Abstract
This study attempted to establish and quantify the connections between parenting, offspring psychosocial adjustment, and the epigenome. The participants, 35 African-American young adults (19 females and 16 males; age = 17 to 29.5 years), represented a subsample of a three-wave longitudinal 15-year study on the developmental trajectories of low-income urban mother-offspring dyads. Mothers were assessed on their perceptions of maternal stress at each wave. Offspring were assessed on their perceptions of maternal parenting at each wave and on their adaptive and maladaptive behavior at the last wave. Genome-wide DNA methylation in peripheral T-lymphocytes at the third wave was assayed using MBD-sequencing. Statistically significant associations were identified between the change in offspring's perception of parenting from middle childhood to adulthood and the DNA methylation in offspring's adult genomes. Specifically, the slope of perceived parental rejection across the three time points was related to an increase in methylation, or a potential downregulation, of 565 genes thought to be involved in the control of a broad spectrum of biological functions generally related to cellular signaling. A subset of these epigenetic marks, clustered in 23 genes, some of which participate in the development and functioning of the CNS, were in turn associated with psychosocial adjustment as captured by interpersonal relationships and emotional self-evaluation. This appears to be one of the first investigations of the modulating role of the methylome in associations between developmental dynamics of parenting throughout the formative years of child and adolescent development and psychosocial adjustment in adulthood.
There is mounting evidence indicating that the relations between parenting and child behavior, emotional well-being, and psychopathology are reciprocal over time (Pardini, 2008). Parenting behavior is subject to short-term and long-term temporal dynamics, with a potential cascading effect on children's psychosocial adjustment later in life exerted not only by type, but also by the stability of the patterns of parent-child interactions (Cox, Mills-Koonce, Propper, & Gariepy, 2010). Despite the growing consensus that both parenting and its lifespan fluctuations impact children's development, the complex multilayered mechanisms behind these transactional effects are yet to be fully understood (Barbot, Crossman, Hunter, Grigorenko, & Luthar, 2014; Pettit & Arsiwalla, 2008). Parents' psychological well-being, characteristics of parenting, and children's perception of the parent-child relationship are interrelated, hampering inferences about the directionality of effects or the disentangling of the role of each of these variables in predicting children's adjustment (Patterson & Fisher, 2002).
A number of factors have been proposed as potential modulators (or modulating factors) that include mediation, moderation, and all other types of nontrivial associations between two variables of interest in the presence of a third variable, in the relation between parenting and child adjustment. At the societal level such factors include the amount and quality of childcare (Romano, Kohen, & Findlay, 2010). At the neurobiological level such factors include low stress reactivity or optimized pre-frontal cortex functioning (Bloomfield, 2011; Rutter, 2012; Silk et al., 2007). In addition, it is acknowledged that parental stress and mental health impact both children's brain and behavior development (Dawson, Ashman, & Carver, 2000), with potential long-term effects on adjustment (Scott, 2012). Despite these and other relevant observations, a framework for understanding the biological underpinnings linking parenting and children's experiences of parent-child relationships with their long-term outcomes on adjustment has not been developed.
During the last decade, epigenetic mechanisms (e.g., histone modifications and DNA methylation) have become central to studies of the modulation between the dynamics of relevant environments and specific phenotypes. Epigenetic mechanisms connecting social environment and behavioral phenotypes are studied within the behavioral epigenetics field (Lester et al., 2011). The basic premises set by this field imply that social environments impact the epigenome in a lasting way, generating persistent and system-wide changes in gene function (McGowan et al., 2009; Suderman et al., 2014; Szyf, 2013; Szyf, McGowan, & Meaney, 2008; Weaver et al., 2004) that, in turn, change relevant physiology (e.g., neuronal plasticity and functioning) and, consequently, affect behavior. However, we do not have a detailed picture of the affected regions of the epigenome and the pathways through which they modulate behavior. This is mainly due to the field's still emerging understanding of (1) the specificities of epigenetic regulation (e.g., DNA methylation) and its role in social experiences, and (2) the specificities of social experiences (e.g., typologies of negative life events) as contexts for epigenome reactivity.
Here, we offer a novel answer to the “old question” regarding what mechanisms modulate the effects of parenting on offspring's long-term adjustment. Our study, grounded in the epigenetic literature, investigates whether epigenetic mechanisms are a candidate for forwarding the effects of parenting to offspring outcomes in adulthood. Specifically, this article reports on associations between parenting behavior, namely offspring's perceived parenting and mothers' perceived parenting stress and the offspring's epigenetic patterns (DNA methylation marks), and the associations of these epigenetic marks with psychosocial adjustment in adulthood. Due to a shortage of knowledge on dynamic associations between the epigenome and social experiences during the human lifespan it is difficult to formulate specific hypotheses. Yet, capitalizing on what is known about the dynamics of parenting and corresponding reflections on the epigenome (Weaver et al., 2004), we hypothesized that indicators of parenting across three distinct developmental periods–middle childhood, adolescence, and young adulthood—might be more sensitive to capturing an association with the changes in the methylome as they would differentiate stable and variable characteristics of parenting. Given the important role of the stability, not only the type, of the patterns of parenting and mother-child interactions in offspring's psychosocial adjustment we investigated the association between the epigenome and the temporal dynamics of parenting over these three periods. Specifically, we investigated whether the stability in comparison to variability of parenting (i.e., the perceived change in parenting over time) may have a different degree of influence on the epigenome.
Method
Participants
The participants in this study were a subsample (35 offspring) of a sample including 361 low-income ethnically diverse urban mother-offspring dyads (see Appendix S1 and Table S1). Mother-offspring dyads were assessed three times over the course of 15 years following the first measurement occasion (T1) in 1996, with the second (T2) and third (T3) occasions following at approximately 5-year intervals.
A cohort of 35 offspring of African-American descent (19 females; age ranging from 17 to 29.5 years; M=22.5±3.3 years) was selected to constrain ethnic diversity, contingent upon participants' willingness to donate biospecimens at T3. All participants provided written consent and a blood sample; ethical approval for the study was obtained from the proper authorities of the institutions involved in this study. Age, gender, and the presence of substance-related and addictive disorders (SRAD) and/or psychiatric problems of the participants were used as covariates in the analyses. Additional demographic information is provided in Appendix S1.
