Abstract
Primary cilia are microtubule-based organelles for sensing of the extracellular milieu and transducing this information into the cell through a variety of molecular signaling pathways. Functioning of the primary cilium has been recently connected to autophagy, a pathway for degradation of cellular components in lysosomes. Autophagy regulates the length of the cilia by removing proteins required for ciliogenesis, a phenomenon that is molecularly different if performed by basal autophagy or when autophagy is induced in response to various stressors. Here we review the current knowledge about the dual interaction between autophagy and ciliogenesis, and discuss the potential role that deregulated ciliary autophagy could have in pathologies with alterations in autophagy and ciliogenesis.
Introduction
Autophagy is a highly conserved intracellular process for the degradation of proteins and organelles in lysosomes [1]. Removal of damaged components by autophagy and continuous turnover of still functional cellular structures to facilitate their renewal is key in the maintenance of cellular homeostasis [2]. Indeed autophagy malfunction leads to the accumulation of altered proteins and organelles that interfere with normal cell functioning and has been linked to pathophysiological conditions such as neurodegenerative diseases, metabolic disorders and cancer [3]. However, the beneficial effect of autophagy goes beyond cellular “cleaning” and resides to a large extent on its ability to recycle back into the cytosol the products resulting from cargo breakdown in lysosomes [4]. Autophagy plays a prominent role in cellular energetics and cell survival during nutrient scarcity [5].
Starvation is a strong selective pressure that has resulted in the evolutionary development of efficient nutrient sensing mechanisms in all organisms. Indeed, during nutrient starvation or under conditions of high metabolic demand (i.e. cellular reprograming, remodeling, differentiation, etc.) cells activate extracellular and intracellular mechanisms directed to sense the energetic defect and provide macromolecules for survival. Receptors and transporters located on the cell surface sense nutrient availability in the extracellular space and transduce the information to the intracellular machinery responsible for energy production [6]. To maximize the sensory functions, most cell types present on their plasma membrane microtubule-based organelles known as primary cilia, which protrude from the cell surface and are enriched in sensing and signaling receptors [7].
Cilia are composed by a central axoneme of nine pairs of microtubules that grow through assembly of tubular components, in a process known as ciliogenesis. The composition of the axoneme differentiates two types of cilia, namely primary cilia (PC) and motile cilia (MC), featuring an extra central pair of microtubules. Both MC and PC are rooted to basal bodies, structures that derive from the centrioles after cell division. However while MC are beating structures with a prominent mechanical role, the PC is a sensory non-motile organelle that clusters receptors and signaling molecules critical for vertebrate development and tissue homeostasis [8,9]. Indeed, defective or absent cilia cause pathologies collectively known as ciliopathies, ranging from polycystic kidney disease (PKD) to respiratory diseases, cognitive impairment disorders and developmental disorders [8]. The ciliary membrane is a continuous yet specialized portion of the plasma membrane enriched in signaling receptors such as components of the Hedgehog (Hh) pathway [10], Wnt signaling pathway [11], olfactory signaling transduction pathway [12], PDGF alpha receptors and calcium channels [13,14], among others. The unique morphology and positioning of the PC allows the activation of these pathways in response to extracellular changes or stressors.
Recent studies show that ciliary signaling pathways are connected to and activate autophagy, and that conversely, autophagy participates in the control of ciliogenesis. In this review, we summarize the current knowledge about the dual interaction between autophagy and the PC, and discuss the pathophysiological implications of altering this interplay.
The primary cilium is required for autophagy activation
Due to its prominent role in the adaptation to nutrient deprivation, autophagy is maximally activated in most cultured cells during the first hours that follow serum removal. In cells that inhibit division by contact, serum removal also induces linear growth of the PC. Despite the parallel temporal activation of both autophagy and ciliogenesis, it was only recently that both processes were linked at the molecular and functional level [15, 16] (Figure 1).
