Abstract
In order to determine whether the deleted in liver cancer-1 (DLC-1) gene is deregulated in non-small-cell lung carcinoma (NSCLC) and to assess the contribution of molecular alterations in DLC-1 to lung carcinogenesis, a total of 84 tissue specimens (30 NSCLC and 30 corresponding adjacent normal tissues; 5 benign tumor and 5 corresponding adjacent normal tissues; and 10 pulmonary bullae and 4 corresponding adjacent normal tissues), were obtained from 45 patients who underwent curative surgical resection. DLC-1 mRNA expression was evaluated by reverse transcription-quantitative polymerase chain reaction (PCR) and its protein level was assessed by western blot analysis. A significant downregulation of DCL-1 at the mRNA and protein levels was observed in NSCLC tissues when compared to benign lung tumors and normal lung tissues (P<0.001). To further determine whether the decreased expression of DLC-1 at the mRNA and protein levels is associated with the methylation of its promoter, methylation-specific PCR was performed following extraction of genomic DNA from the samples. DLC-1 promoter methylation was identified in 7 of the 30 (23.3%) NSCLC tissue samples, but not in the corresponding adjacent normal tissues from NSCLC patients or in lung tissues from non-NSCLC patients. Our data indicated that DLC-1 hypermethylation may play a crucial role in lung carcinogenesis and may be a target for the treatment of NSCLC.
Keywords: expression, methylation, deleted in liver cancer-1, non-small-cell lung carcinomas
Introduction
Lung cancer, with ~80% of the cases comprising non-small-cell lung carcinomas (NSCLCs), is one of the most common types of human cancer worldwide and the most common cause of cancer-related mortality in China (1). Since only 15–20% of the patients survive to 5-years (2), lung cancer cure rates are currently disappointing.
Tumorigenesis is a complex, multistage process, involving numerous epigenetic modifications and genetic alterations. It is essential to characterize the molecular mechanisms underlying tumor formation and progression in order to develop rational approaches to cancer diagnosis and therapy. The inactivation of the tumor suppressor gene deleted in liver cancer-1 (DLC-1), which is induced by promoter methylation, was recently suggested to play a central role in tumorigenesis in several cancer models, including diffuse large B-cell lymphoma (3), gallbladder carcinoma (4), gastric cancer (5), hepatocellular carcinoma (6), breast cancer (7) and multiple myeloma (8). Accumulating evidence indicates that DLC-1 inactivation induced by methylation may also be involved in lung carcinogenesis. It was demonstrated that DLC-1 transcription was significantly reduced or lost in primary tumors and NSCLC cell lines (9), while frequent heterozygous deletion (10) and DNA methylation of DLC-1 (11) was observed in lung cancer.
In this study, we investigated the possible involvement of DLC-1 in NSCLC and analyzed the correlation between aberrant DLC-1 methylation and clinicopathological parameters in Chinese NSCLC patients, with the aim to elucidate the molecular mechanism underlying the development of NSCLC.
Materials and methods
Subjects
A total of 84 tissue specimens were obtained from patients who underwent curative resection at the Department of Thoracic Surgery, China-Japan Friendship Hospital (Beijing, China) between November, 2008 and November, 2009. None of the patients had received radiotherapy or chemotherapy prior to surgery. Tissue specimens were obtained from NSCLC patients (n=30; mean age, 63 years; range, 49–74 years) and from patients with benign lung tumors (n=5; mean age, 56 years; range, 30–61 years) who underwent surgical resection. Normal adjacent lung tissue was available in all the cases. Specimens from patients with pulmonary bullae (n=10; mean age, 28 years; range, 17–63 years) and corresponding normal lung tissue samples (n=4) were also collected. All the normal, benign and malignant tissue samples were immediately snap-frozen and stored at −80°C for analysis. The clinical and histopathological characteristics of all the samples are listed in Table I. To confirm the diagnosis and determine the cellular composition of the tumor, all the samples were carefully reviewed at the Department of Pathology of our hospital. The pathological stage was determined according to the Revised International System for Staging Lung Cancer (12).
