Abstract
The low efficiency of in vitro differentiation of human embryonic stem cells (ESCs) or human induced pluripotent stem cells (iPSCs) into insulin-producing cells thus creates a crucial hurdle for the clinical implementation of human pluripotent stem cells (PSCs). In this study, we investigated the key factors for the differentiation of PSCs into insulin-producing cells. We obtained microarray data of HUES8 and HUES6 from two GeneChips (GPL3921: Affymetrix HT Human Genome U133A Array, GPL570: Affymetrix Human Genome U133 Plus 2.0 Array) in a database of GEO (NCBI), since HUES8 can differentiate into pancreatic cells, while HUES6 hardly demonstrates any differentiation at all. The genes with more than fourfold higher expressions in HUES8 compared to HUES6 included RPS4Y1, DDX3Y, EIF1AY, GREM1, GATA6, and NLGN4Y. Since there were four genes, RPS4Y1, DDX3Y, EIF1AY, and NLGN4Y, on the Y chromosome and HUES8 was a male cell line and HUES6 was a female cell line, we excluded these genes in this study. On the other hand, genes with more than fourfold higher expressions in HUES6 compared to HUES8 included NLRP2, EGR1, and SMC3. We next compared iPSCs derived from pancreatic cells (PiPSCs) and iPSCs derived from fibroblasts (FiPSCs). PiPSCs differentiated into insulin-producing cells more easily than FiPSCs because of their epigenetic memory. The gene expressions of GREM1, GATA6, NLRP2, EGR1, and SMC3 in PiPSCs and FiPSCs were also investigated. The expression level of GREM1 and GATA6 in PiPSCs were higher than in FiPSCs. On the other hand, EGR1, which was lower in HUES8 than in HUES6, was predictably lower in PiPSCs than FiPSCs, while NLRP2 and SMC3 were higher in PiPSCs than FiPSCs. These data suggest that the expression of GATA6 and GREM1 and the inhibition of EGR1 may be important factors for the differentiation of PSCs into insulin-producing cells.
Key words: Embryonic stem cells (ESCs), Induced pluripotent stem cells (iPSCs), GATA6, GREM1, EGR1
INTRODUCTION
Human embryonic stem cells (ESCs) are capable of differentiation into cells from the three embryonic germ layers that constitute the body and, therefore, have been used for regenerative medicine research at many institutions (12,21). However, the use of ESCs remains controversial for ethical reasons,which hinders the clinical uses of ESCs, since ESCs are derived from the inner cell mass of mammalian blastocysts (18). Recently, induced pluripotent stem cells (iPSCs) were generated from somatic cells by the transfection of several transcription factors to human somatic fibroblasts (20). This technical breakthrough has significant implications, since they provide a solution for overcoming the ethical issues associated with ESC derivation from embryos (10).
It has been reported that different human ESC lines express similar markers in the undifferentiated state (11), but marked differences have been observed in the differentiation propensity among various human ESC lines (16). Osafune et al. reported the differentiation potential of 17 human ESC lines, and the Harvard University embryonic stem cell line 8 (HUES8) was found to be the best for pancreatic differentiation, while the HUES3 cell line was the best for cardiomyocyte generation (16). Moreover, HUES8 and HUES6 were the best and the worst cell lines, respectively, for pancreatic differentiation among the 17 identified human ESC lines. Therefore, differences in the gene expressions between HUES8 and HUES6 might be related to the differentiation of the pancreas from PSCs.
On the other hand, it has recently been shown that, following the reprogramming of mouse/human iPSCs, an epigenetic memory is inherited from parental cells, which affects the differentiation capacity of iPSC lines (14,15,17). It has also been shown that the epigenetic memory predisposes iPSCs derived from pancreatic β-cells to differentiate more readily into insulin-producing cells (1). These findings demonstrate that the iPSC phenotype may be influenced by their cells of origin, thereby suggesting that differences in the gene expression between iPSCs derived from pancreatic cells (PiPSCs) and those derived from fibroblasts (FiPSCs) may be related to their differentiation potential.
In this study, we investigated the factors required for the differentiation of PSCs into insulin-producing cells. We first compared the microarray data of the HUES8 and HUES6 cell lines and selected several genes and next compared PiPSCs and FiPSCs with the expressions of the genes that had been selected in the first study.
MATERIALS AND METHODS
Statistical Analysis of Microarray Data
The microarray data of HUES6 (female) and HUES8 (male) were extracted from the homepage of GEO (http://www.ncbi.nlm.nih.gov/geo/). The first set was Affymetrix HT Human Genome U133A Array (platform_id:GPL3921) of HUES6 (geo_accession: GSM 637760) and HUES8 (geo_accession: GSM 637761). The second set was Affymetrix Human Genome U133 Plus 2.0 Array (platform_id:GPL570) of HUES6 (geo_accession: GSM462819, GSM462820, GSM462821) and HUES8 (geo_accession: GSM 310860, GSM 310861, GSM 3108662). The factors were selected based on differences in the gene expressions of more than four times between HUES8 and HUES6 (only GPL570 is a value of p < 0.05) using a Bioconductor (9). A data analysis was then performed using the R statistics software package (http://www.r-project.org/).
Generation of iPSCs From Human Pancreatic Cells
Pancreatic cells (>80% islets) from a human donor (female, aged in her 40s), which were isolated at the University of Alberta as previously described (19) and shipped to Japan, were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum (Thermo Scientific, Kanagawa, Japan), and 0.5% penicillin (Sigma-Aldrich, St. Louis, MO, USA). The cells (1 × 105 cells) were transduced with sendai viral vectors (20 multiplicity of infection; DNAVEC, Ibaraki, Japan) encoding the four pluripotency-inducing factors: octamer-binding transcription factor 4 [OCT4; also known as pituitary-specific positive transcription factor 1 octamer-binding transcription factor 1 Unc-86 (POU) class 5 homeobox 1(POU5F1)], sex-determining region Y box 2 (SOX2), Kruppel-like factor 4 (KLF4), and v-myc avian myelocytomatosis viral oncogene homolog (cMYC). Two days after transduction, the cells were replated on resistant SNL (Sandos Inbred Mice embryo-derived and thioguanine and ouabain cells transformed by neomycin resistance and murine leukemia inhibitory factor) feeder layer (Cosmo Bio Co., Ltd., Tokyo, Japan). The next day, the medium was replaced with a standard ESC culture medium (Repro CELL, Kanagawa, Japan). Three to 4 weeks after transduction, about 10 colonies were visible. The colonies were picked up and then were transferred on an SNL feeder. The cultures were manually passaged at a 1:3–1:6 split ratio every 5–7 days.
