ABSTRACT
Production of proinflammatory cytokines indicative of potent recognition by the host innate immune system has long been recognized as a hallmark of the acute phase of HIV-1 infection. The first components of the machinery by which primary HIV target cells sense infection have recently been described; however, the mechanistic dissection of innate immune recognition and viral evasion would be facilitated by an easily accessible cell line model. Here we describe that reconstituted expression of the innate signaling adaptor STING enhanced the ability of the well-established HIV reporter cell line Tzm-bl to sense HIV infection and to convert this information into nuclear translocation of IRF3 as well as expression of cytokine mRNA. STING-dependent immune sensing of HIV-1 required virus entry and reverse transcription but not genome integration. Particularly efficient recognition was observed for an HIV-1 variant lacking expression of the accessory protein Vpr, suggesting a role of the viral protein in circumventing STING-mediated immune signaling. Vpr as well as STING significantly impacted the magnitude and breadth of the cytokine mRNA expression profile induced upon HIV-1 infection. However, cytoplasmic DNA sensing did not result in detectable cytokine secretion in this cell system, and innate immune recognition did not affect infection rates. Despite these deficits in eliciting antiviral effector functions, these results establish Tzm-bl STING and Tzm-bl STING IRF3.GFP cells as useful tools for studies aimed at dissecting mechanisms and regulation of early innate immune recognition of HIV infection.
IMPORTANCE Cell-autonomous immune recognition of HIV infection was recently established as an important aspect by which the host immune system attempts to fend off HIV-1 infection. Mechanistic studies on host cell recognition and viral evasion are hampered by the resistance of many primary HIV target cells to detailed experimental manipulation. We describe here that expression of the signaling adaptor STING renders the well-established HIV reporter cell line Tzm-bl competent for innate recognition of HIV infection. Key characteristics reflected in this cell model include nuclear translocation of IRF3, expression of a broad range of cytokine mRNAs, and an antagonistic activity of the HIV-1 protein Vpr. These results establish Tzm-bl STING and Tzm-bl STING IRF3.GFP cells as a useful tool for studies of innate recognition of HIV infection.
INTRODUCTION
Virus infection triggers a wide array of immune reactions in the immunocompetent host. Many of these events involve processing of viral proteins into peptides that are presented by major histocompatibility complex (MHC) molecules. The resulting adaptive cellular and humoral immune responses are designed to eliminate productively infected cells and may neutralize infectious virus particles but take several days to weeks to develop. In contrast, innate cell-autonomous immune recognition does not require antigen presentation and allows nonspecialized target cells of an organism to quickly recognize and potentially eliminate incoming virus particles and to limit virus spread (1, 2). The cell-autonomous immune system comprises pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs) to elicit antiviral signaling cascades. Such signal transduction induces antiviral effectors, in particular type I interferons but also other cytokines, to limit virus replication in both infected and uninfected target cells. This response synergizes with intrinsic immune factors whose expression is often induced by interferon (IFN) responses (restriction factors) and that restrict virus replication in acutely infected cells via their direct physical association with viral components (3–5).