Table I.
Characteristics of the investigated patients.
| Characteristics | NSCLC (n=30) | Benign tumors (n=5) | Bullae (n=10) |
|---|---|---|---|
| Gender | |||
| Male | 21 | 3 | 7 |
| Female | 9 | 2 | 3 |
| Age (years) | |||
| <60 | 11 | 2 | 1 |
| ≥60 | 19 | 3 | 9 |
| Histological type | |||
| Adenocarcinoma | 18 | ||
| SCC | 11 | ||
| Others | 1 | ||
| Differentiation | |||
| High/moderate | 15 | ||
| Poor/undifferentiated | 15 | ||
| Stage | |||
| I | 10 | ||
| II | 10 | ||
| III | 10 | ||
| Smoking index | |||
| <400 | 12 | 2 | 10 |
| ≥400 | 18 | 3 | 0 |
NSCLC, non-small-cell lung carcinoma; SCC, squamous cell carcinoma.
The present study was approved by the Institutional Review Board of the China-Japan Friendship Hospital and written informed consent was provided by all the participants.
Evaluation of DLC-1 mRNA using quantitative PCR (qPCR)
DLC-1 mRNA expression was evaluated by reverse transcription (RT)-qPCR. Briefly, total RNA was extracted using the Dr. P kit (BioChain, Newark, CA, USA) according to the manufacturer's instructions. Total RNA (2 µg) was reverse transcribed with the oligo (dT) primer (Invitrogen, Carlsbad, CA, USA) using Moloney Murine Leukemia Virus Reverse Transcriptase (Promega, Madison, WI, USA) in a total reaction volume of 20 µl. Following the RT reaction, sequence-specific PCR primers (Table II) for DLC-1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (both from Invitrogen, Carlsbad, CA, USA) were used for qPCR. DLC-1 was amplified on the same plate under the control of GAPDH, using the Eva QPCR SuperMix kit (BioChain). The data were analyzed with the 2−ΔΔCt method (13) using GADPH as a reference gene. The results are presented as fold-change relative to the GADPH control. A two-fold increased or decreased expression was considered as significant.
Table II.
Primer sequences for DLC-1 and GAPDH in MSP using USP.
| Primers | Sequence, 5′-3′ | PCR product size (bp) |
|---|---|---|
| qDLC-1F | CCG CCT GAG CAT CTA CG | 138 |
| qDLC-1R | ACT ATC CGC TGC ATC CC | |
| GAPDH-F | GAAGGTGAAGGTCGGAGTCA | 226 |
| GAPDH-R | GAAGATGGTGATGGGATTTC | |
| MSP-F | TTTAAAGATCGAAACGAGGGAGCG | 172 |
| MSP-R | CCCAACGAAAAAACCCGACTAACG |
DLC-1, deleted in liver cancer 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PCR, polymerase chain reaction; MSP, methylation-specific PCR; USP, universal sample processing; F, forward; R, reverse.
Western blot analysis
Protein expression was evaluated by western blotting. Briefly, total protein was extracted using the Dr. P kit according to the manufacturer's instructions. The total protein concentration was determined by the BCA Protein Assay kit (Vigorous Instruments, Beijing, China). Equal amounts of total proteins were resolved by SDS-PAGE and subjected to western blotting. The primary antibody was goat polyclonal anti-DLC-1 antibody (dilution 1:800; cat. no. TA302467; OriGene, Rockville, MD, USA). The proteins were transferred from the gels to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked using the Attoglow Western Blot Analysis kit, followed by anti-goat secondary antibody (dilution 1:8,000; cat. no. K3171120; BioChain). The films were scanned and the protein band signal intensity was measured with Adobe Photoshop CS2 (Adobe, San Jose, CA, USA). The relative expression difference of the DLC-1 protein in all the lung tissue samples was calculated compared with the average expression in the normal samples. A two-fold increase or decrease was considered to be a significant expression difference.
