Abstract
Feeder cells are generally required to maintain embryonic stem cells (ESCs)/induced pluripotent stem cells (iPSCs). Mouse embryonic fibroblasts (MEFs) isolated from fetuses and STO mouse stromal cell line are the most widely used feeder cells. The aim of this study was to determine which cells are suitable for establishing iPSCs from human deciduous tooth dental pulp cells (HDDPCs). Primary cultures of HDDPCs were cotransfected with three plasmids containing human OCT3/4, SOX2/KLF4, or LMYC/LIN28 and pmaxGFP by using a novel electroporation method, and then cultured in an ESC qualified medium for 15 days. Emerging colonies were reseeded onto mitomycin C-treated MEFs or STO cells. The colonies were serially passaged for up to 26 passages. During this period, colony morphology was assessed to determine whether cells exhibited ESC-like morphology and alkaline phosphatase activity to evaluate the state of cellular reprogramming. HDDPCs maintained on MEFs were successfully reprogrammed into iPSCs, whereas those maintained on STO cells were not. Once established, the iPSCs were maintained on STO cells without loss of pluripotency. Our results indicate that MEFs are better feeder cells than STO cells for establishing iPSCs. Feeder choice is a key factor enabling efficient generation of iPSCs.
Key words: Deciduous tooth, Dental pulp, Feeder cell, Induced pluripotent stem cells (iPSCs), Mouse embryonic fibroblasts (MEFs), STO cells
INTRODUCTION
Establishment of the human embryonic stem cell (ESC) line was first reported by Thomson et al. (43). Currently, induced pluripotent stem cells (iPSCs) can be obtained from somatic cells by reprogramming these cells using a defined set of reprogramming factors (37,38,55). These ESCs/iPSCs are capable of self-renewal and have the ability to differentiate into various cell types; feeder cells are required to support their growth while maintaining pluripotency (1,30,43,48).
Feeder cells are known to produce growth factors, adhesion molecules, and extracellular matrix components for cell attachment; however, they do not increase the generation frequency of iPSC colonies (4). The most widely used feeders include mouse embryonic fibroblasts (MEFs), an immortalized line established from Santos inbred mouse (SIM) embryonic fibroblasts resistant to 6-thioguanine and ouabain (STO) cells, and several types of human cells. MEFs are primary cells derived from midgestational fetuses, and they can be maintained for a limited number of passages before senescence (11). Since the usefulness of MEFs as feeder cells for maintaining human ESCs was reported by Nichols et al. (22), these cells have been employed to generate iPSCs/ESCs (25,28,37,38,43,55). However, MEFs have some drawbacks. First, it is often difficult to obtain gene-engineered MEFs because their proliferation ability is generally limited in comparison to that of other immortalized cells. Second, isolating MEFs is a laborious and time-consuming process requiring samples from a large number of mice, and the quality of MEFs often differs depending on the cell isolation skills of individuals. STO cells (47) have also proven useful for establishing and maintaining ESCs (3,6,17,18,20,33,34,40). They can continue to proliferate in vitro, but stop their proliferation after treatment with mitotic inhibitors such a mitomycin C (MMC) (21). These properties indicate that STO cells are superior to MEFs for generating gene-engineered ESCs/iPSCs because STO cells do not exhibit senescence, unlike MEFs. Although some laboratories successfully employed STO cells for reprogramming somatic cells from monkeys (24) and humans (23), the overall performance of these cells as feeders for ESC/iPSC culture appears inferior to that of MEFs (2,3,49,51). To overcome the problem of xenocontamination, feeder cells of human origin have also been used to support human ESC growth (1,8,12,15,31,36,52). However, the ability to support undifferentiated growth of human ESCs varies among these feeders (5,32).
In the dental field, little is known regarding which type of feeder cells are suitable for establishing dental cell-derived iPSCs. Tamaoki et al. (42) first reported the successful generation of iPSCs from dental pulp cells (DPCs) isolated from the human wisdom tooth. They employed traditional approaches using retroviral vectors carrying Yamanaka factors and MEFs as feeder cells. Their findings show that DPCs are easy to handle and are highly proliferative in vitro (39). Interestingly, Yan et al. demonstrated that iPSCs could be successfully obtained from three different dental stem/progenitor cells: stem cells from exfoliated deciduous teeth (SHED), stem cells from apical papilla (SCAP), and stem cells from DPCs (53). More importantly, these groups suggested that dental stem cells are more amenable to reprogramming than are dermal fibroblasts (42,53). In preliminary studies, we found that DPCs isolated from deciduous teeth of young children can be easily propagated in vitro. In this study, we found that these cells can be reprogrammed and express several stem cell-specific markers (paper under preparation), similarly to wisdom tooth-derived DPCs (42,53).
Here we report the successful establishment of iPSCs from human deciduous tooth dental pulp cells (HDDPCs) transfected with plasmids carrying cDNAs for Yamanaka factors. We focused on the ability of feeders (MEFs and STO cells) to support reprogramming of HDDPCs and subsequent progression toward HDDPC-derived iPSCs (HDDPC-iPSCs).
MATERIALS AND METHODS
Cell Culture
HDDPCs were collected from patients after obtaining informed consent; the protocols used in this study were approved by the ethical committee for the use and experimentation of the Kagoshima University Graduate School of Medical and Dental Sciences. HDDPCs were isolated as described previously (9), with slight modifications. Pulp tissue from deciduous teeth were removed from the teeth of four young patients (age: 8–10 years; two boys and two girls) and digested in a solution of 3 mg/ml collagenase type I (No. 17100-017; Invitrogen, Carlsbad, CA, USA) and 4 mg/ml dispase (No. 410810077; Roche Applied Science, Basel, Switzerland) for 30–60 min at 37°C. Next, 4 ml of Dulbecco’s modified Eagle’s medium (DMEM; No. 11995-081; Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS; No. SFMB30-2239; Equitech Bio Inc., Kerrville, TX, USA), 50 U/ml penicillin, and 50 mg/ml streptomycin (No. 15140-122; Invitrogen) (DMEM/10% FBS) was then added to stop the digestion reaction. The resulting single cells were seeded onto 60-mm gelatin-coated dishes (Iwaki Glass Co. Ltd., Tokyo, Japan) containing α-modified minimum essential medium (MEMα; No. 135-15175; Wako Pure Chemical Industries, Ltd., Osaka, Japan) with 20% FBS, 100 µM l-ascorbic acid-2-phosphate (No. 323-44822; Wako Pure Chemical Industries Ltd.), 50 U/ml penicillin, and 50 mg/ml streptomycin (MEMα/20% FBS) and were cultured at 37°C in a 5% CO2 atmosphere. After four to six passages, HDDPCs were subjected to iPSC generation. The normal human dermal fibroblast line HDFa (female; No. 106-05a; Cell Applications, Inc., San Diego, CA, USA) was used as a control for reverse transcription (RT)-PCR analysis. The following cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA): STO cells (No. CRL-1503) and PA-1 (No. CRL-1572; female), a teratocarcinoma cell line derived from human ovary (50), which was used as a control for RT-PCR analysis. MEFs isolated from 13.5 days of gestation from outbred CD-1 mice were purchased from Takara Bio Inc. (Shiga, Japan). All cells, except for HDDPCs, were cultured in DMEM/10% FBS at 37°C in a humidified atmosphere of 5% CO2.
Generation of HDDPC-iPSCs
Plasmids (Fig. 1a) carrying Yamanaka factor cDNAs were purchased from Addgene (Cambridge, MA, USA) (26). pCXLE-hOCT3/4-shp53 carries human octamer-binding transcription factor 3/4 (OCT3/4) cDNA and short hairpin RNA (shRNA) for human tumor protein 53 (p53), pCXLE-hUL carries human V-Myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog (L-MYC) and LIN28 cDNAs, and pCXLE-hSK carries human sex-determining region Y box 2 (SOX2) and Krüppel-like factor 4 (KLF4) cDNAs. These cells were propagated in Escherichia coli (DH5α Competent Cells; No. 9057; Takara Bio Inc.), and the DNA was purified using the Qiagen Plasmid Midi Kit (Hilden, Germany).
