we found it was significantly associated with PD risk under the additive, codominant model and dominant model (C vs. T: OR = 1.41, 95% CI: 1.14–1.75, P = 0.001; CC vs. TT: OR = 2.45, 95% CI: 1.52–3.93, P = 0.0002; CT vs. TT: OR = 2.21, 95% CI: 1.38–3.52, P = 0.0009, CC vs. CT+TT: OR = 2.32, 95% CI: 1.49–3.61, P = 0.0002).
Table 2.
Meta-analysis of the association between VDR gene polymorphism and PD
| SNP | Population | Number of studies | Model | Test of association | Heterogeneity | Egger’s test | ||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | I2 (%) | P | ||||
| ApaI | Overall | 4 | T versus G | 1.00 | 0.89–1.12 | 0.97 | 24 | 0.06 |
| Asian | 2 | TT versus GG | 0.93 | 0.73–1.18 | 0.54 | 5 | 0.10 | |
| Caucasian | 2 | TG versus GG | 1.09 | 0.92–1.30 | 0.32 | 7 | 0.43 | |
| TT + TG versus GG | 1.05 | 0.89–1.25 | 0.53 | 23 | 0.27 | |||
| TTversus TG + GG | 0.91 | 0.74–1.13 | 0.40 | 0 | 0.36 | |||
| BsmI | Overall | 4 | A versus G | 0.94 | 0.77–1.15 | 0.53 | 12 | 0.51 |
| Asian | 2 | AA versus GG | 1.09 | 0.69–1.74 | 0.70 | 0 | 0.12 | |
| Caucasian | 2 | AG versus GG | 0.78 | 0.59–1.03 | 0.08 | 28 | 0.37 | |
| AA + AG versus GG | 0.83 | 0.64–1.08 | 0.17 | 24 | 0.39 | |||
| AA versus AG + GG | 1.19 | 0.79–1.80 | 0.40 | 0 | 0.06 | |||
| FokI | Overall | 2 | C versus T | 1.41 | 1.14–1.75 | 0.001 | 0 | – |
| Asian | 1 | CC versus TT | 2.45 | 1.52–3.93 | 0.0002 | 0 | – | |
| Caucasian | 1 | CT versus TT | 2.21 | 1.38–3.52 | 0.0009 | 0 | – | |
| CC + CT versus TT | 2.32 | 1.49–3.61 | 0.0002 | 0 | – | |||
| CC versus CT + TT | 1.31 | 0.98–1.76 | 0.0700 | 0 | – | |||
| TaqI | Overall | 4 | C versus T | 1.03 | 0.85–1.25 | 0.77 | 0 | 0.62 |
| Asian | 2 | CC versus TT | 1.16 | 0.70–1.93 | 0.57 | 0 | 0.94 | |
| Caucasian | 2 | CT versus TT | 0.98 | 0.77–1.26 | 0.89 | 0 | 0.19 | |
| CC + CT versus TT | 1.00 | 0.79–1.27 | 0.99 | 0 | 0.11 | |||
| CC versus CT + TT | 1.18 | 0.74–1.88 | 0.50 | 0 | 0.36 | |||
OR: Odds ratio; CI: Confidence interval; VDR: Vitamin D receptor; PD: Parkinson’s disease; SNP: Single nucleotide polymorphism.
Table 3.