Cell Culture
PiPSCs (induced cells described above) and FiPSCs (201B7, RIKEN BioResource Center, Japan) were maintained on an SNL feeder layer in DMEM-F12 (Sigma-Aldrich), 2 mM l-glutamine (Nacalaitesque, Kyoto, Japan), 1:100 dilution of nonessential amino acid (Life Technologies), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 5 ng/ml basic fibroblast growth factor (bFGF) (Repro CELL), and penicillin/streptomycin (Sigma-Aldrich). For passaging, iPSC colonies were dissociated with dissociation solution for human ESCs/iPSCs (Riken CDB, Kobe, Japan) and split at a ratio between 1:3 and 1:6.
Alkaline Phosphatase Staining
Alkaline phosphatase staining was performed on 15 colonies in 35-mm dishes using the Vector Blue Alkaline Phosphatase Substrate Kit (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions.
Karyotype Analysis
Chromosomal G-band analyses were performed at the Nihon Gene Research Laboratories, Japan. At least 20 metaphases were analyzed.
Quantitative RT-PCR
Total RNA was prepared with the RNeasy Micro Kit (Qiagen, Tokyo, Japan) and treated with RNase free DNase (Qiagen). A total of 500 ng RNA was used for a reverse transcription reaction using the QuantiTect Reverse Transcription Kit (Qiagen). qRT-PCR was performed on the Real-time PCR System (ABI7000) using SYBR Green (both from Life Technologies). The expression levels were normalized to the glycelaldehyde-3-phosphate dehydrogenase (GAPDH). The specific primers are listed in Table 1.
Table 1.
Primer Sequences of Quantitative RT-PCR
| Gene | Sequences (5′ to 3′) |
|---|---|
| ESC-specific genes | |
| OCT3/4 | Forward: GAAACCCACACTGCAGCAGA Reverse: TCGCTTGCCCTTCTGGCG |
| SOX2 | Forward: GGGAAATGGGAGGGGTGCAAAAGAGG Reverse: TTGCGTGAGTGTGGATGGGATTGGTG |
| NANOG | Forward: CTCAGCTACAAACAGGTGAAGAC Reverse: TCCCTGGTGGTAGGAAGAGTAAA |
| REX1 (ZFP42) | Forward: CAGATCCTAAACAGCTCGCAGAAT Reverse: GCGTACGCAAATTAAAGTCCAGA |
| TERT | Forward: CGTACAGGTTTCACGCATGTG Reverse: ATGACGCGCAGGAAAAATGT |
| GAL | Forward: AAACAATATCATGCGCACAATCA Reverse: GGGCACCGGCCTCTTT |
| LEFTY2 | Forward: GCTCAGATGCTGAGCTCTAGTAGGA Reverse: GAAACTCCCAGCTGAAAATGTGT |
| GAPDH | Forward: CCACTCCTCCACCTTTGACG Reverse: ATGAGGTCCACCACCCTGTT |
| Endoderm-specific genes | |
| SOX17 | Forward: AGCCAAGGGCGAGTCCCGTA Reverse: GCCTTCCACGACTTGCCCAGC |
| CXCR4 | Forward: CTGCACCTGTCAGTGGCCGA Reverse: TTGGTGGCGTGGACGATGGC |
| WNT3 | Forward: TCCTCGGCGCCTCTTCTAAT Reverse: CTGTGAGCCCAGAGATGTGT |
| CD117 (KIT) | Forward: ATTCAAGCACAATGGCACGG Reverse: AAGGAGTGAACAGGGTGTGG |
| GATA3 | Forward: CTGGCTCGCAGAATTGCA Reverse: TGGGTACGGCAGAATAAAACG |
| GATA4 | Forward: GTTTTTTCCCCTTTGATTTTTGATC Reverse: AACGACGGCAACAACGATAAT |
| GATA6 | Forward: GGATTGTCCTGTGCCAACTGT Reverse: GGTTCACCCTCGGCGTTT |
| HNF1B | Forward: AACCACCGAAGAGGAAGCAA Reverse: TCGCATCAGTTTGTTCGATGA |
| HNF4A | Forward: CACCTGATGCAGGAACATATGG Reverse: CTGTCCGTTGCTGAGGTGAGT |
| AFP | Forward: GTTGCCAACTCAGTGAGGACAA Reverse: TGATACATAAGTGTCCGATAATAATGTCA |
| Pancreas-specific genes | |
