Abstract
Placental tissue is a biomaterial with remarkable potential for use in regenerative medicine. It has a three-layer structure derived from the fetus (amnion and chorion) and the mother (decidua), and it contains huge numbers of cells. Moreover, placental tissue can be collected without any physical danger to the donor and can be matched with a variety of HLA types. The decidua-derived mesenchymal cells (DMCs) are highly proliferative fibroblast-like cells that express a similar pattern of CD antigens as bone marrow-derived mesenchymal cells (BM-MSCs). Here we demonstrated that induced pluripotent stem (iPS) cells could be efficiently generated from DMCs by retroviral transfer of reprogramming factor genes. DMC-hiPS cells showed equivalent characteristics to human embryonic stem cells (hESCs) in colony morphology, global gene expression profile (including human pluripotent stem cell markers), DNA methylation status of the OCT3/4 and NANOG promoters, and ability to differentiate into components of the three germ layers in vitro and in vivo. The RNA expression of XIST and the methylation status of its promoter region suggested that DMC-iPSCs, when maintained undifferentiated and pluripotent, had three distinct states: (1) complete X-chromosome reactivation, (2) one inactive X-chromosome, or (3) an epigenetic aberration. Because DMCs are derived from the maternal portion of the placenta, they can be collected with the full consent of the adult donor and have considerable ethical advantages for cell banking and the subsequent generation of human iPS cells for regenerative applications.
Keywords: Induced pluripotent stem cells (iPSCs), Decidua, Mesenchymal cells, X-chromosome inactivation
INTRODUCTION
Great breakthroughs in cell reprogramming technology have led to the generation of induced pluripotent stem cells (iPSCs) and dramatic advances in stem cell research (43). To date, iPSCs have been generated efficiently from various accessible human tissues, including dermal fibroblasts (22,27,37,42,47), blood cells (9,10,21), neural stem cells (6,19), mesenchymal stem cells (MSCs) (4,33,35), and keratinocytes (1). It is generally accepted that iPSCs are likely to contribute not only to the realization of regenerative medicine through the use of pluripotent stem cells but also to the elucidation of the molecular pathogenesis of many intractable diseases. The promise of therapies using human iPSCs (hiPSCs) has driven an intense search for good cell sources (34).
Human placenta is a fetal adnexal tissue that contains extraembryonic cells. It is composed of three layers: the amnion and the chorion, which are of fetal origin, and the decidua, which is of maternal origin. Fibroblast-like adherent cells isolated from various components of the fetal adnexa, including the placenta, show multipotency or pluripotency, suggesting that the fetal adnexa represents a new source of human stem cells (3,8,14,46). We recently isolated adherent cells from human term decidua vera, that is, decidua-derived mesenchymal cells (DMCs), and reported some of their properties (18). DMCs are highly proliferative fibroblast-like cells of purely maternal origin, and they express a similar pattern of cluster of differentiation (CD) antigens as bone marrow-derived mesenchymal cells (BM-MSCs). DMCs differentiate well into chondrocytes and moderately into adipocytes, but hardly at all into osteoblasts, in vitro (18). The high-proliferative ability of DMCs makes them relatively easy to prepare and maintain, and their derivation from the maternal portion of the human placenta, which is otherwise discarded, resolves many ethical concerns. These unique properties of DMCs give them several advantages for clinical use, and support their potential as an attractive alternative to allogeneic human stem cells for use in regenerative medicine (15,18).
In this study, we asked whether DMCs could be reprogrammed into hiPSCs and acquire pluripotency. The four reprogramming factors octamer-binding transcription factor 3/4 (OCT3/4), sex-determining region Y box 2 (SOX2), Krüppel-like factor 4 (KLF4), and myelocytomatosis viral oncogene homolog (MYC; OSKM), carried by retroviral vectors (42), were used to reprogram several lines of DMCs. The resulting hiPSCs closely resembled human embryonic stem cells (hESCs) in their morphology, and they expressed hESC markers, which are low or undetectable in native DMCs. These hiPSCs formed embryoid bodies in vitro and teratomas in vivo that contained components of all three germ layers. We think these unique properties of DMCs give them several advantages for clinical use and that the generation of hiPSCs from DMCs could lead to the establishment of hiPSC-banking systems. Such systems would increase the availability of allogeneic hiPSCs, because tissues expressing a wide array of human leukocyte antigen (HLA) variants could be banked for clinical applications.
MATERIALS AND METHODS
Human Tissues and Cells
This study was carried out in accordance with the principles of the Helsinki Declaration, and approvals to use human tissues were obtained from the ethical committee of Osaka National Hospital (No. 72, No. 109, and No. 110). The donor bloods were serologically tested for hepatitis B and C, human immunodeficiency virus (HBs, HCV, HIV), and syphilis. Full-term placental tissues, infant skin samples obtained during plastic surgery, and the results of donor blood tests were collected at the Osaka National Hospital with written informed consent.
Human ESCs (clone KhES1) were obtained from Kyoto University (Kyoto, Japan) (41) and propagated at Keio University in accordance with Japanese guidelines on the utilization of hESCs, under approval of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the ethical committee of Keio University. Human iPSCs (clone 201B7) (42) were obtained from the RIKEN Cell Bank (Tsukuba, Japan).
Human Primary Cell Culture
Human DMCs were propagated from human term decidua vera, in Dulbecco's modified Eagle's medium (DMEM)/F-12 (1:1) supplemented with 10% fetal bovine serum (FBS), HEPES (15 mM), and antibiotic–antimycotic solution (Invitrogen, Carlsbad, CA, USA) at 37°C in 5% CO2 as previously described (18,28). Human amnion-derived epithelial cells (AECs), human amnion-derived mesenchymal cells (AMCs), and human primary BM-MSCs were also propagated as previously described (18). Human primary dermal fibroblast cells (DFBs) were isolated from the toe skin tissue of infants with polydactyly and cultured in DMEM/F-12 (1:1) with 10% fetal bovine serum (FBS).
Generation and Culture of hiPSCs
Four pMX retroviral vectors encoding the reprogramming factors (OSKM) were obtained from Addgene, Inc. (Cambridge, MA, USA) (42). Amphotropic retroviruses were produced by the transfection of Platinum-A retroviral packaging cells (Cell Biolabs, Inc., San Diego, CA, USA) using FuGENE® 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN, USA). Host cells were infected with the retrovirus supernatants supplemented with polybrene (4 mg/ml, Nacalai Tesque, Kyoto, Japan). On day 5 postinfection, the cells were replated on mitomycin C-treated mouse embryonic fibroblast (MEF) feeder cells at a density of 2.5 × 104 cells/6-cm-diameter culture dish or 5 × 105 cells/10-cm culture dish. On the next day, the growth medium was replaced with hESC medium consisting of DMEM/F-12 (1:1) with 20% knockout serum replacement (KSR, Invitrogen)/2-mercaptoethanol (1 mM; Invitrogen)/basic fibroblast growth factor (bFGF; 5ng/ml; Wako Pure Chemical Industries, Ltd., Osaka Japan). The number of alkaline phosphatase (ALP)+ colonies in a 6-cm culture dish was screened and estimated by staining with 1-Step™ NBT/BCIP reagent (Pierce Biotechnology, Rockford, IL, USA), according to the manufacturer's specifications at 28 days postinfection. Concurrently, human ESC-like colonies grown in the 10-cm culture dish were picked up around 30 days after gene induction. The colonies were clonally expanded for further analyses. Human ES cells (KhES1) and hiPSCs (201B7) were also propagated on feeder cells with hESC medium (41).
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated using an RNeasy Mini kit (Qiagen, Valencia, CA, USA), and cDNAs were synthesized using a PrimeScript® First-Strand Synthesis Kit (Takara Bio, Inc., Shiga, Japan), according to the manufacturer's specifications. Quantitative PCR analysis was performed using gene-specific primers (Table 1) with Power SYBR® Green PCR Master Mix and the 7300 real-time PCR system (Applied Biosystems, Foster, CA, USA), and the comparative Ct or standard curve method (20). Total RNA isolated from various human tissues was used for controls (BD Biosciences, San Jose, CA, USA).
Table 1.
Primers Used for RT-PCR and PCR
| Description and Genes | Symbol | Gene ID | Primer Sequence (5′ to 3′) | ||
|---|---|---|---|---|---|
| Transgene detection | |||||
| POU class 5 homeobox 1 | OCT3/4Tg | 5460 | S | CCCCAGGGCCCCATTTTGGTACC | |
| A | TTATCGTCGACCACTGTGCTGCTG | ||||
| SRY (sex-determining region Y)-box 2 | SOX2Tg | 6657 | S | GGCACCCCTGGCATGGCTCTTGGCTC | |
| A | TTATCGTCGACCACTGTGCTGCTG | ||||
| v-Myc myelocytomatosis viral oncogene homolog | MYCTg | 4609 | S | CAACAACCGAAAATGCACCAGCCCCAG | |
| A | TTATCGTCGACCACTGTGCTGCTG | ||||
| Krüppel-like factor 4 | KLF4Tg | 9314 | S | ACGATCGTGGCCCCGGAAAAGGACC | |
| A | TTATCGTCGACCACTGTGCTGCTG | ||||
| Expression of hES markers | |||||
| POU class 5 homeobox 1 | OCT3/4 | 5460 | S | GACAGGGGGAGGGGAGGAGCTAGG | |
| A | CTTCCCTCCAACCAGTTGCCCCAAAC | ||||
| SRY (sex-determining region Y)-box 2 | SOX2 | 6657 | S | GGGAAATGGGAGGGGTGCAAAAGAGG | |
| A | TTGCGTGAGTGTGGATGGGATTGGTG | ||||
| v-Myc myelocytomatosis viral oncogene homolog | MYC | 4609 | S | GCGTCCTGGGAAGGGAGATCCGGAGC | |
| A | TTGAGGGGCATCGTCGCGGGAGGCTG | ||||
| Krüppel-like factor 4 | KLF4 | 9314 | S | ACGATCGTGGCCCCGGAAAAGGACC | |
| A | TGATTGTAGTGCTTTCTGGCTGGGCTCC | ||||
| Nanog homeobox | NANOG | 79923 | S | GCAGAAGGCCTCAGCACCTA | |
| A | GGTTCCCAGTCGGGTTCAC | ||||
| Telomerase reverse transcriptase | TERT | 7015 | S | CTCCATCCTGAAAGCCAAGAA | |
| A | CGAGTCAGCTTGAGCAGGAA | ||||
| X (inactive)-specific transcript(nonprotein coding) | XIST | 7503 | S | TGGCAGGGAGTGCCAGCTCCA | |
| A | GACCAAGGTGCATGGCTGCGGT | ||||
| Expression of endoderm markers | |||||
| Forkhead box A2 | FOXA2 | 3170 | S | TGCTGGTCGTTTGTTGTGG | |
| A | CATGTTGCTCACGGAGGAGTAG | ||||
| GATA binding protein 4 | GATA4 | 2626 | S | CTCTTCAGGCAGTGAGAGCC | |
| A | GGTCCGTGCAGGAATTTGAGG | ||||
| SRY (sex-determining region Y)-box 17 | SOX17 | 64321 | S | CCCATAGTTGGATTGTCAAAACC | |
| A | CACACCCAGGACAACATTTCTTT | ||||
| α-Fetoprotein | AFP | 174 | S | TTGAGAAACCCACTGGAGATGA | |
| A | GTTATGTCTTCCCTCTTCACTTTGG | ||||
| Albumin | ALB | 213 | S | GTTGCATGAGAAAACGCCAGTA | |
| A | AGCATGGTCGCCTGTTCAC | ||||
| Expression of mesoderm markers | |||||
| T, brachyury homolog (mouse) | Brachyury | 6862 | S | TGGAATGCCTGCCCATC | |
| A | CCGTTGCTCACAGACCACA | ||||
| NK2 transcription factor related, locus 5 | NKX2-5 | 1482 | S | CCCCTGGATTTTGCATTCAC | |
| A | CGTGCGCAAGAACAAACG | ||||
| Msh homeobox 1 | MSX1 | 4487 | S | CAGAAGATGCGCTCGTCAAA | |
| A | CGGCTTACGGTTCGTCTTG | ||||
| Collagen, type II, α1 | COL2A1 | 1280 | S | GGAAGAGTGGAGACTACTGGATTGAC | |
| A | TCCATGTTGCAGAAAACCTTCA | ||||
| Expression of neural markers | |||||
| Neurogenin 2 | NEUROG2 | 63973 | S | ATGCCTATTGTCCTGTCCTTCTCT | |
| A | TGACTTCTAACCTGCCCCTCTAAC | ||||
| SRY (sex-determining region Y)-box 1 | SOX1 | 6656 | S | AGCAGTTGTTTCTGGAAGAGTCTGT | |
| A | AGGCCCTTATCCCGGACTAA | ||||
| Paired box 6 | PAX6 | 5080 | S | ACCTGGCTAGCGAAAAGCAA | |
| A | CCCGTTCAACATCCTTAGTTTATCA | ||||
| Expression of astrocyte marker | |||||
| Glial fibrillary acidic protein | GFAP | 2670 | S | ACATCGAGATCGCCACCTAC | |
| A | ACATCACATCCTTGTGCTCC | ||||
| Expression of Epidermis marker | |||||
| Keratin 17 | KRT17 | 3872 | S | AGGAGATTGCCACCTACCG | |
| A | CTTGCCATCCTGGACCTCTT | ||||
| Expression of other genes | |||||
| v-Myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived | MYCL1 | 4610 | S | CGAGAGCCCAAGCGACTCGGAGAA | |
| A | CAGGGGGTCTGCTCGCACCG | ||||
| GLIS family zinc finger 1 | GLIS1 | 148979 | S | TCTGCCCAGCCCACAAGGTTACCA | |
| A | GGCAGCGCTGTGGGGCATGA | ||||
| GLIS family zinc finger 2 | GLIS2 | 84662 | S | CCGCTGTCCGACCTGCAGCA | |
| A | GGCTTCTCACCTGTGTGCGACCG | ||||
| GLIS family zinc finger 3 | GLIS3 | 169792 | S | TGCTCACCAATGGGAAGCCGCG | |
| A | AGCCAAGAGCCCCTTTCCCAGGAT | ||||
| Zinc finger and SCAN domain containing 4 | ZSCAN4 | 201516 | S | TCCACCTGCCTTAGTCCACGTCCA | |
| A | TGGGAGGGTGTCCCCATGTTTGCT | ||||
| Internal control | |||||
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 2597 | S | CCACTTTGTCAAGCTCATTTCCT | |
| A | TCTCTTCCTCTTGTGCTCTTGCT | ||||
| Bisulfite genomic sequencing PCR | |||||
| Nanog homeobox promoter | Bis-NANOG P | 79923 | S | GTTGGGTTTGTTTTTAGGTTTT | |
| A | CATAAAACAACCAACTCAATCC | ||||
| POU class 5 homeobox 1 promoter | Bis-OCT3/4 P | 5460 | S | GTTAAGGTTAGTGGGTGGGATT | |
| A | AACATAAAAAAATCCCCCACA | ||||
| Methylation-specific PCR | |||||
| Unmethylated-XIST promoter | Unmet-XIST P | 7503 | S | TTTTGTTGTAGTGTTTAAGTGGT | |
| A | AACCCACCATATTTTACACTACA | ||||
| Methylated-XIST promoter | Met-XIST P | 7503 | S | TGTCGTAGTGTTTAAGTGGC | |
| A | CCGCCATATTTTACACTACG |
S, sense primer; A, antisense primer.
