Abstract
Background:
Mitochondrial dysfunction is linked to the pathogenesis of Parkinson's disease (PD). However, the precise role of mitochondrial DNA (mtDNA) variations is obscure. On the other hand, mtDNA haplogroups have been inconsistently reported to modify the risk of PD among different population. Here, we try to explore the relationship between mtDNA haplogroups and sporadic PD in a Han Chinese population.
Methods:
Nine single-nucleotide polymorphisms, which define the major Asian mtDNA haplogroups (A, B, C, D, F, G), were detected via polymerase chain reaction-restriction fragment length polymorphism or denaturing polyacrylamide gel electrophoresis in 279 sporadic PD patients and 510 matched controls of Han population.
Results:
Overall, the distribution of mtDNA haplogroups did not show any significant differences between patients and controls. However, after stratification by age at onset, the frequency of haplogroup B was significantly lower in patients with early-onset PD (EOPD) compared to the controls (odds ratio [OR] =0.225, 95% confidence interval [CI]: 0.082–0.619, P = 0.004), while other haplogroups did not show significant differences. After stratification by age at examination, among subjects younger than 50 years of age: Haplogroup B also showed a lower frequency in PD cases (OR = 0.146, 95% CI: 0.030–0.715, P = 0.018) while haplogroup D presented a higher risk of PD (OR = 3.579, 95% CI: 1.112–11.523, P = 0.033), other haplogroups also did not show significant differences in the group.
Conclusions:
Our study indicates that haplogroup B might confer a lower risk for EOPD and people younger than 50 years in Han Chinese, while haplogroup D probably lead a higher risk of PD in people younger than 50 years of age. In brief, particular Asian mtDNA haplogroups likely play a role in the pathogenesis of PD among Han Chinese.
Keywords: Han Chinese, Haplogroups, Mitochondrial DNA, Parkinson's Disease
INTRODUCTION
Parkinson's disease (PD) is a common neurodegenerative disorder which affects about 1.7% of the population over the age of 65 in China.[1] Although the pathological character and clinical features of PD have been clarified, the pathogenesis of idiopathic PD which accounts for 90% of PD is currently unclear.[2]
In the past two decades, increasing evidence suggests that mitochondrial dysfunction is associated with the cause of PD. It has been shown in animal models that initially, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine, an inhibitor of complex I, could give rise to Parkinsonism.[3] In subsequent studies, dysfunction of complex I was found in platelets[4] and substantia nigra[5] of PD patients. The reduction of complex I activity was also detected in cybrids derived from PD patients, pointing to a mitochondrial DNA (mtDNA) impairment.[6]
Interestingly, increasing evidence indicates that a combination of mitochondrial single nucleotide polymorphisms (mtSNPs) might confer susceptibility to PD. MtDNA is a haploid nonrecombining genome. The mtSNPs occur in lineages and are always accompanied by some other polymorphisms. The increased risk of a phenotype could be attributable to a combination of polymorphisms rather than a single polymorphism. According to the available research, the distribution of mtDNA haplogroups varies with geographical regions or population.[7] The classic mtDNA haplogroups includes H, I, J, K, T, U, V, W, X in Europeans, while major mtDNA haplogroups involve A, B, C, D, E, F, G, P, Q, Y, Z in Asians. MtDNA haplogroups J and K, which both share A10398 G, appeared as a protective effect against PD among European population;[8] inconsistent results were found in the English,[9] Italian[10] population; while even a new work denied the association between European mtDNA haplogroups and PD in Spanish.[11] Besides, the reduced risk of PD with haplogroups J, K, and T was mirrored by an increased risk of PD in super-haplogroup HV in Caucasian.[12]
To the best of our knowledge, investigations concerning the role of Asian mtDNA haplogroups in the risk of PD for Han Chinese are few. The Association of Asian mtDNA haplogroups and PD is not clear yet. In this study, we employed nine SNPs representing the major Asian mtDNA haplogroups (A, B, C, D, F, G) to explore the relationships between these six haplogroups and the risk of PD in Han Chinese.
METHODS
Samples
Totally, 279 nonconsanguineous patients were recruited in our study from the Neurology Department in The First Affiliated Hospital of Fujian Medical University and Quanzhou First Hospital from 2007 to 2011. The diagnosis of PD was based on the clinical diagnostic criteria of the UK PD Society Brain Bank[13] and the Chinese PD criteria.[14] The exclusion criteria included: (1) Cases with a family history of PD or other neurodegenerative disorders; (2) cases with secondary Parkinsonism due to trauma, cerebrovascular accident or non-PD neurodegenerative syndromes. The enrolled affected individuals consisted of 174 males and 105 females, with a mean age at onset (AAO) of 58.02 ± 10.73 years (19–83 years), and a mean age at examination (AAE) of 62.18 ± 10.40 years (31–85 years). On the basis of AAO, patients were divided into two groups:[15] early-onset PD (EOPD) with AAO ≤50 years (63 cases, AAE 48.41 ± 8.14 years, 60.3% males) and late-onset PD (LOPD) with AAO >50 years (216 cases, AAE 66.19 ± 7.00 years, 63.0% males). AAE was defined as the age at which the participant was clinically examined and enrolled in the study. AAO was defined as the age at which the patient first noticed the manifestation of PD.