Detection of DLC-1 methylation using methylation-specific PCR
Genomic DNA (500 ng) extracted from tissue samples was modified by sodium bisulfite with the DNA Methylation Detection kit (BioChain) according to the manufacturer's instructions. Modified DNA (7 µl) was used for PCR in a total reaction volume of 25 µl as follows: 35 cycles of melting (30 sec at 95°C), annealing (30 sec at 55°C) and extension (30 sec at 72°C). The 25-µl reaction mixture contained 1X PCR Buffer (Mg2+ Plus; Promega), 200 µM of each dNTP, 0.5 µM forward and 0.5 µM reverse primers (the primer sequences are provided in Table II) and 0.5 U GoTaq® Hot Start Polymerase (Promega). The PCR products were separated in 3% agarose gel supplemented with ethidium bromide. DNA was visualized under ultraviolet illumination.
Statistical analysis
The statistical analysis was performed with Fisher's exact test and Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of DLC-1 at the mRNA and protein level
The DLC-1 expression was significantly reduced in NSCLC compared to that in benign lung tumors and normal lung tissue (both P<0.001; Fig. 1). The DCL-1 mRNA level in the 30 investigated NSCLCs was decreased or undetectable (100%); among these, 26 (86.7%) exhibited less than half of the DCL-1 mRNA levels observed in normal lung tissues. There was no significant difference in DLC-1 expression between normal lung tissues and benign lung tumors (P=0.603). Compared to the unmethylated NSCLC specimens, the DLC-1 expression level in the methylated specimens was significantly reduced (P=0.001 and P<0.05, respectively).
Figure 1.
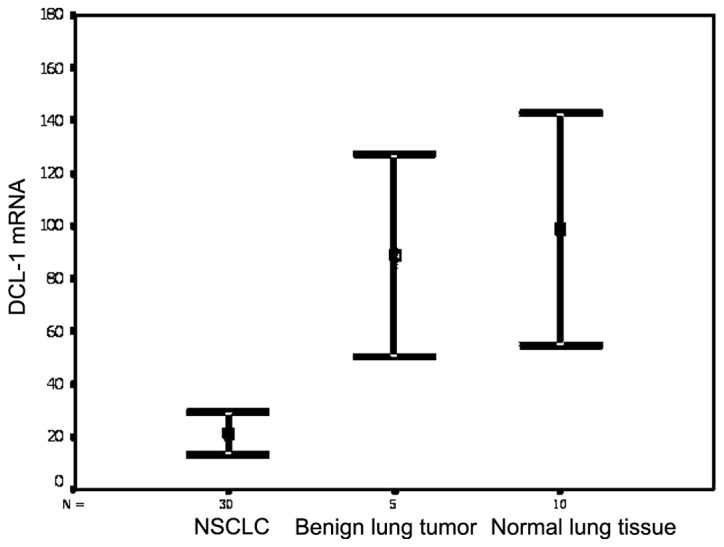
DLC-1 mRNA expression was evaluated by reverse transcription-quantitative polymerase chain reaction. DLC-1, deleted in liver cancer-1; NSCLC, non-small-cell lung carcinoma.
In order to validate the observed reduction of DLC-1 mRNA in NSCLC patients, the protein expression level was evaluated by western blotting. Loss or low expression of the DLC-1 protein was detected in 19 of the 30 (63%) NSCLCs and in 1 of the 5 (20%) benign lung tumor samples (Fig. 2). This result was in accordance with that observed in the DLC-1 mRNA.
Figure 2.