Microarray Analysis
The microarray study was carried out using the GeneChip array (Human Genome U133 Plus 2.0 gene expression array, Affymetrix, Inc., Santa Clara, CA, USA). One hundred nanograms of total RNA was used to synthesize amplified RNA (aRNA) using the 3′ IVT Express Kit, according to the manufacturer's instructions (Affymetrix). After aRNA purification, 15 µg of aRNA was fragmented and hybridized with a preequilibrated GeneChip array at 45°C for 16 h. The GeneChip array was then washed, stained, and scanned according to the manufacturer's instructions. The gene expression data were extracted using Affymetrix Expression Console software, and the data sets were analyzed using GeneSpring GX software (Agilent Technologies, Inc., Santa Clara, CA, USA).
Flow cytometry (FCM) Analysis
Cells were preincubated with Y-27632 (Wako Pure Chemical Industries, Ltd.) for 30 min and then dissociated by trypsin/EDTA (Invitrogen). The dissociated cells were fixed with 4% paraformaldehyde (PFA) for 20 min on ice, washed with PBS, and then reacted with the following primary antibodies (Abs) for 30 min at 4°C: anti-stage-specific embryonic antigen 4 (SSEA-4) Ab (MC-813-70, Chemicon International, Inc., Temecula, CA, USA), anti-SSEA-3 Ab (MC-631, Chemicon), anti-TRA-1-60 Ab (TRA-1-60, Chemicon), or anti-TRA-1-81 Ab (TRA-1-81, Chemicon). After being washed, the cells were incubated with the appropriate secondary Abs (Alexa 488-conjugated anti-mouse IgG, anti-mouse IgM, or anti-rat IgM, Molecular Probes, Invitrogen) for 30 min at 4°C. The stained samples were analyzed by a FACSCalibur™ flow cytometer (BD Biosciences). All stainings were performed with matched-isotype controls.
Karyotype Analysis
Cells were cultured with colcemid (0.06 µg/ml; Invitrogen) at 37°C for 4 h, treated with Y-27632 to prevent apoptosis, and then dissociated with 0.05% trypsin/EDTA. The cells were incubated in KCl solution (75 mM) for 20min and then fixed in Carnoy fluid. The fixed samples were heat-denatured at 95°C for 2 h, incubated in 0.025% trypsin for 10 s, and then stained with Giemsa (Merck, Darmstadt, Germany) for 7 min. The samples were observed with a microscope (Carl Zeiss, Hallbergmoos, Germany), and metaphase samples were analyzed using Ikaros (MetaSystems, Altlussheim, Germany).
Genotyping of Short-Tandem Repeat (STR) Polymorphisms
Genomic DNA (gDNA) was extracted by DNAzol Reagent (Invitrogen). STR loci were analyzed with the Powerplex 16 system (Promega, Madison, WI, USA) using an ABI PRISM3100 Genetic Analyzer (Applied Biosystems) and analyzed by GeneMapper software (Applied Biosystems) following the manufacturer’s instructions (18).
Bisulfite Modification, DNA Sequencing, and Methylation-Specific PCR (MSP)
Genomic DNA was bisulfite-treated with an EZ DNA methylation-Gold Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. The promoter regions of human OCT3/4 and NANOG were amplified with specific primer sets (Table 1) by TaKaRa Taq™ Hot Start Version (Takara Bio, Inc.). The PCR products were subcloned into the pCR®2.1 vector (Invitrogen). Ten clones were sequenced with a universal primer using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems) and analyzed with Sequence analysis software (Applied Biosystems), following the manufacturer's instructions. Episcope® Methylated and Unmethylated HCT116 gDNA (Takara Bio, Inc.) were used as the control human genomic DNAs for the methylation analysis.
In Vitro Differentiation
Cells were incubated with 1 mg/ml collagenase type IV (Invitrogen) for 5 min at 37°C and harvested by scraper. The harvested colonies were broken into properly sized clumps by pipetting and then incubated on noncoated plates in human ES cell medium without bFGF to form embryoid bodies (EBs). After 10 days in floating culture, the EBs that formed were harvested for transcript analysis. Some of the EBs were transferred to gelatin-coated chamber slides (Nalge Nunc International, Rochester, NY, USA) and cultured for another 2 weeks (total 24 days) in DMEM supplemented with 10% FBS at 37°C in 5% CO2.
Teratoma Formation Assay
NOG (non-obese diabetic/Shi-severe combined immunodeficient interleukin 2 receptor γ chain null; NOD/Shi-scid IL2Rγ null) mice (16) aged 6–8 weeks were maintained under specific pathogen-free conditions in the Animal Facility of the Central Institute for Experimental Animals (CIEA) in accordance with the guidelines of the Animal Care Committee at CIEA. All mouse studies were approved by the Animal Care Committee at CIEA. The recipient mice were anesthetized by isoflurane inhalation (Dainippon Pharmaceutical Co., Ltd., Osaka, Japan). Human iPSCs were transplanted into both subcutaneous tissue and kidney capsules. For the subcutaneous transplantation, hiPSCs (1 × 106 cells/0.1 ml of serum-free medium) were injected subcutaneously into the flank. For transplantation into the kidney capsule, the kidney was exteriorized through a dorsal–horizontal incision. A syringe fitted with a 29-G needle with a flattened tip was used. The needle was introduced into the kidney at a site apart from the transplanted region. The kidney was penetrated, and the tip of the needle was held just beneath the renal capsule. The suspension of hiPSCs (1 × 105 cells/10 µl of serum-free medium) was then injected underneath the kidney capsule. The mice were examined daily, and tumors were measured with calipers.
Teratoma samples were dissected and fixed with 4% (v/v) phosphate-buffered formalin, and paraffin-embedded sections were stained with hematoxylin and eosin using routine procedures (H&E staining).
Immunocytochemical and Immunohistochemical Staining
Cultured cells were fixed in 4% PFA, and tissue specimens were permeabilized with 0.1% Triton X-100. Nonspecific reactions were blocked with 10% normal goat serum. The samples were reacted with the following Abs overnight at 4°C: anti-α-fetoprotein (AFP) Ab (SantaCruz, Santa Cruz, CA, USA), anti-cytokeratin19 Ab (clone A53-B/A2, Santa Cruz), anti-desmin Ab (Thermo, Waltham, MA, USA), anti-α-smooth muscle actin (SMA) Ab (clone 1A4, Dako, Glostrup, Denmark), anti-glial fibrillary acidic protein (GFAP) Ab (Sigma, St. Louis, MO, USA), anti-βIII-tubulin Ab (clone TuJ1, Babco, Richmond, CA, USA), anti-vimentin Ab (Chemicon), anti-OCT3/4 Ab (Chemicon), and anti-NANOG Ab (Reprocell, Tokyo, Japan). The samples were then reacted with the appropriate secondary Abs (AlexaFluor®-488-conjugated goat anti-mouse IgG Ab, AlexaFluor®-568-conjugated goat anti-rabbit IgG Ab, Molecular Probes, Invitrogen), and TO-PRO-3® iodide (1 mM, Molecular Probes, Invitrogen) for 1 h at room temperature (RT).
The immunohistochemical staining of teratoma samples was carried out using an automated staining device with the Bond™ Polymer Refine Detection system (Leica Microsystems K.K., Tokyo, Japan) according to the manufacturer's instructions with minor modifications. In brief, 4-μm sections of formalin-fixed, paraffin-embedded tissues were deparaffinized with Bond™ Dewax Solution (Leica Microsystems), and antigen retrieval was performed using Bond™ ER Solution (Leica Microsystems) for 30 min at 100°C. Nonspecific peroxidase activity was quenched by incubating with 0.3% hydrogen peroxide for 5 min. The sections were incubated for 15 min at ambient temperature with the following Abs: anti-cytokeratin Ab (AE1/AE3, Leica Microsystems), anti-neural cell adhesion molecule (NCAM; CD56) Ab (1B6, Nichirei Bioscience, Tokyo, Japan), anti-desmin Ab (D33, Nichirei Bioscience), for 1 h at room temperature. The signals were detected by BondTM Polymer Refine Detection (Leica Microsystems) with 3,3′-diaminobenzidine tetrahydrochloride (DAB; Dojindo Laboratories, Kumamoto, Japan) substrate as the chromogen. The sections were counterstained with hematoxylin.
The immune-stained preparations were examined using a confocal laser scanning microscope (LSM510, Carl Zeiss) or a light microscope (IX70, Olympus, Tokyo, Japan). All stainings were performed with matched isotype controls.
Statistical Analysis
Significant differences in the gene expression levels obtained by qRT-PCR were determined by analysis of variance (ANOVA) with post hoc comparisons. Details are provided in the figure legends.
RESULTS
Phenotype Analysis of Human Placenta-Derived Cells
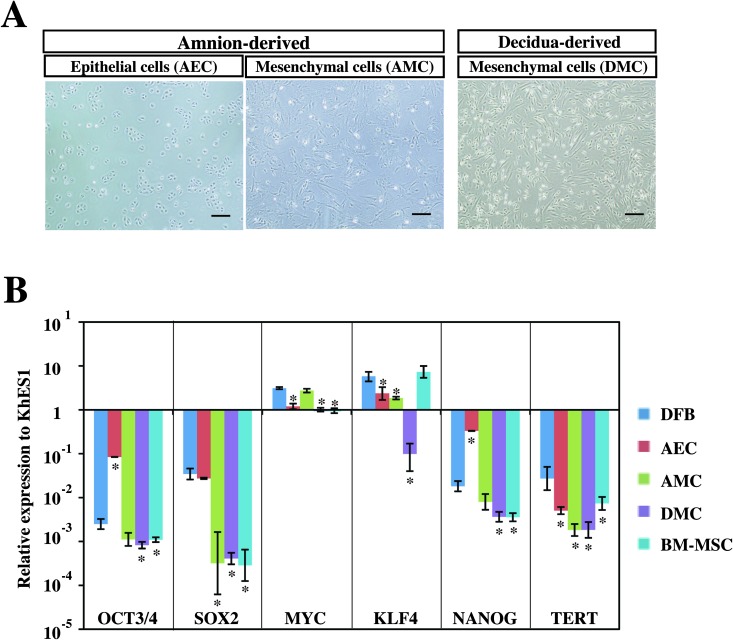
To learn whether hiPSCs could be generated from placenta-derived cells by introducing retroviruses expressing the four reprogramming factors (OSKM), we first propagated three different kinds of somatic cells, DMCs, AECs, and AMCs, from human full-term placenta tissue (Fig.1A) (18) and analyzed their endogenous expression of six pluripotent cell-specific marker genes in comparison with hESCs (KhES1), human primary DFBs, and human primary BM-MSCs (Fig. 1B). Quantitative RT-PCR analysis showed that c-MYC was expressed endogenously in all six cell types at the same level or higher than in hESCs and that the KLF4 levels were about the same in every cell type except DMCs (Fig.1B). In contrast, the expression levels of the other four genes in five cell types (DFBs, AECs, AMCs, DMCs, and BM-MSCs) were significantly lower than in KhES1, to differing degrees (Fig. 1B).