Matched with age and gender, 510 unaffected individuals were randomly selected from the local people. Evaluated by a consultant neurologist, controls had no signs or family history of cognitive or neurological disorders. The healthy group composed 304 males and 206 females, and the mean AAE was 61.97 ± 11.14 years (30–86 years). For a case–control study, controls were also divided into two groups: Control team 1 (CT1) (118 subjects, AAE 47.27 ± 8.97 years, 53.4% males) and control team 2 (CT2) (392 subjects, AAE 66.39 ± 7.25 years, 61.5% males), matched with EOPD and LOPD, respectively.
All the participants were with the same nationality-Han Chinese, the purpose of which was to minimize a race-mixture and avoid a bias of results. Our investigation was approved by the medical ethics committee, and all participants offered informed consents.
Assignment of haplogroups
Nine SNPs were employed in our study to identify the major Asian mtDNA haplogroups. According to MITOMAP (Human Mitochondrial Genome Database; http://www.Mitomap.org) and previous research,[16,17,18] the diagnostic SNPs and corresponding haplogroups are listed in Figure 1.
Figure 1.
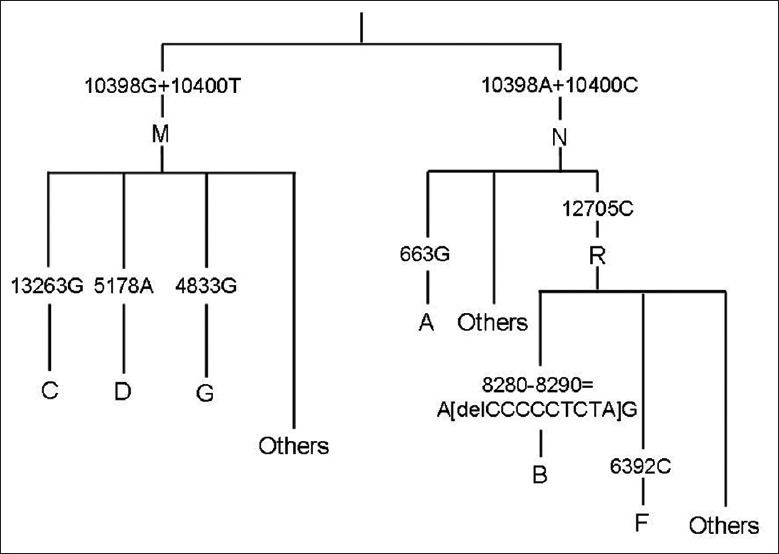
Classification of major Asian mitochondrial DNA haplogoups.
Single-nucleotide polymorphisms genotyping
For all subjects, genomic DNA was extracted from peripheral blood sample using a QIAamp@ DNA kit. Nine mtDNA SNPs classifying different haplogroups were analyzed by polymerase chain reaction-restriction fragment length polymorphism [Table 1]. Briefly, eight pairs of primers were designed based on a revised version of the Cambridge reference sequences[19] in order to amplify specific fragments. Digested by relevant restriction enzymes, the products were electrophoresed through a 2.5% agarose gel. In particular, the SNP 8280–8290 = A[delCCCCCTCTA] G, which is surveyed to determine the haplogroup B, was detected by 8% denaturing polyacrylamide gel electrophoresis without restriction enzyme digestion [Figures 2–4].
Table 1.
Primers and restriction enzymes for defining major Asian mtDNA haplogroups
| SNP | Primer (5’→3’) | Annealing (°C) | Restriction enzymes | Fragment (bp) |
|---|---|---|---|---|
| A10398A+C10400C | F: ACCTGCCACTAATAGTTATGTC | 64 | AluI | 295+20 |
| A10398G+C10400T | R: TGTTGAGGGTTATGAGAGTAGC | 202+93+20 | ||
| A13263G | F: TAGTTGTAGCAGGAATCTTC | 60 | AluI | A: 490 |
| R: GCGATGAGAGTAATAGATAG | G: 316+174 | |||
| C5178A | F: AGCAGTTCTACCGTACAACC | 60 | AluI | C: 383+137 |
| R: ACTTACTGAGGGCTTTGAAG | A: 520 | |||
| A4833G | F: AATAAACCCTCGTTCCACAG | 60 | HhaI | A: 585 |
| R: GTGTTAGTCATGTTAGCTTG | G: 360+225 | |||
| A663G | F: TGGCCACGCACTTAAACAC | 58 | HaeIII | A: 572 |
| R: TGGCACGAAATTGACCAACC | G: 345+227 | |||
| C12705T* | F: TCAGTTCTTCAAATATCTACTGAT | 50 | MboI | C: 158+22 |
| R: TTGTATAGGATGCTTGAATGG | T: 180 | |||
| T6392C* | F: AGGAACAGGTTGAACAGTCTAC | 50 | MunI | T: 118+24 |
| R: ATATTGATAATTGTTGTGATGCA | C: 142 | |||
| 8280–8290=A[delCCCCCTCTA] G | F: AGTTTCATGCCCATCGTCC | 60 | 165/156 | |
| R: ACTGTAAAGAGGTGTTGGTTC |
*The restriction site was detected using a mismatched oligonucleotide. The mismatched nucleotide was underlined. mtDNA: Mitochondrial DNA; SNP: Single-nucleotide polymorphism.