In the case of human immunodeficiency virus type 1 (HIV-1), production of proinflammatory cytokines (“cytokine storm”), indicative of potent recognition by the host innate immune system, has long been recognized as a hallmark of the acute phase of infection, anti-HIV effects of interferon have been described, and key effectors mediating this protection have been identified (6–11). Interferon-induced innate immune responses resulting from cell-autonomous recognition reduce viral replication during acute simian immunodeficiency virus (SIV) infection and attenuate subsequent disease progression (12). Moreover, polymorphisms and expression levels of innate immunity genes, including PRRs and restriction factors, affect HIV transmission rates, replication, and disease progression (13, 14). Finally, the selection of successful transmission-founder viruses with reduced sensitivity to interferon treatment of target cells suggests that evading this response enhances viral fitness in vivo (15, 16). While the relevance and effectors of cell-autonomous recognition of HIV are thus well established, much less is known about the host cell machinery that recognizes HIV infection (17, 18). The molecular events leading to innate recognition of incoming HIV genomes appear to vary remarkably between different types of target cells. In plasmacytoid dendritic cells (DCs), HIV RNA can be sensed by toll-like receptors (TLRs), in particular TLR7, resulting in the production of proinflammatory cytokines (19–22). In contrast, in myeloid cells, DNA products of HIV reverse transcription are recognized by the cyclic GMP-AMP synthase (cGAS) as a cytoplasmic DNA sensor that produces the atypical dicyclic nucleotide cGAMP to activate the central innate signaling adaptor protein STING (23–27). Finally, the interferon gamma inducible protein IFI16 was suggested to act as a cytoplasmic DNA sensor that triggers interferon production and caspase-1-dependent pyroptosis in resting CD4+ T cells and macrophages (28, 29). These responses are complemented by the sensing activities of restriction factors such as Trim5α and CD317/tetherin, which trigger innate signaling cascades upon recognition of incoming or budding viral capsid, respectively (30–33). How these different sensing mechanisms are regulated in space and time, how different effector molecules triggered by these responses affect virus replication, and which additional sensing mechanisms may exist largely remain to be elucidated.
While the action of sensing restriction factors can readily be recapitulated by overexpressing them in cell lines (30–33), the molecular dissection of innate sensing of incoming HIV particles is hampered by donor-to-donor variability, resistance to genetic manipulation, and/or low permissiveness to HIV infection of the primary cell models in which these processes can be observed. We therefore aimed here at establishing and characterizing an easily accessible cell system that mirrors aspects of cell-autonomous immune recognition of HIV-1. We describe the generation of a Tzm-bl-based HIV-1 reporter cell line in which HIV-1 infection is recognized by mechanisms that are in part counteracted by the viral protein Vpr.
MATERIALS AND METHODS
Cells, plasmids, and reagents.
HEK 293T, HeLa, HeLa Kyoto, HeLa S3, and Tzm-bl cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS) and 1% penicillin-streptomycin (all from Invitrogen) (34–38). For the generation of stable cells lines, genes of interest were cloned into pWPI derivatives generated by the modification of the multiple-cloning site (MCS) and replacement of the green fluorescent protein (GFP) in the reporter cistron with a puromycin or neo resistance gene (39), using AscI and BamHI restriction sites. The expression vector for STING was generated by introducing the coding sequence of human STING (human TMEM173, Clone Id 5762441; Open Biosystems) in the pWPI-Puro vector. Tzm-bl CON and Tzm-bl STING cells were generated by transduction of the parental Tzm-bl cells with lentivirus produced in the presence of pWPI-Puro and pWPI-hSTING vectors, respectively, and selection in the presence of 2 μg/ml puromycin (Sigma). After selection, the cells were transduced with lentivirus produced in the presence of the pWPI-Neo IRF3.GFP vector encoding an IRF3.GFP fusion protein. The parental eGFP-IRF3 plasmid was a kind gift from Luis Martinez-Sobrido (Rochester), and Tzm-bl CON IRF3.GFP and Tzm-bl STING IRF3.GFP cells were selected in the additional presence of 600 μg/ml G418. Both cell lines were positively sorted by fluorescence-activated cell sorting (FACS; BD FACSAria III cell sorter) for the expression of GFP to obtain more than 90% of the cells expressing the fusion protein.
The proviral plasmids pHIV-1NL4-3 wt (wt), pHIV-1NL4-3 Vpu stop (ΔVpu), pHIV-1NL4-3 Nef stop (ΔNef), pHIV-1NL4-3 Vif stop (ΔVif), and pHIV-1NL4-3 Env stop Nef stop (ΔΕnv) were kindly provided by Valerie Bosch. When indicated, azidothymidine (AZT; 5 μM) (Sigma-Aldrich), nevirapine (NVP; 5 μM) (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH), raltegravir (RAL; 500 μM), or T20 (50 μM) (the last two kindly provided by Oliver Keppler) was added to the cells 1 h before infection and kept present throughout the experiment to interfere with specific steps of the HIV-1 life cycle.