The expression of the DLC-1 protein in lung tissue was evaluated by western blotting. T, cancer tissue in NSCLC; N, corresponding adjacent normal lung tissue in NSCLC. DLC-1, deleted in liver cancer-1; NSCLC, non-small-cell lung carcinoma. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
DLC-1 methylation detection using methylation-specific PCR (MSP)
To investigate the mechanism underlying the loss of DLC-1 expression at the mRNA and protein levels, the methylation status of the 5′CpG island in the DLC-1 gene was determined using MSP (Fig. 3). DLC-1 promoter methylation was significantly higher in NSCLCs (P=0.011), observed in 23.3% (7/30) of the NSCLC samples, but was not detected in any of the corresponding adjacent normal tissue, benign tumor, or pulmonary bullae samples. By further analyzing the protein expression data in cancer tissues with methylation of DLC-1 DNA, reduced or loss of expression of the DLC-1 protein was observed in 4 of the 7 (57.1%) tumor tissue samples with methylated DLC-1, indicating that the methylation status of the 5′CpG island in the DLC-1 gene may be associated with decreased DLC-1 protein expression in NSCLCs.
Figure 3.
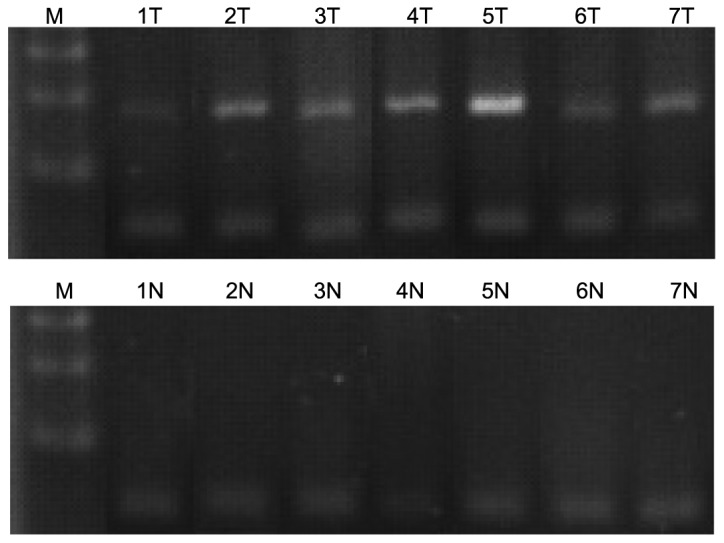
DLC-1 methylation was detected by methylation-specific polymerase chain reaction. T, cancer tissue in NSCLC; N, corresponding adjacent normal lung tissue in NSCLC. DLC-1, deleted in liver cancer-1; NSCLC, non-small-cell lung carcinoma.
DLC-1 methylation and clinicopathological characteristics
The association between the methylation of DLC-1 and clinicopathological characteristics was analyzed in NSCLC patients (Table III). There were no significant correlations of DLC-1 methylation with gender, age, tumor site, differentiation, TNM stage or histological type. However, we observed a tendency for the DLC-1 methylation level in squamous cell carcinomas (5/11) to be higher compared to that observed in adenocarcinomas (2/18) (Chi-square test, likelihood ratio, 0.087). DLC-1 methylation in men also exhibited a tendency for higher incidence compared to that observed in women. The preferential methylation of DLC-1 in male patients and in squamous cell carcinomas prompted us to investigate the association between smoking and DLC-1 methylation in NSCLC. We observed that the incidence of DLC-1 methylation in heavy smoking NSCLC patients [with a smoking index (number of cigarettes smoked per day × years of smoking) of >400] was significantly higher compared to that observed in light smoking (with smoking index <400) and non-smoking patients (Fisher's exact test, P=0.024).
Table III.