Indicators of Parenting
Detailed information on the measures used to assess the indicators of parenting is included in Appendix S1. The Parental Acceptance–Rejection Questionnaire (PARQ; Khaleque & Rohner, 2002) was used to assess participants' perceptions of their relationships with mothers. This self-report questionnaire has four subscales (warmth and affection; hostility and aggression; indifference and neglect; and undifferentiated rejection) and comprises 60 items rated on a 4-point scale from 0 (never true of my mother) to 3 (almost always true of my mother).
Mothers rated their experiences of parenting stress using the Parenting Stress Index – Short Form, PSI-SF (Abidin, 1995), a 36-item self-report measure that uses a 5-point Likert-scale (ranging from 1 = strongly disagree to 5 = strongly agree). The measure yields a total stress score from three subscales with 12 items each, including (1) parental distress; (2) parent-child dysfunctional interactions; and (3) the difficult child subscale.
Two indicators were calculated at each of the three time points: (1) indicators of perceived parental rejection computed as the sum of the hostility-aggression, indifference-neglect, and undifferentiated rejection subscales scores of the PARQ; (2) indicators of maternal stress computed as the sum score of the three aforementioned PSI subscale scores. Finally, scores across all time points were averaged to gauge the general level of perceived parental rejection (Cronbach's α = .60) and maternal stress (Cronbach's α = .90) across the entire study period (see Table S2).
Indicators of Temporal Dynamics of Parenting
Beyond concurrent individual differences in the perception of parental rejection, some children may cope differently with the experience of rejection during different developmental periods. Therefore, we focused on investigating the influences of perceived parental rejection over time (T1-T3) on current epigenetic patterns (T3). Two linear latent growth curve models (Preacher, Wichman, MacCallum, & Briggs, 2008) were specified to derive slope values of parental rejection and maternal stress for subsequent epigenome-wide association study (EWAS) analyses, using a latent variable comprising observed PARQ and PSI subscale scores as indicators for each time point, respectively. This growth curve modeling approach is described in detail in Appendix S1. The factor scores of the slopes were computed (regression method using posterior means) and used as indicators of linear change in parenting, based on perceptions of parental rejection and maternal stress (PARQ-slope and PSI-slope, respectively; see Table S2).
Indicators of Psychosocial Adjustment
Detailed information on the measure used to assess indicators of psychosocial adjustment is included in Appendix S1. The college version of the Behavior Assessment System for Children (BASC-CV; Nowinski, Furlong, Rahban, & Smith, 2007) was used to assess five indicators of psychosocial adjustment: (1) internalizing problems; (2) inattention/hyperactivity; (3) emotional symptoms index; (4) personal adjustment; and (5) a clinical composite. Descriptive statistics on the BASC-CV scores are available in Table S3.
Genome-Wide DNA Methylation Profiling
Detailed information on the approach to genome-wide DNA methylation profiling is included in the Appendix S1. To decrease the variability of DNA methylation profiles due to the high cell-specificity of DNA methylation patterns and differential white blood cell counts among individuals, we focused on a single cell type, T-lymphocytes (CD3). For genome-wide DNA methylation profiling, Methyl-CpG binding domain sequencing (MBD-seq) was utilized, which consists of the capture of methylated DNA (ME-DNA) using the MBD domain of MBD2, and subsequent next generation sequencing of the eluted DNA. The Illumina HiSeq platform was used for the massive parallel sequencing of the ME-DNA; paired-end sequencing (2×75 bp reads) was utilized. MBD-seq outcomes and the results of the alignment are summarized in Table S4.
Each genome-wide profile was represented by a methylation measurement matrix of around 10M 300bp fragments. For the EWAS analyses 480,172 autosomal fragments or markers (ME-markers) that contain the most variation in methylation levels and represent approximately 5% of the genome were used. For details on cutoff criteria for establishing the final list of ME-markers please see Appendix S1.
Epigenome-Wide Association Analyses
To examine the associations between parenting and DNA methylation, we used multiple linear regression analyses. We ran separate ordinary least squares regressions to regress the methylation levels of 480,172 selected ME-markers on the covariates (Table S1) and one of four parenting indicators, that is, the means and slopes of the PARQ and PSI (Table S2). The goal of this approach was to determine the set of ME-markers (thereafter, parenting-associated ME-markers) that were significantly associated with a parenting indicator over and above the effects of potentially confounding variables (i.e., age, gender, and the number of SRAD and psychiatric problems). The p-value associated with the t-test, indicating whether the unstandardized regression coefficient of parenting was significantly different from zero, was adjusted for multiple comparisons using the Benjamini-Hochberg correction (FDR; Benjamini & Hochberg, 1995). Coefficients below .05 according to the FDR-adjusted p-values were retained as statistically significant. Moreover, residuals of the regression models were evaluated for normality using the Shapiro-Wilk test for the markers after the amendment for multiple comparisons. The models that resulted in non-normal distributions of residuals were considered insufficiently robust and disregarded in subsequent analyses.
We then examined whether the resulting sets of parenting-associated ME-markers might be related to psychosocial adjustment. Specifically, linear regression models were fitted using the five BASC-CV composites (Table S3) as dependent variables, methylation levels of parenting-associated ME-markers as independent variables, and the aforementioned as covariates.
Gene-Annotation of the Methylation Fragments and Functional Analysis of Genes
To relate the sets of parenting-associated ME-markers to genes and functional genomic regions, the ME-markers were subsequently annotated to the GRCh37/hg19 human genome assembly, using the bedtools software package (https://github.com/arq5x/bedtools2). Each marker was intersected both within a gene and a 2kb upstream region. To connect the resulting sets of parenting-associated genes to biological functions, as they are defined in Gene Ontology (GO) terms (cellular components, molecular functions and biological processes), the Database for Annotation, Visualization and Integrated Discovery (DAVID) was used (http://david.abcc.ncifcrf.gov). To appraise the activity of the genes and the variability of their expression levels in blood tissue, a co-expression network for the genes was constructed via the WebQTL resource (http://www.genenetwork.org), which combines genetic and phenotypic databases using specific analytic tools. Specifically, the data on the mRNA expression in the blood from Genotype-Tissue Expression (GTEx) project (http://www.gtexportal.org) were used for the network construction.