Figure 1.
Phenotype analysis of human placenta-derived cells. (A) Phase contrast images of amnion-derived epithelial cells (AECs), amnion-derived mesenchymal cells (AMCs), and decidua-derived mesenchymal cells (DMCs). Scale bars: 200 µm. (B) Endogenous transcriptional expression of four reprogramming factors (OCT3/4, SOX2, KLF4, c-MYC) and pluripotent stem cell marker genes (NANOG and TERT). GAPDH was used as the internal control gene, and data are presented as the mean ± SD (n = 3). Statistical significance was determined by Scheffe's test after ANOVA, and the p values between dermal fibroblast cells (DFBs) and other cells are shown (*p < 0.01). See Table 1 for gene definitions. BM-MSCs, bone marrow-derived mesenchymal cells.
Although AECs expressed higher levels of OCT3/4, NANOG, and SOX2 than the other four cell types (Fig.1B) and thus seemed likely to be good candidate cells for generating hiPSCs (17), their growth was too slow (18) for them to be efficiently infected by retroviral vectors, and they were therefore excluded from further examination in this study.
Generation of iPSCs From Human DMCs
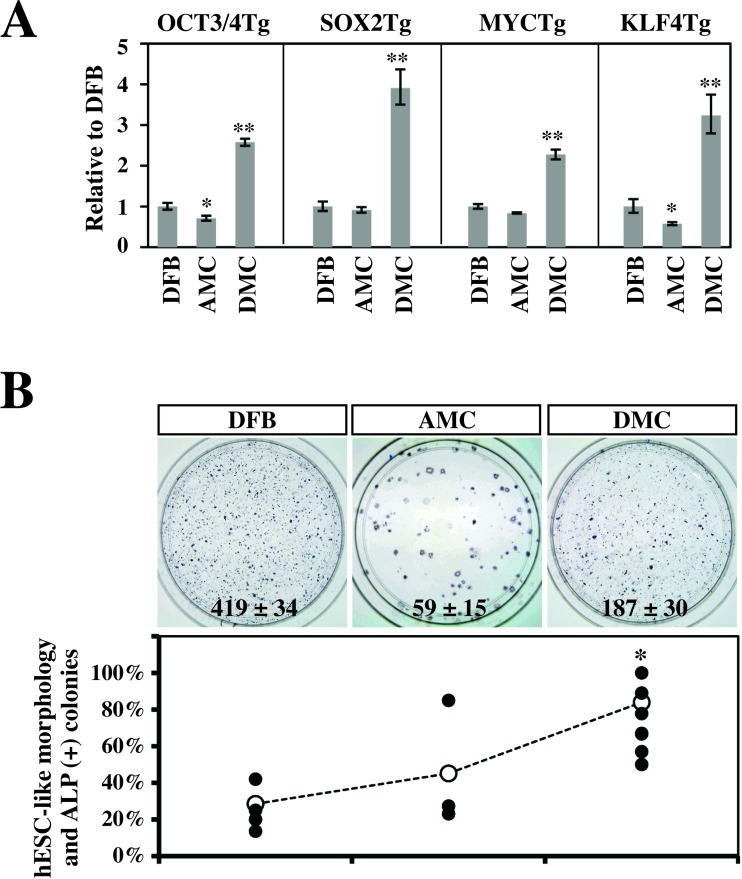
We next examined the gene transduction efficiency by amphotropic retrovirus infection. Three human primary cell types (DFBs, AMCs, and DMCs) were transduced with amphotropic retroviruses encoding the four reprogramming factors (OSKM), and OSKM transgene expression on day 3 postinfection was quantified by qRT-PCR (Fig.2A). The expression of the exogenous reprogramming factors was two to three times higher in DMCs than DFBs (p < 0.01) (Fig. 2A), and AMCs showed a comparable or lower expression than DFBs(p < 0.05). On day 5 postinfection, the cells were split and replated on feeder cultures, and on day 28 postinfection, the reprogramming efficiency of each cell type was estimated by the number of ALP+ (primary) colonies on 6-cm culture dishes. Approximately three times more primary colo-nies were generated from DMCs than AMCs, although the number was still half that generated from DFBs (Fig.2B, top).
Figure 2.
Efficiencies of gene transduction and generation of alkaline phosphatase (ALP)+ colonies. (A) Gene transduction efficiencies of amphotropic retrovirus infection. Efficiencies were estimated on day 3 postinfection by qRT-PCR and are shown in comparison with those in DFBs (mean ± SD, 3 independent experiments). Statistical significance was determined by Scheffe's test after ANOVA (*p < 0.05, **p < 0.01). (B) Generation efficiency of ALP+ colonies. (Top) Representative result of ALP+ primary colonies (blue) emerging from 2.5 × 104 infected cells on a 6-cm culture dish (mean ± SD, 3 independent experiments). (Bottom)Efficiencies of retention of both the human embryonic stem cell (hESC)-like morphology and ALP activity in secondary colonies. Efficiency was determined by dividing the number of secondary colonies by the number of primary hESC-like colonies emerged on two 10-cm dishes. ●Independent results from 5 DFB experiments (DFB01, DFB18, DFB22, DFB37, and DFB44), 3AMC experiments (AMC41 and a duplicate of AMC49), and 12 DMC experiments (DMC41 and duplicates of DMC54, DMC70, DMC71, DMC72, DMC73, DMC75, DMC76, DMC77, DMC83, and DMC92). ?Mean value. Statistical significance was determined by Scheffe's test after ANOVA (*p < 0.01). See Table 1 for gene definitions.
iPSCs of each cell type were generated from two 10-cm dishes containing 1 × 105 cells, infected as described above. The efficiency of hESC-like colony formation was determined by dividing the number of colonies with ESC-like morphology that emerged on the dishes by the number of seeded cells. The efficiency was 0.009 ± 0.002% for DFBs (mean ± SE, five independent experiments with five DFB lines), 0.020 ± 0.008% for AMCs (three independent experiments with two AMC lines), and 0.005 ± 0.001% for DMCs (12 experiments with eleven DMC lines), respectively.
We also examined the cellular properties of the morphologically hESC-like colonies generated on 10-cm culture dishes, as candidates for further analysis. After they were subcultured, approximately 90% of the colonies derived from primary colonies of DMCs retained both their hESC-like morphology and positive ALP activity after the second passage, whereas only 40% of the colonies from AMCs and DFBs met both conditions (Fig. 2B, bottom). Based on these findings, we selected DMCs as the first cells to use for iPSC generation.
Cellular Properties of DMC-hiPSCs
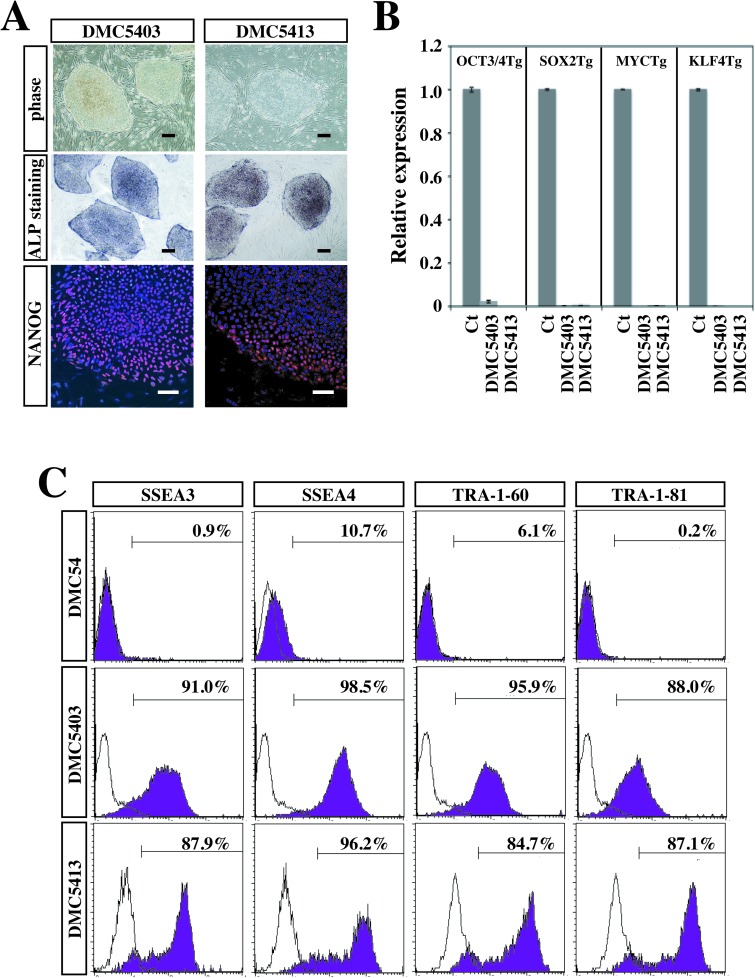
The DMC-hiPSC clones that retained their hESC-like morphology and positive ALP activity after several passages were also confirmed to express NANOG protein by immunocytochemistry (Fig. 3A). Expression analysis of the four transgenes by qRT-PCR analysis showed that each transgene was almost completely suppressed in two representative DMC-hiPSC clones (DMC5403 and DMC5413) compared with those on day 3 postinfection (Fig. 3B). To evaluate the protein levels of hESC markers, we analyzed two established clones and their parental DMCs (DMC54) by FCM. This analysis revealed that the two established clones highly expressed hESC-specific surface antigens (SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81), whereas the parental DMCs showed little or no expression of these antigens, in comparison with the isotypic control antibody samples (Fig. 3C).
Figure 3.
Characterization of decidua-derived mesenchymal cell-derived human induced pluripotent stem cells (DMC-hiPSCs). (A)Representative results of morphology and cytochemical analyses. Morphology (top), ALP staining (middle), and Nanog protein staining (bottom) of DMC-hiPSCs. Scale bar: 200 µm (morphology and ALP staining) and 50 µm (Nanog staining). (B) Expression of the four exogenous transgenes (OSKM) in DMC-hiPSC clones (DMC5403 and DMC5413) relative to infected cells extracted on day3 postinfection (mean ± SD, 3 independent experiments). (C) Flow cytometry analysis of hESC-specific cell surface markers [stage-specific embryonic antigen 3 (SSEA3), SSEA4, TRA-1-60, and TRA-1-81]. The shaded histograms represent the distribution of cells stained by the respective antibodies, and the open histograms show the isotype control staining.
Karyotyping, Genotyping, and Promoter Methylation Analysis
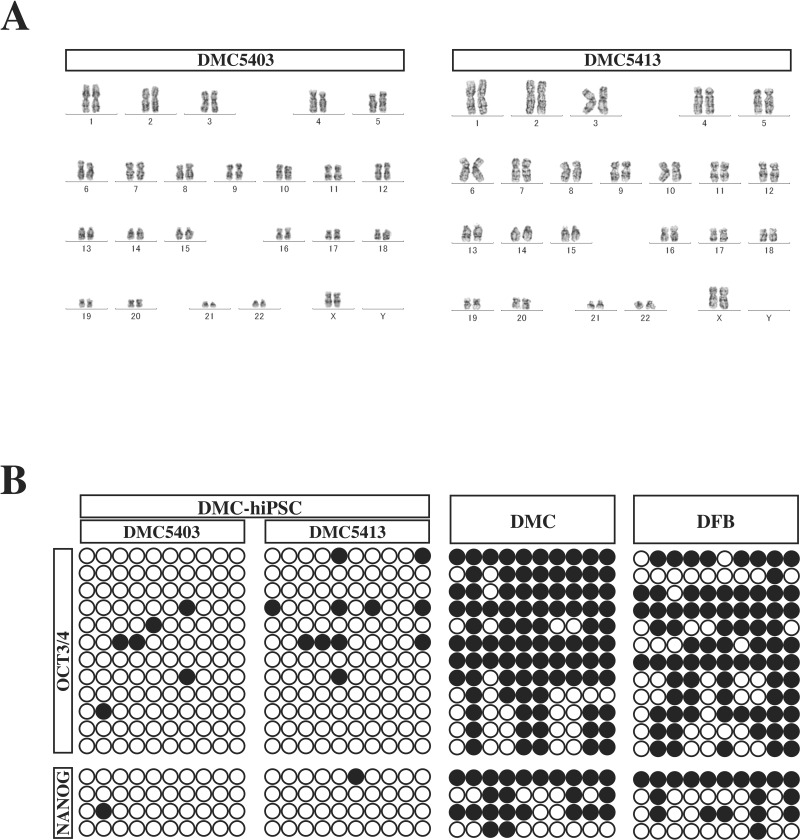
Karyotyping by G band staining showed that both DMC-hiPSC clones had a normal female karyotype (46, XX) (Fig. 4A). Analysis of the methylation state of the OCT3/4 and NANOG promoters revealed that most of the CpG sites in the DMC-hiPSC clones were unmethylated, while those of the parental DMCs were highly methylated (Fig. 4B). Genotyping by STR-PCR showed a complete match of STRs between the parental DMCs and the two established clones (Table 2). These findings confirmed that the two DMC-hiPSC clones were derived from the parental DMCs and that their epigenetic status had been reprogrammed efficiently.