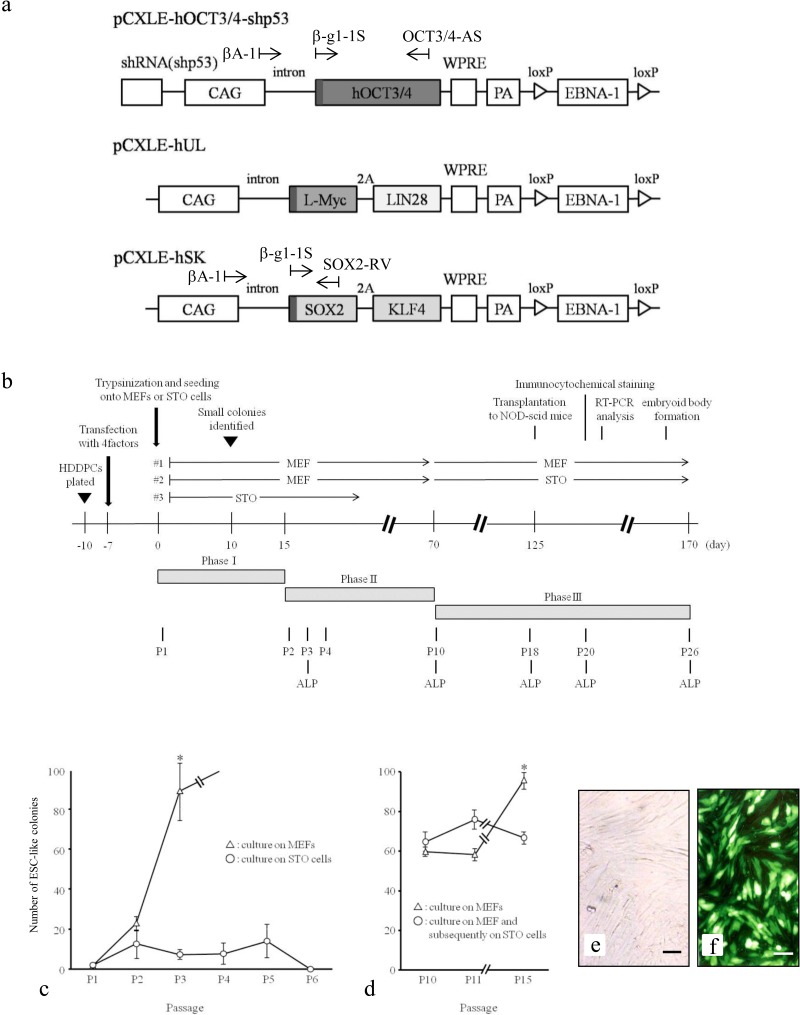
Figure 1.
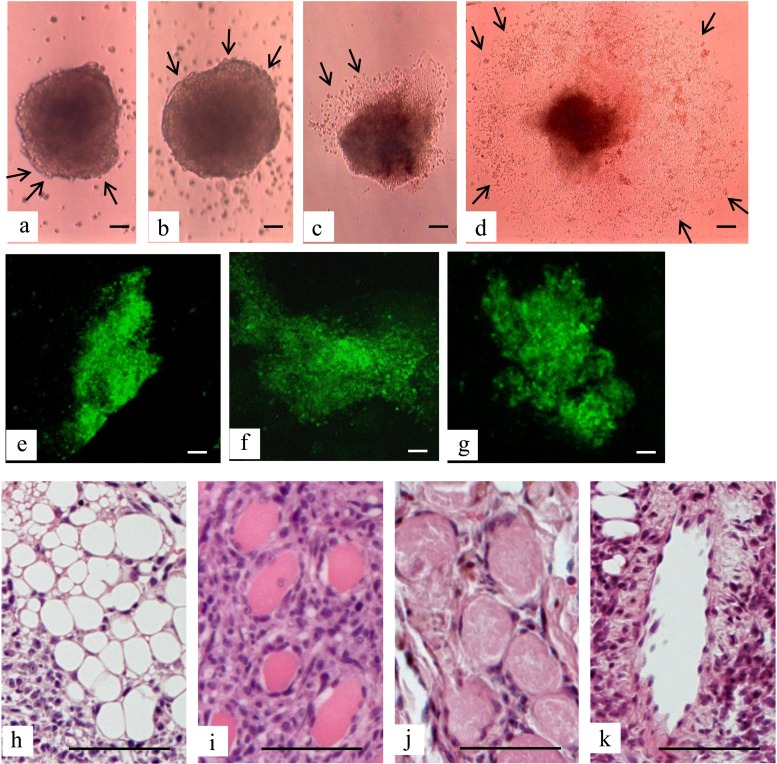
Generation of human deciduous teeth dental pulp cell induced pluripotent stem cells (HDDPC-iPSCs). (a) Plasmid vectors used for reprogramming. The location of each primer is denoted above the construct. shRNA(shp53): short hairpin RNA for tumor protein 53 (p53); CAG, cytomegalovirus enhancer + chicken β-actin promoter; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; PA, poly(A) sites; EBNA-1, Epstein–Barr nuclear antigen 1. (b) Reprogramming protocol of HDDPCs. HDDPCs were plated (day −10) in the absence of a feeder layer and transfected (day −7) with three different plasmid vectors. Seven days after transfection, the emerging colonies were then reseeded onto mouse embryonic fibroblasts (MEFs) or STO cells (an immortalized line established from mouse Santos inbred mouse (SIM) embryonic fibroblasts resistant to 6-thioguanine and ouabain). This first passage of embryonic stem cell (ESC)-like colonies was designated as passage 1 (P1). Staining for alkaline phosphatase (ALP) activity was performed on P3, P10, P18, P20, and P26. NOD-Scid, nonobese diabetic-severe combined immunodeficient. (c, d) Determination of colony formation efficiency. Approximately 17 days after transfection, several foci were observed. The number of growing colonies appeared to be the same regardless of feeder type, but after P1, the number of ESC-like colonies seeded onto MEFs increased at least up to P5; however, the number of cells seeded onto STO cells did not (c). In contrast, the number of growing colonies appeared to be the same between those continuously grown on MEFs and those grown on MEFs and then grown on STO cells at P10 (d). (e, f) HDDPCs before (e) and after (f) transfection. When enhanced green fluorescent protein (EGFP)-derived fluorescence was inspected 1 day after transfection, more than 70% of the cells were successfully transfected, as shown in (f). Bars: 50 µm.
The study was conducted in accordance with the guidelines of the Ethics Committee of the Kagoshima University Graduate School of Medical and Dental Sciences to derive and culture the iPSC lines. For transfection, HDDPCs (5 × 104) were electroporated using a Neon® microporation system (Invitrogen) in 100 µl of R-buffer (Invitrogen) containing 1 µg of pCXLE-hOCT3/4-shp53, 1 µg of pCXLE-hUL, 1 µg of pCXLE-hSK, and 0.5 µg of pmaxGFP [a green fluorescent protein (GFP) indicator plasmid for monitoring transfection efficiency; Lonza GmbH, Cologne, Germany] under electric condition No. 4 (one electrical pulse at 1,600 V and 20 ms pulse). The electroporated cells were then seeded onto three wells of a gelatin-coated 24-well plate (Iwaki Glass Co. Ltd.) without feeder containing DMEM/20% FBS. One day after transfection, cells were inspected for green fluorescence under UV illumination to confirm that cells had been successfully transfected. The cells were further cultivated in the same medium. Medium changes were performed every day or every 2 days. Seven days after transfection, cells in the 24-well plate were trypsinized and subsequently reseeded onto MMC-treated (No. M4287, Sigma-Aldrich, St. Louis, MO, USA) MEFs or STO cells in a 60-mm gelatin-coated dish with human ESC culture medium iPSellon (No. 007001; Cardio, Kobe, Japan) supplemented with 5 ng/ml recombinant human basic fibroblast growth factor (bFGF; Wako Pure Chemical Industries, Ltd.), as the first passage (P1) (Fig. 1b). Fifteen days after seeding onto feeder cells, the dish containing emerging small ESC-like colonies was washed once with phosphate-buffered saline (PBS) without Ca2+ and Mg2+, incubated with PBS containing 10 mg/ml collagenase IV (No. 17104-019; Invitrogen), 1 M CaCl2/PBS, 20% Knockout Serum Replacement (KSR; No. 10828-028; Invitrogen), and 0.25% trypsin (No. 15090-046; Invitrogen) at 37°C for approximately 5 min, and then reseeded onto new feeder cells in a 60-mm gelatin-coated dish, which was designated as P2. Six to 8 days after reseeding, growing colonies were again dissociated using the method described above, split to 1:5 and reseeded onto new feeder cells in a 60-mm gelatin-coated dish, which was designated as P3. Similar passages were performed until P26 (Fig. 1b). The medium was changed every day. Seventy-seven days after transfection (corresponding to P10) (Fig. 1b), some ESC-like colonies grown on MEFs were transferred onto MMC-treated STO cells to examine whether STO cells could support growth and maintain pluripotency of HDDPC-iPSCs (#2 of Fig. 1b).