Correlation of DLC-1 methylation and mRNA and protein expression with clinical and histological variables in NSCLC patients.
| Methylation | mRNA | Protein | ||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | Present | Absent | P-value | (mean ± SE) | P-value | Normal | Reduced | P-value |
| Age (years) | 1 | 0.502 | 1 | |||||
| <60 | 9 | 2 | 24.6±7.3 | 4 | 7 | |||
| ≥60 | 14 | 5 | 19.0±4.7 | 7 | 12 | |||
| Smoking index | 0.024 | 0.042 | 0.442 | |||||
| <400 | 12 | 0 | 30.7±6.7 | 3 | 9 | |||
| ≥400 | 11 | 7 | 14.6±4.3 | 8 | 10 | |||
| Histological type | 0.071 | 0.399 | 0.432 | |||||
| Adenocarcinoma | 6 | 5 | 16.5±6.5 | 5 | 6 | |||
| SCC | 16 | 2 | 23.8±5.3 | 5 | 13 | |||
| TNM stage | 0.657 | 0.592 | 0.425 | |||||
| I/II | 16 | 4 | 19.5±4.5 | 6 | 14 | |||
| III | 7 | 3 | 24.1±7.8 | 5 | 5 | |||
| Differentiation | 1 | 0.883 | 1 | |||||
| High/moderate | 3 | 0 | 19.2±7.4 | 1 | 2 | |||
| Poor/undifferentiated | 20 | 7 | 21.2±4.3 | 10 | 17 | |||
DLC-1, deleted in liver cancer-1; NSCLC, non-small-cell lung carcinoma; SCC, squamous cell carcinoma; SE, standard error; TNM, tumor node metastastis.
Discussion
In the present study, DLC-1 expression at the mRNA level was evaluated by RT-qPCR and at the protein level by western blotting. Significant downregulation of DCL-1 at the mRNA as well as the protein level was observed in NSCLC tissues when compared to benign lung tumors and normal lung tissues (P<0.001). To further investigate whether the reduced expression of DLC-1 at the mRNA and protein levels is associated with the methylation on its promoter, methylation-specific PCR was performed following extraction of genomic DNA from the samples. DLC-1 promoter methylation was identified in 7 of the 30 (23.3%) tumor tissue samples, but not in corresponding adjacent normal tissue samples from NSCLC patients or in lung tissues from non-NSCLC patients. Our data are consistent with those of previous studies (9,11,14). The level of expression of DLC-1 in methylated specimens was significantly reduced compared to that in unmethylated NSCLC samples. The majority of the methylated tumors exhibited a reduction or a loss of expression of the DLC-1 protein. Promoter methylation has been hypothesized to be one of the key mechanisms responsible for the inactivation of the DLC-1 gene in NSCLCs (15). Our data further support this hypothesis.
DLC-1 methylation was observed in 4 of 20 (20%) stage I–II tumors and the percentage of methylation did not increase significantly in more advanced-stage tumors, suggesting that DLC-1 methylation may occur at a relatively early stage during the multistep process of lung carcinogenesis. However, the pathobiological significance of DLC-1 methylation in multistage carcinogenesis may differ between squamous cell carcinomas and adenocarcinomas. DLC-1 methylation appears to be a relatively early event in adenocarcinomas, since its incidence was 14% in stage I tumors but did not significantly change in more advanced-stage tumors. By contrast, DLC-1 methylation appears to be a late event in squamous cell carcinomas, since its incidence increased significantly as tumor stage advanced from I to III (Chi-square test, P=0.015 and P<0.05). By contrast, methylation of the differentially expressed in adenocarcinoma of the lung-1 gene was reported to be an early event in squamous cell carcinomas but a late event in adenocarcinomas (16). The finding that 27.3% (3/11) of NSCLCs exhibited methylation with normal expression of the DLC-1 protein suggests that DLC-1 methylation is an early event during lung cancer development. In addition, the expression of DLC-1 was significantly reduced in NSCLC and was marginally lower in benign lung tumors. Therefore, methylation of DLC-1 appears to be a useful biomarker for the early diagnosis of NSCLC.
Tobacco smoking is the main etiologic factor associated with lung carcinoma and an estimated 86% of lung carcinoma deaths are attributed to smoking (17). Although there is no direct evidence that smoking induces DNA hypermethylation, it was recently demonstrated that there is an association of DNA methylation with exposure to the carcinogens in tobacco.