Results
The present study conceptualizes epigenetic marks as potential modulators of the relation between parenting and long-term psychosocial adjustment. Results are presented in three sections: (1) for the behavior analyses, the association between perceived parental rejection and adult psychosocial outcomes; (2) for the EWAS analyses, the identification of regions in the methylome that are associated with different indicators of parenting; and (3) for the combined behavior-methylome analyses, the association between the methylome markers identified in (2) and indicators of psychosocial adjustment.
Perceived Parental Rejection is Associated with Adult Psychosocial Outcomes
As expected, parenting variables were related to offspring psychosocial adjustment. Specifically, the increase in offspring's perception of parental rejection over time (PARQ-slope) was related to higher levels (all p-values < .001) of internalizing problems (r = .59), inattention/hyperactivity (r = .52), emotional problems (r = .62), and clinical symptoms (r = .55); and lower levels (r = -.48, p < .01) of personal adjustment in adulthood. The correlations between psychosocial adjustment and the average level of perceived parental rejection across all three time points showed lower values but similar magnitudes: internalizing problems (r = .50), inattention/hyperactivity (r = .49), emotional problems (r = .54), clinical symptoms (r = .52), and personal adjustment (r = -.50); all p-values < .01. Thus, there were significant associations between perceived parental rejection (mean and slope) and adult outcomes, whereas maternal stress (PSI-slope and averaged PSI scores) was not significantly associated with offspring's adult outcomes.
DNA Methylation is Associated with Changes in Perceived Parental Rejection
The EWAS did not show significant associations between the epigenetic patterns and the indicators of parenting (PARQ and PSI), when PARQ and PSI measurements were considered separately for each of three developmental periods and were averaged across the three time points. Also, there were no significant associations observed between the linear change of maternal stress (PSI-slope) and the ME-markers. However, there were 818 significant (p < .05, after multiple-testing corrections) associations between ME-markers and the indicators of linear change in perceived parental rejection, PARQ-slope. The resulting ME-markers and statistics for the association with the PARQ-slope are shown in Table S5. The age, gender, and mental-health indicators, used as covariates in the regression models, did not show any significant contribution to the variability of the methylation levels of these 818 ME-markers (see Figure S1).
Most of the 818 ME-markers (787 or approximately 96%) were positively related to the PARQ-slope. Moreover, mean methylation levels of the parenting-associated markers and the PARQ-slope yielded positive correlations (Figure 1A&B). Specifically, a one unit increase in the PARQ-slope was associated with a 0.11 unit increase in mean methylation levels across the 818 ME-markers. However, we interpret this effect cautiously because much of the effect was due to three cases, which show slope values of about 5, while most of the 35 individuals clustered in the range of -2 to 2 on the PARQ-slope (Figure 1A). While values this high are possible and, perhaps, true to the assumed biological mechanism, they are discrepant with the remaining cluster of values and thus exert a certain degree of leverage on the fit of the regression model. Although we believe that these are legitimate cases, this finding may indicate that the nature of the studied sample is such that variables are often distributed non-normally. Without the three cases in question, adding the PARQ-slope to the model still accounted for a significant amount of variance in the mean methylation level over and above the contribution of the covariates (F(1, 26) = 4.48, p = .04, Cohen's f2 = 0.17). Moreover, a significant association between the PARQ-slope and DNA methylation was confirmed even after removing the three outliers (see Figure 1B), supporting the robustness of the association between change in perception of parental rejection and DNA methylation in specific genomic regions. Notably, the 818 ME-markers are widely distributed across the entire genome. Gene-annotation of these marks indicated that about 70% of them are allied with 565 different genes (see Table S6); they are located within the sequence or the 2kb region upstream of these genes. Full results of the functional analysis of these 565 genes are presented in Table S7; a summary of this analysis (e.g., GO categories significantly overrepresented among these genes) is provided in Table 1. The GO annotation demonstrated that the list is especially enriched in genes controlling functions such as GTPase activity and its regulation, ATP-binding, and metal ion binding. These molecular functions are involved in a broad spectrum of biological processes and pathways, such as development, the cell cycle, and metabolic processes.
Figure 1.
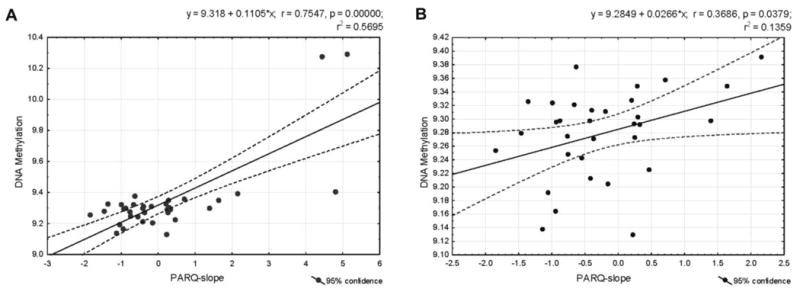
Linear regression model that relates the score of dynamic change in perceived parental rejection (PARQ-slope) and the mean methylation levels of 818 parenting-associated markers, or 300bp MBD-seq fragments for the entire sample of 35 individuals (A) and for 32 samples, with the exclusion of extreme cases (B). Figure 1A shows that the sample contains three extreme cases. One case had an unusually high PARQ-slope value (4.82) with average DNA methylation compared to the rest of the sample (i.e., exerting a leverage on the regression line). Two cases showed discrepancies as reflected in high PARQ-slope values (5.12 and 4.45) and high DNA methylation. A set of diagnostic analyses of the regression of mean methylation on PARQ-slope values and the covariates was performed to identify any concerns with these extreme cases. Results showed (1) no concerning leverage of mean methylation on the fitted values as indicated by hat values (values were 0.48, 0.46, and 0.46; mean hat value was 0.17); (2) no unusual large or small residuals as indicated by most studentized residuals within the ± 2 range (largest studentized residual was 2.64 with a Bonferroni-adjusted p-value of .47); (3) no major influence on the regression coefficients of PARQ-slope as indicated by a plot of residuals against leverage (Cook's d values were 0.87, 0.54, and 0.84). Given the small sample size, we have tested and evaluated other types of robust regression models using high breakdown point (M- or MM-) estimators as these models tend to be less vulnerable to unusual data. Estimators such as “least trimmed squares” and “least median of squares” are alternatives to ordinary least squares regression that account better for the effect of extreme values. However, these methods led to problems of convergence in many of the regression models we tested. In robust regression using M-estimators, Wald-type inference of the significance of coefficients typically requires larger samples—due to the unreliable asymptotic covariance matrix in small samples—than the data available in this study. Thus, although we were aware of the potential influence of these cases on the regression, we decided to utilize ordinary least squares regression but cautiously interpret the results. Figure 1B shows that the association between the PARQ-slope and the mean of ME-markers is high and significant within the sample after removing the three extreme cases. The y-axis (mean DNA-methylation) has been rescaled to better illustrate the linear interrelation.