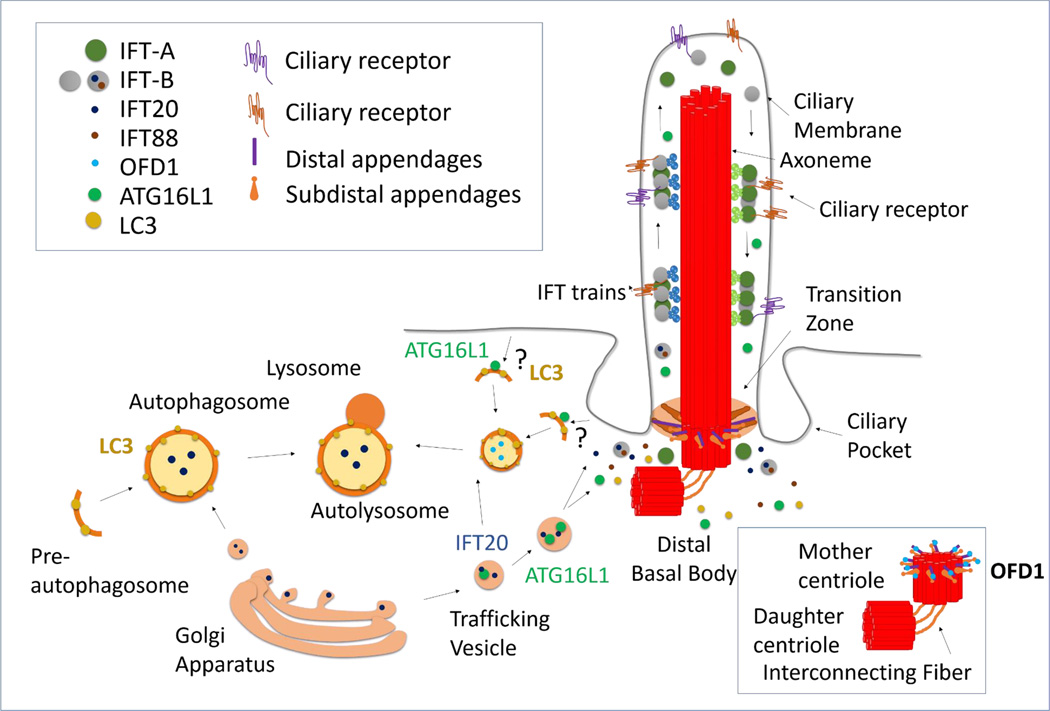
Figure 1. Molecular interaction between of autophagy and ciliogenesis.
Growth of primary cilia (PC) from the mother centriole is attained through continuous bidirectional intraflagellar trafficking (IFT) of proteins such as IFT20. Ciliogenesis is prevented by OFD1, a repressor that localizes at the distal appendages of the mother centriole (capping of the distal appendages by OFD1 is shown in the right boxed area). Activation of ciliogenesis or ciliary Hedgehog signaling induces autophagy. During activation of autophagy, autophagy related proteins (ATG) proteins organize into functional complexes to form a preautophagosome membrane, which surrounds the cargo and then seals into double membrane vesicles (autophagosomes). Autophagosome content is degraded upon fusion with lysosomes. In ciliary-induced autophagy, the pre-autophagosomal marker ATG16L1 trafficks in IFT20-containing vesicles to the ciliary base, where autophagosomes may form from either the plasma membrane or the ciliary pocket.
The first connection between autophagy and the PC came from the realization that autophagy activation upon starvation requires the presence of a functional PC [15]. PC growth and function depends on the continuous trafficking of structural, regulatory and signaling molecules in and out of the ciliary axoneme mediated by the intraflagellar transport (IFT) system [17]. Downregulation or loss of anterograde components of the IFT machinery (IFT88 and IFT20) - known to impair ciliogenesis [18]- also prevents full activation of autophagy in response to starvation [15]. Indeed, starvation-induced increase in autophagosome formation depends on the activation of ciliary Hh signaling upon serum removal [15]. The molecular mechanisms connecting ciliary Hh signaling with autophagy induction are still poorly understood, but recruitment of several autophagy-related proteins (ATG) towards the ciliary base during serum starvation has proven necessary [15]. In fact, while several ATG proteins are constitutively present in this location others, such as ATG16L1 - one of the autophagy proteins involved in the early events of the autophagosome membrane formation [19] - are only recruited to the basal body upon serum removal, where they traffic in vesicles known to participate in IFT (IFT20 containing cytosolic vesicles) (Figure 1) [15]. This sharing of molecular components between autophagy and ciliary trafficking systems highlights the possibility of additional common functions, beyond the described adaptation to starvation. The Hh-dependent recruitment of ATGs involved in the first steps of autophagy to the basal body suggests that the ciliary base could be a niche for autophagosome formation. The single nature of the PC (one per cell) is in clear contrast with other sites of autophagosome biogenesis such as ER, mitochondria or late endosomes, where multiple foci of biogenesis can occur at once [20]. However, the ciliary pocket is an active spot for endocytosis [21] suggesting that vesicle formation can occur from this region. Furthermore, the ciliary membrane is in continuum with the plasma membrane, which can also contribute to the formation of autophagosomes [22] in coordinated manner with the endosomal system [23]. Therefore, it is tantalizing to hypothesize that upon ciliary signaling autophagosomes could form out of membranes derived from the endocytic activity of the ciliary pocket (Figure 1). Indeed, the concept that autophagy initiation sites within the cell vary according to the cargo and initiation stimulus has recently emerged from the better characterization of selective autophagy [24].