In human NSCLCs, it has been demonstrated that the hypermethylation of the p16/cyclin-dependent kinase inhibitor 2A (18,19), Ras association domain family 1 isoform A (20) and tumor suppressor in lung cancer 1/immunoglobulin superfamily member 4 genes (21) was significantly associated with tobacco smoking, indicating that the carcinogens in tobacco smoke induce DNA hypermethylation. Similar to other published reports, our data demonstrated that aberrant methylation of DLC-1 is associated with exposure to smoke and correlated to long-term smoking habit (20,22). Although only few adenocarcinoma patients were smokers, the two methylated adenocarcinoma patients were heavy smokers. There were no significant differences between former smokers (had quit smoking >12 months prior to tumor development) and current smokers (smoked within the 12 months preceding tumor development), although a trend towards a higher methylation frequency among smokers was noted. The incidence of DLC-1 methylation in tumors was significantly higher among heavy smokers. In particular, methylation was detected in the 3 former smokers, whereas methylation rates did not differ between current and former smokers (4/10 vs. 3/9, respectively). These data indicate that aberrant methylation is not only correlated to current smoke exposure, but continues for a long period of time following smoking cessation. Smoking cessation reduces the subsequent risk of lung cancer and quitting smoking before middle age eliminates >90% of the risk attributable to tobacco (23).
The mechanism through which the carcinogens in cigarette smoke cause DLC-1 methylation in NSCLC remains unclear. Smoking exposure may induce significant and sustained lung inflammation (24), which was found to be significantly associated with aberrant DNA methylation (25). Inflammation may increase CpG island methylation. Furthermore, smoking increases the activity of DNA methyltransferase (26), which drives the de novo hypermethylation of susceptible loci (27). These direct or indirect mechanisms may be involved in the enhanced DNA methylation observed in the NSCLC of heavy smokers.
In conclusion, DLC-1 promoter methylation was detected in 7 of 30 (23.3%) primary NSCLCs. Furthermore, our results indicated that DLC-1 promoter hypermethylation is a potential molecular marker for the early detection of lung cancer. This methylation is associated with heavy tobacco smoking and may play a role in lung carcinogenesis. The promoter of the DLC-1 gene is likely one of the important targets of methylation by smoking during the development or progression of NSCLCs. Our results also indicated that epigenetic silencing of DLC-1 in Chinese NSCLC patients may be used as a biomarker for NSCLC diagnosis and a therapeutic option.
Acknowledgements
The authors would like to thank the Department of Pathology of China-Japan Friendship Hospital for specimen collection and BioChain Science-Technology Inc., for assisting with the experiments.
Glossary
Abbreviations
- NSCLCs
non-small-cell lung carcinomas
- DLC-1
deleted in liver cancer-1
- MSP
methylation-specific polymerase chain reaction
References
- 1.Song F, He M, Li H, Qian B, Wei Q, Zhang W, Chen K, Hao X. A cancer incidence survey in Tianjin: the third largest city in China-between 1981 and 2000. Cancer Causes Control. 2008;19:443–450. doi: 10.1007/s10552-007-9105-6. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Pike BL, Greiner TC, Wang X, Weisenburger DD, Hsu YH, Renaud G, Wolfsberg TG, Kim M, Weisenberger DJ, Siegmund KD, Ye W, Groshen S, Mehrian-Shai R, Delabie J, Chan WC, Laird PW, Hacia JG. DNA methylation profiles in diffuse large B-cell lymphoma and their relationship to gene expression status. Leukemia. 2008;22:1035–1043. doi: 10.1038/leu.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García P, Manterola C, Araya JC, Villaseca M, Guzman P, Sanhueza A, Thomas M, Alvarez H, Roa JC. Promoter methylation profile in preneoplastic and neoplastic gallbladder lesions. Mol Carcinog. 2009;48:79–89. doi: 10.1002/mc.20457. [DOI] [PubMed] [Google Scholar]