Table 1.
Summary of the functional annotation of 565 parenting-associated genes performed using the DAVID annotation tool.
| Function | GO term | Genes Count | Genes % | p-value | Fold Enrichment | FDR-adjusted p-value* |
|---|---|---|---|---|---|---|
| GTPase regulator activity | GO:0030695∼GTPase regulator activity | 26 | 6.30 | 3.68E-06 | 2.89 | .0018 |
| GO:0060589∼nucleoside-triphosphatase regulator activity | 26 | 6.30 | 5.44E-06 | 2.82 | .0014 | |
| GO:0005083∼small GTPase regulator activity | 16 | 3.87 | 0.001195 | 2.62 | .0488 | |
| ATP binding | GO:0032559∼adenyl ribonucleotide binding | 57 | 13.80 | 5.44E-05 | 1.71 | .0091 |
| GO:0005524∼ATP binding | 55 | 13.32 | 1.39E-04 | 1.67 | .0139 | |
| GO:0030554∼adenyl nucleotide binding | 58 | 14.04 | 1.19E-04 | 1.65 | .0148 | |
| GO:0001883∼purine nucleoside binding | 58 | 14.04 | 1.78E-04 | 1.62 | .0148 | |
| GO:0001882∼nucleoside binding | 58 | 14.04 | 2.13E-04 | 1.62 | .0152 | |
| Metal ion binding | GO:0046872∼metal ion binding | 119 | 28.81 | 6.38E-04 | 1.29 | .0392 |
| GO:0043169∼cation binding | 119 | 28.81 | 9.40E-04 | 1.28 | .0461 | |
| GO:0005509∼calcium ion binding | 36 | 8.72 | 0.001177 | 1.76 | .0503 | |
| Associated with the long qt syndrome** | 5 | 1.21 | 5.74E-05 | 21.48 | .0066 | |
Notes.
The FDR-adjusted p-values (≤ .05) were used as inclusion criteria to trim the overrepresented term lists. Fold enrichment of each gene group is estimated in comparison to the base set of genes in the DAVID databases (19,235 genes).
The long QT syndrome—a rare inherited heart condition related to the violation of ventricular fibrillation. The enrichment in this functional group of genes (5 of 12 known associates including two candidate genes, KCNQ1 and KCNH2) is considered an unexpected finding. We cannot speculate about the causal association between these genes and parenting. Most likely, the association was obtained because of the presence of the disease in the study cohort, known to be a population with a high risk and frequency of cardio-vascular disorders (an assumption we could not confirm due to a lack of medical records), and a possible causal relation between the syndrome and abnormal methylation of the candidate genes. This might be an issue for future investigation of the candidate-genes for the disorder.
The Epigenetic Links between Parenting and Psychosocial Adjustment
After multiple-testing corrections, 38 of the 818 ME-markers showed significant (p < .05) and negative relations with the BASC-CV personal adjustment composite (Table S5). Specifically, lower levels in personal adjustment were associated with higher methylation levels while controlling for age, gender and the number of SRAD and psychiatric problems. In a model with the mean level of methylation among the 38 identified markers as an independent variable in the regression (Figure 2), the results showed that DNA methylation was significantly and negatively related to the BASC-CV personal adjustment composite (B = -26.80, SE = 7.58, β = -.74, p = .002), but the association between the PARQ-slope and BASC-CV personal adjustment composite was not significant over and above the other variables in the model (B = 0.15, SE = 2.07, β = .02, p = .943).
Figure 2.
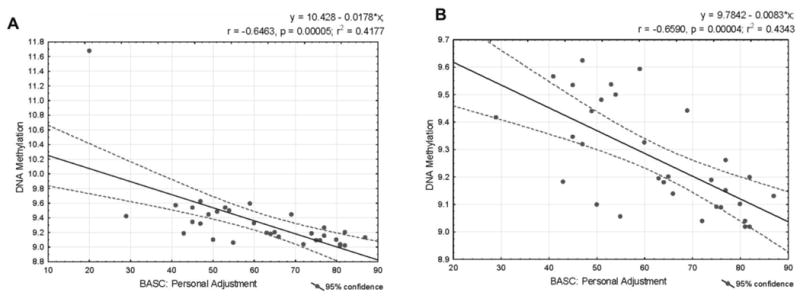
Linear regression model that describes the relationship between the Personal Adjustment composite score and the average methylation level of 38 methylation markers that were associated with both Personal Adjustment and the PARQ-slope. Relationships were established for the entire studied sample (A) and for the sample without any cases with extremely high methylation levels (B). Both plots show high negative associations between the variables.
Although no other associations with the BASC-CV composite or individual subscales survived corrections for multiple testing, we found suggestive associations prior to p-value corrections. Indicators of social stress, anxiety, and hyperactivity were related to methylation levels for 31 ME-markers (with average uncorrected p-values across all ME-markers of .003, .003, and .006, respectively). Indicators of depression and attention problems were associated with 443 and 337 ME-markers (with average uncorrected p-values across all 443 and 337 ME-markers of 1.33e-04 and 4.85e-04), respectively. The lowest set of FDR-adjusted p-values was .07 and .08 for depression and attention problems, respectively.