Figure 4.
Genomic DNA analysis of DMC-hiPSCs. (A) Representative G-banding karyotype analysis of DMC-hiPSCs (DMC5403 and DMC5413). The DMC-hiPSC clones showed a normal karyotype at passage 30 and 38, respectively. (B) DNA methylation profile of the OCT3/4 and NANOG promoters in the DMC-hiPSCs, parental DMCs, and DFBs. ?Unmethylated CpGs; ●methylated CpGs.
Table 2.
Short-Tandem Repeat (STR) Genotyping
| Locus/Clone | DMC54 | DMC5403 | DMC5413 | UCB54 |
|---|---|---|---|---|
| Penta_E | 18, 21 | 18, 21 | 18, 21 | 14, 18 |
| D18S51 | 14, 16 | 14, 16 | 14, 16 | 14, 15 |
| D21S11 | 31.2, 33.2 | 31.2, 33.2 | 31.2, 33.2 | 31.2, 33.2 |
| TH01 | 9, 9 | 9, 9 | 9, 9 | 7, 9 |
| D3S1358 | 15, 16 | 15, 16 | 15, 16 | 14, 15 |
| FGA | 19, 22 | 19, 22 | 19, 22 | 19, 23 |
| TPOX | 8, 9 | 8, 9 | 8, 9 | 8, 11 |
| D8S1179 | 16, 16 | 16, 16 | 16, 16 | 10, 16 |
| vWA | 14, 16 | 14, 16 | 14, 16 | 16, 16 |
| Amelogenin | X, X | X, X | X, X | X, Y |
| Penta_D | 11, 15 | 11, 15 | 11, 15 | 11, 12 |
| CSF1PO | 11, 12 | 11, 12 | 11, 12 | 11, 11 |
| D16S539 | 9, 12 | 9, 12 | 9, 12 | 9, 9 |
| D7S820 | 8, 11 | 8, 11 | 8, 11 | 11, 12 |
| D13S317 | 8, 8 | 8, 8 | 8, 8 | 8, 11 |
| D5S818 | 11, 13 | 11, 13 | 11, 13 | 8, 11 |
Fifteen polymorphic STR DNA loci plus Amelogenin (AMEL) for sex chromosomes were analyzed.
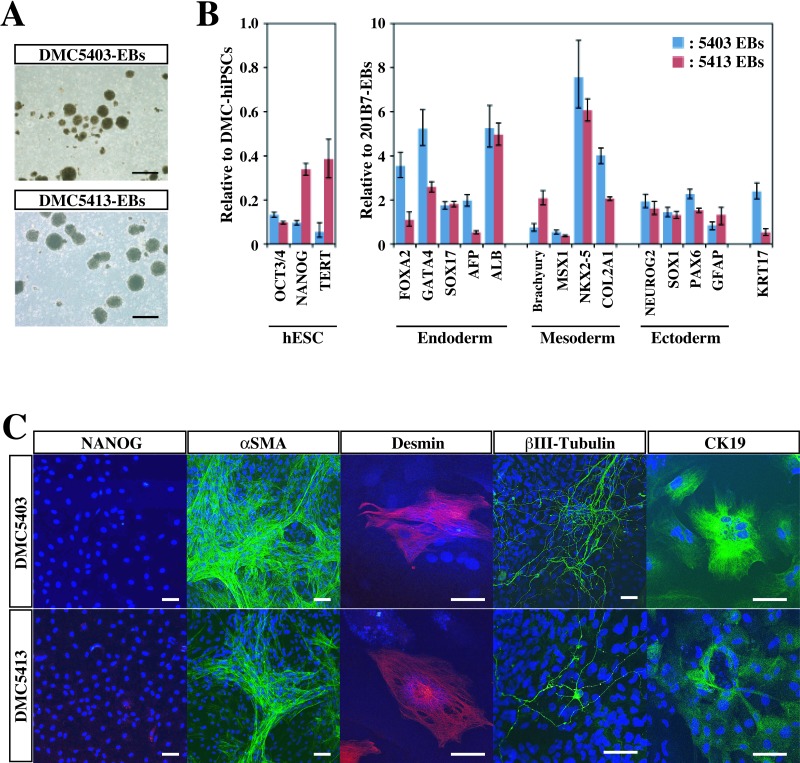
In Vitro Differentiation of DMC-hiPSCs
To confirm that the DMC-hiPSCs were pluripotent in vitro, we performed EB formation assays. The DMC-hiPSCs formed EBs in a 10-day suspension culture (Fig.5A), and qRT-PCR analysis showed the downregulated expression of pluripotent stem cell marker genes [hESCs: OCT3/4, NANOG, and telomerase reverse transcriptase (TERT)] (Fig. 5B). The expression of differentiation-specific marker genes of the three germ layers [endoderm: forkhead box A2 (FOXA2), SOX17, AFP, and albumin (ALB); mesoderm: GATA binding protein 4 (GATA4), Brachyury, Mut S homolog (msh) homeobox 1 (MSX1), and NK2 transcription factor related, locus 5 (NKX2-5); ectoderm: neurogenin 2 (NEUROG2), SOX1, and paired box 6 (PAX6); epidermis: keratin 17 (KRT17)] was observed in these EBs (Fig. 5B). The expression levels of the differentiation-specific marker genes were equal to or surpassed those in EBs formed from 201B7 hiPSCs (Fig. 5B).
Figure 5.
In vitro differentiation of DMC-hiPSCs. (A) Embryoid bodies (EBs) derived from two DMC-hiPSC clones on day 10. Scale bar: 200 μm. (B, left) Expression of human pluripotent stem cell marker genes in the same EBs on day 10 in suspension culture compared with their corresponding DMC-hiPSCs. (Right) Differentiation-specific marker gene expression relative to that of EBs derived from 201B7. (C) Immunofluorescence staining of differentiated cells in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) for an additional 2 weeks. Scale bars: 50 μm. See Table 1 for gene definitions.
To confirm the progression of differentiation, we replated the 10-day-cultured EBs on gelatin-coated chamber slides, cultured them in 10% FBS-containing medium for an additional 2 weeks, and then examined the cells by immunocytochemistry on day 24 of total in vitro differentiation. On day 24, the cells in EBs had differentiated into various adherent cells, most of which expressed very little NANOG (Fig. 5C), although some expressed endoderm markers (CK19), mesoderm markers (desmin, αSMA), and ectoderm markers (βIII-tubulin, GFAP) at various levels (Fig. 5C). These results indicated that the DMC-hiPSCs were fully pluripotent in vitro.
In Vivo Differentiation of DMC-hiPSCs
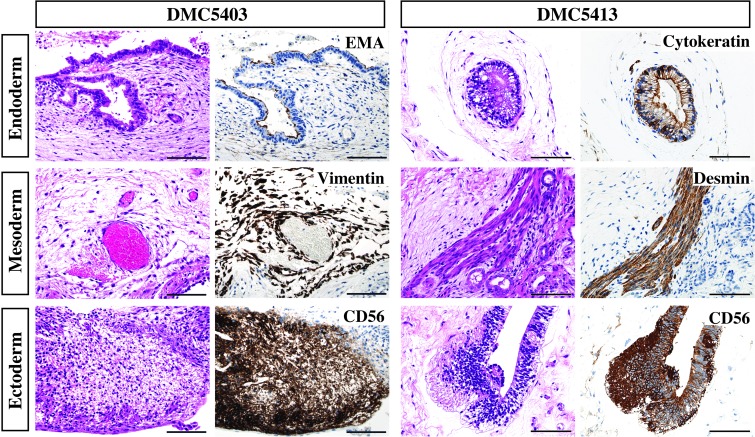
The pluripotency of the DMC-hiPSCs was also evaluated by an in vivo teratoma formation assay. DMC-hiPSCs formed tumor masses in NOG mice after several months, and these tumor masses contained histological components of the three germ layers. Some tumors showed neuroepithelial-like structures that expressed human NCAM (ectoderm) (Fig. 6). The tumors also showed blood vessels and fibrous stroma that were positive for human vimentin, muscle-like structures that expressed desmin (mesoderm), and gut-like epithelium that was positive for epithelial membrane antigen (EMA) or cytokeratin (endoderm) (Fig. 6). Thus, the DMC-hiPSCs formed teratomas showing in vivo pluripotency, which indicated that the DMC-hiPSCs met the criteria for hiPSCs.
Figure 6.
In vivo differentiation of DMC-hiPSCs. Teratomas derived from two DMC-hiPSCs (DMC5403, DMC5413) were examined by H&E and immunohistochemical staining. Teratomas from both DMC-hiPSCs contained histological components of the three germ layers, including neuroepithelial structures expressing human neural cell adhesion molecule (NCAM) (CD56, ectoderm), blood vessels and fibrous stroma-like structures expressing human vimentin or muscle-like structures expressing desmin (mesoderm), and gut-like epithelium expressing epithelial membrane antigen (EMA) or AE1 and AE3 (endoderm). Scale bars: 100 μm.
Gene Expression Profile of DMC-hiPSCs
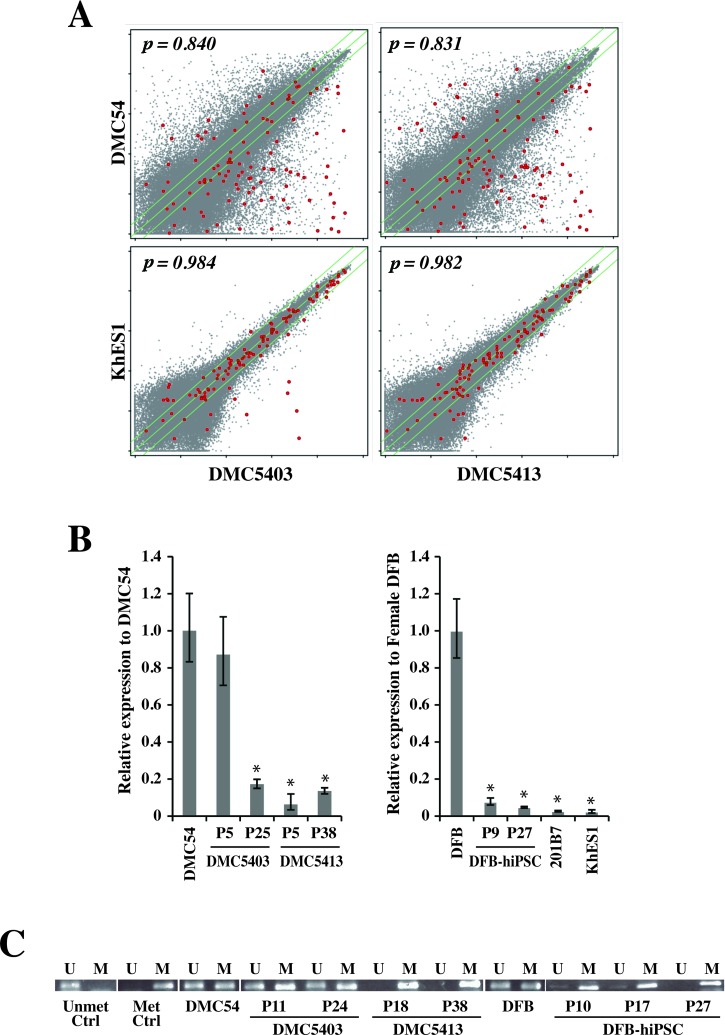
To further characterize the DMC-hiPSCs at the molecular level, we examined the genome-wide gene expression profile of two DMC-hiPSCs in comparison to those of hESCs and the parental DMCs. Microarray analysis verified that there was a large difference in the transcriptome profile between the DMC-hiPSCs and parental DMCs and, in contrast, a strong similarity between that of the DMC-hiPSCs and KhES1 (Fig. 7A). Many undifferentiated ES cell marker genes (2), such as OCT3/4 (POU5F1), SOX2, NANOG, TERT, FOXD3, growth differentiation factor 3 (GDF3), zinc finger protein 42homolog (ZFP42/reduced expression protein 1 [REX-1]), and teratocarcinoma-derived growth factor 1 (TDGF1), were expressed in the DMC-iPSCs at the same level as in the hESCs (Table 3), although these transcripts were not detected in the parental DMCs.
Figure 7.
Transcriptional expression analysis of DMC-hiPSCs. (A) Scatter plots comparing the DNA microarray analysis data between DMC-iPSCs and parental DMCs (top) and between DMC-hiPSCs and human ES cells (KhES1) (bottom). The diagonal lines indicate twofold changes between the two samples, and the Pearson's coefficients are shown in the scatter plots. Red dots indicate data from probe sets for characterizing undifferentiated stem cells (see Table 1). (B) qRT-PCR analysis of X (inactive)-specific transcript (XIST) gene expression in female cells. Data are presented as the value relative to parental DMCs or DFBs. (C) Methylation-specific PCR was performed to examine the methylation status of the XIST promoter. The PCR products were specific for the methylated (M) or unmethylated (U) genome.