Figure 2.
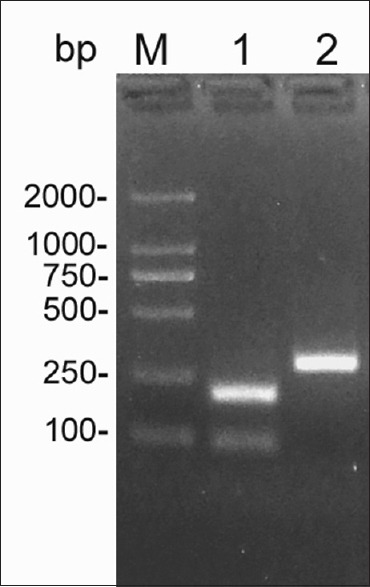
Polymerase chain reaction-restriction fragment length polymorphism products of mitochondrial DNA single nucleotide polymorphism A10398G and C10400T analyzed with agarose gel electrophoresis. Lane M: D2000 marker; Lane 1: 10398G and 10400T; Lane 2: 10398A and10400C.
Figure 4.
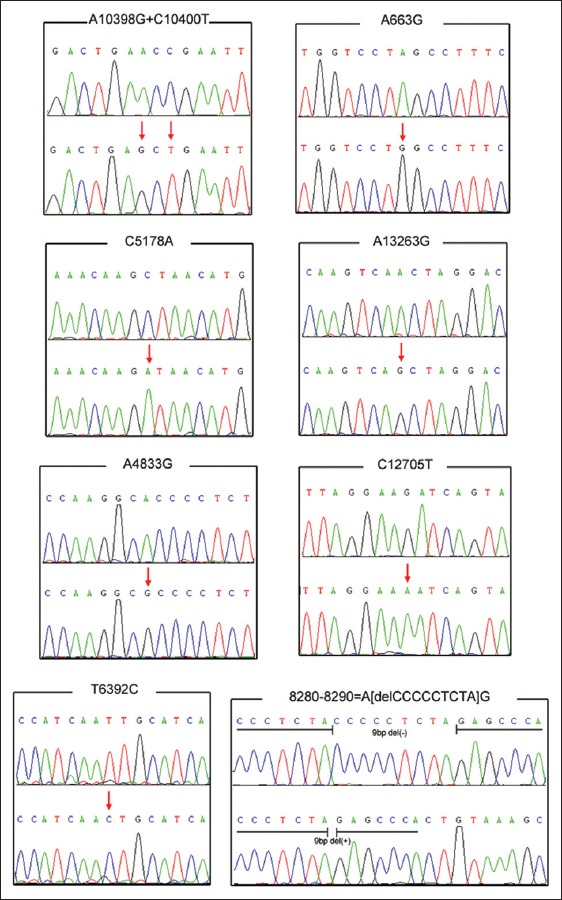
Direct sequencing of the mitochondrial DNA fragments to confirm the alleles of each single nucleotide polymorphism. *Forward sequencing for all of the polymerase chain reaction-amplified fragments except the one encompassing C12705T which was detected by reverse sequencing.
Figure 3.