- 5.Kim TY, Jong HS, Song SH, Dimtchev A, Jeong SJ, Lee JW, Kim TY, Kim NK, Jung M, Bang YJ. Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene. 2003;22:3943–3951. doi: 10.1038/sj.onc.1206573. [DOI] [PubMed] [Google Scholar]
- 6.Wong CM, Lee JM, Ching YP, Jin DY, Ng IO. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63:7646–7651. [PubMed] [Google Scholar]
- 7.Teramoto A, Tsukuda K, Yano M, Toyooka S, Dote H, Doihara H, Shimizu N. Less frequent promoter hypermethylation of DLC-1 gene in primary breast cancers. Oncol Rep. 2004;12:141–144. [PubMed] [Google Scholar]
- 8.Song YF, Xu R, Zhang XH, Chen BB, Chen Q, Chen YM, Xie Y. High-frequency promoter hypermethylation of the deleted in liver cancer-1 gene in multiple myeloma. J Clin Pathol. 2006;59:947–951. doi: 10.1136/jcp.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan BZ, Jefferson AM, Baldwin KT, Thorgeirsson SS, Popescu NC, Reynolds SH. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- 10.Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, Singer S, Kuehnel F, Wigler M, Powers S, Zender L, Lowe SW. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, Hattenhorst UE, Hofmann HS, Silber RE, Burdach S, Hansen G. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005;41:1223–1236. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S, Sánchez-Carbayo M. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. doi: 10.1186/1479-5876-8-86. doi: 10.1186/1479-5876-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD, Rha SY, Tan J, Hsieh WS, Ambinder RF, Lin X, Chan AT, Tao Q. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–944. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 16.Kikuchi S, Yamada D, Fukami T, Masuda M, Sakurai-Yageta M, Williams YN, Maruyama T, Asamura H, Matsuno Y, Onizuka M, Murakami Y. Promoter methylation of DAL-1/4.1B predicts poor prognosis in non-small cell lung cancer. Clin Cancer Res. 2005;11:2954–2961. doi: 10.1158/1078-0432.CCR-04-2206. [DOI] [PubMed] [Google Scholar]
- 17.Shopland DR, Eyre HJ, Pechacek TF. Smoking-attributable cancer mortality in 1991: is lung cancer now the leading cause of death among smokers in the United States? J Natl Cancer Inst. 1991;83:1142–1148. doi: 10.1093/jnci/83.16.1142. [DOI] [PubMed] [Google Scholar]
- 18.Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61:3419–3424. [PubMed] [Google Scholar]
- 19.Jarmalaite S, Kannio A, Anttila S, Lazutka JR, Husgafvel-Pursiainen K. Aberrant p16 promoter methylation in smokers and former smokers with nonsmall cell lung cancer. Int J Cancer. 2003;106:913–918. doi: 10.1002/ijc.11322. [DOI] [PubMed] [Google Scholar]
- 20.Kim DH, Kim JS, Ji YI, Shim YM, Kim H, Han J, Park J. Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer Res. 2003;63:3743–3746. [PubMed] [Google Scholar]
- 21.Kikuchi S, Yamada D, Fukami T, Maruyama T, Ito A, Asamura H, Matsuno Y, Onizuka M, Murakami Y. Hypermethylation of the TSLC1/IGSF4 promoter is associated with tobacco smoking and a poor prognosis in primary nonsmall cell lung carcinoma. Cancer. 2006;106:1751–1758. doi: 10.1002/cncr.21800. [DOI] [PubMed] [Google Scholar]
- 22.Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, Shimizu N, Shimizu K, Lee H, Chen CY, Fong KM, Gilcrease M, Roth JA, Minna JD, Gazdar AF. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 23.Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R. Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ. 2000;321:323–329. doi: 10.1136/bmj.321.7257.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 25.Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, Tamura G, Saito D, Sugimura T, Ichinose M, Ushijima T. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 26.Belinsky SA, Nikula KJ, Baylin SB, Issa JP. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci USA. 1996;93:4045–4050. doi: 10.1073/pnas.93.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vertino PM, Yen RW, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/MCB.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]