Table 3.
Gene Expression Data of the Probe Sets in the Affymetrix Human Genome U133 Plus 2.0 Array for the Characterization of Undifferentiated Stem Cells
| Flag a s | Fold Change Versus DMC54 | Fold Change Versus KhES1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe Set ID | UniGene ID | Gene Title | Symbol | DMC54 | DMC5403 | DMC5413 | KhES1 | DMC5403 | DMC5413 | DMC5403 | DMC5413 | |
| 1552982_a_at | Hs.1755 | Fibroblast growth factor 4 | FGF4 | A | A | A | A | 1.8 | 2.6 | 1.5 | 2.2 | |
| 1554776_at | Hs.335787 | Zinc finger protein 42 homolog (mouse) | ZFP42 | A | P | P | P | 84.4 | 75.3 | –1.2 | –1.4 | |
| 1554777_at | Hs.335787 | Zinc finger protein 42 homolog (mouse) | ZFP42 | A | P | P | P | 126.1 | 220.4 | –1.5 | 1.2 | |
| 1555271_a_at | Hs.492203 | Telomerase reverse transcriptase | TERT | A | A | A | M | 4.1 | 3.0 | 1.1 | –1.2 | |
| 1560469_at | Hs.33446 | Nuclear receptor subfamily 5, group A, member 2 | NR5A2 | A | P | A | A | 33.0 | 2.7 | 11.4 | –1.1 | |
| 1569689_s_at | Hs.302352 | γ-Aminobutyric acid (GABA) A receptor, β3 | GABRB3 | A | A | A | A | 1.6 | –1.3 | 1.6 | –1.3 | |
| 201005_at | Hs.114286 | CD9 molecule | CD9 | P | P | P | P | 2.2 | 2.8 | 1.1 | 1.4 | |
| 201315_x_at | Hs.709321 | Interferon-induced transmembrane protein 2 (1-8D) | IFITM2 | P | P | P | P | –1.6 | –1.2 | 1.5 | 1.9 | |
| 201578_at | Hs.723993 | Podocalyxin-like | PODXL | P | P | P | P | 8.7 | 7.8 | 1.2 | 1.1 | |
| 201601_x_at | Hs.458414 | Interferon-induced transmembrane protein 1 (9-27) | IFITM1 | P | P | P | P | 3.9 | 4.5 | 1.7 | 1.9 | |
| 202575_at | Hs.405662 | Cellular retinoic acid binding protein 2 | CRABP2 | A | P | P | P | 7.5 | 25.7 | –2.7 | 1.3 | |
| 204053_x_at | Hs.500466 | Phosphatase and tensin homolog | PTEN | P | P | P | P | –1.3 | –1.3 | 1.0 | 1.0 | |
| 204054_at | Hs.500466 | Phosphatase and tensin homolog | PTEN | P | P | P | P | –2.2 | –2.1 | –1.0 | 1.0 | |
| 204271_s_at | Hs.82002 | Endothelin receptor type B | EDNRB | A | P | P | P | 596.2 | 435.3 | 2.2 | 1.6 | |
| 204273_at | Hs.82002 | Endothelin receptor type B | EDNRB | A | P | P | P | 514.5 | 609.4 | 2.8 | 3.3 | |
| 204535_s_at | Hs.631513 | RE1-silencing transcription factor | REST | A | A | A | A | –3.2 | –1.1 | –1.1 | 2.7 | |
| 204863_s_at | Hs.532082 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6ST | P | A | A | A | –11.8 | –7.5 | –1.4 | 1.1 | |
| 204864_s_at | Hs.532082 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6ST | P | A | A | P | –10.1 | –1.2 | –7.3 | 1.1 | |
| 205051_s_at | Hs.479754 | v-Kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | KIT | P | P | P | P | 9.1 | 10.8 | –1.6 | –1.4 | |
| 205603_s_at | Hs.226483 | Diaphanous homolog 2 (Drosophila) | DIAPH2 | P | P | P | P | 29.9 | 17.6 | 1.3 | –1.3 | |
| 205726_at | Hs.226483 | Diaphanous homolog 2 (Drosophila) | DIAPH2 | P | P | P | P | 4.2 | 4.1 | 1.0 | –1.0 | |
| 205850_s_at | Hs.302352 | γ-Aminobutyric acid (GABA) A receptor, β3 | GABRB3 | A | P | P | P | 44.4 | 50.6 | –1.4 | –1.2 | |
| 205876_at | Hs.133421 | Leukemia inhibitory factor receptor α | LIFR | A | P | A | P | 3.4 | 1.5 | –1.0 | –2.3 | |
| 206012_at | Hs.520187 | Left-right determination factor 2 | LEFTY2 | A | P | P | P | 970.9 | 465.7 | 1.1 | –1.9 | |
| 206268_at | Hs.724790 | Left-right determination factor 1 | LEFTY1 | A | P | P | P | 342.1 | 155.5 | 1.2 | –1.8 | |
| 206286_s_at | Hs.385870 | Teratocarcinoma-derived growth factor 1 teratocarcinoma-derived growth factor 3, pseudogene | TDGF1 | A | P | P | P | 12405.4 | 9524.5 | 1.4 | 1.1 | |
| TDGF3 | ||||||||||||
| 206701_x_at | Hs.82002 | Endothelin receptor type B | EDNRB | A | P | P | P | 197.9 | 108.6 | 2.7 | 1.5 | |
| 206783_at | Hs.1755 | Fibroblast growth factor 4 | FGF4 | A | P | P | P | 24.5 | 31.9 | 1.1 | 1.5 | |
| 206805_at | Hs.252451 | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | SEMA3A | P | P | P | P | 2.4 | 1.2 | 1.3 | –1.4 | |
| 207199_at | Hs.492203 | Telomerase reverse transcriptase | TERT | A | P | P | P | 3.0 | 3.9 | –1.8 | –1.4 | |
| 207466_at | Hs.278959 | Galanin prepropeptide | GAL | A | P | P | P | 28.6 | 12.6 | 1.5 | –1.5 | |
| 207545_s_at | Hs.654609 | Numb homolog (Drosophila) | NUMB | P | P | P | P | 1.1 | 1.2 | 1.9 | 2.0 | |
| 207742_s_at | Hs.586460 | Nuclear receptor subfamily 6, group A, member 1 | NR6A1 | A | A | A | A | 11.4 | 19.9 | –2.2 | –1.2 | |
| 208275_x_at | Hs.458406 | Undifferentiated embryonic cell transcription factor 1 | UTF1 | A | P | P | P | 13.0 | 29.6 | 1.1 | 2.5 | |
| 208286_x_at | Hs.632482 | POU class 5 homeobox 1 | POU5F1 | A | P | P | P | 1925.3 | 2025.3 | 1.1 | 1.2 | |
| POU class 5 homeobox 1B | POU5F1B | |||||||||||
| POU class 5 homeobox 1 pseudogene 3 | POU5F1 p3 | |||||||||||
| POU class 5 homeobox 1 pseudogene 4 | POU5F1 P4 | |||||||||||
| 208337_s_at | Hs.33446 | Nuclear receptor subfamily 5, group A, member 2 | NR5A2 | A | P | P | P | 37.8 | 26.4 | 1.0 | –1.4 | |
| 208343_s_at | Hs.33446 | Nuclear receptor subfamily 5, group A, member 2 | NR5A2 | A | P | P | P | 153.3 | 39.3 | 1.4 | –2.7 | |
| 208378_x_at | Hs.37055 | Fibroblast growth factor 5 | FGF5 | P | A | A | A | –9.8 | –55.8 | 1.9 | –3.0 | |
| 208500_x_at | Hs.546573 | Forkhead box D3 | FOXD3 | A | A | A | A | 5.5 | 14.1 | –7.9 | –3.1 | |
| 209073_s_at | Hs.654609 | Numb homolog (Drosophila) | NUMB | P | P | P | P | –1.2 | 1.1 | 1.1 | 1.4 | |
| 210002_at | Hs.514746 | GATA binding protein 6 | GATA6 | P | A | A | A | –38.1 | –4.8 | –1.3 | 6.0 | |
| 210174_at | Hs.33446 | Nuclear receptor subfamily 5, group A, member 2 | NR5A2 | A | P | P | P | 28.8 | 9.6 | 2.3 | –1.3 | |
| 210310_s_at | Hs.37055 | Fibroblast growth factor 5 | FGF5 | P | A | A | A | –127.2 | –102.7 | –1.4 | –1.1 | |
| 210311_at | Hs.37055 | Fibroblast growth factor 5 | FGF5 | P | A | M | A | –4.5 | –6.8 | 1.6 | 1.0 | |
| 210391_at | Hs.586460 | Nuclear receptor subfamily 6, group A, member 1 | NR6A1 | A | A | A | A | 1.5 | 4.1 | –1.5 | 1.7 | |
| 210392_x_at | Hs.586460 | Nuclear receptor subfamily 6, group A, member 1 | NR6A1 | A | M | A | A | 6.7 | 6.1 | 1.2 | 1.1 | |
| 210560_at | Hs.184945 | Gastrulation brain homeobox 2 | GBX2 | A | A | A | A | 4.5 | 10.3 | 3.6 | 8.4 | |
| 210761_s_at | Hs.86859 | Growth factor receptor-bound protein 7 | GRB7 | A | P | P | P | 1.6 | 3.2 | –1.3 | 1.5 | |
| 211000_s_at | Hs.532082 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6ST | P | A | A | A | –8.2 | –7.5 | 1.3 | 1.5 | |
| 211402_x_at | Hs.586460 | Nuclear receptor subfamily 6, group A, member 1 | NR6A1 | A | A | A | A | 13.6 | 10.7 | –1.0 | –1.3 | |
| 211711_s_at | Hs.500466 | Phosphatase and tensin homolog | PTEN | P | P | P | P | –1.8 | –2.2 | 1.0 | –1.2 | |
| 212195_at | Hs.532082 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6ST | P | P | P | P | –17.9 | –15.2 | 1.2 | 1.4 | |
| 212196_at | Hs.532082 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6ST | P | P | P | P | –4.2 | –3.7 | 1.1 | 1.3 | |
| 212920_at | Hs.631513 | RE1-silencing transcription factor | REST | P | P | P | P | 1.5 | 2.4 | –1.1 | 1.4 | |
| 213721_at | Hs.518438 | SRY (sex-determining region Y)-box 2 | SOX2 | A | P | P | P | 72.0 | 53.6 | 1.1 | –1.3 | |
| 213722_at | Hs.518438 | SRY (sex-determining region Y)-box 2 | SOX2 | A | A | A | A | 9.6 | 2.9 | 1.4 | –2.3 | |
| 214022_s_at | Hs.458414 | Interferon-induced transmembrane protein 1 (9-27) | IFITM1 | P | P | P | P | 5.7 | 6.1 | 1.7 | 1.8 | |
| 214178_s_at | Hs.518438 | SRY (sex-determining region Y)-box 2 | SOX2 | A | A | A | A | 4.3 | 3.3 | 1.4 | 1.1 | |
| 214218_s_at | Hs.730656 | X (inactive)-specific transcript (nonprotein coding) | XIST | P | P | P | P | –1.0 | –60.2 | 80.1 | 1.4 | |
| 214240_at | Hs.278959 | Galanin prepropeptide | GAL | A | P | P | P | 3871.9 | 2443.0 | 1.6 | –1.0 | |
| 217246_s_at | Hs.226483 | Diaphanous homolog 2 (Drosophila) | DIAPH2 | A | A | A | P | 1.4 | 1.2 | –1.1 | –1.3 | |
| 218048_at | Hs.720384 | COMM domain containing 3 | COMMD3 | P | P | P | P | –5.1 | –9.7 | 1.3 | –1.4 | |
| 218847_at | Hs.35354 | Insulin-like growth factor 2 mRNA binding protein 2 | IGF2BP2 | P | P | P | P | 2.2 | 2.8 | 1.1 | 1.5 | |
| 219177_at | Hs.718510 | BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) | BRIX1 | P | P | P | P | 1.6 | 2.8 | –1.1 | 1.6 | |
| 219735_s_at | Hs.156471 | Transcription factor CP2-like 1 | TFCP2L1 | A | P | P | P | 3.5 | 2.7 | 1.2 | –1.1 | |
| 219823_at | Hs.86154 | Lin-28 homolog (C. elegans) | LIN28 | A | P | P | P | 25980.2 | 23501.4 | 1.1 | 1.0 | |