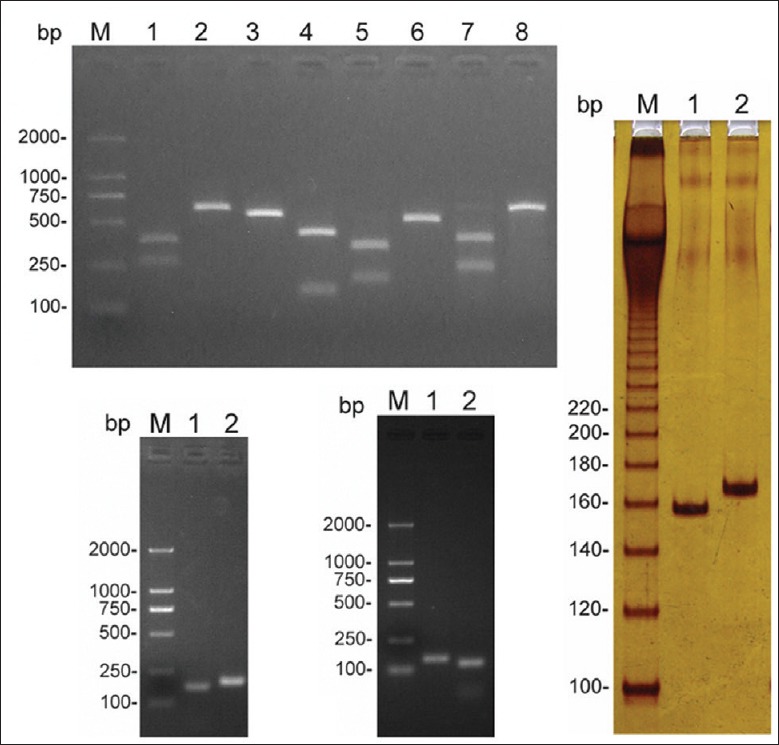
Polymerase chain reaction-restriction fragment length polymorphism analysis of mitochondrial DNA single nucleotide polymorphism A663G, C5178A, A13263G, A4833G, C12705T, T6392C, and 8280–8290 = A[delCCCCCTCTA] G. (a) Lane M: D2000 marker; Lane 1: 663G; Lane 2: 663A; Lane 3: 5178A; Lane 4: 5178C; Lane 5: 13263G; Lane 6: 13263A; Lane 7: 4833G; Lane 8: 4833A; (b) Lane M: D2000 marker; Lane 1: 12705C; Lane 2: 12705T; (c) Lane M: D2000 marker; Lane 1: 6392C; Lane 2: 6392T; (d) Lane M: Marker-20-bp Ladder; Lane 1: 9-bp Del (+); Lane 2: 9-bp Del (−).
Statistical analysis
All statistical analyses were performed using SPSS software, version 13.0 (SPSS Inc, Chicago, USA). Statistical significance was established at P < 0.05 and based on two-tailed 5% level. To compare the AAE and gender between the two groups, we adopted an independent-samples t-test and Chi-square test, respectively. As appropriate, we used two-tailed Fisher's exact test or χ2 test with Yates’ correction to assess the differences in frequency of each haplogroup between PD cases and unaffected subjects, while each haplogroup was compared to all other haplogroups pooled into one group. To evaluate the risk of PD for each haplogroup or mtSNPs, we calculated odds ratios (ORs) and associated 95% confidence intervals (CIs) by binary logistic regression analysis. Meanwhile, to adjust for potential confounding, AAE and gender were also brought into the analysis as covariates.
RESULTS
Baseline
No significant differences were found in AAE (t = 0.255, P = 0.798) or gender (χ2 = 0.574, P = 0.449) between patients and healthy controls. Furthermore, each case-control group was statistically comparable in both AAE and gender (EOPD vs. CT1: t = 0.842, P = 0.401; χ2 = 0.799, P = 0.371. LOPD vs. CT2: t = −0.335, P = 0.738; χ2 = 0.130, P = 0.718).
Distribution of mitochondrial DNA haplogroups
The distribution of the six major Asian mtDNA haplogroups is listed in Table 2. In the healthy controls, the distribution of haplogroup A, B, C, D, F, and G accounted for 4.3%, 21.2%, 3.7%, 13.9%, 15.7%, and 2.9%, respectively, which echoes similar results with MITOMAP and previous reports.[20] In addition, 39.1% of patients and 38.2% of healthy controls were assigned to the “others” group, which could not be classified to any of the six representative Asian haplogroups. However, no significant difference in the distribution of mtDNA haplogroups was found between the PD patients and controls (χ2 = 3.872, P = 0.694). Similarly, we did not find any difference after stratification by gender (males: χ2 = 2.927, P = 0.818; females: Fisher's exact test, P = 0.669) [Table 3].
Table 2.
Distribution of mtDNA haplogroups between PD patients and controls, n (%)
| Haplogroup* | PD (n = 279) | Control (n = 510) |
|---|---|---|
| A | 16 (5.7) | 22 (4.3) |
| B | 46 (16.5) | 108 (21.2) |
| C | 8 (2.9) | 19 (3.7) |
| D | 43 (15.4) | 71 (13.9) |
| F | 47 (16.8) | 80 (15.7) |
| G | 10 (3.6) | 15 (2.9) |
| Others | 109 (39.1) | 195 (38.2) |
*Chi-square test for the overall haplogroup distribution: χ2 = 3.872, P = 0.694. mtDNA: Mitochondrial DNA; PD: Parkinson’s disease.
Table 3.