Subgroup analysis of the association between the VDR gene polymorphism and PD
| SNP | Model | Asian | Caucasian | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | I2 (%) | OR | 95% CI | P | I2 (%) | ||
| ApaI | T versus G | 0.96 | 0.85–1.09 | 0.53 | 55 | 1.15 | 0.90–1.47 | 0.25 | 0 |
| TT versus GG | 0.82 | 0.62–1.09 | 0.17 | 0 | 1.35 | 0.82–2.21 | 0.24 | 5 | |
| TG versus GG | 1.07 | 0.89–1.30 | 0.46 | 66 | 1.21 | 0.76–1.91 | 0.42 | 7 | |
| TT + TG versus GG | 1.02 | 0.85–1.23 | 0.81 | 67 | 1.26 | 0.82–1.93 | 0.30 | 0 | |
| TT versus TG + GG | 0.82 | 0.63–1.05 | 0.11 | 0 | 1.17 | 0.81–1.70 | 0.40 | 0 | |
| BsmI | A versus G | 0.88 | 0.62–1.25 | 0.47 | 69 | 0.97 | 0.76–1.24 | 0.26 | 0 |
| AA versus GG | 1.89 | 0.55–6.46 | 0.31 | 0 | 1.00 | 0.61–1.65 | 1.00 | 0 | |
| AG versus GG | 0.74 | 0.50–1.11 | 0.14 | 76 | 0.82 | 0.55–1.20 | 0.30 | 0 | |
| AA + AG versus GG | 0.80 | 0.55–1.18 | 0.26 | 75 | 0.86 | 0.60–1.24 | 0.42 | 0 | |
| AA versus AG + GG | 2.03 | 0.59–6.98 | 0.26 | 0 | 1.12 | 0.72–1.73 | 0.62 | 0 | |
| FokI | C versus T | 1.34 | 1.04–1.72 | – | – | 1.61 | 1.09–2.40 | – | – |
| CC versus TT | 2.23 | 1.26–3.95 | – | – | 3.00 | 1.27–7.08 | – | – | |
| CT versus TT | 2.10 | 1.19–3.69 | – | – | 2.50 | 1.06–5.64 | – | – | |
| CC + CT versus TT | 2.16 | 1.26–3.70 | – | – | 2.68 | 1.21–5.90 | – | – | |
| CC versus CT + TT | 1.24 | 0.88–1.75 | – | – | 1.53 | 0.87–2.69 | – | – | |
| TaqI | C versus T | 0.98 | 0.71–1.35 | 0.90 | 0 | 1.06 | 0.83–1.36 | 0.64 | 7 |
| CC versus TT | – | – | – | 0 | 1.16 | 0.70–1.93 | 0.57 | 0 | |
| CT versus TT | 0.98 | 0.71–1.36 | 0.90 | 0 | 0.99 | 0.77–1.26 | 0.89 | 55 | |
| CC + CT versus TT | 0.98 | 0.71–1.36 | 0.90 | 0 | 1.03 | 0.72–1.46 | 0.88 | 51 | |
| CC versus CT + TT | – | – | – | 0 | 1.18 | 0.74–1.88 | 0.50 | 0 | |
OR: Odds ratio; CI: Confidence interval; VDR: Vitamin D receptor; PD: Parkinson’s disease; SNP: Single nucleotide polymorphism.
We further performed a subgroup analysis according to ethnicity. Consistently, ethnicity-specific analysis revealed no association between ApaI, BsmI or TaqI polymorphisms and PD risk while a significant association between FokI (C/T) polymorphisms and PD was observed in both Asian and Caucasian population [Table 3]. A sensitivity analysis was additionally conducted in which one study at a time was removed. The results showed no significant alterations in pooled OR and 95% CI values, indicating that our analysis is statistically robust.
Heterogeneity and publication bias
No between-study heterogeneity was found in analyses of the ApaI, BsmI or TaqI polymorphisms in the combined population. Visual inspection of funnel plots did not find any evidence of funnel plot asymmetry [Figure 2]. Statistical result of Egger's test till did not show publication bias among studies [Table 2].
Figure 2.
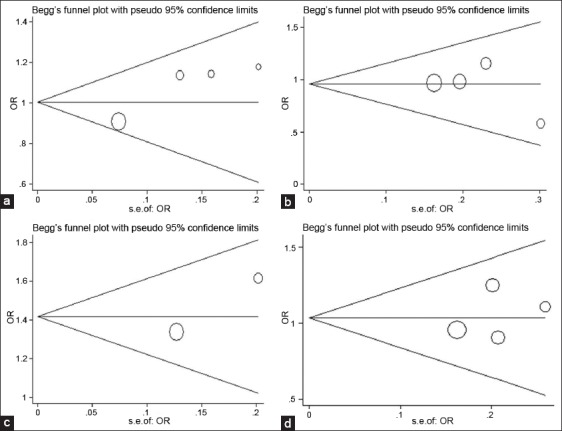
Funnel plots for vitamin D receptor gene polymorphisms and Parkinson's disease ([a] T vs. G of ApaI; [b] A vs. G of BsmI; [c] C vs. T of FokI; [d] C vs. T of TaqI).