| 220053_at | Hs.86232 | Growth differentiation factor 3 | GDF3 | A | P | P | P | 15.8 | 22.7 | –1.1 | 1.3 | |
| 220184_at | Hs.635882 | Nanog homeobox | NANOG | A | P | P | P | 15134.8 | 13855.2 | 1.4 | 1.3 | |
| 220668_s_at | Hs.643024 | DNA (cytosine-5-)-methyltransferase 3β | DNMT3B | P | P | P | P | 99.7 | 82.3 | 1.2 | –1.1 | |
| 221728_x_at | Hs.730656 | X (inactive)-specific transcript (nonprotein coding) | XIST | P | P | A | A | –1.3 | –88.8 | 1945.5 | 27.6 | |
| 222176_at | Hs.500466 | Phosphatase and tensin homolog | PTEN | A | A | A | A | –3.7 | –7.5 | –3.4 | –6.9 | |
| 223121_s_at | Hs.481022 | Secreted frizzled-related protein 2 | SFRP2 | A | P | P | P | 258.7 | 375.6 | –1.1 | 1.3 | |
| 223122_s_at | Hs.481022 | Secreted frizzled-related protein 2 | SFRP2 | A | P | P | P | 1264.8 | 14.76.7 | –1.4 | –1.2 | |
| 223963_s_at | Hs.35354 | Insulin-like growth factor 2 mRNA binding protein 2 | IGF2BP2 | P | M | P | A | 1.3 | 1.4 | –1.2 | –1.1 | |
| 224588_at | Hs.730656 | X (inactive)-specific transcript (nonprotein coding) | XIST | P | P | A | A | –1.3 | –176.7 | 355.0 | 2.7 | |
| 224589_at | Hs.730656 | X (inactive)-specific transcript (nonprotein coding) | XIST | P | P | A | A | –2.5 | –187.9 | 7.8 | –9.6 | |
| 224590_at | Hs.730656 | X (inactive)-specific transcript (nonprotein coding) | XIST | P | P | A | A | –5.7 | –128.8 | 34.4 | 1.5 | |
| 225363_at | Hs.500466 | Phosphatase and tensin homolog | PTEN | P | P | P | P | –1.9 | –3.0 | 1.1 | –1.4 | |
| 225571_at | Hs.133421 | Leukemia inhibitory factor receptor α | LIFR | P | P | P | P | 14.0 | 14.3 | 1.3 | 1.3 | |
| 225575_at | Hs.133421 | Leukemia inhibitory factor receptor α | LIFR | P | P | P | P | 6.4 | 6.5 | 1.4 | 1.4 | |
| 227642_at | Hs.156471 | Transcription factor CP2-like 1 | TFCP2L1 | A | P | P | P | 29.3 | 33.4 | 1.4 | 1.6 | |
| 227671_at | Hs.730656 | X (inactive)-specific transcript (nonprotein coding) | XIST | P | P | A | A | –1.0 | –66.6 | 44.9 | –1.4 | |
| 227690_at | Hs.302352 | γ-Aminobutyric acid (GABA) A receptor, β3 | GABRB3 | A | P | P | P | 133.3 | 161.2 | 1.0 | 1.2 | |
| 227771_at | Hs.133421 | Leukemia inhibitory factor receptor α | LIFR | P | P | P | P | 2.9 | 1.8 | 1.1 | –1.5 | |
| 227830_at | Hs.302352 | γ-Aminobutyric acid (GABA) A receptor, β3 | GABRB3 | A | P | P | P | 121.6 | 129.7 | 1.2 | 1.2 | |
| 228038_at | Hs.518438 | SRY (sex-determining region Y)-box 2 | SOX2 | A | P | P | P | 8739.3 | 7576.9 | 1.2 | 1.1 | |
| 229282_at | Hs.514746 | GATA binding protein 6 | GATA6 | M | M | P | A | 1.7 | 1.6 | 3.1 | 3.0 | |
| 229341_at | Hs.156471 | Transcription factor CP2-like 1 | TFCP2L1 | A | A | A | A | 1.2 | 2.9 | –3.9 | –1.6 | |
| 229724_at | Hs.302352 | γ-Aminobutyric acid (GABA) A receptor, β3 | GABRB3 | A | P | P | P | 51.7 | 53.0 | 1.2 | 1.3 | |
| 230462_at | Hs.654609 | Numb homolog (Drosophila) | NUMB | P | P | P | P | 2.1 | 1.6 | –1.4 | –1.9 | |
| 230916_at | Hs.370414 | Nodal homolog (mouse) | NODAL | A | P | P | P | 26.2 | 27.7 | 1.0 | 1.1 | |
| 231798_at | Hs.248201 | Noggin | NOG | P | P | P | P | –17.0 | –13.5 | 1.2 | 1.5 | |
| 233254_x_at | Hs.500466 | Phosphatase and tensin homolog | PTEN | A | A | A | A | 1.1 | –1.6 | –1.1 | –2.0 | |
| 233314_at | Hs.500466 | Phosphatase and tensin homolog | PTEN | A | P | A | P | 17.2 | 2.4 | 1.0 | –7.2 | |
| 233317_at | Hs.114286 | CD9 molecule | CD9 | A | A | A | A | 8.2 | 1.9 | 1.4 | –3.0 | |
| 233322_at | Hs.114286 | CD9 molecule | CD9 | A | A | A | A | 2.1 | 1.9 | 5.7 | 5.0 | |
| 234474_x_at | Hs.532082 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6ST | A | A | A | A | –9.9 | –15.9 | –1.8 | –2.9 | |
| 234967_at | Hs.532082 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6ST | A | A | A | A | –2.8 | –3.0 | –4.4 | –4.8 | |
| 235446_at | Hs.724376 | X (inactive)-specific transcript (nonprotein coding) | XIST | A | A | A | A | 1.5 | –1.1 | 1.2 | –1.4 | |
| 236930_at | Hs.654609 | Numb homolog (Drosophila) | NUMB | A | P | P | P | 2.3 | 2.1 | –1.6 | –1.7 | |
| 237896_at | Hs.370414 | Nodal homolog (mouse) | NODAL | A | P | P | P | 6.3 | 5.6 | 1.1 | –1.0 | |
| 241609_at | Hs.546573 | Forkhead box D3 | FOXD3 | A | A | A | A | 2.9 | 7.7 | 2.0 | 5.3 | |
| 241612_at | Hs.546573 | Forkhead box D3 | FOXD3 | A | P | P | P | 758.4 | 868.1 | 1.0 | 1.2 | |
| 242622_x_at | Hs.500466 | Phosphatase and tensin homolog | PTEN | A | A | A | A | 1.2 | –1.1 | –2.6 | –3.6 | |
| 243161_x_at | Hs.335787 | Zinc finger protein 42 homolog (mouse) | ZFP42 | A | P | P | P | 8492.4 | 8389.2 | –1.3 | –1.3 | |
| 243712_at | Hs.730656 | X (inactive)-specific transcript (nonprotein coding) | XIST | A | A | A | A | 1.3 | 1.9 | –1.8 | –1.2 | |
| 244163_at | Hs.252451 | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | SEMA3A | P | P | P | P | 1.2 | –1.2 | 1.7 | 1.2 | |
| 244849_at | Hs.252451 | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | SEMA3A | P | P | A | P | 1.8 | –1.4 | 1.3 | –1.9 | |
Flag indicates whether a transcript is present (P), marginal (M), or absent (A), as assigned by the Detection Algorithm.
Among the 108 probe sets in the Human Genome U133 plus 2.0 Array for characterizing undifferentiated stem cells (2), the expression of the X (inactive)-specific transcript (nonprotein coding) gene (XIST) in one of the DMC-iPSCs (DMC5403) was more than 10 times that in the hESCs and was the same as in the parental DMCs (Table 3). Quantitative RT-PCR analysis for the XIST gene expression showed that the expansion and continuous passage of DMC-iPSCs led to the loss of XIST transcription in clone DMC5403 (Fig. 7B). The other DMC-iPSC clone (DMC5413) and a female DFB-iPS clone expressed XIST at a very low level at the beginning of culture (less than 10 passages) and after many passages (no less than 27 passages) (Fig. 7B). The methylation status of the XIST promoter region was assessed by methylation-specific PCR. Genomic DNA extracted from the parental female cells and DMC-hiPSC clone 5403 produced similar amounts of PCR products with both methylated and unmethylated DNA-specific primer sets. In contrast, the methylated DNA-specific primer set produced almost all the amplified products from the other clone, 5413, and the female DFB-hiPSCs (Fig. 7C).
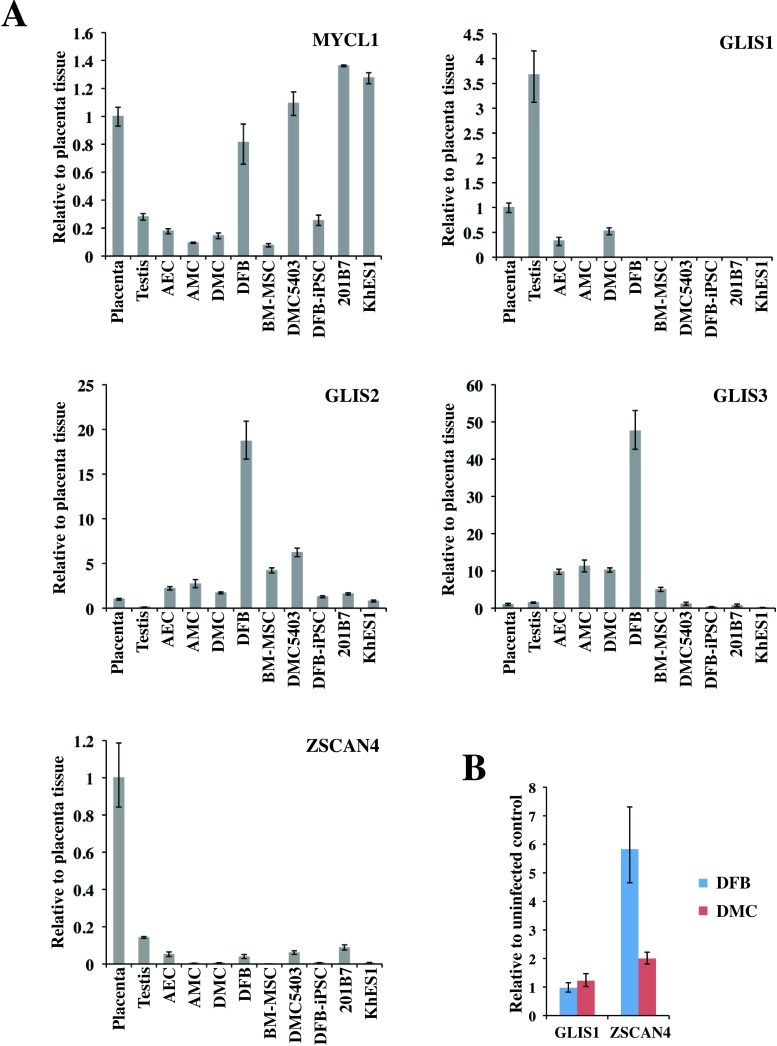
Recently, new key molecules for efficient reprogramming, lung carcinoma-derived Myc (MYCL1) and glioma-associated oncogene similar protein [GLIS] family zinc finger (GLIS1), were reported (24,36). We evaluated the expression of these genes and of other GLI-related Krüppel-like zinc finger genes, GLIS2 and GLIS3, in several cell lines and tissues (Fig. 8A). Quantitative RT-PCR analysis showed that MYCL1 was expressed in all the primary cells, with the DFBs expressing three to four times the level observed in the placenta-derived cells or BM-MSCs. GLIS1 expression was detected in the testis at a high level and in the placenta at a moderate level. Of the primary cells, a low level of GLIS1 expression was detected in only the AECs and DMCs; no significant expression of GLIS1 was detected in the hESCs or hiPSCs. On the other hand, significant amounts of GLIS2 and GLIS3 were expressed in the placenta-derived cells, DFBs, and BM-MSCs, while very low levels were expressed in pluripotent stem cells, testis, and placenta (Fig. 8A). Seventeen factors that can substitute for Klf4 have been identified, although they have much lower efficiencies in iPSC generation (24). Among these factors, we evaluated Zinc finger and SCAN domain containing 4 (Zscan4), which is expressed exclusively in mouse late two-cell embryos and embryonic stem cells (7). ZSCAN4 expression was detected in five primary cultured cells at very low levels, but was roughly five times higher in AECs and DFBs than in AMCs, DMCs, or BM-MSCs (Fig. 8A). We also examined the expression of GLIS1 and ZSCAN4 in DFBs and DMCs 3 days after the transduction of OSKM. Under this condition, we observed an increase in ZSCAN4 but not in GLIS1 expression in both DFBs and DMCs (Fig. 8B).
Figure 8.
Transcriptional expression of novel factors for efficient reprogramming. (A) Endogenous transcriptional expression of MYCL1 (top left), GLIS1 (top right), GLIS2 (middle left), GLIS3 (middle right), and ZSCAN4 (bottom left). GAPDH was used as the internal control gene, and data are presented relative to the values from placenta tissue and as the mean ± SD (n = 3). (B) Expression of GLIS1 and ZSCAN4 3 days after the transduction of reprogramming factor genes. Resuslts are shown in comparison to individual uninfected control cells. See Table 1 for gene definitions.