Distribution of mtDNA haplogroups after stratification by gender
| Subjects | Haplogroup (n, %) | ||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | F | G | Others | |
| Males* | |||||||
| PD (n=174) | 12 (6.9) | 31 (17.8) | 5 (2.9) | 29 (16.7) | 27 (15.5) | 8 (4.6) | 62 (35.6) |
| Control (n=304) | 14 (4.6) | 64 (21.1) | 8 (2.6) | 46 (15.1) | 46 (15.1) | 9 (3.0) | 117 (38.5) |
| Females† | |||||||
| PD (n=105) | 4 (3.8) | 15 (14.3 | 3 (2.9) | 14 (13.3) | 20 (19.0) | 2 (1.9) | 47 (44.8) |
| Control (n=206) | 8 (3.9) | 44 (21.4) | 11 (5.3) | 25 (12.1) | 34 (16.5) | 6 (2.9) | 78 (37.9) |
*Chi-square test: χ2 = 2.927, P = 0.818; †Fisher’s exact test: P = 0.669. mtDNA: Mitochondrial DNA; PD: Parkinson’s disease.
Significant differences regarding age at onset and age at examination
After stratification by AAO, a significant difference was found in the distribution of haplogroups between the EOPD and CT1 groups (Fisher's exact test: P = 0.023). This was especially true for the frequency of haplogroup B, which was significantly lower in patients with EOPD compared to CT1 (χ2 = 8.665, P = 0.003; OR = 0.225, 95% CI: 0.082–0.619, P = 0.004) [Table 4]. Interestingly, between LOPD and CT2, this difference disappeared (χ2 = 0.998, P = 0.986) [Table 5]. Stratified by AAE, haplogroup B also showed a lower frequency in PD cases who are younger than 50 years (χ2 = 5.066, P = 0.024; OR = 0.146, 95% CI: 0.030–0.715, P = 0.018), while haplogroup D contained a higher proportion of these cases (χ2 = 5.522, P = 0.019; OR = 3.579, 95% CI: 1.112–11.523, P = 0.033) [Table 6].
Table 4.
Distribution of mtDNA haplogroups after stratification by AAO (EOPD vs. CT1)
| Haplogroup* | EOPD (n = 63) n (%) | CT1 (n = 118) n (%) | P† | Logistic regression analysis | ||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | ||||
| A | 4 (6.3) | 5 (4.2) | 0.792 | 1.665 | 0.424–6.534 | 0.465 |
| B‡ | 5 (7.9) | 31 (26.3) | 0.003 | 0.225 | 0.082–0.619 | 0.004 |
| C | 1 (1.6) | 3 (2.5) | 1.000 | 0.674 | 0.067–6.755 | 0.737 |
| D | 14 (22.2) | 14 (11.9) | 0.066 | 2.083 | 0.918–4.726 | 0.079 |
| F | 14 (22.2) | 18 (15.3) | 0.242 | 1.701 | 0.765–3.779 | 0.192 |
| G | 2 (3.2) | 1 (0.8) | 0.577 | 3.271 | 0.286–37.452 | 0.341 |
| Others | 23 (36.5) | 46 (39.0) | 0.744 | 0.905 | 0.477–1.718 | 0.760 |
*Fisher’s exact test for the overall haplogroup distribution: P = 0.023; †P values for 2 × 2 tables; ‡P < 0.05. mtDNA: Mitochondrial DNA; AAO: Age at onset; EOPD: Early-onset Parkinson’s disease; CT1: Control team 1; OR: Odds ratio; CI: Confidence interval.
Table 5.
Distribution of mtDNA haplogroups after stratification by AAO (LOPD vs. CT2)
| Haplogroup* | LOPD (n = 216) n (%) | CT2 (n = 392) n (%) |
|---|---|---|
| A | 12 (5.6) | 17 (4.3) |
| B | 41 (19.0) | 77 (19.6) |
| C | 7 (3.2) | 16 (4.1) |
| D | 29 (13.4) | 57 (14.5) |
| F | 33 (15.3) | 62 (15.8) |
| G | 8 (3.7) | 14 (3.6) |
| Others | 86 (39.8) | 149 (38.0) |
*Chi-square test for the overall haplogroup distribution: χ2 = 0.998, P = 0.986. mtDNA: Mitochondrial DNA; AAO: Age at onset; LOPD: Late- onset Parkinson’s disease; CT2: Control team 2.
Table 6.
Distribution of mtDNA haplogroups after stratification by AAE <50
| Haplogroup* | PD (n = 32) n (%) | Control (n = 63) n (%) | P† | Logistic regression analysis | ||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | ||||
| A | 3 (9.4) | 2 (3.2) | 0.428 | 3.037 | 0.459–20.117 | 0.249 |
| B‡ | 2 (6.3) | 16 (25.4) | 0.024 | 0.146 | 0.030–0.715 | 0.018 |
| C | 0 (0) | 3 (4.8) | 0.526 | 0.000 | 0.000–NA|| | 0.999 |
| D§ | 9 (28.1) | 6 (9.5) | 0.019 | 3.579 | 1.112–11.523 | 0.033 |
| F | 5 (15.6) | 9 (14.3) | 1.000 | 1.449 | 0.413–5.086 | 0.563 |
| G | 0 | 0 | – | – | – | – |
| Others | 13 (40.6) | 27 (42.9) | 0.835 | 0.953 | 0.393–2.313 | 0.916 |
*Fisher’s exact test for the overall haplogroup distribution: P = 0.032; †P values for 2 × 2 tables; ‡P < 0.05; §P < 0.05; ||NA: Not applicable; mtDNA: Mitochondrial DNA; AAE: Age at examination; OR: Odds ratio; CI: Confidence interval; PD: Parkinson’s disease.