As per the genome annotation analysis, out of 38 ME-markers associated with both the PARQ-slope and BASC-CV personal adjustment 11 were annotated within intergenic regions with unknown regulatory functions, 4 markers were localized within transcription factor (TF) binding sites, and the rest of the markers were related to 23 genes (Table 2). The analysis of the mRNA expression of these genes in blood tissue (based on data from the GTEx project, http://www.gtexportal.org) indicated high co-expression (i.e., co-variability in their expression levels) across individuals (Figure 3). This observation suggests system-level functionality that might arise from co-regulatory and/or co-functional relations between these genes. The annotation of these genes' biological functions within the GO categories pointed out that almost half (10 of 23) of them are directly involved in the control of neuronal development and CNS functioning (Table 2).
Table 2.
List of 38 ME-markers that are significantly associated with both the linear change in perceived parental rejection (PARQ-slope) and psychosocial adjustment (BASC-CV, personal adjustment composite).
| ME-marker, start position | Regression coefficients | Genome Annotation | ||
|---|---|---|---|---|
|
| ||||
| β | p-value | Gene/Locus | GO: Biological Process | |
|
| ||||
| chr1:33998550 | -.461 | .039 | CSMD2 | integral to membrane |
| chr1:113202150 | -.513 | .080 | CAPZA1 | cell motion, cytoskeleton organization; actin filament-based process |
| chr1:203954250 | -.473 | .039 | TF binding site | |
| chr2:4183550 | -.502 | .030 | ||
| chr2:11147750 | -.555 | .030 | ||
| chr2:54325850 | -.544 | .030 | ACYP2 | phosphate-containing compound metabolic process |
| chr2:125652050 | -.518 | .056 | CNTNAP5 | cell adhesion |
| chr3:183525750 | -.676 | .030 | YEATS2-AS1 | |
| chr3:196318950 | -.634 | .035 | ||
| chr4:1198750 | -.480 | .031 | SPON2 | cell morphogenesis; cell motion; immune response; cell adhesion; axonogenesis |
| chr4:76533250 | -.469 | .044 | CDKL2 | protein phosphorylation; signal transduction; reproductive developmental process |
| chr6:170496250 | -.665 | .030 | ||
| chr7:27141650 | -.523 | .030 | HOXA2 | regulation of transcription; cell morphogenesis; cell motion; axonogenesis; neuron differentiation |
| chr7:100843550 | -.684 | .000 | MOGAT3 | glycerol, alditol and polyol metabolic process; lipid biosynthetic process |
| chr7:101454050 | -.638 | .030 | ||
| chr7:130522550 | -.291 | .208 | ||
| chr8:108555250 | -.492 | .030 | ||
| chr8:123888850 | -.621 | .029 | ZHX2 | regulation of transcription, mRNA catabolic process; regulation of neuron differentiation |
| chr8:142389850 | -.534 | .030 | lncRNA | |
| chr9:99251950 | -.659 | .030 | HABP4 | regulation of transcription; platelet degranulation and activation; blood coagulation |
| chr9:140928850 | -.557 | .091 | CACNA1B | calcium ion transport; synaptic transmission; neurotransmitter secretion; locomotory behavior |
| chr10:88476150 | -.492 | .039 | LDB3 | muscle alpha-actinin binding; zinc ion binding |
| chr11:1673750 | -.477 | .030 | MOB2 | stimulates the autophosphorylation and kinase activity of STK38 and STK38L |
| chr11:69269150 | -.479 | .033 | ||
| chr13:49796550 | -.580 | .030 | ||
| chr13:10823895 | -.496 | .030 | FAM155A | |
| chr14:10404775 | -.506 | .030 | APOPT1; | intrinsic apoptotic signaling pathway |
| KLC1 | microtubule motor activity and tubulin binding | |||
| chr16:88448350 | -.480 | .030 | ||
| chr17:64550 | -.492 | .031 | RPH3AL | exocytosis; response to drug; glucose homeostasis; regulation of G-protein signaling pathway |
| chr17:4755050 | -.660 | .019 | MINK1 | protein phosphorylation; intracellular signaling cascade; synaptic transmission; cell-cell adhesion |
| chr17:43309250 | -.601 | .030 | FMNL1 | substrate-dependent cell migration; regulation of cell shape; cortical actin cytoskeleton organization |
| chr17:75864350 | -.504 | .030 | ||
| chr18:61489650 | -.568 | .030 | ||
| chr19:55813850 | -.483 | .039 | BRSK1 | intracellular signaling cascade, neurotransmitter secretion; neuron morphogenesis; axonogenesis |
| chr20:47087850 | -.505 | .030 | TF binding site | |
| chr21:44715450 | -.487 | .064 | TF binding site | |
| chr22:36874350 | -.598 | .030 | TXN2 | electron transport chain; response to internal and external stimulus; response to axon injury |
| chr22:50422050 | -.546 | .030 | TF binding site | |
Notes. ME-markers are presented in the order of their genomic localization. The EWAS showed that all 38 ME-markers were significantly associated with the PARQ-slope and the BASC-CV personal adjustment composite. A series of regression analyses was performed to test the modulating role of these markers in the relationship between the PARQ-slope and the BASC-CV personal adjustment composite. Regression coefficients and p-values (adjusted for multiple comparisons using FDR method) for the 38 ME-markers were estimated in 38 separate regressions of the BASC-CV personal adjustment composite on one ME-marker and the PARQ slope, controlling for age, gender and the number of SRAD and psychiatric problems. p-values > .05 are italicized. Genes directly related to CNS development and functioning, as defined by GO categories, are marked in bold.
Figure 3.