| PDX1 | Forward: GATACTGGATTGGCGTTGTTTG Reverse: TCCCAAGGTGGAGTGCTGTAG |
| HNF6 | Forward: TGCGCAACCCCAAACC Reverse: TCCACATCCTCCGGAAGGT |
| NKX6.1 | Forward: GCCTGTACCCCTCATCAAGGA Reverse: AAGTGGGTCTCGTGTGTTTTCTC |
| INS | Forward: ACGAGGCTTCTTCTACACACCC Reverse: TCCACAATGCCACGCTTCTGCA |
| PAX6 | Forward: GCTTCACCATGGCAAATAACCT Reverse: GGCAGCATGCAGGAGTATGA |
| SOX9 | Forward: CCCATGTGGAAGGCAGATG Reverse: GAAGGTTAACTGCTGGTGTTCTGA |
| Genes selected by a microarray analysis | |
| GREM1 | Forward: CCCCCCGCCAGACAAG Reverse: TTGCACCAGTCTCGCTTCAG |
| EGR1 | Forward: TTTCACGTCTTGGTGCCTTTT Reverse: TCCCTCACAATTGCACATGTC |
| NLRP2 | Forward: CTTTGAGGAAACCACTGTGCAA Reverse: AACTGAACGGAGGGATGGAA |
| SMC3 | Forward: TTGCTCTGATTTTTGCCATTCA Reverse: CATCCAGAGCCTGGTCAATTTC |
OCT3/4, octamer-binding transcription factor 3/4 [also known as pituitary-specific positive transcription factor 1 octamer-binding transcription factor 1 Unc-86 (POU) class 5 homeobox 1(POU5F1)]; SOX2: sex-determining region Y box 2; REX1, reduced expression protein 1 (Zinc finger protein 42; ZFP42); TERT, telomerase reverse transcriptase; GAL:, galanin/galanin-message-associated peptide (GMAP) pre-propeptide; LEFTY2, left-right determination factor 2; GAPDH, glyceraldehyde 3-phospahte dehydrogenase; CXCR4, chemokine C-X-C motif receptor 4; WNT3, wingless-type MMTV integration site family, member 3; CD117, cluster of differentiation 117 (also known as v-kit Hardy–Zuckerman 4 feline sarcoma viral oncogene homolog, KIT); GATA3, guanine–adenine–thymine–adenine-binding protein 3; HNF1B, hepatocyte nuclear factor 1B; AFP, αfeto-protein; PDX1, pancreatic and duodenal homeobox 1; NKX6.1, NK6 homeobox 1; INS, insulin; PAX6, paired box 6; GREM1, Gremlin 1 [also known as differential screening-selected gene aberrative in neuroblastoma (DAN) family bone morphogenetic protein (BMP) antagonist]; EGR1, early growth response 1; NLRP2, nucleotide-binding domain and leucine-rich repeat containing gene (NLR) family, pyrin domain containing 2; SMC3, structural maintenance of chromosomes 3.
RESULTS
Comparison of the Gene Expressions of Human ESC Lines by the Microarray Data
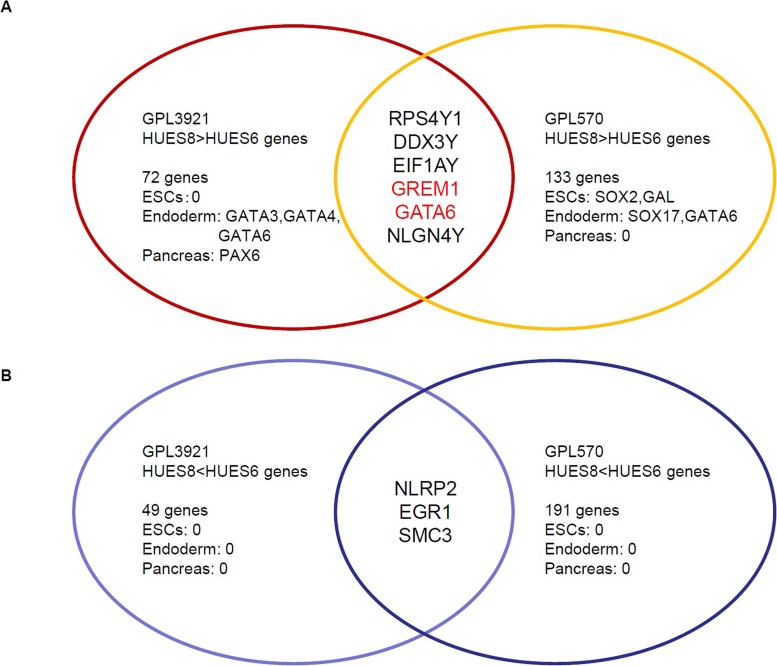
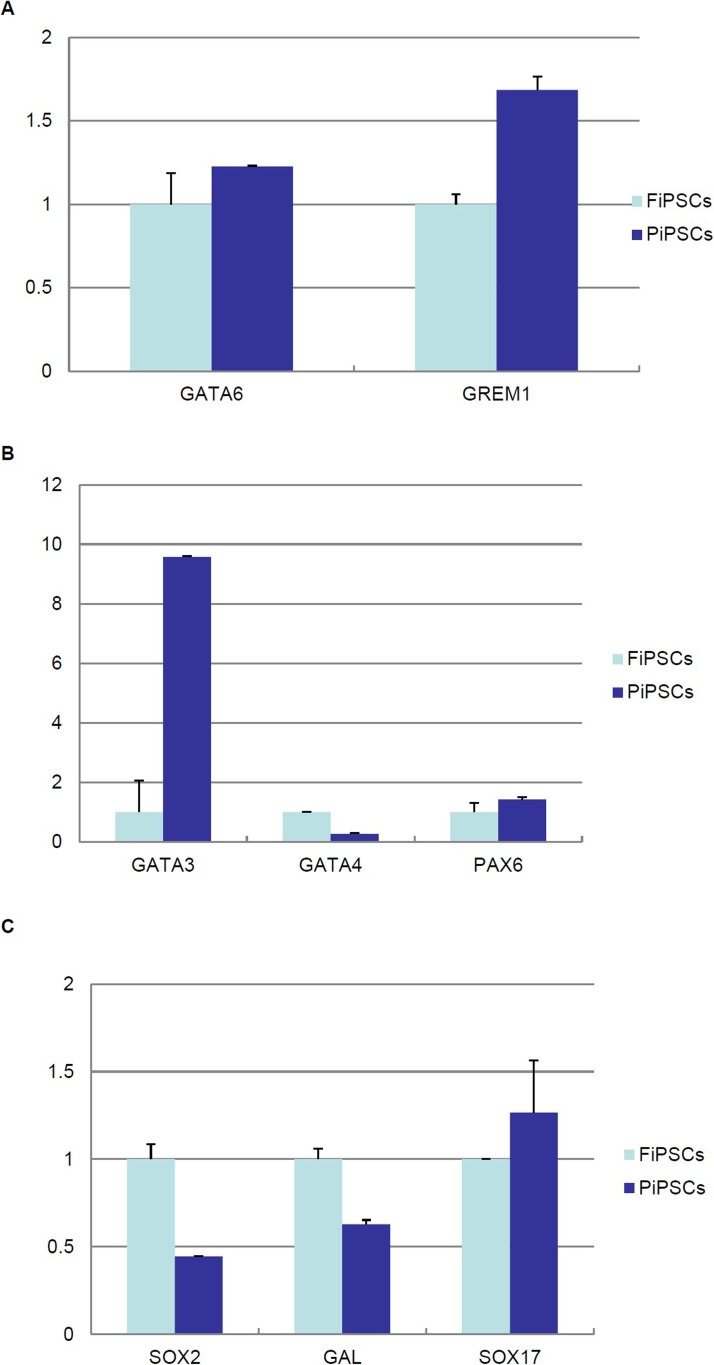
Osafune et al. reported the differentiation potential of 17 human ESC lines, and the male HUES8 and female HUES6 were found to be the best and the worst lines, respectively, for pancreatic differentiation (16). We obtained the microarray data of HUES8 and HUES6 from two GeneChips (GPL3921: Affymetrix HT Human Genome U133A Array, GPL570: Affymetrix Human Genome U133 Plus 2.0 Array) in a database of GEO (NCBI). Seventy-two genes on GPL3921 and 133 genes on GPL 570 were expressed more than fourfold higher in HUES8 than in HUES6. The matched genes of these two chips were ribosomal protein S4, Y-linked 1 (RPS4Y1), DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked (DDX3Y), eukaryotic translation initiation factor 1A, Y-linked (EIF1AY), Gremlin 1 [GREM1 also known as differential screening-selected gene aberrative in