DISCUSSION
DMCs Are a Promising Human Allogeneic Cell Type for Generating hiPSCs
In this study, we demonstrated that hiPSCs were efficiently generated from DMCs, which were derived from the maternal components of placental tissue. The DMC-hiPSCs maintained a normal karyotype and showed equivalent characteristics to hESCs in colony morphology, global gene expression profile (including human pluripotent stem cell markers), and DNA methylation status of the OCT3/4 and NANOG promoters, although these iPSC clones showed minor differences from each other in gene expression patterns and in the methylation status of promoter regions. In addition, the DMC-hiPSCs were able to differentiate into components of the three germ layers in vitro and in vivo. These data indicated that the DMC-hiPSC clones acquired full pluripotency equal to that of hiPSCs derived from DFBs or other cell sources and that the generation of iPSCs from DMCs is feasible.
The overall efficiency of reprogramming DMCs was almost the same as for DFBs, the most commonly used primary cell type for generating hiPSCs. However, we found several differences between DMCs and DFBs in the present study. In comparison with DFBs, DMCs formed about half the number of ALP+ colonies (Fig. 2B, top). However, about 90% of the hESC-like colonies from DMCs were fully reprogrammed, while fewer than 40% of those from DFBs were successfully reprogrammed (Fig. 2B, bottom). Consequently, the overall reprogramming efficiency of DMCs was comparable to that of DFBs. There are various possible reasons for this higher reprogramming efficiency of DMCs. First, given that DMCs expressed the amphotropic retrovirus-introduced transgenes at two to three times the level expressed by DFBs (Fig. 2A), DMCs may be particularly compatible with this gene transduction system. Second, the endogenous gene expression profile of DMCs might result in improved reprogramming efficiency. Many previous reports have indicated that OSKM are important for reprogramming cells to iPSCs and that the endogenous expression of any of the four factors can reduce the number of genes needed for reprogramming (6,19,35). Furthermore, recently reported key molecules for efficient reprogramming, MYCL1 and GLIS1 (24,36), were expressed by the DMCs.
Our quantitative RT-PCR analysis showed that, among the four reprogramming factors (OSKM), only c-MYC was expressed in DMCs at the same level as in hESCs, similar to other placenta-derived cells and DFBs (Fig. 1). However, expression of the GLIS1 gene was detected only in the testis, placenta, and their derivative cells, the AECs and DMCs (Fig. 8). The transduction of GLIS1 along with OCT3/4, KLF4, and SOX2 markedly increases the expression of several genes that enhance iPSC generation, including lin-28 homolog A (C. elegans; LIN28A) (47), NANOG (38), neuroblastoma-derived Myc (MYCN), and MYCL1 (31), and the number of fully reprogrammed cells (24). The low endogenous expression of GLIS1 in the DMCs, combined with the efficient retroviral gene transduction, might contribute to the higher efficiency of DMC reprogramming.
On the other hand, the level of MYCL1 expression in the three placenta-derived cell types was low compared to that in DFBs (Fig. 8A). It is well known that DFBs can be reprogrammed efficiently by three (OSK) or fewer factors, that is, c-MYC is not required (30). Although we tried several times to reprogram the DMCs using OSK, we could not generate hiPSCs (unpublished results). The lower endogenous expression of MYCL1 in the DMCs compared with DFBs might explain the reduced efficiency of DMC reprogramming when c-MYC is omitted.
We also evaluated the expression of the GLIS1 homologs GLIS2 and GLIS3 in several cell lines and tissues (Fig. 8). GLIS2 and GLIS3 were hardly expressed in pluripotent stem cells (hESCs and hiPSCs), placenta, or testis (Fig. 8). In contrast, placenta-derived cells (AECs, AMCs, DMCs), BM-MSCs, and DFBs expressed GLIS2 and GLIS3; in particular, DFBs highly expressed both GLIS1 homologs (Fig. 8). Although the significance of GLIS2 and GLIS3 in generating hiPSCs is not yet clear, these differences in GLI-related Krüppel-like zinc finger genes might result in different efficiencies of hiPSC generation from various cell sources. Another factor, Zscan4, was previously shown to facilitate the reprogramming of MEFs (12). Unlike in MEFs, we detected an increase in the intrinsic expression of ZSCAN4 in human DMCs and DFBs when the reprogramming factors were overexpressed; however, ZSCAN4’s contribution to iPSC generation has yet to be elucidated. Zscan4 was shown to be required only for the first few days of iPSC formation and to mediate the activation of genes detected only in preimplantation embryos (1-cell to blastocyst) (12). Besides GLIS1 and ZSCAN4, other preimplantation-specific genes that are expressed in blastomeres and have reprogramming activity in nuclear transplantation (5) should be considered for improving the generation of iPSCs.
Molecular Characteristics of DMC-hiPSCs
The global expression profile of DMC-iPSC clones, which retained a hESC-like morphology after several passages, verified that there was a strong similarity between DMC-hiPSCs and KhES1 (p > 0.98) (Fig. 7A). Despite the similarities, one of the genes for characterizing undifferentiated ES cells (2), XIST, was highly expressed in one of the DMC-iPSC clones (DMC5403), that is, at more than 10 times the level in female hESCs (KhES1), and at the same level as in the parental DMCs. In female cells, one of the two X-chromosomes is transcriptionally silenced through a process called X-chromosome inactivation (XCI); this process depends on the large noncoding RNA XIST (45). In the mouse system, the key pluripotency factors Nanog, Oct3/4, and Sox2 bind within Xist’s intron 1 and cooperate to repress the Xist transcript in undifferentiated embryonic stem (ES) cells; therefore, both X-chromosomes remain active (Xa) (32). In the process of reprogramming female somatic cells to miPSCs, the inactivated X-chromosome (Xi) is reactivated in concert with the activation of pluripotency factors (25,40). On the other hand, the X-chromosomes of female hESCs have a variable epigenetic status (2,39), and many hESC lines display XCI in the undifferentiated state. Studies of the epigenetic state of the X-chromosomes in hiPSCs have led to two conflicting conclusions. One study showed that a hiPSC clone, unlike miPSCs, was reprogrammed without undergoing X-reactivation and maintained the XCI of the mother fibroblast cell (44). Another study showed that complete X-reactivation was achieved in some hiPSC lines and that subsequent X-inactivation occurred during differentiation (26).
We briefly examined the epigenetic state by assessing the XIST expression and performing MSP analysis of the XIST promoter region. In agreement with the expectation that XIST would be silenced on Xa and actively expressed from Xi, the results indicated that Xi/Xa was present in the parental female cells and in the early phase of one DMC-hiPSC clone (DMC5403 at passage 5). In contrast, Xa/Xa was present in the other DMC-hiPSC clone (DMC5413) and in DFB-hiPSCs (Fig. 7B). DMC5403 cells from multiply passaged cultures did not express the XIST gene, although the XIST promoter region in half of the chromosomes remained unmethylated. The silencing of XIST expression in the presence of an inactive X is often observed in both undifferentiated and differentiated cell states (39,44). Overall, regardless of the epigenetic state of the X-chromosome, the DMC-iPSC clones exhibited genome-wide expression profiles similar to those of hESCs and formed differentiated tissues containing all three germ layers in vitro and in vivo (early passage, Figs. 5–7) (late passage, data not shown). Thus, not just the X-chromosome but the overall epigenetic state of iPSCs is an important quality and safety concern in clinical applications of hiPSCs and remains to be fully addressed.
DMC-hiPSCs May Be Useful as Allogeneic hiPSCs in Regenerative Medicine
Extraembryonic tissues such as the umbilical cord and placenta have been proposed as attractive human cell sources for use in regenerative medicine. In addition, human amnion-derived cells, which are of fetal origin, were reported as a promising cell source for efficient hiPSC generation (4,29,48). In this study, we used DMCs isolated from the decidua membrane, which is a maternal portion of the placenta (18). DMCs exhibit a typical fibroblast-like morphology and have a high proliferative potential for over 30 population doublings, which is better than that of BM-MSCs (18). They strongly express the mesenchymal cell marker vimentin, but not cytokeratin 19 or HLA-G, and FCM analysis shows a resemblance between DMCs and BM-MSCs. In vitro, the DMCs showed good differentiation into chondrocytes and moderate differentiation into adipocytes, but scant evidence of osteogenesis, compared with BM-MSCs. These findings indicated that DMCs are mesenchymal cells of purely maternal origin and unique cells that have MSC-like properties but differ from BM-MSCs. It was previously reported that mesenchymal cells derived from the human term decidua are multipotent, and these decidua-derived mesenchymal cells were suggested to be useful for regenerative medicine (11,13,23). However, in the mouse system, MSCs purified from adult tissue are useful for producing high-quality iPSCs (33). In addition, iPSCs have been generated efficiently from various somatic stem cells, including MSCs (4,33,35) and neural stem cells (6,19). In this context, the generation of iPSCs from immature somatic stem cells is an important option that needs to be thoroughly examined.
Here, we showed that DMCs can be efficiently reprogrammed into pluripotent stem cells. Although DMCs are not superior to BM-MSCs in their multipotency and might have some risk of genomic abnormalities accumulated during aging, their greater proliferative ability means that their cultivation might require less maintenance, and their derivation from the maternal portion of the human fetal adnexal tissues, which are otherwise discarded, would resolve many ethical concerns associated with the use of embryonic stem cells. The high success rate of DMC isolation from tissues stored more than 24 h indicates that it may be feasible to develop a system for collecting and banking fetal adnexal tissues; such a bank would provide tissues of wide HLA variation to multiple and even remote hospitals (18). We also previously reported that the pericellular matrix prepared from DMCs (PCM-DM) is as potent a substrate for the growth and pluripotency of hESCs as Matrigel (28). Furthermore, we have confirmed that hiPSCs can be generated on PCM-DM by a retroviral gene transduction procedure (Fukusumi et al., submitted).
To be sure, many concerns about the feasibility and potential risks of cell-based therapies using iPSCs have been recognized, and many scientific and technical issues remain to be elucidated before iPSC technology can be applied to regenerative medicine. Since patient safety is the foremost consideration, extensive exploration and testing of iPSCs will be required before this technology can be considered for clinical use.
Taken together, these findings indicate that DMCs have several advantages for clinical use and that the generation of hiPSCs from DMCs might enable the establishment of hiPSC-banking systems. Such systems could increase the availability of allogeneic hiPSCs that have broad HLA variations and thus increase the feasibility of their use in clinical applications.
ACKNOWLEDGMENTS
We thank Dr. Chiaki Ban (Department of Obstetrics and Gynecology Osaka National Hospital) and Ms.Chika Teramoto and Ms. Ai Takada (Osaka National Hospital) for support in collecting the human placenta tissues. We also thank Mr. Kenji Kawai and Ms. Miyuki Kuromuma (CIEA) for support in the teratoma formation assay. This study was supported by the Project for the Realization of Regenerative Medicine, the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, the Cooperative Link of Unique Science and Technology for Economy Revitalization (CLUSTER) project from MEXT, Japan, and the Research on New Drug Development, Health and Labour Sciences Research Grants, the Ministry of Health, Labour and Welfare (MHLW) of Japan. The authors declare no conflicts of interest.