To determine the reprogramming efficiency of HDDPCs, the number of ESC-like colonies from 5 × 104 HDDPCs that had been transfected with reprogramming factors was determined 22 days (P2) after transfection. The number of ES-like colonies was also determined for subsequent passages (P3 to P6 and P10 to P15). The data were plotted as graphs as the average of three examinations (as shown in Fig. 1c, d). The determination of ES-like colony formation efficiency was subjected to statistical analysis.
Alkaline Phosphatase (ALP) and Immunocytochemical Staining
To detect ALP activity, the Leukocyte Alkaline Phosphatase Kit (No. ALP-TK1; Sigma-Aldrich) was used. HDDPC-iPSCs were plated onto a well of the Lab-Tek® Chamber Slide™ System (No. 177399; Nalge Nunc International, Penfield, NY, USA) into which MEFs had been seeded. During staining, the cells were fixed with 4% paraformaldehyde (PFA; Sigma-Aldrich) for 10 min at room temperature and subjected to cytochemical staining following the manufacturer’s instructions.
For immunocytochemical staining using ES markers, cells fixed with 4% PFA were permeabilized with 0.05% Triton X-100 (Sigma-Aldrich), if necessary, and were blocked with 10% normal goat serum (NGS; Invitrogen). Cells were stained with the primary antibodies OCT3/4 (1:400; clone 10H11.2, No. MAB4401; Merck Millipore, Billerica, MA, USA), stage-specific embryonic antigen-1 (SSEA-1) (1:500; No. Ab16285; Abcam Inc., Cambridge, MA, USA), SSEA-3 (1:500; No. MAB4303; Merck Millipore), TRA-1-60 (1:500; MAB4360; Merck Millipore), forkhead box protein A2 (FOXA2; 1:250; No. ab5074; Abcam Inc.), βIII tubulin (antibody clone TUJ1; 1:250; No. ab14545; Abcam Inc.), or smooth muscle actin (SMA; 1:250; No. ab124438; Abcam Inc.). The secondary antibodies used were Alexa Fluor 568-conjugated goat anti-mouse IgM (1:400; Invitrogen), fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibodies (1:400; Invitrogen), Alexa Fluor 488-conjugated donkey anti-goat IgG H&L (1:250; No. ab150129; Abcam Inc.), FITC-conjugated rat monoclonal SB84a anti-mouse IgG2a γ chain (1:250; No. ab79092; Abcam Inc.), or Alexa Fluor 488-conjugated goat anti-rabbit IgG H&L (1:250; No. ab150077; Abcam Inc.). Nuclear staining was performed using 6-diamidino-2-phenylindole (DAPI; 1.5 µg/ml; No. H-1200; Vector Laboratories Inc, Burlingame, CA, USA).
Fluorescence was examined using an Olympus BX60 fluorescence microscope. Microphotographs were obtained using a digital camera (FUJIX HC-300/OL; Fuji Film, Tokyo, Japan) attached to the fluorescence microscope and printed using a Mitsubishi digital color printer (CP700DSA; Mitsubishi, Tokyo, Japan).
PCR-Based Detection of Transgene and RT-PCR
Genomic DNA and total RNA from each sample were extracted using the DNA/RNA Mini Kit (No. 80204; Qiagen), which allows for simultaneous purification of DNA and RNA.
To identify expression of endogenous and exogenous (transgene) target mRNA by RT-PCR, RT was first performed using the First-Strand cDNA Synthesis Kit (No. 18080-051; Invitrogen). The resulting cDNAs were then PCR amplified from undiluted cDNA samples (1 µl) in a total volume of 20 μl using AmpliTaq Gold® 360 Master Mix (No. 4398881; Applied Biosystems, Foster City, CA, USA). PCR was performed for 38 cycles of 30-s denaturation at 95°C, 30-s annealing at 58°C, and 60-s extension at 72°C in a PC708 thermal cycler (Astec, Fukuoka, Japan). A negative, no-template control (designated as −RT) was included for each reaction. In addition, cDNA from the human dermal fibroblast cell line HDFa was used as a negative control. cDNA from human PA-1 teratocarcinoma cells was used as a positive control. Information regarding PCR primers used is listed in Table 1. The products (5 µl) were then analyzed by electrophoresis in 2% agarose gels (Nacalai Tesque, Inc., Kyoto, Japan) and visualized after staining with ethidium bromide (1 µg/ml; No. E1510; Sigma-Aldrich).
Table 1.
Primer Sets Used for PCR Analysis
| Target mRNAor DNA | Primer Name: Sequence (5′–3′) | Size (bp) | Reference |
|---|---|---|---|
| Endogenous OCT3/4 mRNA | OCT3/4-S (forward): ATT TCA CCA GGC CCC CGG CT OCT3/4-AS (reverse): GCT GAT CTG CTG CAG TGT GGG T |
822 | 16 |
| Endogenous SOX2 mRNA | SOX2-S (forward): AGG ACC AGC TGG GCT ACC CG SOX2-AS (reverse): GGC GCC GGG GAG ATA CAT GC |
320 | 29 |
| Endogenous KLF4 mRNA | KLF4-S (forward): CGT GCT GAA GGC GTC GCT GA KLF4-AS (reverse): GGG TGC ACG AAG AGA CCG CC |
173 | 10 |
| Endogenous NANOG mRNA | N-S (forward): CCT CCA GCA GAT GCA AGA ACT C N-AS (reverse): GTA AAG GCT GGG GTA GGT AGG TG |
172 | 46 |
| Endogenous ALP mRNA | ALP-S (forward): TGG CCC CCA TGC TGA GTG ACA C ALP-AS (reverse): TGG CGC AGG GGC ACA GCA GAC |
160 | 27 |
| Endogenous GAPDH mRNA | GAPDH-S (forward): GGC GAT GCT GGC GCT GAG TA GAPDH-AS (reverse): ACA GTT TCC CGG AGG GGC CA |
325 | 14 |
| Exogenous OCT3/4 mRNA | βA-1 (forward): TCT GAC TGA CCG CGT TAC TCC CAC A OCT3/4-AS (reverse): GCT GAT CTG CTG CAG TGT GGG T |
836 | 13,16 |
| Exogenous SOX2 mRNA | βA-1 (forward): TCT GAC TGA CCG CGT TAC TCC CAC A SOX2-AS (reverse): GGC GCC GGG GAG ATA CAT GC |
945 | 13,16 |
| Transgene OCT3/4 DNA | β-gl-1S (forward): TGT GCT GTC TCA TCA TTT TGG CAA A OCT3/4-AS (reverse): GCT GAT CTG CTG CAG TGT GGG T |
787 | 16,45 |
| Transgene SOX2 DNA | β-gl-1S (forward): TGT GCT GTC TCA TCA TTT TGG CAA A SOX2-AS (reverse): GGC GCC GGG GAG ATA CAT GC |
880 | 29,45 |
Exogenous octamer-binding transcription factor 3/4 (OCT3/4) and sex-determining region Y box 2 (SOX2) mRNA are derived from the transgenes pCXLE-hOCT3/4-shp53 and pCXLE-hSK, respectively. Similarly, exogenous OCT3/4 and SOX2 DNA correspond to the integrated pCXLE-hOCT3/4-shp53 and pCXLE-hSK in human deciduous tooth dental pulp cell-derived induced pluripotent stem cells (HDDPC-iPSCs), respectively. KLF4, Kruppel-like factor 4; ALP, alkaline phosphatase, tissue-nonspecific isozyme; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
To identify transgenes integrated into the host genome, genomic DNA (approximately 0.5 µg) was subjected to PCR. To identify pCXLE-hOCT3/4-shp53, the primer set β-gl-1S and OCT3/4-AS (Fig. 1a and Table 1) was used. Similarly, the primer set β-gl-1S and SOX2-RV (Fig. 1a and Table 1) was used to identify pCXLE-hSK. PCR conditions were the same as those used for RT-PCR.
In Vitro differentiation
To induce embryoid body formation, pieces of HDDPC-iPSC colonies at P25 (#1 of Fig. 1b) were dissected mechanically using a pipette tip under a stereomicroscope and then seeded onto an ultralow attachment 30-mm dish (No. MS-9035X; Sumitomo Bakelite Co., Ltd., Tokyo, Japan) with DMEM/10% FBS. Ten days after cultivation, emerging embryoid bodies were transferred onto gelatin-coated 30-mm dish (Iwaki Glass Co. Ltd.) and cultured for another 10 days in DMEM/10% FBS to allow enhanced differentiation into various cell types.