The type of autophagy modulated by PC and the mechanisms behind this regulation seem to be context-dependent. Thus, recent studies in a paradigm of kidney epithelial cell differentiation have demonstrated a positive regulatory effect of ciliogenesis on basal autophagy [25]. In this context, the decreased basal autophagy resulting from a block in ciliogenesis can be restored by suppressing mTOR signaling, a well-characterized endogenous repressor of autophagy [25], whereas a similar intervention cannot overcome the defect in starvation-induced autophagy in fibroblasts lacking PC [15]. These divergences suggest that the mechanisms behind cilia-mediated autophagy are stimulus- and/or cell type dependent and could originate from a diversification in the receptor composition of the ciliary membrane according to the cell type or cell conditions. Furthermore, the same signal may have different impacts on autophagy depending on where it originates. Thus, while Hh signaling from the cilium stimulates autophagy [15], other reports have demonstrated that Hh inhibits autophagy in rapidly dividing cells that presumably lack PC [26]. It is possible that the closer proximity to the nucleus of the PC-originating signaling may favor the recruitment of downstream mediators different from those deployed when Hh signaling originates from other cellular locations.
Autophagy regulates ciliogenesis
In recent years, changes in ciliary growth have been directly related to changes in autophagy activity. However, the complexity of this regulatory role has originated conflicting reports whereby blockage of autophagy can either increase or decrease ciliary length. Part of these opposite effects arise from the fact that autophagy degrades proteins that contribute to ciliary growth [15] as well as regulatory proteins that block ciliogenesis [16].
Under nutrient-rich conditions, basal autophagy prevents continuous PC growth by direct degradation of IFT20. However, upon serum removal, IFT proteins are required for ciliogenesis, and thus spared from degradation. This switch from basal to induced autophagy promotes selective degradation of OFD1 and subsequent PC growth [16]. Although OFD1 protein localization at the distal end of the centrioles is necessary for appendage formation [27] and ciliogenesis [28], there is a second pool of OFD1 at centriolar satellites (electron-dense granules of about 70–100 nm in diameter around centrosomes that contain regulatory proteins of centrosome and cilia) that seems to instead suppress ciliogenesis [16] (Figure 2).
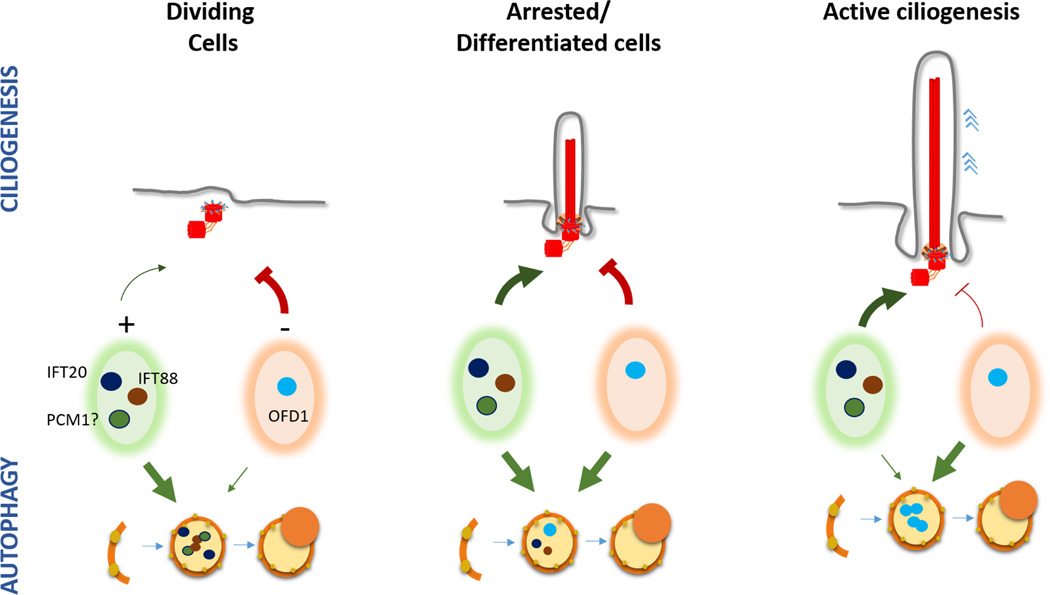
Figure 2. The complex role of autophagy in the regulation of ciliogenesis.