DISCUSSION
Although the multifactorial nature of PD is well-recognized, genetic factors are considered the significant determinants. Numerous genes have been studied in connection with PD, and the VDR gene has been one of the research hotspots recently.[22] It is suggested that genetic polymorphisms in the VDR gene alter the affinity of the receptor for vitamin D, which may be an environmentally modifiable factor in the pathogenesis of PD. Given the potential association between VDR and PD, VDR polymorphisms affecting its expression have been analyzed to determine their potential effects on PD. The human VDR gene contains eight exons and seven introns spanning >75 kb DNA on chromosome 12q12.[23] To date, over 60 polymorphisms have been identified for VDR genes. These subtle DNA sequence variations, which often occur in the population, have a modest but real biological effect.[24] Among these, ApaI, BsmI, FokI, and TaqI polymorphisms have been most intensively investigated in relation to PD as well as other neurodegenerative diseases. However, controversial results have been obtained.
To establish the association of VDR ApaI, BsmI, FokI, and TaqI polymorphisms with risk of PD, we performed a meta-analysis of seven studies (including 2034 PD cases and 2432 controls). We failed to detect any association between Apal, Bsml, Taql polymorphisms and PD susceptibility in all four genetic models. Ethnicity-specific analysis additionally revealed no association between them. A previous study suggested strong linkage disequilibrium for BsmI, ApaI, and TaqI RFLPs,[11] which may explain the high degree of consistency between these three polymorphisms and their link to PD susceptibility. These three polymorphisms are located at the 3’end of the VDR gene neighboring the 3’UTR region, which do not result in changes in the predicted amino acid sequence of the VDR.[25] However, one thing we should pay attention to is that the serum vitamin D level, which most of the included studies did not measure, may influence the effect size of a VDR polymorphism. In a study analyzing association between VDR polymorphisms and incidence of Type I diabetes mellitus, the OR of the risk allele increasing with higher vitamin D levels in the studied population.[26] A recent report that the positive association between VDR genetic variants and multiple sclerosis was found in studies conducted in regions at lower latitudes but rather than higher latitudes.[27] We know that the duration of sun exposure can change active vitamin D levels. Therefore, it is important to include measurement of serum vitamin D levels in future studies to better characterize the genetic effects of VDR polymorphisms in PD.
Our data suggest that FokI (C/T) polymorphism is significantly associated with PD and the C allele may increase the risk of PD. The FokI polymorphism is located in exon 2 at the 5’ coding region of the gene. The functional FokI polymorphism is the most important start codon polymorphism in VDR gene.[28] Previous studies by Gross et al.[29] suggested that VDR encoded by the C allele is three amino acids shorter than that encoded by the T allele. Differences in length may result in altered VDR function.[25,30] However, since there are only two studies on the FokI (C/T) gene polymorphism have been documented, one in an Asian and the other in a Caucasian population. Although the results from both investigations were consistent, data should be carefully interpreted because of the relatively small sample sizes. Additional studies with larger sample sizes are necessary to verify the role of FokI polymorphisms in PD.
Several limitations of our investigation merit comment. First, the number of cases and controls involved in the meta-analysis were limited, which could increase the probability of false positives or false negatives. Second, although publication bias among studies were not detected, and the quality of our studies was good (NOS≥6), the possibility of bias could not be eliminated because of the relatively small sample size. Third, factors such as gene-gene or gene-environment interactions may influence the gene-disease factor. However, the lack of individual data from the included studies limited further evaluation of these potential interactions.