In Vivo differentiation
The animal experiment protocols were approved by the Animal Care and Use Committee of the Kagoshima University Graduate School of Medical and Dental Sciences. For teratoma formation (n = 3), HDDPC-iPSCs (2 × 106) at P18 (#1 of Fig. 1b) were mixed with Cellmatrix Type I-A (No. 631-00651; Wako Pure Chemical Industries, Ltd.), as described by the manufacturer. This mixture (100 µl/site) was then injected subcutaneously into immune-compromised mice (8-week-old, male) [non-obese diabetic/severe combined immunodeficient (NOD/Shi-scid); Kyudo Co., Ltd., Tosu, Saga, Japan]. Seven to 8 weeks posttransplantation, teratomas were harvested and fixed with 4% PFA at 4°C for 4 days. Tissue sections were embedded in paraffin and stained with hematoxylin and eosin (Sigma-Aldrich).
Karyotype Analysis
Karyotyping of HDDPC-iPSCs was performed by Chromocenter Inc. (Tottori, Japan).
Statistical Analysis
The data obtained from experiment 1 were subjected to statistical analysis using SPSS® 15.0 J for Windows software (SPSS Inc., Chicago, IL, USA). The determination of ESC-like colony formation efficiency was subjected to statistical analysis. Statistical significance was determined by Welch’s t-test. Values of p < 0.05 were considered statistically significant.
RESULTS
Generation of HDDPC-iPSCs
To generate iPSCs, we used one line termed MM among the four HDDPC lines established from each independent patient, since this line exhibited the highest degree of proliferation (data not shown). MM cells were cotransfected with plasmids, each of which included OCT3/4, L-MYC/LIN28, or SOX2/KLF4 cDNAs at a ratio of 1:1:1, together with the reporter construct pmaxGFP using the Neon® microporation system. When cells were inspected for fluorescence 1 day after transfection, transfection efficiency was more than 70%, as shown in Figure 1f. Seven days after transfection, cells were reseeded onto MEFs or STO cells (Fig. 2). Seventeen days posttransfection, small colonies with ESC-like morphology were observed on both types of feeders (Fig. 2a, g). From 5 × 104 MM cells, we obtained approximately three ESC-like colonies [efficiency, ∼0.006% (3/5 × 104); P1 in Fig. 1c]. Further independent experiments yielded similar results (data not shown). Twenty-two days posttransfection, we seeded the P1 cells onto new feeder cells as P2. Five to 7 days after seeding as P2, emerging colonies exhibited a flat shape with an irregular periphery and a rough surface (arrows in Fig. 2b, h). This morphological property was common, regardless of the feeder type (Fig. 2b, h). These growing colonies were next passaged onto new feeder cells, and then passaging was continued until P26. During this period, the undifferentiated state of the colonies was microscopically monitored daily with periodic testing for expression of one of the stem cell markers, ALP.
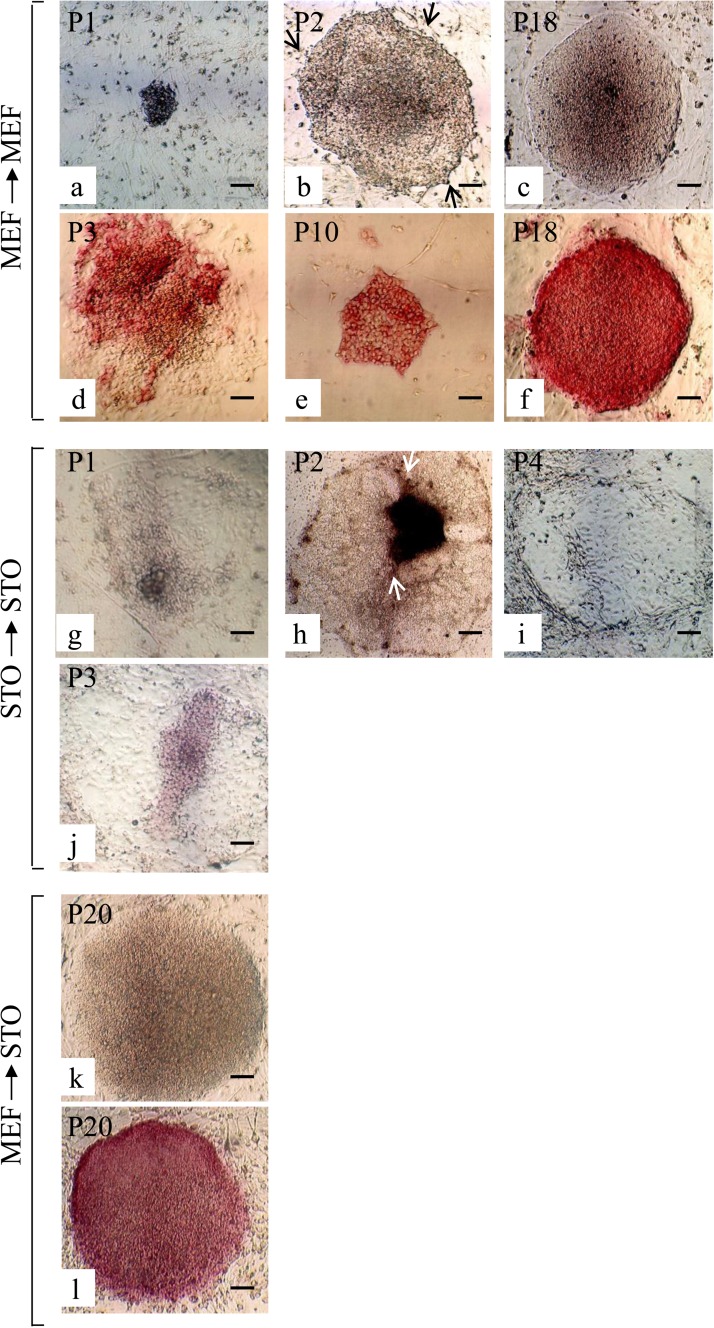
Figure 2.
ALP activity of ESC-like colonies. Seven days after transfection with Yamanaka factors, HDDPCs were reseeded onto MEFs or STO cells. Emerging ESC-like colonies grown on MEFs were then serially passaged onto the same feeder cells (a–f). Similarly, ESC-like colonies grown on STO cells were subjected to serial passage (g–j). In some cases, the ESC-like colonies grown on MEFs were seeded onto STO cells at P10 and then were serially passaged using the same feeder (k, l). (a–c, g–i, k) Photographed under phase contrast microscopy; (d–f, j, l) cytochemical staining for ALP activity. Scale bars: 50 µm.
ESC-Like Colonies Can be Maintained in the Presence of MEFs, but not STO Cells
As described above, ESC-like colonies in the initial stage of reprogramming (P2 to P10) maintained on MEFs exhibited colony morphology with irregular borders, but after P10, they exhibited structural characteristics typical for undifferentiated ESC colonies, such as tightly packed colonies with a smooth outline and cell surface, as shown in Figure 2c. Staining with ALP was correlated with these observed morphologies. ESC-like colonies maintained on MEFs by P3 exhibited relatively low levels of ALP activity and were often mosaic (Fig. 2d), while those maintained at P10 and thereafter were uniform and showed a high degree of ALP activity (Fig. 2e, f). In contrast, ESC-like colonies maintained on STO cells failed to form clear colonies during passaging (Fig. 2g–i) and finally ceased to proliferate by P6 (Fig. 1c). Concomitantly, ALP activity in the colonies remained very low (Fig. 2j). These findings indicate that MEFs, but not STO cells, can support the initial growth of iPSC colonies, which may have been undergoing reprogramming.
To test whether STO cells can support the growth of established iPSCs, HDDPC-iPSCs at P10 that had been grown on MEFs were transferred to the STO cells. As shown in Figure 2k, ESC-like colonies grew well and exhibited smooth edges when inspection was performed on P26. High ALP activity was observed in these colonies (Fig. 2l). These findings suggest that STO cells can support the growth and pluripotency of iPSCs, which have been established as a continuous cultured cell line.