Autophagy has been shown both to enhance and to decrease ciliogenesis. This contrasting effect may depend on the cell type, cellular conditions and likely also on the type of autophagy and the cargo targeted by the autophagic process. Based on the published studies reporting autophagic degradation of both ciliogenesis inhibitors (such as OFD1) and activators (such as IFT20, IFT88 and likely also PCM1) we propose that changes in the ratio of degradation by autophagy of these two pool of regulators could determine the final ciliogenesis rate.
These studies indicate that basal and inducible autophagy have different roles in the control of ciliogenesis, and the switch in cargo suggests participation of different molecular players. In this regard, Skp2 E3 ligase has emerged as one possible candidate by controlling the extent of mTORC1 inhibition over autophagy and thus, modulating PC length [29]. In nutrient-rich conditions activation of autophagy is prevented by recruitment of active mTORC1 to the lysosomal surface [30]. However, to avoid hyperactivation of mTORC1 by the continuous exposure to nutrients, Skp2 limits the amount of TORC1 recruited to lysosomes through ubiquitination of the machinery that usually mediates this recruitment [29]. If Skp2 is depleted, the persistent TORC1-dependent autophagy inhibition leads to increased ciliary length, whereas overexpression of Skp2 results in PC shortening [29]. Basal Skp2 levels or activity could in part determine the observed differences in PC length under normal conditions among the different cell types in a tissue.
The complexity of the contribution of autophagy to the regulation of ciliary length makes any attempt to target autophagy in PC-related diseases premature. Thus, cells exposed to several chemical compounds detected to induce autophagy also show longer PC, although in this case longer cilia originate from reduced disassembly rather than from increased growth [31,32]. Although the increase in autophagy observed with these drugs could be secondary to the longer PC or to their effect in other cellular process, the idea that autophagy could regulate PC length both at the level of growth or at the level of disassembly is attractive. Adding complexity to the autophagy/PC interplay, the consequences of autophagy inhibition on PC growth have been shown to be dependent in part on the ability of different cell types to engage proteasome degradation upon autophagy blockage, although the specific proteins targeted by the proteasome under these conditions are yet to be identified [25].