neuroblastoma (DAN) family bone morphogenetic protein (BMP) antagonist], guanine–adenine–thymine–adenine-binding protein 6 (GATA6), and neuroligin 4, Y-linked (NLGN4Y). Since four of these genes, RPS4Y1, DDX3Y, EIF1AY, and NLGN4Y, are on the Y chromosome, and HUES8 is a male cell line, while HUES6 is a female cell line, we excluded these genes in the following study. We also investigated ESC-specific genes, endoderm-specific genes, and pancreas-specific genes as shown in Table 2 (2,3,4,20,22) in 72 genes on GPL3921 and 133 genes on GPL 570. There were no ESC-specific genes, three endodermal-specific genes (GATA3, GATA4, and GATA6), and one pancreas-specific gene (paired box 6; PAX6) out of the 72 genes on GPL3921. There were two ESC-specific genes [SOX2 and galanin/galanin-message-associated peptide (GMAP) pre-propeptide (GAL)], two endodermal-specific genes (SOX17 and GATA6), and no pancreas-specific genes in 133 genes on GPL 570 (Fig. 1A).
Table 2.
Human ESC-Specific Genes, Endoderm-Specific Genes, Pancreas-Specific Genes
| ESC-Specific Genes | Endoderm-Specific Genes | Pancreas-Specific Genes |
|---|---|---|
| OCT3/4 (POU5F1) | WNT3 | INSULIN |
| SOX2 | CD117 | PDX1 |
| NANOG | GATA4 | PAX6 |
| REX1 (ZFP42) | SOX17 | NKX6.1 |
| LEFTY2 | HNF4 | SOX9 |
| TERT | CXCR4 | HNF6 |
| GAL | HNF1B | |
| AFP | ||
| GATA6 | ||
| GATA3 |
Figure 1.
Microarray analysis. (A) Genes that are expressed more than fourfold higher in HUES8 compared to HUES6 in two gene chips (GPL3921 and GPL570). (B) Genes that are expressed more than fourfold higher in HUES6 compared to HUES8. ESCs, embryonic stem cells; GATA3, guanine–adenine–thymine–adenine-binding protein 3; PAX6, paired box 6; RPS4Y1, ribosomal protein S4, Y-linked 1; DDX3Y, DEAD (Asp-Glu-Ala-Asp) box helicase 3; EIF1AY, eukaryotic translation initiation factor 1A, Y-linked, GREM1, Gremlin 1 [also known as differential screening-selected gene aberrative in neuroblastoma (DAN) family bone morphogenetic protein (BMP) antagonist]; NLGN4Y, neuroligin 4, Y-linked; SOX2, sex-determining region Y box 2; GAL, galanin/galanin-message-associated peptide pre-propeptide (GMAP); NLRP2, nucleotide-binding domain and leucine-rich repeat containing gene (NLR) family, pyrin domain containing 2; EGR1, early growth response 1; SMC3, structural maintenance of chromosomes 3.
Forty-nine genes on GPL3921 and 191 genes on GPL 570 were expressed more than fourfold higher in HUES6 than in HUES8. The matched genes of these two chips were nucleotide-binding domain and leucine-rich repeat containing gene (NLR) family, pyrin domain containing 2 (NLRP2), early growth response 1 (EGR1), and structural maintenance of chromosomes 3 (SMC3). We also investigated the ESC-specific genes, endoderm-specific genes, and pancreas-specific genes as shown in Table 2 (2,3,4,20,22) in 49 genes on GPL3921 and 191 genes on GPL 570. There were no ESC-specific genes, endodermal-specific genes, or pancreas-specific genes out of the 49 genes on GPL3921 or out ofthe 191 genes on GPL 570 (Fig. 1B).