ESC-Like Colonies Express ESC Markers
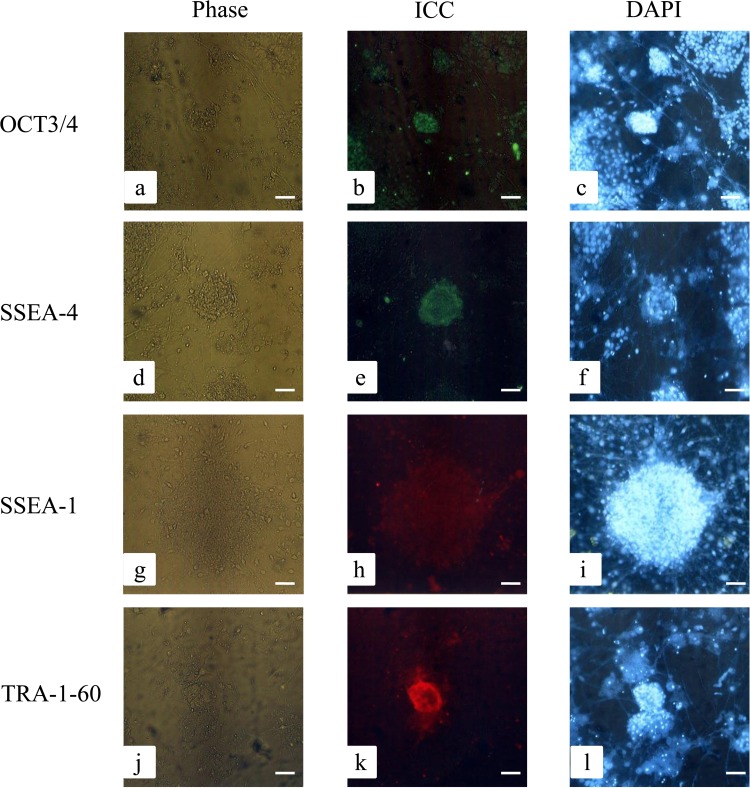
Next, we performed immunocytochemical staining to determine whether the resulting ESC-like colonies maintained continuously on MEFs over P18 could express ESC-specific proteins. As expected, cells were reactive to the antibodies produced against OCT3/4 (Fig. 3a–c), SSEA-4 (Fig. 3d–f), and TRA-1-60 (Fig. 3j–l), but not to those against SSEA-1 (Fig. 3g–i).
Figure 3.
Immunostaining of ESC-like colonies. Immunocytochemical staining (ICC) of ESC-like colonies showed reactivity against octamer-binding transcription factor 3/4 (OCT3/4), stage-specific embryonic antigen 4 (SSEA-4), and TRA-1-60, but not against SSEA-1. Nuclei were stained with 6-diamidino-2-phenylindole (DAPI). Phase, photographs taken using a phase-contrast microscope. Scale bars: 50 μm.
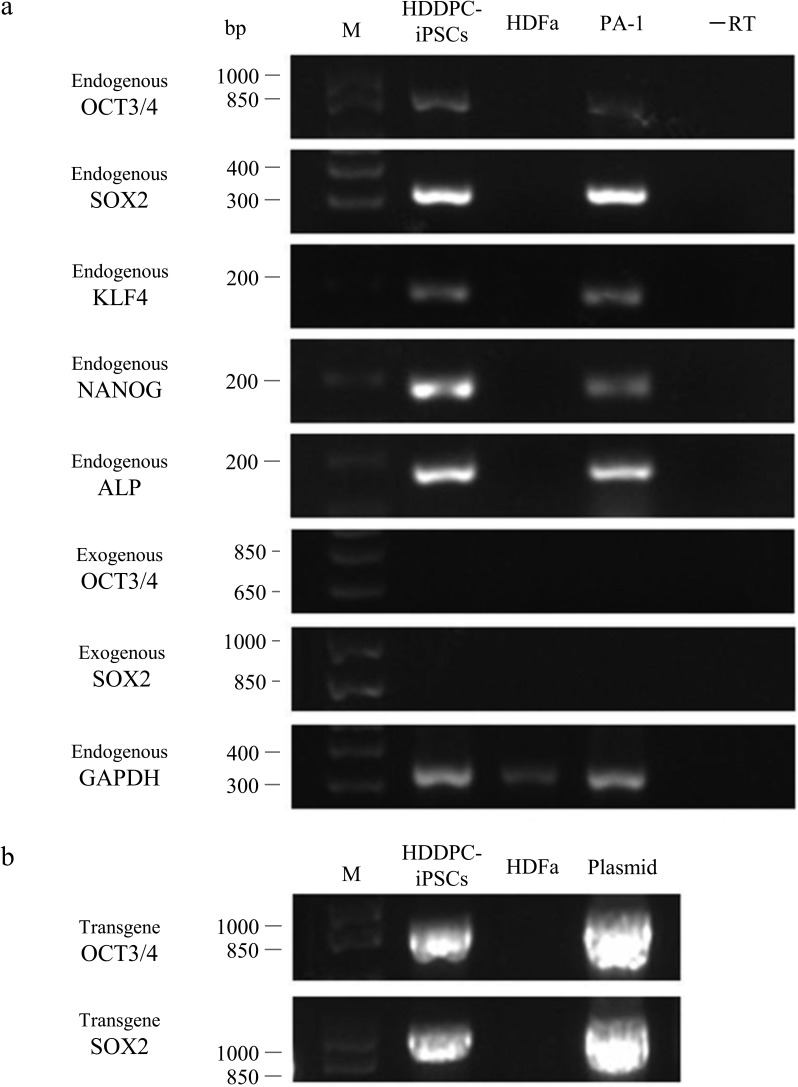
RT-PCR demonstrated that the transcript derived from the integrated pCXLE-hOCT3/4-shp53 or pCXLE-hSK plasmid was undetectable (exogenous OCT3/4 and exogenous SOX2 in Fig. 4a). As expected, ESC-like colonies expressed endogenous stem cell-specific marker mRNAs, including OCT3/4, SOX2, KLF4, NANOG, and ALP (Fig. 4a).
Figure 4.
Gene expression profiling of ESC-like colonies. (a) RT-PCR analysis performed to detect the expression of endogenous genes [OCT3/4, sex-determining region Y box 2 (SOX2), Krüppel-like factor 4 (KLF4), NANOG, and ALP] in HDDPC-iPSCs, HDFa (negative control), and PA-1 (positive control). Expression of OCT3/4 and SOX2 mRNA from the exogenous construct was also assessed using primer sets βA-1/OCT3/4-AS (for OCT3/4) and βA-1/SOX2-RV (for SOX2). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. −RT indicates the negative control (PCR with no RT product). M, 100-bp ladder markers. (b) PCR analysis of genomic DNA to detect the presence of integrated transgenes using the primer sets β-gl-1S/OCT3/4-AS (for pCXLE-hOCT3/4-shp53) and β-gl-1S/SOX2-RV (for pCXLE-hSK). As positive controls, plasmid DNA (5 ng) was concomitantly subjected to PCR and loaded.
When genomic DNA isolated from 10 iPSC colonies was subjected to PCR, all tested colonies contained transgenes (at least pCXLE-hOCT3/4-shp53 and pCXLE-hSK) in their genomes (Fig. 4b).
Embryoid Body Formation and In Vitro Differentiation
The formation of embryoid bodies can be a good indicator of the ability of ESCs/iPSCs to differentiate in vitro. We cultured HDDPC-iPSCs at P25 on a nonadherent dish in DMEM/10% FBS for up to 10 days. The cells formed aggregates with differentiated cells on their surfaces (arrows in Fig. 5a, b). The resulting aggregates were next cultured on an adherent tissue culture dish to examine whether the outgrowth of differentiated cells from the aggregates occurs. As expected, outgrowth occurred approximately 2 days after attachment to the substratum (arrows in Fig. 5c), and it was further accelerated over additional days (arrows in Fig. 5d). The presence of differentiated cells from three germ layers (endoderm, mesoderm, and ectoderm) was confirmed by immunocytochemical staining of differentiated cells 10 days after attachment of embryoid bodies (derived from HDDPC-iPSCs at P25) to the substratum using typical endodermal (FOXA2) (Fig. 5e), ectodermal (antibody clone TUJ1) (Fig. 5f), and mesodermal (SMA) (Fig. 5g) markers.
Figure 5.
Teratoma formation. (a, b) Embryoid bodies formed from the ESC-like cells 2 (a) and 10 (b) days after floating culture. Note the formation of differentiated cells (arrows) at the surface of embryoid bodies. (c, d) In vitro outgrowth of embryoid bodies 2 (c) and 10 (d) days after transfer of floating embryoid bodies to the adhesive substratum. Note that extensive outgrowth of differentiated cells (arrows) from the center of a colony is observed along with extension of cultural period. (e–g) Immunocytochemical staining of cells 10 days after in vitro differentiation of embryoid bodies from HDDPC-iPSCs at P25 by typical endodermal (forkhead box protein A2; FOXA2, e), ectodermal (βIII tubulin; antibody clone TUJ1, f) and mesodermal (smooth muscle actin; SMA, g) markers. (h–k) Hematoxylin–eosin staining of sections of solid tumors generated after transplantation of HDDPC-iPSCs at P18 into immunocompromised mice. Bars: 50 µm.