Overall, all these studies demonstrate that autophagy controls PC length both positively and negatively depending on the context, and highlight the need for a fine-tune regulation of this opposite effect of autophagy to assure rapid adaptation of ciliary length to the cellular requirements.
Cilia related autophagy and disease
Although the interplay between cilia and autophagy is still a young finding whose molecular details require more in depth characterization, the connection between both processes has already contributed to enlightening the pathogenic mechanisms of several human disorders (Figure 3).
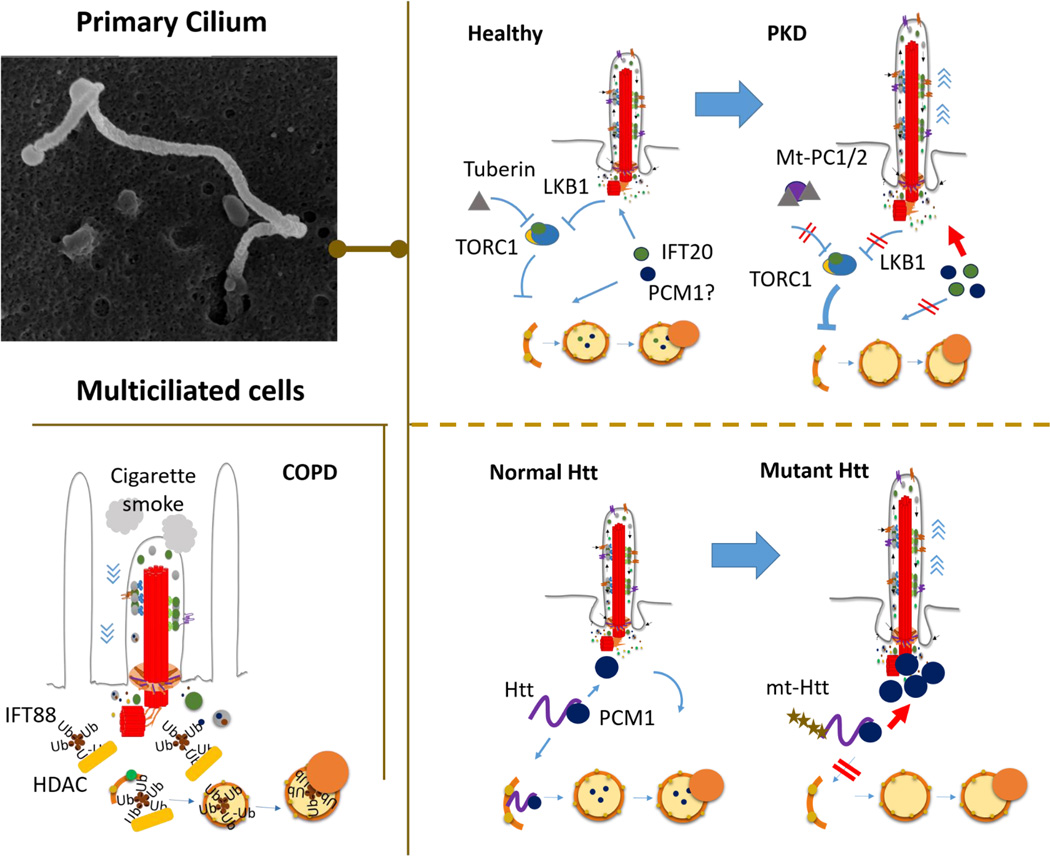
Figure 3. Human diseases in the crossroad between primary cilia and autophagy.
Malfunction of both motile cilia (MC) and primary cilia (PC) in different pathological conditions has been recently connected to autophagy. Top left: scanning electron microscopy of primary cilia in mouse embryonic fibroblasts. Bottom left: Autophagy has been shown to contribute to shortening of MC in chronic obstructive respiratory disease (COPD) through HDAC-mediated degradation of intraflagellar (IFT) proteins required for ciliary growth and function. Cigarette smoke triggers this event by inducing global polyubiquitination. Right: Autophagy may contribute to the abnormally long PC observed in polycystic kidney disease (PKD) (top) and Huntington’s disease (HD) (bottom). In PKD, the observed exacerbation of TORC1 signaling may repress degradation of ciliary components by autophagy such as IFT20 and maybe PCM1, and contribute to enhanced ciliogenesis. In the case of HD, ciliary malfunction may originate from the disruption in the patients of the dual role of huntingtin (Htt) in selective autophagy and trafficking of ciliogenesis regulators, such as PCM1.
Ciliophagy and chronic obstructive pulmonary disease
The first connection between autophagy of cilia and disease was established for degradation of motile cilia (MC) [33]. Defects in MC structure and function associate with disease in epithelia containing systems. In the respiratory tract, cigarette smoking, the most common cause for chronic obstructive pulmonary disease (COPD), leads to impairment of mucociliary clearance and shortening of the epithelial cilia. Autophagy contributes to this shortening through the direct degradation of modified ciliary components, a process now termed ciliophagy [34]. Cigarette smoking triggers ciliophagy upon inducing global protein ubiquitination, with aggregation of ciliary proteins such as IFT88, that are then degraded through HDAC6-mediated autophagy [33]. Indeed, loss of autophagy protects from cigarette smoking-induced cilia shortening in mice [33]. Notably, this is the first evidence that cilia can activate damage signals to trigger autophagy as part of the celllular stress response.