Generation of iPSCs From Human Pancreatic Cells
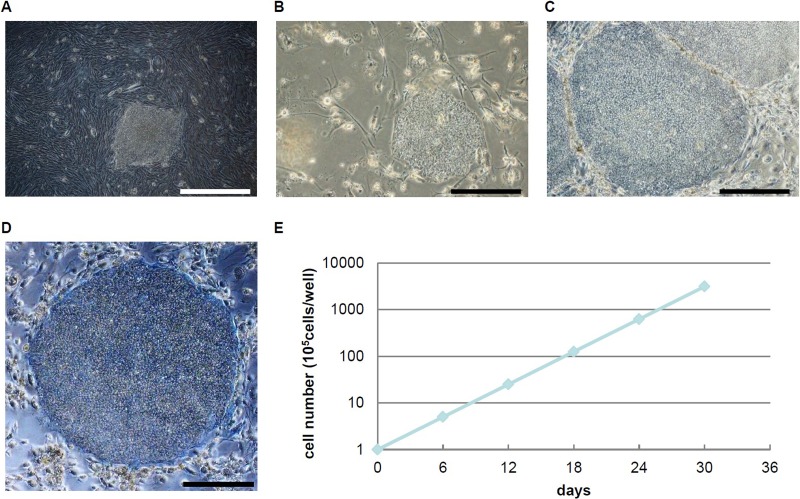
Epigenetic memory has been shown to predispose iPSCs derived from pancreatic β-cells to differentiate more readily into insulin-producing cells (1). These findings suggest that differences in the gene expression between PiPSCs and FiPSCs may be related to their differentiation potential. In this study, 201B7 cells (20) were used as FiPSCs. To generate PiPSCs, human pancreatic cells (more than 80% islets) were cultured and then transduced with four sendai viral vectors encoding the reprogramming factors OCT4, SOX2, KLF4, and c-MYC. Four weeks after transduction, 10 colonies were visible. The colonies were picked up and then were transferred on an SNL feeder (Fig. 2A). After three to four passages, the PiPSC lines showed a typical ESC-like morphology (Fig. 2B), and they could be maintained beyond 20 passages (Fig. 2C). PiPSCs expressed alkaline phosphatase (Fig. 2D) and divided actively without showing any growth inhibition (Fig. 2E).
Figure 2.
Generation of iPSCs from human pancreatic cells.(A) PiPSCs; passage 0. (B) PiPSCs; passage 4. (C) PiPSCs; passage 20. (D) Alkaline phosphatase staining of PiPSCs (passage 20). Scale bars: 500 μm. (F) Growth curve of PiPSCs.
Karyotype of PiPSCs
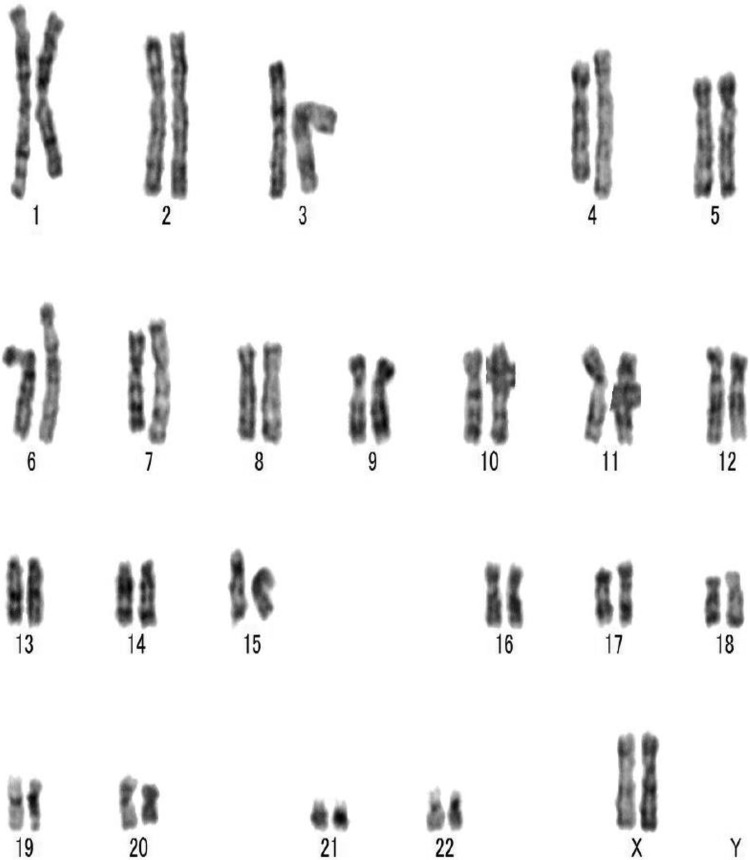
In some cases, chromosomal abnormalities have been shown to exist in human ESC lines and human iPSC lines. We investigated the karyotype of PiPSCs. PiPSCs exhibit a normal diploid karyotype of 46XX chromosomes (Fig. 3).
Figure 3.
Karyotype of PiPSCs. PiPSCs exhibit a normal diploid karyotype of the 46XX chromosomes.