Teratoma Formation
To test whether the HDDPC-iPSCs maintained on MEFs can form teratomas in vivo, we transplanted cells at P18 (#1 of Fig. 1b) under the skin of immune-compromised mice. Generation of solid tumors at the transplanted region (3/4 grafted) was noted less than 1.5 months after grafting. Histological examination of the resulting tumors revealed that the tumors contained several types of differentiated cells derived from three germ layers, including adipose tissue (mesoderm) (Fig. 5h), skeletal muscle (mesoderm) (Fig. 5i), keratin-containing epidermal tissue (ectoderm) (Fig. 5j), and columnar structure (endoderm) (Fig. 5k).
Cytogenetic Analysis
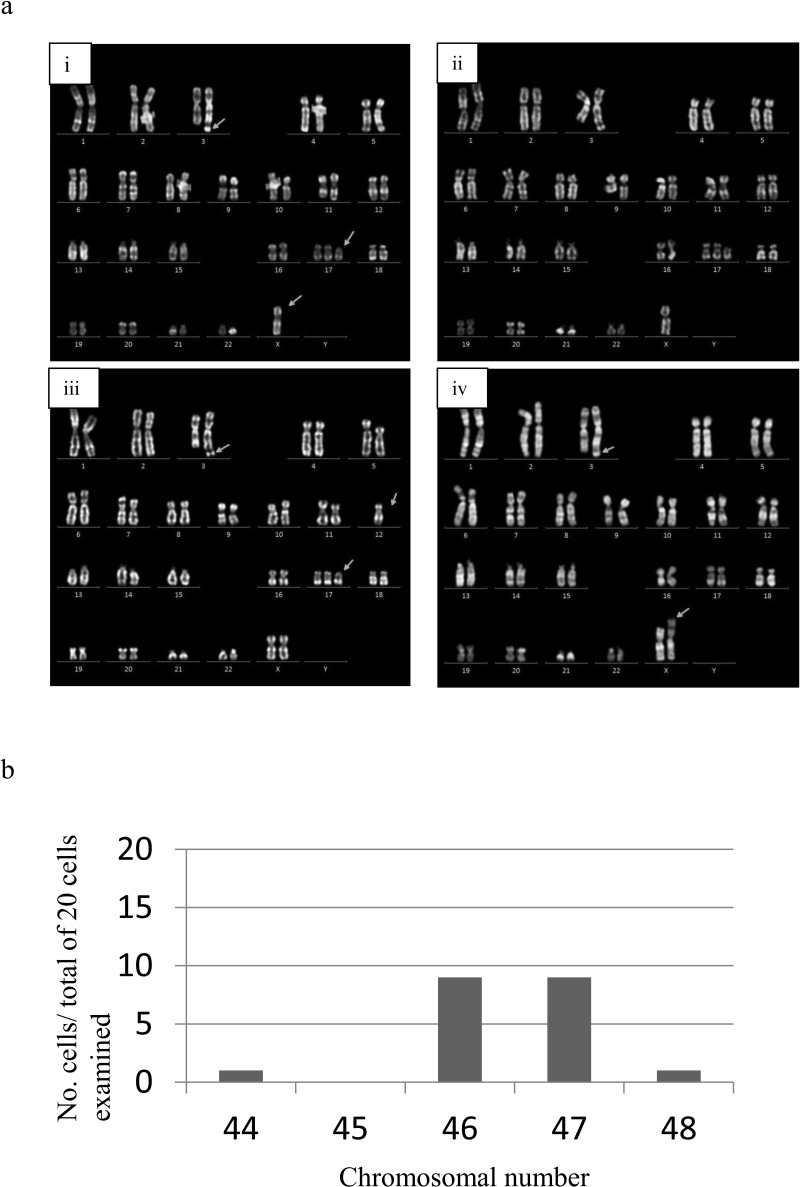
Cytogenetic analysis of HDDPC-iPSCs (P15) revealed abnormal karyotypes (Fig. 6a). Of 20 cells examined, there were typical karyotypes exemplified by 47,XX,add(3)(q29),+del(17)(p11) (50%) (Fig. 6a–i), 46,XX,add(3)(q29),−8,+del(17)(p11) (10%) (Fig. 6a-ii), 46,XX,add(3)(q29),−12,+del(17)(p11) (10%) (Fig. 6a-iii), 46,X,add(3)(q29),+del(17)(p11) (20%) (Fig. 6a-iv), and 46,X,der(X)add(p2?),add(3)(q29) (10%; not shown) by images of G-band staining of HDDPC-iPSCs. The chromosomal number ranged between 46 and 47 (Fig. 6b). We observed the presence of chromosomal number 44 and 48 (Fig. 6b), but in this case the slides might have had excessive spreading of the chromosomes and consequently generation of artifacts.
Figure 6.
Karyotype analysis. (a) Abnormal karyotypes in HDDPC-iPSCs. A total of 20 cells were examined. Arrows indicate chromosomal abnormality including addition or deletion of chromosomal fragments. (b) Distribution pattern of chromosomal number obtained after karyotype analysis.
DISCUSSION
The best feeder cell type for establishing HDDPC-derived iPSCs remains unknown, although both MEFs and STO cells have been shown to be effective for establishing ESCs. Therefore, we examined these two types of cells and found that MEFs are superior to STO cells for supporting the growth of “young” iPSCs, which do not appear to be fully reprogrammed.
What is the difference between MEFs and STO cells in their abilities to support the growth of young iPSCs? Recent studies have revealed a difference in the quality and amounts of secreted substances between MEF and STO cells. For example, mesenchymal stem cells derived from human ESCs (hESC-MSCs) secreted bFGF, one of the key factors in maintaining human ESCs (2,5,44,51), whereas STO cells did not (15). CF-1 cells, MEFs derived from 13.5-day fetuses of CF-1 outbred mice, expressed 10-fold more activin A, a key factor (2,5,44,51), than did STO cells (41). Notably, MEFs produce transforming growth factor-β (TGF-β), wingless-type mouse mammary tumor virus (MMTV) integration site family members (WNTs), and bone morphogenetic protein 4 (BMP4), in addition to bFGF and activin A (5,19,35), which are also important for ESC proliferation and for maintaining its pluripotency. Interestingly, Lee et al. demonstrated that STO cells overexpressing human bFGF can readily support the pluripotency of ESCs (18). Furthermore, STO cells genetically engineered to express E-cadherin, a cell–cell adhesion molecule expressed abundantly in ESCs, exhibited better performance in maintaining a pluripotent state of ESCs than did parental STO cells (11). These data clearly indicate that MEFs significantly can produce more growth factors that are beneficial for ESC growth compared to STO cells.
During reprogramming of somatic cells into iPSCs, at least three phases appear to exist, as shown in Figure 1b: (i) Phase I, during which the first ESC-like colony appears after transduction with a set of reprogramming factors, and the growth of ESC-like colonies is supported by their nontransfected parental cells; (ii) Phase II, during which the outline of ESC-like colony is often irregular, and the surface of the colony is not smooth, suggesting incomplete reprogramming; and (iii) Phase III, during which the outline of ESC-like colony is distinct and the surface of the colony is smooth, suggesting complete reprogramming. ESC-like colonies in Phase II likely require specific factors (i.e., activin A and bFGF) to guide their growth and pluripotency. In the present study, we found that STO cells were unable to support the growth of iPSCs during this phase (Fig. 1c). In contrast, ESC-like colonies in Phase III do not appear to require these factors to the same extent as do iPSCs in Phase II (Fig. 1d). This may be why STO cells were able to support the growth of iPSCs in Phase III.
It is important to create iPSCs in which no exogenous DNA has been integrated because the integrated component may activate oncogenic genes or inhibit the normal function of the cell itself. Thus, a vector-free induction system for iPSC generation has been developed (7,54). The plasmids used in this study are thought to be hard to integrate (26). However, we observed the presence of at least two vector components (pCXLE-hOCT3/4-shp53 and pCXLE-hSK) in the genome of the resulting iPSC clones tested (see Fig. 4b). Fortunately, we did not detect a transcript generated from the integrated pCXLE-hOCT3/4-shp53 or pCXLE-hSK transgene (see Fig. 4a). This may be due to silencing of the integrated transgenes. A lack of gene expression from the integrated transgenes would have allowed HDDPC-iPSCs to differentiate into various types of cells under in vitro and in vivo conditions.
Finally, we succeeded in producing iPSCs from one HDDPC line. The other four lines were also successfully reprogrammed using the same method (unpublished results). Furthermore, we succeeded in generating iPSCs from other tissues such as human MSCs and fibroblasts using the same method employed here (unpublished results). Notably, the overall efficiency for HDDPC-iPSC generation was 0.006% (30/5 × 104), which appeared approximately 10-fold lower than that of other iPSCs produced via retroviral transduction (∼0.06% = ∼30/5 × 104) (4). This low efficiency may be due to differences in the transfection methods used. We employed a cotransfection approach using plasmids in which simultaneous uptake of three plasmids by a cell is a prerequisite for iPSC induction. Such simultaneous uptake may not have been frequent in this system.