REFERENCES
- 1. Aasen T.; Raya A.; Barrero M. J.; Garreta E.; Consiglio A.; Gonzalez F.; Vassena R.; Bilic J.; Pekarik V.; Tiscornia G.; Edel M.; Boue S.; Izpisua Belmonte J.C. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26(11):1276–1284; 2008. [DOI] [PubMed] [Google Scholar]
- 2. Adewumi O.; Aflatoonian B.; Ahrlund-Richter L.; Amit M.; Andrews P. W.; Beighton G.; Bello P. A.; Benvenisty N.; Berry L. S.; Bevan S.; Blum B.; Brooking J.; Chen K. G.; Choo A. B. H.; Churchill G. A.; Corbel M.; Damjanov I.; Draper J. S.; Dvorak P.; Emanuelsson K.; Fleck R. A.; Ford A.; Gertow K.; Gertsenstein M.; Gokhale P. J.; Hamilton R. S.; Hampl A.; Healy L. E.; Hovatta O.; Hyllner J.; Imreh M. P.; Itskovitz-Eldor J.; Jackson J.; Johnson J. L.; Jones M.; Kee K.; King B.L.; Knowles B. B.; Lako M.; Lebrin F.; Mallon B. S.; Manning D.; Mayshar Y.; McKay R. D. G.; Michalska A.E.; Mikkola M.; Mileikovsky M.; Minger S. L.; Moore H. D.; Mummery C. L.; Nagy A.; Nakatsuji N.; O'Brien C. M.; Oh S. K. W.; Olsson C.; Otonkoski T.; Park K.-Y.; Passier R.; Patel H.; Patel M.; Pedersen R.; Pera M. F.; Piekarczyk M. S.; Pera R. A. R.; Reubinoff B. E.; Robins A. J.; Rossant J.; Rugg-Gunn P.; Schulz T. C.; Semb H.; Sherrer E. S.; Siemen H.; Stacey G. N.; Stojkovic M.; Suemori H.; Szatkiewicz J.; Turetsky T.; Tuuri T.; van den Brink S.; Vintersten K.; Vuoristo S.; Ward D.; Weaver T. A.; Young L. A.; Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25(7):803–816; 2007. [DOI] [PubMed] [Google Scholar]
- 3. Barlow S.; Brooke G.; Chatterjee K.; Price G.; Pelekanos R.; Rossetti T.; Doody M.; Venter D.; Pain S.; Gilshenan K.; Atkinson K. Comparison of human placenta- and bone marrow-derived multipotent mesenchymal stem cells. Stem Cells Dev. 17:1095–1107; 2008. [DOI] [PubMed] [Google Scholar]
- 4. Cai J.; Li W.; Su H.; Qin D.; Yang J.; Zhu F.; Xu J.; He W.; Guo X.; Labuda K.; Peterbauer A.; Wolbank S.; Zhong M.; Li Z.; Wu W.; So K. F.; Redl H.; Zeng L.; Esteban M. A.; Pei D. Generation of human induced pluripotent stem cells from umbilical cord matrix and amniotic membrane mesenchymal cells. J. Biol. Chem. 285:11227–11234; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Egli D.; Sandler V. M.; Shinohara M. L.; Cantor H.; Eggan K. Reprogramming after chromosome transfer into mouse blastomeres. Curr. Biol. 19:1403–1409; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eminli S.; Utikal J.; Arnold K.; Jaenisch R.; Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells 26:2467–2474; 2008. [DOI] [PubMed] [Google Scholar]
- 7. Falco G.; Lee S. L.; Stanghellini I.; Bassey U. C.; Hamatani T.; Ko M. S. Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 307:539–550; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fukuchi Y.; Nakajima H.; Sugiyama D.; Hirose I.; Kitamura T.; Tsuji K. Human placenta-derived cells have mesenchymal stem/progenitor cell potential. Stem Cells 22:649–658; 2004. [DOI] [PubMed] [Google Scholar]
- 9. Haase A.; Olmer R.; Schwanke K.; Wunderlich S.; Merkert S.; Hess C.; Zweigerdt R.; Gruh I.; Meyer J.; Wagner S.; Maier L. S.; Han D. W.; Glage S.; Miller K.; Fischer P.; Scholer H. R.; Martin U. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell 5:434–441; 2009. [DOI] [PubMed] [Google Scholar]
- 10. Hanna J.; Markoulaki S.; Schorderet P.; Carey B. W.; Beard C.; Wernig M.; Creyghton M. P.; Steine E. J.; Cassady J. P.; Foreman R.; Lengner C. J.; Dausman J. A.; Jaenisch R. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133:250–264; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayati A. R.; Nur Fariha M. M.; Tan G. C.; Tan A.E.; Chua K. Potential of human decidua stem cells for angiogenesis and neurogenesis. Arch. Med. Res. 42:291–300; 2011. [DOI] [PubMed] [Google Scholar]
- 12. Hirata T.; Amano T.; Nakatake Y.; Amano M.; Piao Y.; Hoang H. G.; Ko M. S. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci. Rep. 2:208; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang Y. C.; Yang Z. M.; Chen X. H.; Tan M. Y.; Wang J.; Li X. Q.; Xie H. Q.; Deng L. Isolation of mesenchymal stem cells from human placental decidua basalis and resistance to hypoxia and serum deprivation. Stem Cell Rev. 5:247–255; 2009. [DOI] [PubMed] [Google Scholar]
- 14. Igura K.; Zhang X.; Takahashi K.; Mitsuru A.; Yamaguchi S.; Takashi T. A. Isolation and characterization of mesenchymal progenitor cells from chorionic villi of human placenta. Cytotherapy 6:543–553; 2004. [DOI] [PubMed] [Google Scholar]
- 15. In't Anker P. S.; Scherjon S. A.; Kleijburg-van der Keur C.; de Groot-Swings G. M.; Claas F. H.; Fibbe W. E.; Kanhai H. H. Isolation of mesenchymal stem cells of fetal or maternal origin from human placenta. Stem Cells 22:1338–1345; 2004. [DOI] [PubMed] [Google Scholar]
- 16. Ito M.; Hiramatsu H.; Kobayashi K.; Suzue K.; Kawahata M.; Hioki K.; Ueyama Y.; Koyanagi Y.; Sugamura K.; Tsuji K.; Heike T.; Nakahata T. NOD/SCID/gamma(c)(null) mouse: An excellent recipient mouse model for engraftment of human cells. Blood 100:3175–3182; 2002. [DOI] [PubMed] [Google Scholar]
- 17. Izumi M.; Pazin B. J.; Minervini C. F.; Gerlach J.; Ross M. A.; Stolz D. B.; Turner M. E.; Thompson R. L.; Miki T. Quantitative comparison of stem cell marker-positive cells in fetal and term human amnion. J. Reprod. Immunol. 81:39–43; 2009. [DOI] [PubMed] [Google Scholar]
- 18. Kanematsu D.; Shofuda T.; Yamamoto A.; Ban C.; Ueda T.; Yamasaki M.; Kanemura Y. Isolation and cellular properties of mesenchymal cells derived from the decidua of human term placenta. Differentiation 82:77–88; 2011. [DOI] [PubMed] [Google Scholar]
- 19. Kim J. B.; Sebastiano V.; Wu G.; Arauzo-Bravo M. J.; Sasse P.; Gentile L.; Ko K.; Ruau D.; Ehrich M.; van den Boom D.; Meyer J.; Hubner K.; Bernemann C.; Ortmeier C.; Zenke M.; Fleischmann B. K.; Zaehres H.; Scholer H. R. Oct4-induced pluripotency in adult neural stem cells. Cell 136:411–419; 2009. [DOI] [PubMed] [Google Scholar]
- 20. Livak K. J.; Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408; 2001. [DOI] [PubMed] [Google Scholar]
- 21. Loh Y. H.; Agarwal S.; Park I. H.; Urbach A.; Huo H.; Heffner G. C.; Kim K.; Miller J. D.; Ng K.; Daley G. Q. Generation of induced pluripotent stem cells from human blood. Blood 113:5476–5479; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lowry W. E.; Richter L.; Yachechko R.; Pyle A. D.; Tchieu J.; Sridharan R.; Clark A. T.; Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. USA 105:2883–2888; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Macias M. I.; Grande J.; Moreno A.; Dominguez I.; Bornstein R.; Flores A. I. Isolation and characterization of true mesenchymal stem cells derived from human term decidua capable of multilineage differentiation into all 3 embryonic layers. Am. J. Obstet. Gynecol. 203:e495; 2010. [DOI] [PubMed] [Google Scholar]
- 24. Maekawa M.; Yamaguchi K.; Nakamura T.; Shibukawa R.; Kodanaka I.; Ichisaka T.; Kawamura Y.; Mochizuki H.; Goshima N.; Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis 1. Nature 474:225–229; 2011. [DOI] [PubMed] [Google Scholar]
- 25. Maherali N.; Sridharan R.; Xie W.; Utikal J.; Eminli S.; Arnold K.; Stadtfeld M.; Yachechko R.; Tchieu J.; Jaenisch R.; Plath K.; Hochedlinger K. Directly reprogrammed fibroblasts showglobalepigeneticremodeling andwidespreadtissuecontribution. Cell Stem Cell 1:55–70; 2007. [DOI] [PubMed] [Google Scholar]
- 26. Marchetto M. C. N.; Carromeu C.; Acab A.; Yu D.; Yeo G. W.; Mu Y.; Chen G.; Gage F. H.; Muotri A. R. A Model for neural development and treatment of rett syndrome using human induced pluripotent stem cells. Cell 143:527–539; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meissner A.; Wernig M.; Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 25:1177–1181; 2007. [DOI] [PubMed] [Google Scholar]
- 28. Nagase T.; Ueno M.; Matsumura M.; Muguruma K.; Ohgushi M.; Kondo N.; Kanematsu D.; Kanemura Y.; Sasai Y. Pericellular matrix of decidua-derived mesenchymal cells: A potent human-derived substrate for the maintenance culture of human ES cells. Dev. Dyn. 238:1118–1130; 2009. [DOI] [PubMed] [Google Scholar]
- 29. Nagata S.; Toyoda M.; Yamaguchi S.; Hirano K.; Makino H.; Nishino K.; Miyagawa Y.; Okita H.; Kiyokawa N.; Nakagawa M.; Yamanaka S.; Akutsu H.; Umezawa A.; Tada T. Efficient reprogramming of human and mouse primary extraembryonic cells to pluripotent stem cells. Genes Cells 14:1395–1404; 2009. [DOI] [PubMed] [Google Scholar]
- 30. Nakagawa M.; Koyanagi M.; Tanabe K.; Takahashi K.; Ichisaka T.; Aoi T.; Okita K.; Mochiduki Y.; Takizawa N.; Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26:101–106; 2008. [DOI] [PubMed] [Google Scholar]
- 31. Nakagawa M.; Takizawa N.; Narita M.; Ichisaka T.; Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA 107:14152–14157; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Navarro P.; Chambers I.; Karwacki-Neisius V.; Chureau C.; Morey C.; Rougeulle C.; Avner P. Molecular coupling of xist regulation and pluripotency. Science 321:1693–1695; 2008. [DOI] [PubMed] [Google Scholar]
- 33. Niibe K.; Kawamura Y.; Araki D.; Morikawa S.; Miura K.; Suzuki S.; Shimmura S.; Sunabori T.; Mabuchi Y.; Nagai Y.; Nakagawa T.; Okano H.; Matsuzaki Y. Purified mesenchymal stem cells are an efficient source for iPS cell induction. PLoS One 6:e17610; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nori S.; Okada Y.; Yasuda A.; Tsuji O.; Takahashi Y.; Kobayashi Y.; Fujiyoshi K.; Koike M.; Uchiyama Y.; Ikeda E.; Toyama Y.; Yamanaka S.; Nakamura M.; Okano H. Grafted human-induced pluripotent stem cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc. Nat. Acad. Sci. USA 108:16825–16830; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oda Y.; Yoshimura Y.; Ohnishi H.; Tadokoro M.; Katsube Y.; Sasao M.; Kubo Y.; Hattori K.; Saito S.; Horimoto K.; Yuba S.; Ohgushi H. Induction of pluripotent stem cells from human third molar mesenchymal stromal cells. J. Biol. Chem. 285:29270–29278; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okita K.; Matsumura Y.; Sato Y.; Okada A.; Morizane A.; Okamoto S.; Hong H.; Nakagawa M.; Tanabe K.; Tezuka, K.-i.; Shibata T.; Kunisada T.; Takahashi M.; Takahashi J.; Saji H.; Yamanaka S. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8:409–412; 2011. [DOI] [PubMed] [Google Scholar]
- 37. Park I. H.; Zhao R.; West J. A.; Yabuuchi A.; Huo H.; Ince T. A.; Lerou P. H.; Lensch M. W.; Daley G. Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451:141–146; 2008. [DOI] [PubMed] [Google Scholar]
- 38. Silva J.; Nichols J.; Theunissen T. W.; Guo G.; van Oosten A. L.; Barrandon O.; Wray J.; Yamanaka S.; Chambers I.; Smith A. Nanog is the gateway to the pluripotent ground state. Cell 138:722–737; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silva S. S.; Rowntree R. K.; Mekhoubad S.; Lee J. T. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Nat. Acad. Sci. USA 105:4820–4825; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stadtfeld M.; Maherali N.; Breault D. T.; Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2:230–240; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suemori H.; Yasuchika K.; Hasegawa K.; Fujioka T.; Tsuneyoshi N.; Nakatsuji N. Efficient establishment of human embryonic stem cell lines and long-term maintenance with stable karyotype by enzymatic bulk passage. Biochem. Biophys. Res. Commun. 345:926–932; 2006. [DOI] [PubMed] [Google Scholar]
- 42. Takahashi K.; Tanabe K.; Ohnuki M.; Narita M.; Ichisaka T.; Tomoda K.; Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872; 2007. [DOI] [PubMed] [Google Scholar]
- 43. Takahashi K.; Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676; 2006. [DOI] [PubMed] [Google Scholar]
- 44. Tchieu J.; Kuoy E.; Chin M. H.; Trinh H.; Patterson M.; Sherman S. P.; Aimiuwu O.; Lindgren A.; Hakimian S.; Zack J. A.; Clark A. T.; Pyle A. D.; Lowry W. E.; Plath K. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell 7:329–342; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wutz A.; Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5:695–705; 2000. [DOI] [PubMed] [Google Scholar]
- 46. Yen B. L.; Huang H. I.; Chien C. C.; Jui H. Y.; Ko B. S.; Yao M.; Shun C. T.; Yen M. L.; Lee M. C.; Chen Y. C. Isolation of multipotent cells from human term placenta. Stem Cells 23:3–9; 2005. [DOI] [PubMed] [Google Scholar]
- 47. Yu J.; Vodyanik M. A.; Smuga-Otto K.; Antosiewicz-Bourget J.; Frane J. L.; Tian S.; Nie J.; Jonsdottir G.A.; Ruotti V.; Stewart R.; Slukvin I. I.; Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920; 2007. [DOI] [PubMed] [Google Scholar]
- 48. Zhao H. X.; Li Y.; Jin H. F.; Xie L.; Liu C.; Jiang F.; Luo Y. N.; Yin G. W.; Wang J.; Li L. S.; Yao Y. Q.; Wang X. H. Rapid and efficient reprogramming of human amnion-derived cells into pluripotency by three factors OCT4/SOX2/NANOG. Differentiation 80:123–129; 2010. [DOI] [PubMed] [Google Scholar]