Virus production and infection.
For production of lentiviral vectors, 2 × 106 HEK 293T cells were seeded per 6-cm dish 24 h before transfection (40) and transfected using Pei (Sigma) with 20 μg of pWPI-Puro or pWPI-hSTING, 20 μg of pPAX2, and 6.5 μg of vesicular stomatitis virus G protein plasmid (pMD2.G). Virus supernatants were harvested after 48 h, filtered through 0.45-μm-pore-size filters, and used immediately for transduction of seeded Tzm-bl cells.
Stocks of infectious HIV-1 were generated by transfection of proviral HIV-1 plasmids into HEK 293T cells using Pei (Sigma) (41). Three days posttransfection, culture supernatants were harvested and filtered through 0.45-μm-pore-size filters, and virus stocks were concentrated by ultracentrifugation through a 20% sucrose cushion (28,000 rpm, 1.5 h). Virion-associated reverse transcriptase (RT) activity was determined by a Sybr green I PCR enhanced RT assay (SG-PERT) (42). If not mentioned otherwise, the quantity of virus used for infection was equivalent to an SG-PERT count of 1.9 × 108 pU/ml.
Cell surface receptor levels and toxicity measurement.
For the quantification of cell surface receptor levels or cell viability, Tzm-bl, Tzm-bl CON, and Tzm-bl STING cells were incubated with peridinin chlorophyll protein (PerCP)/Cy5.5–anti-human CD4 antibody (BioLegend), allophycocyanin (APC)–mouse anti-human CD184 (CXCR4), and fluorescein isothiocyanate (FITC)–mouse anti-human CD195 (both from BD Pharmingen) or 7-aminoactinomycin D (7-AAD) (2.5 μg/ml) (BD Pharmingen), respectively, for 1 h. Following several washing steps, cells were analyzed on a FACSCalibur flow cytometer, and data were analyzed with FlowJo (TreeStar) or Cyflogic (CyFlo Ltd.) software.
Immunofluorescence microscopy.
Tzm-bl CON, STING, CON IRF3.GFP, or STING IRF3.GFP cells grown on coverglasses were infected with HIV-1 WT or HIV-1 ΔVpr. At 48 hpi (hpi), the cells were fixed with 3% paraformaldehyde (PFA) prior to permeabilization and saturation in phosphate-buffered saline (PBS) containing 0.3% Triton X-100 and 5% FCS. When staining for endogenous IRF3, the primary rabbit anti-IRF3 (1:200, cell signaling) antibody was applied overnight, followed by several washing steps and incubation with goat anti-rabbit Alexa Fluor 488 and Hoechst 33258 stain (1 ng/μl) for 1 h. Cells were incubated for 30 min with Hoechst 33258 stain (1 ng/μl) prior to mounting. Images were recorded with an Olympus IX81 microscope or on a spinning-disc UltraVIEW Vox microscope (PerkinElmer) equipped with a Hamamatsu ORCA-Flash4.0 V2 camera and analyzed with ImageJ software.
Quantification of nuclear IRF3 localization.
Cells stained for endogenous IRF3 or expressing IRF3.GFP were labeled with Hoechst stain and subjected to microscopic analysis to determine the frequency of cells that display translocation of IRF3 to the nucleus. Based on their characteristic appearance in the Hoechst stain channel, mitotic cells were excluded from the analysis. Nonmitotic cells were grouped into cells with detectable IRF3 signal in the nucleus and cells in which nuclei were devoid of an IRF3 signal. At least 150 cells per condition were counted. To assess the magnitude of IRF3 translocation, the relative nuclear fluorescence intensity of IRF3 per cell was calculated by delimiting the cell and nucleus contour on a single plane of confocal images and calculating the ratio of the integrated fluorescence signal in the nucleus to that of the total cell. For illustration of IRF3 localization, profile plots of the total cell IRF3 signal along a defined cross section of a single confocal plane are presented.
Cytokine quantification.