Mitochondrial single nucleotide polymorphism at positions 10398 and 10400
According to previous studies, mtDNA 10398G seems to be a protecting factor for PD patients,[7,21] and is usually closely linked to mtDNA 10400T.[18,22,23] We detected the polymorphisms of 10398G and 10400T, but no significant difference was found between groups (46.2% vs. 44.7%, χ2 = 0.171, P = 0.680; data not shown).
DISCUSSION
To date, the relationship between Asian mtDNA haplogroups and the risk of PD remains unsubstantiated. According to MITOMAP and previous reports,[18,20] carriers of mtDNA haplogroup A, B, C, D, F, G are in the majority in the Asian population. Here, we focused on these six haplogroups in a population from Southern China, with the proportions of each haplogroup in our control group being similar to those listed in MITOMAP. Consonant with prior research,[22] the frequency of haplogroup B and F shows a tendency to decrease from Southern to Northern Chinese population while those of A and D increase, which also corresponds to our data. In other words, the control group can be considered representative of the general population in Southern China.
For each PD patient, AAO and AAE were significant, with the former indicating the occurrence of first PD symptoms and the latter meaning early diagnosis and therapy. In southern Han Chinese, haplogroup B seems to be a protective factor for whose AAO or AAE are under 50 years while haplogroup D leads a susceptibility to PD for whose AAE is under 50 years. In addition, mtDNA SNPs 10398G and 10400T do not confer any protective effect upon our patients.
Mitochondrial DNA haplogroup B has been established for East Asian population as one of the distinctive mitochondrial lineages, which derives from R, a sub-haplogroup under N. One of the most characteristics of haplogroup B is a 9-bp deletion 8280–8290 = A[delCCCCCTCTA] G, which is in a noncoding (NC) region of 25-bp lengths between cytochrome c oxidase subunit II and tRNALys. As reported, the haplogroup B is associated with a risk of exacerbating acute mountain sickness in southwestern Han Chinese.[24] In Taiwanese people, the intergenic 9-bp deletion can be seen in high prevalence in MELAS or MERRF patients.[25] Recently, it has been reported that even mtDNA synonymous polymorphisms, which do not result in the substitution of amino acids, experience selection pressure, and indicate functional relevance.[26] These findings suggest the possibility that the 9-bp deletion in NC region might be subtly functional. We propose that the 9-bp deletion located in the small region between two functional genes may affect the intergenic structure, leading to an abnormal molecular conformation which could cause a protective function against EOPD. On the other hand, as haplogroup B subordinates to R, individuals with haplogroup B also carry the 12705C that encodes for Ile in the NADH dehydrogenase 5, which is a mitochondrial subunit of complex I. Since the effect of single causal variation is possibly enhanced or subdued by combining with a particular mtDNA polymorphism.[27] The 9-bp deletion possibly cooperates with other mtSNP, such as 12705C, to reduce the risk of developing EOPD.
Mitochondrial DNA haplogroup D, a sub-cluster of haplogroup M, is one of the prevalent lineages in East Asian population and is defined by C5178A. The SNP changes amino acids from Leu to Met in ND2, which is also a mitochondrial subunit of complex I. Several studies have nominated Mt5178A as a candidate genetic marker of longevity,[28] as well as a protector against certain adult-onset diseases[29,30,31] in the Japanese population. Nevertheless, the relationship between C5178A and PD still lacks clarification among the same population. It is the case that certain mtDNA variations are representative for specific races or geographical regions.[32,33] In Han Chinese, the protective effect against some adult-onset diseases has not been detected in 5178A, and on the contrary, our research suggests that 5178A is a predisposition to PD in people younger than 50 years of age. Specifically, it proved the probable relationship between the haplogroups D and PD in the Han population. In addition, as a sub-cluster of haplogroup M, haplogroup D possesses 10398G and 5178A, makes a possible influence on the function of complex I with an integrative effect for different age brackets. The potential role of haplogroup D in younger patients may be based on several elements: Such as oxidative stress, cell apoptosis, and respiratory chain complex activities.[7] By the way, in AAO <50 year, haplogroup D did not present a higher frequency. We believe that the difference between the two results come from the different methods of stratification (AAE/AAO). Certainly, a larger sample and age-matched study will be necessary to explore this influence in the future.