Expression of ESC-Specific Genes, Endoderm-Specific Genes, and Pancreas-Specific Genes in PiPSCs and FiPSCs
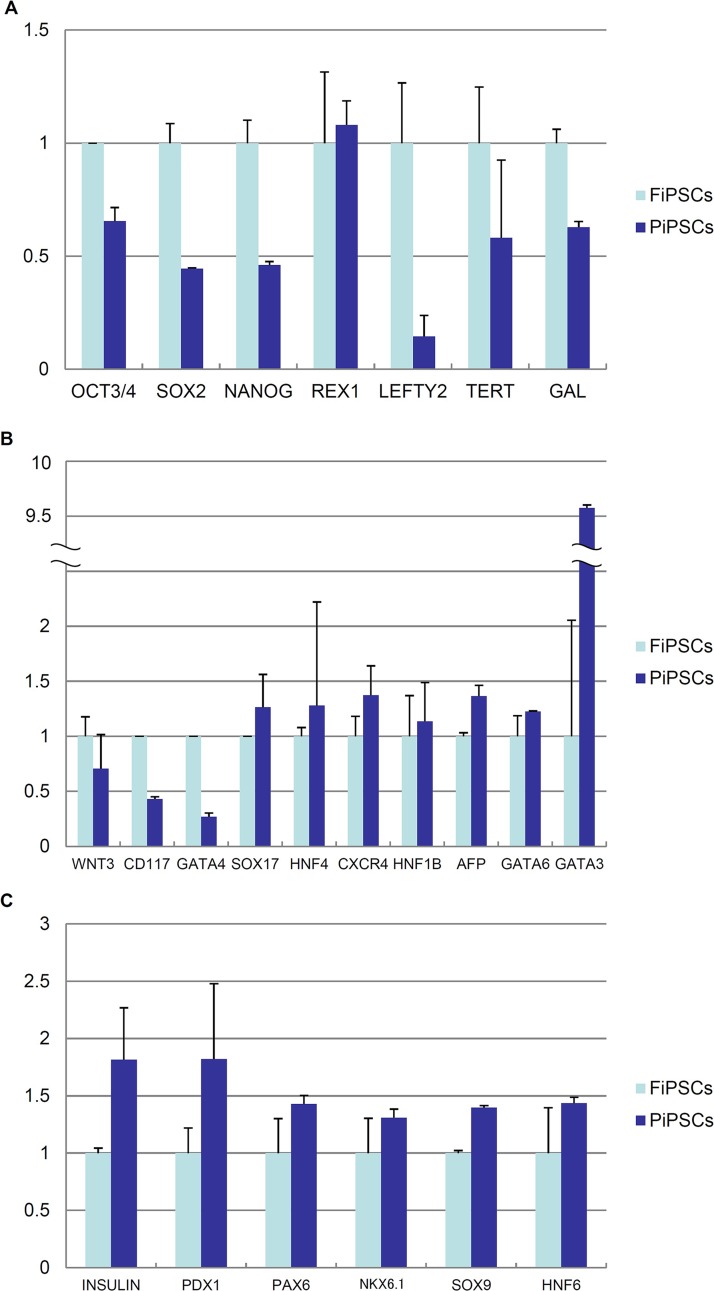
An expression analysis of ESC-specific genes, endoderm-specific genes, and pancreas-specific genes (Table 2) in PiPSCs and FiPSCs was conducted using quantitative RT-PCR (qRT-PCR). ESC-specific genes, endoderm-specific genes, and pancreatic-specific genes are shown in Table 2. The expression of ESC-specific genes in PiPSCs was relatively lower than that in FiPSCs, except for the reduced expression protein 1 (REX1 also known as zinc finger protein 42; ZFP42) gene (Fig. 4A). For endoderm-specific genes, the expressions of wingless-type mouse mammary tumor virus (MMTV) integration site family, member 3 (WNT3), cluster of differentiation 117 (CD117, also known as v-kit Hardy–Zuckerman 4 feline sarcoma viral oncogene homolog, KIT), and GATA4, which are known to be early endodermal markers in PiPSCs, were relatively lower than those observed in FiPSCs, while the expression of other endodermal-specific genes in PiPSCs was relatively higher than that in FiPSCs. In particular, the expression of GATA3 in PiPSCs was about nine times higher than that in FiPSCs (Fig. 4B). The expression of pancreas-specific genes in PiPSCs was relatively higher than that of FiPSCs (Fig. 4C). These data suggest that, following the reprogramming of PiPSCs, an epigenetic memory is thus inherited from the pancreatic cells.
Figure 4.
Gene expression in FiPSCs and PiPSCs. (A) A quantitative RT-PCR analysis of ESC-specific genes in FiPSCs and PiPSCs. (B) A quantitative RT-PCR analysis of endodermal-specific genes in FiPSCs and PiPSCs. (C) A quantitative RT-PCR analysis of pancreas-specific genes in FiPSCs and PiPSCs. The data are expressed as the genes-to-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) ratio, with that of FiPSCs arbitrarily set at 1 (n = 3). OCT3/4, octamer-binding transcription factor 3/4 [also known as pituitary-specific positive transcription factor 1 octamer-binding transcription factor 1 Unc-86 (POU) class 5 homeobox 1(POU5F1)]; REX1, reduced expression protein 1 (Zinc finger protein 42; ZFP42); LEFTY2, left-right determination factor 2; TERT, telomerase reverse transcriptase; WNT3, wingless-type MMTV integration site family, member 3; CD117, cluster of differentiation 117 (also known as v-kit Hardy–Zuckerman 4 feline sarcoma viral oncogene homolog, KIT); HNF4, hepatocyte nuclear factor 4; CXCR4, chemokine C-X-C motif receptor 4; AFP, αfeto-protein; PDX1, pancreatic and duodenal homeobox 1; NKX6.1, NK6 homeobox 1.
Expression of Genes Selected by Microarray Analysis in PiPSCs and FiPSCs
An expression analysis of the genes selected by microarray analysis (Fig. 1) in PiPSCs and FiPSCs was conducted using qRT-PCR. GREM1 and GATA6 genes, which both had a higher expression in HUES8 than in HUES6 for both GPL3921 and GPL570, were expressed at higher levels in PiPSCs than FiPSCs (Fig. 5A). For three genes, in which the expression was higher in HUES8 than HUES6 only on GPL3921, GATA3, and PAX6, the expressions were relatively higher in PiPSCs than FiPSCs, while GATA4 was lower in PiPSCs than FiPSCs (Fig. 5B). For three genes in which the expression was higher in HUES8 than HUES6 only on GPL570, SOX17 was relatively higher in PiPSCs than FiPSCs, while SOX2 and GAL were lower in PiPSCs than in FiPSCs (Fig. 5C).
Figure 5.
Expression analysis of the genes selected by a microarray analysis (HUES8>HUES6) in PiPSCs and FiPSCs.(A) A quantitative RT-PCR analysis of the GATA6 and GREM1 genes, in which the expression was higher in HUES8 than in HUES6 on both GPL3921 and GPL570, in FiPSCs and PiPSCs. (B) A quantitative RT-PCR analysis of GATA3, GATA4, and PAX6 genes, in which the expression was higher in HUES8 than HUES6 on only GPL3921, in FiPSCs and PiPSCs. (C) A quantitative RT-PCR analysis of SOX2, GAL, SOX17 genes, in which expression was higher in HUES8 than HUES6 on only GPL570, in FiPSCs and PiPSCs. The data are expressed as the genes-to-GAPDH ratio, with that of FiPSCs arbitrarily set at 1 (n = 3).