Unfortunately, the karyotype of HDDPC-iPSCs we established here was abnormal with addition or deletion of some chromosomal fragments (see Fig. 6). It is clear that this is not ascribed to the chromosomal abnormality of parental cells (data not shown). It remains unknown why such this abnormality generates in our iPSCs. We are now investigating possible chromosomal abnormality using other established iPSC clones.
Direct reprogramming of dental cells has been recently reported by Tamaoki et al. and Yan et al. (42,53). Other than these two reports, few studies have reported the successful generation of iPSCs in the dental field. Our successful establishment of iPSCs from HDDPCs isolated from young patients appears to be the first. Our present finding that MEFs are suitable for obtaining HDDPC-iPSCs may be helpful for researchers attempting to isolate iPSCs from primary dental cells.
ACKNOWLEDGMENTS
We thank Dr. Akihiro Umezawa (Department of Reproductive Biology and Pathology, National Research Institute for Child Health and Development, Tokyo, Japan) for his support during iPSC preparation. This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sports, Culture, and Technology of Japan (23792364, 23792439 and 25293418). Issei Saitoh: conception and design, collection and assembly data, data analysis and interpretation, manuscript writing; Emi Inada: collection and assembly data, data analysis and interpretation; Yoko Iwase: provision of study material or patients; Hirofumi Noguchi: conception and design and approval of manuscript; Tomoya Murakami: data analysis and interpretation; Miki Soda: data analysis and interpretation; Naoko Kubota: collection and assembly data; Hiroko Hasegawa: partial financially support; Eri Akasaka: provision of study material or patients; Yuko Matsumoto: partial financially support; Kyoko Oka: data analysis and interpretation; Youichi Yamasaki: laboratory and financial support, approval of manuscript; Haruaki Hayasaki: laboratory and financial support, approval of manuscript; Masahiro Sato: conception and design, laboratory and financial support, manuscript writing. The authors declare no conflicts of interest.
REFERENCES
- 1. Amit M.; Margulets V.; Segev H.; Shariki K.; Laevsky I.; Coleman R.; Itskovitz-Eldor J. Human feeder layers for human embryonic stem cells. Biol. Reprod. 68(6):2150–2156; 2003. [DOI] [PubMed] [Google Scholar]
- 2. Beattie G. M.; Lopez A. D.; Bucay N.; Hinton A.; Firpo M. T.; King C. C.; Hayek A. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells 23(4):489–495; 2005. [DOI] [PubMed] [Google Scholar]
- 3. Brook F. A.; Gardner R. L. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA 94(11):5709–5712; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen M.; Sun X.; Jiang R.; Shen W.; Zhong X.; Liu B.; Qi Y.; Huang B.; Xiang A. P.; Ge J. Role of MEF feeder cells in direct reprogramming of mousetail-tip fibroblasts. Cell Biol. Int. 33(12):1268–1273; 2009. [DOI] [PubMed] [Google Scholar]
- 5. Eiselleova L.; Peterkova I.; Neradil J.; Slaninova I.; Hampl A.; Dvorak P. Comparative study of mouse and human feeder cells for human embryonic stem cells. Int. J. Dev. Biol. 52(4):353–363; 2008. [DOI] [PubMed] [Google Scholar]
- 6. Evans M. J.; Kaufman M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292(5819):154–156; 1981. [DOI] [PubMed] [Google Scholar]
- 7. Fusaki N.; Ban H.; Nishiyama A.; Saeki K.; Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85(8):348–362; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Genbacev O.; Krtolica A.; Zdravkovic T.; Brunette E.; Powell S.; Nath A.; Caceres E.; McMaster M.; McDonagh S.; Li Y.; Mandalam R.; Lebkowski J.; Fisher S. J. Serum-free derivation of human embryonic stem cell lines on human placental fibroblast feeders. Fertil. Steril. 83(5):1517–1529; 2005. [DOI] [PubMed] [Google Scholar]
- 9. Gronthos S.; Mankani M.; Brahim J.; Robey P. G.; Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 97(25):13625–13630; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoffmeyer K.; Raggioli A.; Rudloff S.; Anton R.; Hierholzer A.; Del Valle I.; Hein K.; Vogt R.; Kemler R. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336(6088):1549–1554; 2012. [DOI] [PubMed] [Google Scholar]
- 11. Horie M.; Ito A.; Kiyohara T.; Kawabe Y.; Kamihira M. E-cadherin gene-engineered feeder systems for supporting undifferentiated growth of mouse embryonic stem cells. J. Biosci. Bioeng. 110(5):582–587; 2010. [DOI] [PubMed] [Google Scholar]
- 12. Hovatta O.; Mikkola M.; Gertow K.; Stromberg A. M.; Inzunza J.; Hreinsson J.; Rozell B.; Blennow E.; Andang M.; Ahrlund-Richter L. A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Hum. Reprod. 18(7):1404–1409; 2003. [DOI] [PubMed] [Google Scholar]
- 13. Kawarabayashi T.; Shoji M.; Sato M.; Sasaki A.; Ho L.; Eckman C. B.; Prada C. M.; Younkin S. G.; Kobayashi T.; Tada N.; Matsubara E.; Iizuka T.; Harigaya Y.; Kasai K.; Hirai S. Accumulation of beta-amyloid fibrils in pancreas of transgenic mice. Neurobiol. Aging 17(2):215–222; 1996. [DOI] [PubMed] [Google Scholar]
- 14. Kondo S.; Kubota S.; Mukudai Y.; Nishida T.; Yoshihama Y.; Shirota T.; Shintani S.; Takigawa M. Binding of glyceraldehyde-3-phosphate dehydrogenase to the cis-acting element of structure-anchored repression in ccn2 mRNA. Biochem. Biophys. Res. Commun. 405(3):382–387; 2011. [DOI] [PubMed] [Google Scholar]
- 15. Lee E. J.; Kang H. J.; Lee H. N.; Kang S. K.; Kim K. H.; Lee S. W.; Lee G.; Park Y. B.; Kim H. S. New culture system for human embryonic stem cells: Autologous mesenchymal stem cell feeder without exogenous fibroblast growth factor 2. Differentiation 83(1):92–100; 2012. [DOI] [PubMed] [Google Scholar]
- 16. Lee J.; Kim H. K.; Rho J. Y.; Han Y. M.; Kim J. The human OCT-4 isoforms differ in their ability to confer self-renewal. J. Biol. Chem. 281(44):33554–33565; 2006. [DOI] [PubMed] [Google Scholar]
- 17. Lee K. H.; Chuang C. K.; Guo S. F.; Tu C. F. Simple and efficient derivation of mouse embryonic stem cell lines using differentiation inhibitors or proliferation stimulators. Stem Cells Dev. 21(3):373–383; 2012. [DOI] [PubMed] [Google Scholar]
- 18. Lee W. Y.; Kim J.; Gil C. H.; Lee J. H.; Song H.; Kim J. H.; Chung H. M. Maintenance of human pluripotent stem cells using 4SP-hFGF2-secreting STO cells. Stem Cell Res. 7(3):210–218; 2011. [DOI] [PubMed] [Google Scholar]
- 19. Lim J. W.; Bodnar A. Proteome analysis of conditioned medium from mouse embryonic fibroblast feeder layers which support the growth of human embryonic stem cells. Proteomics 2(9):1187–1203; 2002. [DOI] [PubMed] [Google Scholar]
- 20. Martin G. R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78(12):7634–7638; 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin G. R.; Evans M. J. Differentiation of clonal lines of teratocarcinoma cells: Formation of embryoid bodies in vitro. Proc. Natl. Acad. Sci. USA 72(4):1441–1445; 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nichols J.; Zevnik B.; Anastassiadis K.; Niwa H.; Klewe-Nebenius D.; Chambers I.; Scholer H.; Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95(3):379–391; 1998. [DOI] [PubMed] [Google Scholar]
- 23. Ohta S.; Imaizumi Y.; Okada Y.; Akamatsu W.; Kuwahara R.; Ohyama M.; Amagai M.; Matsuzaki Y.; Yamanaka S.; Okano H.; Kawakami Y. Generation of human melanocytes from induced pluripotent stem cells. PLoS One 6(1):e16182; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okamoto S.; Takahashi M. Induction of retinal pigment epithelial cells from monkey iPS cells. Invest. Ophthalmol. Vis. Sci. 52(12):8785–8790; 2011. [DOI] [PubMed] [Google Scholar]