In our study, 10398G and 10400T seem to not be associated with the risk of PD. Although the function of these SNPs remains unclear, some investigations have found that 10398G shows a protective effect on PD in certain population,[7,21] a conclusion not arrived at in other studies.[9,10,11,34,35] MtDNA 10398G and 10400T are located in the ND3 gene, which encodes one of the seven subunits constituting complex I. A10398G leads a substitution of Thr with Ala, while C10400T causes a synonymous mutation with Thr. As described in several studies,[18,22,23] the 10398G and 10400T are usually tightly linked. Since the effect of a single variation can possibly be modified by other existing mtDNA or a nuclear gene polymorphism,[27] we decided to analyze the coexistent A10398G and C10400T together in order to avoid bias. From our data, we failed to find any significant association between the SNPs and PD in Han Chinese, which is consistent with the results of similar studies carried out in other population.[9,10,11] In a previous report,[36] single common variation did not seem to be PD-deterministic because of the overall modifying effect by other SNPs. As the typical SNPs for super-haplogroup M and found at a large frequency in our samples, the 10398G and 10400T may just be a marker for mtDNA haplogroup but not a genetic factor involved in PD among Han Chinese.
A strength of our study is that the control group is representative of the general population in Southern China, and the ethnic backgrounds of the subjects are consistent. As for limitations, a similar percentage of patients and controls could not be classified to any of the six major Asian haplogroups. The samples pooled in the “others” category might be derived from some particular haplogroups which have not been incorporated to our research, such as M7-M10,[20] or other rare haplogroups, such as Y or P.
In summary, we speculate that mtDNA haplogroup B might confer a lower risk for EOPD and people younger than 50 years in Han Chinese, while haplogroup D probably lead a higher risk of PD in people younger than 50 years of age. Our research reveals that particular Asian mtDNA haplogroups likely play a part in the pathogenesis of PD among Han Chinese. Certainly, a case–control study with larger samples and further biochemical analysis will be warranted to clarify the exact function of mtDNA haplogroups in PD among Han Chinese.
ACKNOWLEDGMENTS
This work was supported by the grant 81322017 from the National Natural Science Foundation of China, grant NCET-13–0736 from Program for New Century Excellent Talents in University, National Key Clinical Specialty Discipline Construction Program and Key Clinical Specialty Discipline Construction Program of Fujian.
Footnotes
Edited by: Ya-Lin Bao
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Zhang ZX, Roman GC, Hong Z, Wu CB, Qu QM, Huang JB, et al. Parkinson's disease in China: Prevalence in Beijing, Xian, and Shanghai. Lancet. 2005;365:595–7. doi: 10.1016/S0140-6736(05)17909-4. [DOI] [PubMed] [Google Scholar]
- 2.Thomas B, Beal MF. Parkinson's disease. Hum Mol Genet. 2007;16 Spec No 2:R183–94. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 3.Nicklas WJ, Youngster SK, Kindt MV, Heikkila RE. MPTP, MPP+ and mitochondrial function. Life Sci. 1987;40:721–9. doi: 10.1016/0024-3205(87)90299-2. [DOI] [PubMed] [Google Scholar]
- 4.Parker WD, Jr, Boyson SJ, Parks JK. Abnormalities of the electron transport chain in idiopathic Parkinson's disease. Ann Neurol. 1989;26:719–23. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- 5.Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson's disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 6.Swerdlow RH, Parks JK, Miller SW, Tuttle JB, Trimmer PA, Sheehan JP, et al. Origin and functional consequences of the complex I defect in Parkinson's disease. Ann Neurol. 1996;40:663–71. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- 7.Yan MH, Wang X, Zhu X. Mitochondrial defects and oxidative stress in Alzheimer disease and Parkinson disease. Free Radic Biol Med. 2013;62:90–101. doi: 10.1016/j.freeradbiomed.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–11. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyle A, Foltynie T, Tiangyou W, Lambert C, Keers SM, Allcock LM, et al. Mitochondrial DNA haplogroup cluster UKJT reduces the risk of PD. Ann Neurol. 2005;57:564–7. doi: 10.1002/ana.20417. [DOI] [PubMed] [Google Scholar]
- 10.Ghezzi D, Marelli C, Achilli A, Goldwurm S, Pezzoli G, Barone P, et al. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson's disease in Italians. Eur J Hum Genet. 2005;13:748–52. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- 11.Fachal L, Mosquera-Miguel A, Pastor P, Ortega-Cubero S, Lorenzo E, Oterino-Durán A, et al. No evidence of association between common European mitochondrial DNA variants in Alzheimer, Parkinson, and migraine in the Spanish population. Am J Med Genet B Neuropsychiatr Genet. 2015;168B:54–65. doi: 10.1002/ajmg.b.32276. [DOI] [PubMed] [Google Scholar]