CONCLUSION
Data from our meta-analysis suggest that the ApaI, BsmI, and TaqI gene polymorphisms are not significantly associated with PD while the FokI (C/T) polymorphism may increase PD risk. Given the limitations mentioned above, further studies with more specific common information and larger sample sizes are warranted. Furthermore, additional investigations are required to determine the biological mechanisms underlying the observed associations and their potential relationship with PD.
Footnotes
Edited by: Li-Shao Guo
Source of Support: This work was supported by a grant from the Natural Science Foundation of Shandong Province, China (No. ZR2012CM028).
Conflict of Interest: None declared.
REFERENCES
- 1.Kempster PA, Hurwitz B, Lees AJ. A new look at James Parkinson's essay on the shaking palsy. Neurology. 2007;69:482–5. doi: 10.1212/01.wnl.0000266639.50620.d1. [DOI] [PubMed] [Google Scholar]
- 2.Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide BM, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: The PDGene database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evatt ML, DeLong MR, Kumari M, Auinger P, McDermott MP, Tangpricha V, et al. High prevalence of hypovitaminosis D status in patients with early Parkinson disease. Arch Neurol. 2011;68:314–9. doi: 10.1001/archneurol.2011.30. [DOI] [PubMed] [Google Scholar]
- 4.Knekt P, Kilkkinen A, Rissanen H, Marniemi J, Sääksjärvi K, Heliövaara M. Serum vitamin D and the risk of Parkinson disease. Arch Neurol. 2010;67:808–11. doi: 10.1001/archneurol.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcion E, Wion-Barbot N, Montero-Menei CN, Berger F, Wion D. New clues about vitamin D functions in the nervous system. Trends Endocrinol Metab. 2002;13:100–5. doi: 10.1016/s1043-2760(01)00547-1. [DOI] [PubMed] [Google Scholar]
- 6.Eyles DW, Smith S, Kinobe R, Hewison M, McGrath JJ. Distribution of the vitamin D receptor and 1 alpha-hydroxylase in human brain. J Chem Neuroanat. 2005;29:21–30. doi: 10.1016/j.jchemneu.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Burne TH, McGrath JJ, Eyles DW, Mackay-Sim A. Behavioural characterization of vitamin D receptor knockout mice. Behav Brain Res. 2005;157:299–308. doi: 10.1016/j.bbr.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Bouillon R, Carmeliet G, Daci E, Segaert S, Verstuyf A. Vitamin D metabolism and action. Osteoporos Int. 1998;8 Suppl 2:S13–9. doi: 10.1007/pl00022727. [DOI] [PubMed] [Google Scholar]
- 9.Cai Q, Chandler JS, Wasserman RH, Kumar R, Penniston JT. Vitamin D and adaptation to dietary calcium and phosphate deficiencies increase intestinal plasma membrane calcium pump gene expression. Proc Natl Acad Sci U S A. 1993;90:1345–9. doi: 10.1073/pnas.90.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler MW, Burt A, Edwards TL, Zuchner S, Scott WK, Martin ER, et al. Vitamin D receptor gene as a candidate gene for Parkinson disease. Ann Hum Genet. 2011;75:201–10. doi: 10.1111/j.1469-1809.2010.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–56. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 13.Zintzaras E, Ioannidis JP. HEGESMA: Genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 14.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JS, Kim YI, Song C, Yoon I, Park JW, Choi YB, et al. Association of vitamin D receptor gene polymorphism and Parkinson's disease in Koreans. J Korean Med Sci. 2005;20:495–8. doi: 10.3346/jkms.2005.20.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv Z, Tang B, Sun Q, Yan X, Guo J. Association study between vitamin d receptor gene polymorphisms and patients with Parkinson disease in Chinese Han population. Int J Neurosci. 2013;123:60–4. doi: 10.3109/00207454.2012.726669. [DOI] [PubMed] [Google Scholar]