- 25. Okita K.; Ichisaka T.; Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 448(7151):313–317; 2007. [DOI] [PubMed] [Google Scholar]
- 26. Okita K.; Matsumura Y.; Sato Y.; Okada A.; Morizane A.; Okamoto S.; Hong H.; Nakagawa M.; Tanabe K.; Tezuka K.; Shibata T.; Kunisada T.; Takahashi M.; Takahashi J.; Saji H.; Yamanaka S. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8(5):409–412; 2011. [DOI] [PubMed] [Google Scholar]
- 27. Orimo H.; Goseki-Sone M.; Sato S.; Shimada T. Detection of deletion 1154–1156 hypophosphatasia mutation using TNSALP exon amplification. Genomics 42(2):364–366; 1997. [DOI] [PubMed] [Google Scholar]
- 28. Park J. H.; Kim S. J.; Oh E. J.; Moon S. Y.; Roh S. I.; Kim C. G.; Yoon H. S. Establishment and maintenance of human embryonic stem cells on STO, a permanently growing cell line. Biol. Reprod. 69(6):2007–2014; 2003. [DOI] [PubMed] [Google Scholar]
- 29. Pierantozzi E.; Gava B.; Manini I.; Roviello F.; Marotta G.; Chiavarelli M.; Sorrentino V. Pluripotency regulators in human mesenchymal stem cells: Expression of NANOG but not of OCT-4 and SOX-2. Stem Cells Dev. 20(5):915–923; 2011. [DOI] [PubMed] [Google Scholar]
- 30. Rajala K.; Hakala H.; Panula S.; Aivio S.; Pihlajamaki H.; Suuronen R.; Hovatta O.; Skottman H. Testing of nine different xeno-free culture media for human embryonic stem cell cultures. Hum. Reprod. 22(5):1231–1238; 2007. [DOI] [PubMed] [Google Scholar]
- 31. Richards M.; Fong C. Y.; Chan W. K.; Wong P. C.; Bongso A. Human feeders support prolonged undifferentiated growth of human inner cell masses and embryonic stem cells. Nat. Biotechnol. 20(9):933–936; 2002. [DOI] [PubMed] [Google Scholar]
- 32. Richards M.; Tan S.; Fong C. Y.; Biswas A.; Chan W. K.; Bongso A. Comparative evaluation of various human feeders for prolonged undifferentiated growth of human embryonic stem cells. Stem Cells 21(5):546–556; 2003. [DOI] [PubMed] [Google Scholar]
- 33. Saitoh I.; Sato M.; Iwase Y.; Inada E.; Nomura T.; Akasaka E.; Yamasaki Y.; Noguchi H. Generation of mouse STO feeder cell lines that confer resistance to several types of selective drugs. Cell Med. 3(1–3):97–102; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shamblott M. J.; Axelman J.; Wang S.; Bugg E. M.; Littlefield J. W.; Donovan P. J.; Blumenthal P. D.; Huggins G. R.; Gearhart J. D. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc. Natl. Acad. Sci. USA 95(23):13726–13731; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soh B. S.; Song C. M.; Vallier L.; Li P.; Choong C.; Yeo B. H.; Lim E. H.; Pedersen R. A.; Yang H. H.; Rao M.; Lim B. Pleiotrophin enhances clonal growth and long-term expansion of human embryonic stem cells. Stem Cells 25(12):3029–3037; 2007. [DOI] [PubMed] [Google Scholar]
- 36. Stojkovic P.; Lako M.; Stewart R.; Przyborski S.; Armstrong L.; Evans J.; Murdoch A.; Strachan T.; Stojkovic M. An autogeneic feeder cell system that efficiently supports growth of undifferentiated human embryonic stem cells. Stem Cells 23(3):306–314; 2005. [DOI] [PubMed] [Google Scholar]
- 37. Takahashi K.; Tanabe K.; Ohnuki M.; Narita M.; Ichisaka T.; Tomoda K.; Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5):861–872; 2007. [DOI] [PubMed] [Google Scholar]
- 38. Takahashi K.; Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676; 2006. [DOI] [PubMed] [Google Scholar]
- 39. Takeda T.; Tezuka Y.; Horiuchi M.; Hosono K.; Iida K.; Hatakeyama D.; Miyaki S.; Kunisada T.; Shibata T.; Tezuka K. Characterization of dental pulp stem cells of human tooth germs. J. Dent. Res. 87(7):676–681; 2008. [DOI] [PubMed] [Google Scholar]
- 40. Talbot N. C.; Powell A. M.; Garrett W. M. Spontaneous differentiation of porcine and bovine embryonic stem cells (epiblast) into astrocytes or neurons. In Vitro Cell Dev. Biol. Anim. 38(4):191–197; 2002. [DOI] [PubMed] [Google Scholar]
- 41. Talbot N. C.; Sparks W. O.; Powell A. M.; Kahl S.; Caperna T. J. Quantitative and semiquantitative immunoassay of growth factors and cytokines in the conditioned medium of STO and CF-1 mouse feeder cells. In Vitro Cell Dev. Biol. Anim. 48(1):1–11; 2012. [DOI] [PubMed] [Google Scholar]
- 42. Tamaoki N.; Takahashi K.; Tanaka T.; Ichisaka T.; Aoki H.; Takeda-Kawaguchi T.; Iida K.; Kunisada T.; Shibata T.; Yamanaka S.; Tezuka K. Dental pulp cells for induced pluripotent stem cell banking. J. Dent. Res. 89(8):773–778; 2010. [DOI] [PubMed] [Google Scholar]
- 43. Thomson J. A.; Itskovitz-Eldor J.; Shapiro S. S.; Waknitz M. A.; Swiergiel J. J.; Marshall V. S.; Jones J. M. Embryonic stem cell lines derived from human blastocysts. Science 282(5391):1145–1147; 1998. [DOI] [PubMed] [Google Scholar]
- 44. Vallier L.; Alexander M.; Pedersen R. A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118(Pt 19):4495–4509; 2005. [DOI] [PubMed] [Google Scholar]
- 45. van Ooyen A. J.; de Boer H. A.; Ab G.; Gruber M. Specific inhibition of ribosomal RNA synthesis in vitro by guanosine 3′ diphosphate, 5′ diphosphate. Nature 254(5500):530–531; 1975. [DOI] [PubMed] [Google Scholar]
- 46. Wang X.; Wang J.; Huang V.; Place R. F.; Li L. C. Induction of NANOG expression by targeting promoter sequence with small activating RNA antagonizes retinoic acid-induced differentiation. Biochem. J. 443(3):821–828; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ware L. M.; Axelrad A. A. Inherited resistance to N- and B-tropic murine leukemia viruses in vitro: Evidence that congenic mouse strains SIM and SIM.R differ at the Fv-1 locus. Virology 50(2):339–348; 1972. [DOI] [PubMed] [Google Scholar]
- 48. Wobus A. M.; Boheler K. R. Embryonic stem cells: Prospects for developmental biology and cell therapy. Physiol. Rev. 85(2):635–678; 2005. [DOI] [PubMed] [Google Scholar]
- 49. Wobus A. M.; Holzhausen H.; Jakel P.; Schoneich J. Characterization of a pluripotent stem cell line derived from a mouse embryo. Exp. Cell Res. 152(1):212–219; 1984. [DOI] [PubMed] [Google Scholar]
- 50. Xia S. L.; Zhang X.; Jing N. H. [The induction and differentiation of a human teratocarcinoma cell line (PA-1) in vitro]. Shi Yan Sheng Wu Xue Bao. 28(4):397–407; 1995. [PubMed] [Google Scholar]
- 51. Xiao L.; Yuan X.; Sharkis S. J. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells 24(6):1476–1486; 2006. [DOI] [PubMed] [Google Scholar]
- 52. Xu C.; Jiang J.; Sottile V.; McWhir J.; Lebkowski J.; Carpenter M. K. Immortalized fibroblast-like cells derived from human embryonic stem cells support undifferentiated cell growth. Stem Cells 22(6):972–980; 2004. [DOI] [PubMed] [Google Scholar]
- 53. Yan X.; Qin H.; Qu C.; Tuan R. S.; Shi S.; Huang G. T. iPS cells reprogrammed from human mesenchymal-like stem/progenitor cells of dental tissue origin. Stem Cells Dev. 19(4):469–480; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu J.; Hu K.; Smuga-Otto K.; Tian S.; Stewart R.; Slukvin I. I.; Thomson J. A. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324(5928):797–801; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu J.; Vodyanik M. A.; Smuga-Otto K.; Antosiewicz-Bourget J.; Frane J. L.; Tian S.; Nie J.; Jonsdottir G. A.; Ruotti V.; Stewart R.; Slukvin II; Thomson J. A. Induced pluripotent stem cell lines derived from human somatic cells. Science 318(5858):1917–1920; 2007. [DOI] [PubMed] [Google Scholar]