- 12.Hudson G, Nalls M, Evans JR, Breen DP, Winder-Rhodes S, Morrison KE, et al. Two-stage association study and meta-analysis of mitochondrial DNA variants in Parkinson disease. Neurology. 2013;80:2042–8. doi: 10.1212/WNL.0b013e318294b434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: A clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55:181–4. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XD. Diagnostic criteria and differential diagnosis of Parkinson's disease. Chin J Psychiatry. 1985;18:256. [Google Scholar]
- 15.Tanner CM, Ottman R, Goldman SM, Ellenberg J, Chan P, Mayeux R, et al. Parkinson disease in twins: An etiologic study. JAMA. 1999;281:341–6. doi: 10.1001/jama.281.4.341. [DOI] [PubMed] [Google Scholar]
- 16.Torroni A, Schurr TG, Yang CC, Szathmary EJ, Williams RC, Schanfield MS, et al. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics. 1992;130:153–62. doi: 10.1093/genetics/130.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torroni A, Miller JA, Moore LG, Zamudio S, Zhuang J, Droma T, et al. Mitochondrial DNA analysis in Tibet: Implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol. 1994;93:189–99. doi: 10.1002/ajpa.1330930204. [DOI] [PubMed] [Google Scholar]
- 18.Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–71. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 20.Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet. 2002;70:635–51. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huerta C, Castro MG, Coto E, Blázquez M, Ribacoba R, Guisasola LM, et al. Mitochondrial DNA polymorphisms and risk of Parkinson's disease in Spanish population. J Neurol Sci. 2005;236:49–54. doi: 10.1016/j.jns.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 22.Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, et al. The emerging limbs and twigs of the East Asian mtDNA tree. Biol Evol. 2002;19:1737–51. doi: 10.1093/oxfordjournals.molbev.a003996. Erratum in: Mol Biol Evol 2003;20:162. [DOI] [PubMed] [Google Scholar]
- 23.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–30. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 24.Li FX, Ji FY, Zheng SZ, Yao W, Xiao ZL, Qian GS. MtDNA haplogroups M7 and B in southwestern Han Chinese at risk for acute mountain sickness. Mitochondrion. 2011;11:553–8. doi: 10.1016/j.mito.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu CS, Cheng WL, Chen YY, Ma YS, Pang CY, Wei YH. High prevalence of the COII/tRNA (Lys) intergenic 9-bp deletion in mitochondrial DNA of Taiwanese patients with MELAS or MERRF syndrome. Ann N Y Acad Sci. 2005;1042:82–7. doi: 10.1196/annals.1338.058. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Wang K, Rodova M, Esteves R, Berry D, Lezi E, et al. Polymorphic variation in cytochrome oxidase subunit genes. J Alzheimers Dis. 2010;21:141–54. doi: 10.3233/JAD-2010-100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swerdlow RH, Weaver B, Grawey A, Wenger C, Freed E, Worrall BB. Complex I polymorphisms, bigenomic heterogeneity, and family history in Virginians with Parkinson's disease. J Neurol Sci. 2006;247:224–30. doi: 10.1016/j.jns.2006.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K. Mitochondrial genotype associated with longevity. Lancet. 1998;351:185–6. doi: 10.1016/S0140-6736(05)78211-8. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Taniyama M, Suzuki Y, Katagiri T, Ban Y. Association of the mitochondrial DNA 5178A/C polymorphism with maternal inheritance and onset of type 2 diabetes in Japanese patients. Exp Clin Endocrinol Diabetes. 2001;109:361–4. doi: 10.1055/s-2001-17407. [DOI] [PubMed] [Google Scholar]
- 30.Takagi K, Yamada Y, Gong JS, Sone T, Yokota M, Tanaka M. Association of a 5178C-->A (Leu237Met) polymorphism in the mitochondrial DNA with a low prevalence of myocardial infarction in Japanese individuals. Atherosclerosis. 2004;175:281–6. doi: 10.1016/j.atherosclerosis.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Ohkubo R, Nakagawa M, Ikeda K, Kodama T, Arimura K, Akiba S, et al. Cerebrovascular disorders and genetic polymorphisms: Mitochondrial DNA5178C is predominant in cerebrovascular disorders. J Neurol Sci. 2002;198:31–5. doi: 10.1016/s0022-510x(02)00055-2. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–8. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–6. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 34.Otaegui D, Paisán C, Sáenz A, Martí I, Ribate M, Martí-Massó JF, et al. Mitochondrial polymporphisms in Parkinson's disease. Neurosci Lett. 2004;370:171–4. doi: 10.1016/j.neulet.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Chen CM, Kuan CC, Lee-Chen GJ, Wu YR. Mitochondrial DNA polymorphisms and the risk of Parkinson's disease in Taiwan. J Neural Transm. 2007;114:1017–21. doi: 10.1007/s00702-007-0658-z. [DOI] [PubMed] [Google Scholar]
- 36.Swerdlow RH. Does mitochondrial DNA play a role in Parkinson's disease. A review of cybrid and other supportive evidence? Antioxid Redox Signal. 2012;16:950–64. doi: 10.1089/ars.2011.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]