A role for autophagy in ciliopathies
Ciliopathies are a broad group of disorders ranging from developmental disorders to polycystic kidney diseases (PKD), characterized by defects in PC structure and functioning. Mutations in the PKD1 and PKD2 genes, which encode polycystin-1 (PC1) and polycystin-2 (PC2) proteins respectively, lead to autosomal dominant forms of PKD (ADPKD). PC1 and PC2 form an ion channel in the cilium that allows the compartmentalized regulation of ciliary calcium without altering the cytoplasmic calcium levels [35,13,14]. The role of PC on autophagy regulation makes likely that disrupted PC function in PKD may result in autophagic defects. Indeed, reduced autophagy flux has been observed in PKD animal models [36], and although autophagy malfunction has not been demonstrated yet in ADPKD patients, several aberrant signaling pathways implicated in the disease are well-known autophagy modulators [37]. Among them, aberrant mTOR activation has proven to contribute to pathogenesis in these patients since rapamycin reduces the size of the polycystic kidneys [38]. Increased mTOR signaling can originate from the aberrant interaction of mutant PC1 with tuberin, one of the major negative regulators of mTOR [38]. In addition, because bending of the PC inhibits mTORC1 activity through activation of Lkb1 at the base of the cilia, it is also possible that loss of this physiological function of PC in ADPKD patients contributes to their increased mTOR signaling [39]. Sustained mTOR activation in ADPKD patients should presumably impair their ability to induce autophagy. In this respect, although the longer PC in the disease occur in part through changes in protein synthesis mediated by mTORC1 activation [40], in light of the newly discovered role of autophagy in ciliogenesis, the sustained inhibitory effect of mTORC1 on autophagy may also contribute to aberrantly increased PC length in the patients.
Impairment of autophagy and ciliogenesis in Huntington´s disease
Although still lacking direct experimental support, a possible connection between defective PC structures in Huntington’s Disease (HD) and the described autophagy dysfunction in this disease has been recently proposed [41]. The widely recognized autophagy malfunction in HD originates at different levels, including abnormal autophagy induction, failure to selectively recognize the autophagic cargo as well as altered trafficking of the autophagic vesicles [42, 43]. The first two defects have been recently attributed to loss of physiological function of huntingtin (Htt) - the protein mutated in this disease - on autophagy. Htt stabilizes the simultaneous interaction of the autophagy receptor p62 with the cargo and the autophagy protein LC3, and it also contributes to direct activation of selective autophagy by neutralizing the inhibitory effect of mTOR on this pathway [44].
Htt mutation had been previously linked to altered ciliogenesis, resulting in longer and malfunctional PC in the HD brain. This ciliary elongation originates from an abnormally enhanced interaction between mutant Htt and its partner protein HAP1 [45]. The complex Htt/HAP1 usually participates in regulating cellular trafficking, including that of the pericentriolar material 1 protein (PCM1) to the centrosome. Mutation in Htt results in PCM1 accumulation at the centrosome and increased ciliogenesis [45]. The recent involvement of Htt in selective autophagy and the fact that other ciliogenesis mediators undergo degradation by autophagy [15,16] opens the attractive possibility that increased centrosomal accumulation of PCM1 in the in the context of mutant Htt results from its failure to undergo selective autophagy. In this case, PCM1 could join IFT20 and OFD1 as part of the pool of ciliogenesis-related proteins under the control of autophagy.
Conclusions and future directions
Despite common stimulus and time frame activation, the dual interplay between autophagy and ciliogenesis has only been characterized recently. As increasing number of signaling receptors are being discovered on the ciliary membrane, it is likely that additional pathways besides Hh and mTORC1 participate in the regulation of autophagy through the PC. Different ciliary pathways could trigger different types of autophagy, and the possibility that some of them negatively regulate autophagy is also conceivable. Furthermore, cilia-mediated autophagy could also be regulated by cellular events that interfere with the physical presence of the PC, such as the pre-mitotic resorption of PC during the cell cycle. How these events affect autophagy remains unknown. Conversely, since basal and inducible autophagy contribute differentially to ciliogenesis, it would be interesting to know if specific autophagy receptors or adaptors contribute to promote the degradation of ciliary activators or repressors.
An enticing consequence of the PC/autophagy interplay is the possibility of using PC signaling as therapeutic target in diseases with defective autophagy, such as neurodegeneration or metabolic disorders. Similarly, modulation of autophagy could be of therapeutic value in ciliopathies. However, before these interventions can be implemented, future studies should be directed towards the molecular characterization of the dual interplay between autophagy and ciliogenesis, as well as to study the role of cilia-mediated autophagy in physiological tissue homeostasis.