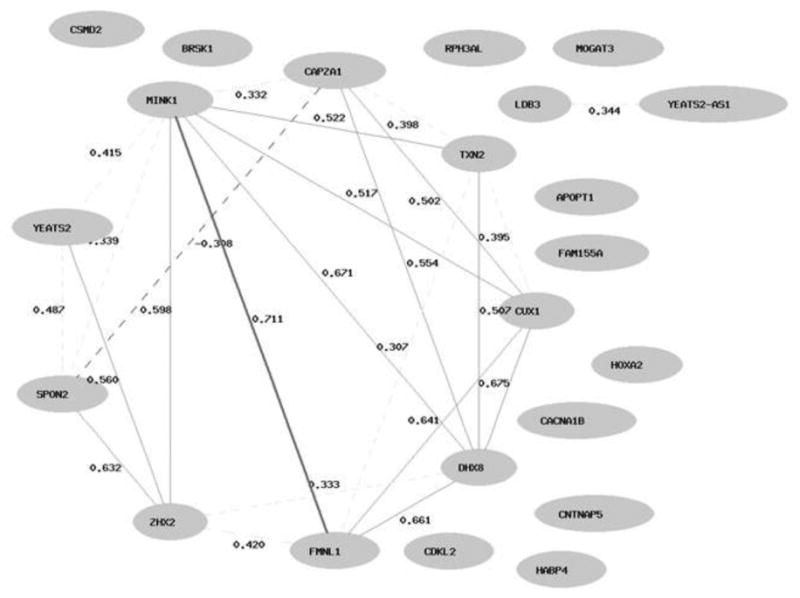
The tissue (blood) specific co-expression network for the 23 genes associated with both the PARQ-slope and the BASC-CV Personal Adjustment, constructed based on Genotype-Tissue Expression (GTEx) project database (GTExConsortium, 2013) via a WebQTL resource (http://www.genenetwork.org). Interactions for absolute correlations above .50 are shown; Pearson pairwise correlations are represented. The nodes contain the symbols of genes. Notably, 12 of these 23 genes are known to be expressed in blood cells, and 9 of 12 active genes showed a high co-variability in their expression levels among individuals.
To investigate the modulating role of the 38 ME-markers in the relation between the change in parental rejection and psychosocial adjustment, a series of 38 separate regressions of the BASC-CV composite on each of the ME-markers and the PARQ slope were conducted, controlling for age, gender and the number of SRAD and psychiatric problems. The results showed that 33 out of the 38 ME-markers related to 20 genes were significantly associated with the BASC-CV personal adjustment over and above the effect of the covariates (Table 2).
Discussion
Although the epigenetic literature has provided evidence for the presence of significant associations between negative early experiences and long-lasting alterations in epigenetic patterns (McGowan et al., 2009; Suderman et al., 2014; Naumova et al., 2012; Weaver et al., 2004; Borghol et al., 2012), there are few studies on the connection between the epigenome and family environment in the context of mother-child relationships, as well as studies on the connection between the epigenome and the change (or stability) of patterns in these relationships. Moreover, there is a gap in knowledge on the role of alterations and modifications in the epigenome driven by early experiences in relation to long-term behavioral outcomes. In our study we attempted to provide a comprehensive picture of the potential modulating role of the epigenome, namely whole genome DNA methylation patterns, in established relations between perceived characteristics of parenting (in particular, stability and variation in parenting across time) and adult psychosocial outcomes.
Genome-wide DNA methylation patterns were assessed when participants were adults. The availability of data on perception of parenting and perceived maternal stress at three distinct developmental periods (middle childhood, adolescence, and young adulthood) allowed us to evaluate the connections between parenting, the epigenome, and adult behavior outcomes in a longitudinal study. Specifically, we evaluated the associations between epigenetic patterns and parenting indicators at different time points, averaged across all developmental stages, and with changes in the perception of parenting over time, from middle childhood to young adulthood.
The results of the EWAS showed that the trace left on the epigenome by subjective experiences (as reflected in perception) of parenting at each of three studied time points (from middle childhood to young adult) is better captured not through stable (i.e., absolute value) but dynamic (i.e., fluctuation between timepoints) characteristics of parenting. This finding may indicate that change in negative perception of parenting over time may exert an accumulating effect on the epigenome. We hope that other larger-scale studies will provide additional evidence supporting this observation.
Specifically, significant associations were obtained between the PARQ-slope and 818 ME-markers located mostly in genes controlling GTPase activity, ATP-binding, and metal ion binding, which are involved in a broad spectrum of biological processes and pathways. GTPases control differentiation during cell division, the synthesis of proteins and their translocation through membranes, and the activation of transmembrane receptors. ATP-binding plays an important role in many metabolic and cellular processes, including mitochondrial energy metabolism, DNA synthesis, transcription activation, and cell signaling. In complicity, ion- and ATP-binding play a main role in the activation of signal receptors and, as a consequence, in signal transduction. Of particular interest is that all of these functional groups are related to intra- and inter-cellular signal transduction. Similar findings (i.e., alterations in the methylation of genes involved in the control of key cell signaling pathways) have been reported in association with adverse experiences early in life, such as child abuse (McGowan et al., 2009; Suderman et al., 2014), maternal deprivation (Meaney & Szyf, 2005; Naumova et al., 2012; Weaver et al., 2004), disadvantaged socio-emotional position in childhood (Borghol et al., 2012), and others.
Besides the possible common effects on intra and inter-cellular processes across different tissues, the enriched GO categories might be described in terms of cell-specific effects of the epigenetic alterations. Thus, the enrichment in GO categories related to the metabolism of purines (GTP and ATP) may indicate a disturbance in immune system functioning. There is evidence (Scheele, Marks, & and Boss, 2007) that small GTPases (GO:0005083; Table 1) play an important role in maintaining normal immune system regulation of the functions of T-cells, along with B-lymphocytes and dendritic cells. Disturbances in purine nucleotide metabolism, such as a decrease in the activity of enzymes involved in these metabolic pathways and increase in the A/G ratio in the lymphocytes may cause important malfunctions of the immune system to the point of developing serious disorders such as chronic lymphocytic leukemia (Carlucci et al., 1997). Thus, although preliminary, this finding contributes to the growing literature substantiating the connection between negative early environment and lifespan health outcomes. Specifically, it suggests that the modulating role of the epigenome might be carried out, at least in part, through the alterations of the functioning of the immune system, which, in turn, trigger a cascade of negative organismic events resulting in poor developmental outcomes.