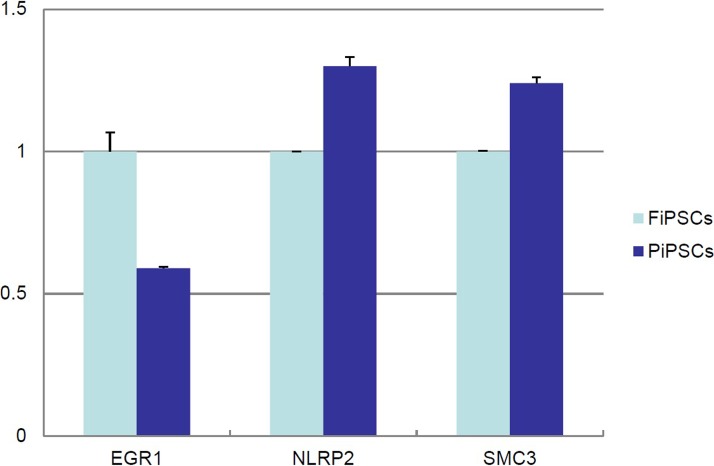
For three genes in which the expression was lower in HUES8 than HUES6 on both GPL3921 and GPL570, EGR1 was lower in PiPSCs than FiPSCs, while NLRP2 and SMC3 were unexpectedly higher in PiPSCs than FiPSCs (Fig. 6).
Figure 6.
Expression analysis of the genes selected by a microarray analysis (HUES8<HUES6) in PiPSCs and FiPSCs. A quantitative RT-PCR analysis of EGR1, NLRP2, and SMC3 genes, in which the expression was lower in HUES8 than in HUES6 on both GPL3921 and GPL570, in FiPSCs and PiPSCs. The data are expressed as the genes-to-GAPDH ratio, with that of FiPSCs arbitrarily set at 1 (n = 3).
These data suggest that expression of GREM1 and GATA6 and the suppression of EGR1 may explain why these cells can be differentiated to insulin-producing cells more readily.
DISCUSSION
Based on a report regarding the differences in the differentiation induction efficiency among 17 ESC lines (16), we analyzed the microarray data ofthe HUES8 and HUES6 cell lines. The data that we obtained suggest that a high expression of GREM1 and GATA6, and the suppression of EGR1, NLRP2, and SMC3 might be important factors for the differentiation of ESCs into pancreatic cells. We next analyzed the expression of these genes in PiPSCs and FiPSCs based on epigenetic memory of iPSCs (1). The data for iPSCs suggest that a high expression of GREM1 and GATA6 and the suppression of EGR1 could play an important role in the differentiation of iPSCs into pancreatic cells. GATA6 is well known to play an important role in the development of the pancreas (5,13). On the other hand, there are few reports that have investigated the relationship between GREM1 and pancreas development. Dodge et al. analyzed the gene expression profiles of three different stages during in vitro islet generation: namely, the initial adherent, expanded, and differentiated stages. In the expanded and differentiated stages, the expression of the GREM1 gene was upregulated (6). GREM1 is a member of a BMP (osteogenic protein) antagonist family. Since the BMP signal is important for liver development, its inhibition by GREM1 might therefore be an important factor for pancreas development.
The protein encoded by the EGR1 gene belongs to the EGR family of C2H2-type zinc-finger proteins. It is a nuclear protein, and it functions as a transcriptional regulator. One group reported EGR1 to regulate the transcription of the insulin gene (7) and the Pdx1 gene (8). Since the relationship between EGR1 and pancreas development is unknown, we therefore plan to investigate the role of EGR1 in pancreas development in a future study.
In conclusion, the expression of GREM1 and the inhibition of EGR1 might therefore be important factors in the differentiation of PSCs into insulin-producing cells. Further study of these genes is needed to generate insulin-producing cells with glucose sensitivity from ES/iPS cells.
ACKNOWLEDGMENTS
This work was supported in part by the Japan Society for the Promotion of Science and the Ministry of Health, Labour and Welfare. The authors declare no conflicts of interest.
REFERENCES
- 1. Bar-Nur O.; Russ H. A.; Efrat S.; Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived fromhuman pancreatic islet beta cells. Cell Stem Cell 9:17–23; 2011. [DOI] [PubMed] [Google Scholar]
- 2. Cheng X.; Ying L.; Lu L.; Galvão A. M.; Mills J. A.; Lin H. C.; Kotton D. N.; Shen S. S.; Nostro M. C.; Choi J. K.; Weiss M. J.; French D. L.; Gadue P. Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell 10:371–384; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D’Amour K. A.; Agulnick A. D.; Eliazer S.; Kelly O. G.; Kroon E.; Baetge E. E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23:1534–1541; 2005. [DOI] [PubMed] [Google Scholar]
- 4. D’Amour K. A.; Bang A. G.; Eliazer S.; Kelly O. G.; Agulnick A. D.; Smart N. G.; Moorman M. A.; Kroon E.; Carpenter M. K.; Baetge E. E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 24:1392–1401; 2006. [DOI] [PubMed] [Google Scholar]
- 5. Decker K.; Goldman D. C.; Grasch C. L.; Sussel L. Gata6 is an important regulator of mouse pancreas development. Dev. Biol. 298:415–429; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dodge R.; Loomans C.; Sharma A.; Bonner-Weir S. Developmental pathways during in vitro progression of human islet neogenesis. Differentiation 77:135–147; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eto K.; Kaur V.; Thomas M. K. Regulation of insulin gene transcription by the immediate-early growth response gene Egr-1. Endocrinology 147:2923–2935; 2006. [DOI] [PubMed] [Google Scholar]