Acknowledgments
Research in our groups is supported by grants from Initiative of Excellence of the University of Bordeaux and the Ligue Européenne Contre la Maladie d’Alzheimer (OP) and the National Institutes of Health National Institute on Aging (AMC) and awards from The Rainwaters Foundation and The Beatrice and Roy Backus Foundation and a generous gift from Robert and Renee Belfer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Zhang H, Baehrecke EH. Eaten alive: Novel insights into autophagy from multicellular model systems. Trends in cell biology. 2015;25(7):376–387. doi: 10.1016/j.tcb.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005;12:1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 3.Choi AMK, Ryter SW, Levine B. Autophagy in human health and disease. New England Journal of Medicine. 2013;368(7):651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 4.Shen HM, Mizushima N. At the end of the autophagic road: An emerging understanding of lysosomal functions in autophagy. Trends in biochemical sciences. 2014;39(2):61–71. doi: 10.1016/j.tibs.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16(8):461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 6.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517(7534):302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachury MV. How do cilia organize signalling cascades? Philos Trans R Soc Lond B Biol Sci. 2014;369(1650) doi: 10.1098/rstb.2013.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen ST, Pedersen LB, Schneider L, Satir P. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 2007;8(2):97–109. doi: 10.1111/j.1600-0854.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- 9.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nature Reviews Genetics. 2010;5:331. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briscoe J, Therond PP. The mechanisms of hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes JM, Katsanis N. Ciliary function and wnt signal modulation. Curr Top Dev Biol. 2008;85:175–195. doi: 10.1016/S0070-2153(08)00807-7. [DOI] [PubMed] [Google Scholar]
- 12.McEwen DP, Jenkins PM, Martens JR. Olfactory cilia: Our direct neuronal connection to the external world. Curr Top Dev Biol. 2008;85:333–370. doi: 10.1016/S0070-2153(08)00812-0. [DOI] [PubMed] [Google Scholar]
- 13.DeCaen PG, Delling M, Vien TN, Clapham DE. Direct recording and molecular identification of the calcium channel of primary cilia. Nature. 2013;504(7479):315–318. doi: 10.1038/nature12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504(7479):311–314. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502(7470):194–200. doi: 10.1038/nature12639. ••Identification of the regulatory role of primay cilia on starvation-induced autophagy and of the selective degradation of IFT20 in the negative control of ciliogenesis by basal autophagy.
- 16. Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. Autophagy promotes primary ciliogenesis by removing ofd1 from centriolar satellites. Nature. 2013;502(7470):254–257. doi: 10.1038/nature12606. ••Identification of the first negative regulator of ciliogenesis whose degradation by autophagy promotes ciliogenesis.
- 17. Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426(6962):83–87. doi: 10.1038/nature02061. •Identification of the involvement of IFT in hedgehog sinaling from the cilia
- 18.Follit JA, Xu F, Keady BT, Pazour GJ. Characterization of mouse ift complex b. Cell Motil Cytoskeleton. 2009;66(8):457–468. doi: 10.1002/cm.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell research. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Current opinion in cell biology. 2013;25(4):455–460. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Benmerah A. The ciliary pocket. Current opinion in cell biology. 2013;25(1):78–84. doi: 10.1016/j.ceb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nature cell biology. 2010;12(8):747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puri C, Renna M, Bento Carla F, Moreau K, Rubinsztein David C. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154(6):1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sica V, Galluzzi L, Bravo-San Pedro JM, Izzo V, Maiuri MC, Kroemer G. Organelle-specific initiation of autophagy. Mol Cell. 2015;59(4):522–539. doi: 10.1016/j.molcel.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Livingston MJ, Su Y, Dong Z. Reciprocal regulation of cilia and autophagy via the mtor and proteasome pathways. Autophagy. 2015;11(4):607–616. doi: 10.1080/15548627.2015.1023983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jimenez-Sanchez M, Menzies FM, Chang YY, Simecek N, Neufeld TP, Rubinsztein DC. The hedgehog signalling pathway regulates autophagy. Nat Commun. 2012;3:1200. doi: 10.1038/ncomms2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singla V, Romaguera-Ros M, Garcia-Verdugo JM, Reiter JF. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev Cell. 2010;18(3):410–424. doi: 10.1016/j.devcel.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, Dolle P, Franco B. Oralfacial- digital type i protein is required for primary cilia formation and left-right axis specification. Nat Genet. 2006;38(1):112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- 29. Jin G, Lee SW, Zhang X, Cai Z, Gao Y, Chou PC, Rezaeian AH, Han F, Wang CY, Yao JC, Gong Z, et al. Skp2-mediated raga ubiquitination elicits a negative feedback to prevent amino-acid-dependent mtorc1 hyperactivation by recruiting gator1. Mol Cell. 2015;58(6):989–1000. doi: 10.1016/j.molcel.2015.05.010. •Demonstration of the mechanism that prevents hyperactivation of mTor during continous nutrient exposure
- 30. Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-rag complex targets mtorc1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. •First description of the association of mTOR with lysosomes for functional purposes
- 31.Kim ES, Shin JH, Park SJ, Jo YK, Kim JS, Kang IH, Nam JB, Chung DY, Cho Y, Lee EH, Chang JW, et al. Inhibition of autophagy suppresses sertraline-mediated primary ciliogenesis in retinal pigment epithelium cells. PLoS One. 2015;10(2):e0118190. doi: 10.1371/journal.pone.0118190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin JH, Bae DJ, Kim ES, Kim HB, Park SJ, Jo YK, Jo DS, Jo DG, Kim SY, Cho DH. Autophagy regulates formation of primary cilia in mefloquine-treated cells. Biomol Ther (Seoul) 2015;23(4):327–332. doi: 10.4062/biomolther.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, Cervo M, Yao H, Chung AL, Mizumura K, An CH, et al. Histone deacetylase 6–mediated selective autophagy regulates copd-associated cilia dysfunction. The Journal of clinical investigation. 2013;123(12) doi: 10.1172/JCI69636. 0-0. ••First data linking the degradation of ciliary components by autophagy to pathological conditions.