Due to a broad spectrum of biological processes controlled by genes that contain parenting-related methylation events, we can assume a direct or indirect involvement of these ME-markers in shaping offspring's phenotype in the broadest sense, including behavioral phenotype. To investigate epigenetic factors as potential modulators of behavioral phenotypes examined through the prism of characteristics of adaptive and maladaptive behavior and psychological adjustment using the BASC-CV, we applied a set of multiple regression models to delineate associations between 818 parenting-associated ME-markers and the BASC-CV composite and individual subscales. Out of 818 parenting-related ME-markers, 38 showed significant negative association with the BASQ-CV personal adjustment, and 33 of them passed conservative tests of a modulating role of their methylation states in the association between the temporal dynamics in perceived parenting from middle childhood to young adulthood and long-term behavioral outcomes, namely personal adjustment in adulthood. Most of these 33 ME-markers are related to 20 genes with known biological functions, including the genes directly involved in the control of brain development and functioning, and genes associated with various brain disorders, such as neuronitis (HOXA2, MINK1, SPON2, and ZHX2), neuropathy and schizophrenia (CSMD2; LDB3), and others.
It is important to note that we observed a number of suggestive (statistically borderline significant) associations between almost half of these 818 markers and such behavior outcomes as anxiety, hyperactivity, depression, and attention problems. These findings, first, are consistent with the literature, which provides evidence associating psychiatric and neurodevelopmental disorders, such as depression and autism spectrum disorders, to alterations in DNA methylation (Dempster et al., 2014; Siniscalco, Cirillo, Bradstreet, & Antonucci, 2013). And, second, these observations indicate that the link between parenting-associated ME-markers and behavior outcomes might be even more extensive, and that more associations might be detected in future studies.
Limitations and Future Directions
Several limitations in this study outline the need for further work. First, with regard to methodology, there is an insufficiency of current analytical techniques for conducting EWAS. The small sample size decreased the power to detect small effects and precluded the use of robust high breakdown point estimators or non-parametric approaches more suitable for accounting for the presence of participants with extreme values (i.e., perceptions of change in parenting). Although the method used for genome-wide profiling of the methylome (MBD-seq) has been proven to be highly informative and cost-efficient, it captures only the genome regions with high densities of methylated CpGs. This introduces some constraints on detecting methylation levels of low-density regions or single methylation events that might be related to a phenotype. Moreover, although some way to correct for multiple comparisons was needed, perhaps the strategy used was overly conservative. It is possible that any positive findings that did not survive correction for multiple testing may resurface when different methods are applied (e.g., Bayesian adjustment through the assignment of prior probabilities to models).
Second, due to the low starting material, namely the amount of T-cells derived from a blood sample, we were not able to obtain RNA. A lack of data on the transcriptional activity of genes whose methylation statuses were found to be associated with negative parenting does not allow us to conclude, only to assume, that the found methylation marks may cause changes in gene activity. Therefore, the functional relation between DNA methylation and the expression levels of those genes needs to be examined in further research involving a parallel analysis of methylation and transcription profiles in T-cells.
Third, due to a lack of longitudinal epigenetic data we could not address important questions concerning the dynamics of epigenetic alterations associated with family environment— how to detect the most critical and sensitive developmental stages when changes occur, and how to explore their stability during development. Given well-established statements on the potentially reversible nature of DNA methylation changes, it is possible that by examining adult genomes we detected only especially stable changes that remain over the long-term. At the same time we might have missed alterations that are short-lived but crucial at earlier stages of development. Also, longitudinal data on medical records and data on various facets of development will be needed if we are to determine the effects of DNA methylation alterations on developmental cascades and health outcomes during child development. All these questions hopefully should be addressed by future EWASs focused on longitudinal investigations of the epigenome at different stages of development in the context of early environment, taking into account potential confounding variables, such as data on prenatal, early postnatal and other environmental factors.
Conclusions
Despite some limitations, our study provides evidence supporting an association between early social environment and the epigenome. Namely, the results of the study provide initial evidence of the differential associations between the offspring's epigenome and different facets of parenting. We found stronger associations with indicators of perceived parenting than with maternal reports of parenting stress. This suggests that changes in perceived parental rejection have a stronger association with the offspring's epigenome than measures of maternal parenting difficulties. However, since significant correlations were observed between both constructs that were self-reported by the participating offspring (i.e., personal adjustment and perceived maternal parenting), this finding highlights the possibility of shared method variance that should be explored in future studies by including other-reported indicators of parenting and/or adult psychosocial outcomes in a multi-trait, multi-method framework. The results of this study also imply that individual differences in the dynamics of offspring perceptions of parenting are crucial for understanding the factors of family environment that leave their mark on the epigenome.
In conclusion, this study presents a new longitudinal and multi-phenotypic approach to investigating associations between family environment (e.g., parenting), epigenome (e.g., methylation), and long-term outcomes (e.g., psychosocial maladjustment). Together, the results contribute to a growing body of literature in the field of behavioral epigenetics on the association of parenting, methylome, and behavior, wherein the methylome may play a role as a connecting link or mediating mechanism between negative parenting and diverse facets of long-term psychosocial and behavioral adaptation.
Supplementary Material
Acknowledgments
This research was supported by NIDA DA010726, R01-DA11498 and R01-DA14385 (PI: S. S. Luthar). Grantees undertaking such projects are encouraged to express freely their professional judgment. This article, therefore, does not necessarily reflect the position or policies of the NIH, and no official endorsement should be inferred. We thank the families for their participation and willingness to provide blood samples. We also thank Mei Tan for her editorial efforts and valuable input to the discussion of this manuscript.