- 8. Eto K.; Kaur V.; Thomas M. K. Regulation of pancreas duodenum homeobox-1 expression by early growth response-1. J. Biol. Chem. 282:5973–5983; 2007. [DOI] [PubMed] [Google Scholar]
- 9. Gentleman R. C.; Carey V. J.; Bates D. M.; Bolstad B.; Dettling M.; Dudoit S.; Ellis B.; Gautier L.; Ge Y.; Gentry J.; Hornik K.; Hothorn T.; Huber W.; Iacus S.; Irizarry R.; Leisch F.; Li C.; Maechler M.; Rossini A. J.; Sawitzki G.; Smith C.; Smyth G.; Tierney L.; Yang J. Y.; Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5:R80; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inoue H.; Yamanaka S. The use of induced pluripotent stem cells in drug development. Clin. Pharmacol. Ther. 89:655–661; 2011. [DOI] [PubMed] [Google Scholar]
- 11. International Stem Cell Initiative; Adewumi O.; Aflatoonian B.; Ahrlund-Richter L.; Amit M.; Andrews P. W.; Beighton G.; Bello P. A.; Benvenisty N.; Berry L. S.; Bevan S.; Blum B.; Brooking J.; Chen K. G.; Choo A. B.; Churchill G. A.; Corbel M.; Damjanov I.; Draper J. S.; Dvorak P.; Emanuelsson K.; Fleck R. A.; Ford A.; Gertow K.; Gertsenstein M.; Gokhale P. J.; Hamilton R. S.; Hampl A.; Healy L. E.; Hovatta O.; Hyllner J.; Imreh M. P.; Itskovitz-Eldor J.; Jackson J.; Johnson J. L.; Jones M.; Kee K.; King B. L.; Knowles B. B.; Lako M.; Lebrin F.; Mallon B. S.; Manning D.; Mayshar Y.; McKay R. D.; Michalska A. E.; Mikkola M.; Mileikovsky M.; Minger S. L.; Moore H. D.; Mummery C. L.; Nagy A.; Nakatsuji N.; O’Brien C. M.; Oh S. K.; Olsson C.; Otonkoski T.; Park K. Y.; Passier R.; Patel H.; Patel M.; Pedersen R.; Pera M. F.; Piekarczyk M. S.; Pera R. A.; Reubinoff B. E.; Robins A. J.; Rossant J.; Rugg-Gunn P.; Schulz T. C.; Semb H.; Sherrer E. S.; Siemen H.; Stacey G. N.; Stojkovic M.; Suemori H.; Szatkiewicz J.; Turetsky T.; Tuuri T.; van den Brink S.; Vintersten K.; Vuoristo S.; Ward D.; Weaver T. A.; Young L. A.; Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25:803–816; 2007. [DOI] [PubMed] [Google Scholar]
- 12. Itskovitz-Eldor J.; Schuldiner M.; Karsenti D.; Eden A.; Yanuka O.; Amit M.; Soreq H.; Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 6:88–95; 2000. [PMC free article] [PubMed] [Google Scholar]
- 13. Ketola I.; Otonkoski T.; Pulkkinen M. A.; Niemi H.; Palgi J.; Jacobsen C. M.; Wilson D. B.; Heikinheimo M. Transcription factor GATA-6 is expressed in the endocrine andGATA-4 in the exocrine pancreas. Mol. Cell. Endocrinol. 226:51–57; 2004. [DOI] [PubMed] [Google Scholar]
- 14. Kim K.; Doi A.; Wen B.; Ng K.; Zhao R.; Cahan P.; Kim J.; Aryee M. J.; Ji H.; Ehrlich L. I.; Yabuuchi A.; Takeuchi A.; Cunniff K. C.; Hongguang H.; McKinney-Freeman S.; Naveiras O.; Yoon T. J.; Irizarry R. A.; Jung N.; Seita J.; Hanna J.; Murakami P.; Jaenisch R.; Weissleder R.; Orkin S. H.; Weissman I. L.; Feinberg A. P.; Daley G. Q. Epigenetic memory in induced pluripotent stem cells. Nature 467:285–290; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohi Y.; Qin H.; Hong C.; Blouin L.; Polo. J. M.; Guo T.; Qi Z.; Downey S. L.; Manos P. D.; Rossi D. J.; Yu J.; Hebrok M.; Hochedlinger K.; Costello J. F.; Song J. S.; Ramalho-Santos M. Incomplete DNA methylation underlies a transcriptional memory of somatic cells inhuman iPS cells. Nat. Cell Biol. 13:541–549; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Osafune K.; Caron L.; Borowiak M.; Martinez R. J.; Fitz-Gerald C. S.; Sato Y.; Cowan C. A.; Chien K. R.; Melton D. A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26:313–315; 2008. [DOI] [PubMed] [Google Scholar]
- 17. Polo J. M.; Liu S.; Figueroa M. E.; Kulalert W.; Eminli S.; Tan K. Y.; Apostolou E.; Stadtfeld M.; Li Y.; Shioda T.; Natesan S.; Wagers A. J.; Melnick A.; Evans T.; Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28:848–855; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robertson J. A. Human embryonic stem cell research: Ethical and legal issues. Nat. Rev. Genet. 2:74–78; 2001. [DOI] [PubMed] [Google Scholar]
- 19. Shapiro A. M.; Lakey J. R.; Ryan E. A.; Korbutt G. S.; Toth E.; Warnock G. L.; Kneteman N. M.; Rajotte R. V. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N. Engl. J. Med. 343:230–238; 2000. [DOI] [PubMed] [Google Scholar]
- 20. Takahashi K.; Tanabe K.; Ohnuki M.; Narita M.; Ichisaka T.; Tomoda K.; Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872; 2007. [DOI] [PubMed] [Google Scholar]
- 21. Thomson J. A.; Itskovitz-Eldor J.; Shapiro S. S.; Waknitz M. A.; Swiergiel J. J.; Marshall V. S.; Jones J. M. Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147; 1998. [DOI] [PubMed] [Google Scholar]
- 22. Van Hoof D.; D’Amour K. A.; German M. S. Derivation of insulin-producing cells fromhuman embryonic stem cells. Stem Cell Res. 3:73–87; 2009. [DOI] [PubMed] [Google Scholar]