- 34.Cloonan SM, Lam HC, Ryter SW, Choi AM. "Ciliophagy": The consumption of cilia components by autophagy. Autophagy. 2014;10(3):532–534. doi: 10.4161/auto.27641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cebotaru V, Cebotaru L, Kim H, Chiaravalli M, Boletta A, Qian F, Guggino WB. Polycystin-1 negatively regulates polycystin-2 expression via the aggresome/autophagosome pathway. J Biol Chem. 2014;289(10):6404–6414. doi: 10.1074/jbc.M113.501205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belibi F, Zafar I, Ravichandran K, Segvic AB, Jani A, Ljubanovic DG, Edelstein CL. Hypoxia-inducible factor-1alpha (hif-1alpha) and autophagy in polycystic kidney disease (pkd) Am J Physiol Renal Physiol. 2011;300(5):F1235–F1243. doi: 10.1152/ajprenal.00348.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravichandran K, Edelstein CL. Polycystic kidney disease: A case of suppressed autophagy? Semin Nephrol. 2014;34(1):27–33. doi: 10.1016/j.semnephrol.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Shillingford JM, Murcia NS, Larson CH, Low SH, Hedgepeth R, Brown N, Flask CA, Novick AC, Goldfarb DA, Kramer-Zucker A, Walz G, et al. The mtor pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci U S A. 2006;103(14):5466–5471. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boehlke C, Kotsis F, Patel V, Braeg S, Voelker H, Bredt S, Beyer T, Janusch H, Hamann C, Godel M, Muller K, et al. Primary cilia regulate mtorc1 activity and cell size through lkb1. Nature cell biology. 2010;12(11):1115–1122. doi: 10.1038/ncb2117. •First data indicating that the primary cilium regulates the central pathway mTORC1 through Lkb1.
- 40.Yuan S, Li J, Diener DR, Choma MA, Rosenbaum JL, Sun Z. Target-of-rapamycin complex 1 (torc1) signaling modulates cilia size and function through protein synthesis regulation. Proc Natl Acad Sci U S A. 2012;109(6):2021–2026. doi: 10.1073/pnas.1112834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaliszewski M, Knott AB, Bossy-Wetzel E. Primary cilia and autophagic dysfunction in huntington/'s disease. Cell Death Differ. 2015;22(9):1413–1424. doi: 10.1038/cdd.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin DD, Ladha S, Ehrnhoefer DE, Hayden MR. Autophagy in huntington disease and huntingtin in autophagy. Trends Neurosci. 2015;38(1):26–35. doi: 10.1016/j.tins.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 43. Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in huntington's disease. Nat Neurosci. 2010;13(5):567–576. doi: 10.1038/nn.2528. •First connection between huntingtin and abnormal recognition of cargo by autophagy
- 44. Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, Cuervo AM, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nature cell biology. 2015;17(3):262–275. doi: 10.1038/ncb3101. •Identification of a dual physiological function for huntingtin in selective autophagy
- 45. Keryer G, Pineda JR, Liot G, xE raldine, Kim J, Dietrich P, Benstaali C, Smith K, Cordeli xE, et al. Ciliogenesis is regulated by a huntingtin-hap1-pcm1 pathway and is altered in huntington disease. The Journal of Clinical Investigation. 2011;121(11):4372–4382. doi: 10.1172/JCI57552. •First data supporting that mutant huntingtin alters ciliogenesis through abnormal interaction with ciliary components