References
- Abidin RR. Parenting Stress Index (PSI) manual. 3rd. Charlottesville, VA: Pediatric Psychology Press; 1995. [Google Scholar]
- Barbot B, Crossman E, Hunter SR, Grigorenko EL, Luthar SS. Reciprocal Influences Between Maternal Parenting and Child Adjustment in a High-risk Population: A Five-Year Cross-Lagged Analysis of Bidirectional Effects. American Journal of Orthopsychiatry. 2014;85(5):567–580. doi: 10.1037/ort0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;B57:289–300. [Google Scholar]
- Bloomfield FH. Epigenetic modifications may play a role in the developmental consequences of early life events. 2011;3:348–355. doi: 10.1007/s11689-011-9096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, et al. Associations with early-life socio-economic position in adult DNA methylation. The International Journal of Epidemiology. 2012;41:62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlucci F, Rosi F, Di Pietro C, Marinello E, Pizzichini M, Tabucchi A. Purine nucleotide metabolism: specific aspects in chronic lymphocytic leukemia lymphocytes. Biochimica et Biophysica Acta. 1997;1360:203–210. doi: 10.1016/s0925-4439(96)00077-4. [DOI] [PubMed] [Google Scholar]
- Cox MJ, Mills-Koonce R, Propper C, Gariepy JL. Systems theory and cascades in developmental psychopathology. Development and Psychopathology. 2010;22:497–506. doi: 10.1017/S0954579410000234. [DOI] [PubMed] [Google Scholar]
- Dawson G, Ashman SB, Carver LJ. The role of early experience in shaping behavioral and brain development and its implications for social policy. Development and Psychopathology. 2000;12:695–712. doi: 10.1017/s0954579400004089. [DOI] [PubMed] [Google Scholar]
- Dempster EL, Wong CC, Lester KJ, Burrage J, Gregory AM, Mill J, et al. Genome-wide Methylomic Analysis of Monozygotic Twins Discordant for Adolescent Depression. Biololical Psychiatry. 2014;76(12):977–983. doi: 10.1016/j.biopsych.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleque A, Rohner RP. Reliability of measures assessing the pancultural association between perceived parental acceptance-rejection and psychological adjustment: A meta-analysis of cross-cultural and intracultural studies. Journal of Cross-Cultural Psychology. 2002;33:87–99. [Google Scholar]
- Lester BM, Tronick E, Nestler E, Abel T, Kosofsky B, Kuzawa CW, et al. Behavioral epigenetics. Annals of of the New York Academy of Sciences. 2011;1226:14–33. doi: 10.1111/j.1749-6632.2011.06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthar SS, Sexton CC. Maternal drug abuse versus maternal depression: Vulnerability and resilience among school-age and adolescent offspring. Development and Psychopathology. 2007;19(1):205–225. doi: 10.1017/S0954579407070113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature Neuroscience. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome. Dialogues in Clinical Neuroscience. 2005;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén LK, Muthén BO. User's Guide. Seventh Edition. Los Angeles, CA: Muthén & Muthén; 2012. Mplus: Statistical Analysis With Latent Variables. [Google Scholar]
- Naumova OY, Lee M, Koposov R, Szyf M, Dozier M, Grigorenko EL. Differential patterns of whole-genome DNA methylation in institutionalized children and children raised by their biological parents. Development and Psychopathology. 2012;24:143–155. doi: 10.1017/S0954579411000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowinski LA, Furlong MJ, Rahban R, Smith SR. Initial reliability and validity of the BASC-2, SRP, College version. Journal of Psychoeducational Assessment. 2007;26(2):156–167. [Google Scholar]
- Pardini DA. Novel insights into longstanding theories of bidirectional parent-child influences: introduction to the special section. Journal of Abnormal Child Psychology. 2008;36:627–631. doi: 10.1007/s10802-008-9231-y. [DOI] [PubMed] [Google Scholar]
- Patterson GR, Fisher PA. Recent developments in our understanding of parenting: Bidirectional effects, causal models, and the search for parsimony. In: Bornstein MH, editor. Handbook of parenting. Vol. 5. Mahwah, NJ: Lawrence Erlbaum; 2002. pp. 59–88. Practical issues in parenting. [Google Scholar]
- Pettit GS, Arsiwalla DD. Commentary on special section on “bidirectional parent-child relationships”: the continuing evolution of dynamic, transactional models of parenting and youth behavior problems. Journal of Abnormal Child Psychology. 2008;36:711–718. doi: 10.1007/s10802-008-9242-8. [DOI] [PubMed] [Google Scholar]
- Preacher KJ, Wichman AL, MacCallum RC, Briggs NE. Latent growth curve modeling. Thousand Oaks, CA: Sage Publications; 2008. [Google Scholar]
- Robins LN, Cottler L, Bucholtz K, Compton W. Diagnostic Interview Schedule for DSM-IV. St. Louis, MO: Washington University; 1995. [Google Scholar]
- Romano E, Kohen D, Findlay LC. Associations among child care, family, and behavior outcomes in a nation-wide sample of preschool-aged children. International Journal of Behavioral Development. 2010;34:427–440. [Google Scholar]
- Rutter M. Achievements and challenges in the biology of environmental effects. Proceedings of the National Academy of Sciences. 2012;109:17149–17153. doi: 10.1073/pnas.1121258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele JS, Marks RE, Boss GR. Signaling by small GTPases in the immune system. Immunological Reviews. 2007;218:92–101. doi: 10.1111/j.1600-065X.2007.00530.x. [DOI] [PubMed] [Google Scholar]
- Scott S. Parenting quality and children's mental health: biological mechanisms and psychological interventions. Current Opinion in Psychiatry. 2012;25:301–306. doi: 10.1097/YCO.0b013e328354a1c5. [DOI] [PubMed] [Google Scholar]
- Silk JS, Vanderbilt-Adriance E, Shaw DS, Forbes EE, Whalen DJ, Ryan ND, et al. Resilience among children and adolescents at risk for depression: Mediation and moderation across social and neurobiological contexts. Development and Psychopathology. 2007;19:841–865. doi: 10.1017/S0954579407000417. [DOI] [PubMed] [Google Scholar]
- Siniscalco D, Cirillo A, Bradstreet JJ, Antonucci N. Epigenetic findings in autism: new perspectives for therapy. International Journal of Environmental Research and Public Health. 2013;10(9):4261–4273. doi: 10.3390/ijerph10094261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman MNB, Pappas JJ, Pinto Pereira SM, Pembrey M, Hertzman C, et al. Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Medical Genomics. 2014;7(13) doi: 10.1186/1755-8794-1187-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. DNA methylation, behavior and early life adversity. Journal of Genetics and Genomics. 2013;40:331–338. doi: 10.1016/j.jgg.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Szyf M, McGowan P, Meaney MJ. The social environment and the epigenome. Environmental and Molecular Mutagenesis. 2008;49:46–60. doi: 10.1002/em.20357. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