- 17.Han X, Xue L, Li Y, Chen B, Xie A. Vitamin D receptor gene polymorphism and its association with Parkinson's disease in Chinese Han population. Neurosci Lett. 2012;525:29–33. doi: 10.1016/j.neulet.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 18.Liu HX, Han X, Zheng XP, Li YS, Xie AM. Association of vitamin D receptor gene polymorphisms with Parkinson disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2013;30:13–6. doi: 10.3760/cma.j.issn.1003-9406.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Lin CH, Chen KH, Chen ML, Lin HI, Wu RM. Vitamin D receptor genetic variants and Parkinson's disease in a Taiwanese population. Neurobiol Aging. 2014;35:1212e11–3. doi: 10.1016/j.neurobiolaging.2013.10.094. [DOI] [PubMed] [Google Scholar]
- 20.Török R, Török N, Szalardy L, Plangar I, Szolnoki Z, Somogyvari F, et al. Association of vitamin D receptor gene polymorphisms and Parkinson's disease in Hungarians. Neurosci Lett. 2013;551:70–4. doi: 10.1016/j.neulet.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Petersen MS, Bech S, Christiansen DH, Schmedes AV, Halling J. The role of vitamin D levels and vitamin D receptor polymorphism on Parkinson's disease in the Faroe Islands. Neurosci Lett. 2014;561:74–9. doi: 10.1016/j.neulet.2013.12.053. [DOI] [PubMed] [Google Scholar]
- 22.Peterson AL. A review of vitamin D and Parkinson's disease. Maturitas. 2014;78:40–4. doi: 10.1016/j.maturitas.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 23.McDonnell DP, Pike JW, O’Malley BW. The vitamin D receptor: A primitive steroid receptor related to thyroid hormone receptor. J Steroid Biochem. 1988;30:41–6. doi: 10.1016/0022-4731(88)90074-x. [DOI] [PubMed] [Google Scholar]
- 24.Köstner K, Denzer N, Müller CS, Klein R, Tilgen W, Reichrath J. The relevance of vitamin D receptor (VDR) gene polymorphisms for cancer: A review of the literature. Anticancer Res. 2009;29:3511–36. [PubMed] [Google Scholar]
- 25.Zmuda JM, Cauley JA, Ferrell RE. Molecular epidemiology of vitamin D receptor gene variants. Epidemiol Rev. 2000;22:203–17. doi: 10.1093/oxfordjournals.epirev.a018033. [DOI] [PubMed] [Google Scholar]
- 26.Ponsonby AL, Pezic A, Ellis J, Morley R, Cameron F, Carlin J, et al. Variation in associations between allelic variants of the vitamin D receptor gene and onset of type 1 diabetes mellitus by ambient winter ultraviolet radiation levels: A meta-regression analysis. Am J Epidemiol. 2008;168:358–65. doi: 10.1093/aje/kwn142. [DOI] [PubMed] [Google Scholar]
- 27.Smolders J, Peelen E, Thewissen M, Menheere P, Tervaert JW, Hupperts R, et al. The relevance of vitamin D receptor gene polymorphisms for vitamin D research in multiple sclerosis. Autoimmun Rev. 2009;8:621–6. doi: 10.1016/j.autrev.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Ferrari S, Rizzoli R, Manen D, Slosman D, Bonjour JP. Vitamin D receptor gene start codon polymorphisms (FokI) and bone mineral density: Interaction with age, dietary calcium, and 3’-end region polymorphisms. J Bone Miner Res. 1998;13:925–30. doi: 10.1359/jbmr.1998.13.6.925. [DOI] [PubMed] [Google Scholar]
- 29.Gross C, Krishnan AV, Malloy PJ, Eccleshall TR, Zhao XY, Feldman D. The vitamin D receptor gene start codon polymorphism: A functional analysis of FokI variants. J Bone Miner Res. 1998;13:1691–9. doi: 10.1359/jbmr.1998.13.11.1691. [DOI] [PubMed] [Google Scholar]
- 30.Arai H, Miyamoto K, Taketani Y, Yamamoto H, Iemori Y, Morita K, et al. A vitamin D receptor gene polymorphism in the translation initiation codon: Effect on protein activity and relation to bone mineral density in Japanese women. J Bone Miner Res. 1997;12:915–21. doi: 10.1359/jbmr.1997.12.6.915. [DOI] [PubMed] [Google Scholar]