The amounts of cytokines and chemokines present in cell culture supernatants were determined by Eve Technologies Corporation using a Discovery Assay (human cytokine array/chemokine array 42-plex). Results are expressed in picograms of cytokines/chemokines per milliliter according to the company protein standard.
Virion infectivity assays.
Tzm-bl CON and STING cells were seeded in a 96-well format (5 × 103 cells/well) 1 day before infection. The infectivity of HIV-1 was determined 48 h after infection by analysis of firefly luciferase activity (43). For detection of productively infected cells, cells were fixed with 3% PFA and incubated for 30 min with an FITC-conjugated anti-p24 antibody (Beckman Coulter) prior to analysis on a FACSCalibur cytometer. Data were analyzed with Cyflogic (CyFlo Ltd.) software.
Quantitative PCR (qPCR).
Total RNA was extracted using the NucleoSpin RNA II kit (Macherey-Nagel) and reverse transcribed using the SuperScript One-Step RT-PCR system (Life Technologies) according to the manufacturers' instructions. cDNA levels were determined by using the SYBR green PCR master mix (Life Technologies), and reactions were performed on an ABI PRISM 7500 sequence detection system (Applied Biosystems) using the following program: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was used for normalization of input RNA. RT-PCR data were analyzed by using the ΔΔCT method. The following primers were used: alpha interferon (IFN-α) forward primer (Fwd), 5′-GCCTCGCCCTTTGCTTTACT-3′, and IFN-α reverse primer (Rev), 5′-CTGTGGGTCTCAGGGAGATCA-3′; IFN-β Fwd, 5′-CGCCGCATTGACCATCTA-3′, and IFN-β Rev, 5′-GACATTAGCCAGGAGGTTCTC-3′; IFN-γ Fwd, 5′-CTTCCACAGGATCACTGTGTACCT-3′, and IFN-γ Rev, 5′-TTCTGCTCTGACCACCTCCC-3′; interleukin-1α (IL-1α) Fwd, 5′-TGGTAGTAGCAACCAACGGGA-3′, and IL-1α Rev, 5′-ACTTTGATTGAGGGCGTCATTC-3′; IL-1β Fwd, 5′-ACAGATGAAGTGCTCCTTCCA-3′, and IL-1β Rev, 5′-GTCGGAGATTCGTAGCTGGAT-3′; IL-2 Fwd, 5′-ATGAGACAGCAACCATTGTAGAATTT-3′, and IL-2 Rev, 5′-CACTTAATTATCAAGTCAGTGTTGAGATGA-3′; IL-4 Fwd 5′-AGATCATCGGCATTTTGAACG-3′, and IL-4 Rev, 5′-TTTGGCACATCCATCTCCG-3′; IL-5 Fwd, 5′-AGCTGCCTACGTGTATGCCA-3′, and IL-5 Rev, 5′-GCAGTGCCAAGGTCTCTTTCA-3′; IL-6 Fwd, 5′-CCAGAAACCGCTATGAAGTTCC-3′, and IL-6 Rev, 5′-TCACCAGCATCAGTCCCAAG-3′; IL-8 Fwd, 5′-ATGACTTCCAAGCTGGCCGTGGCT-3′, and IL-8 Rev, 5′-TCTCAGCCCTCTTCAAAAACTTCTC-3′; IL-10 Fwd, 5′-GACTTTAAGGGTTACCTGGGTTG-3′, and IL-10 Rev, 5′-TCACATGCGCCTTGATGTCTG-3′; IL-13 Fwd, 5′-GCTTATTGAGGAGCTGAGCAACA-3′, and IL-13 Rev, 5′-GGCCAGGTCCACACTCCATA-3′; tumor necrosis factor alpha (TNF-α) Fwd, 5′-CCTCTCTCTAATCAGCCCTCTG-3′, and TNF-α Rev, 5′-GAGGACCTGGGAGTAGATGAG-3′; transforming growth factor beta (TGF-β) Fwd, 5′-CCCGAAGCGGACTACTATGC-3′, and TGF-β Rev 5′-ATAGATGGCGTTGTTGCGGT-3′; interferon-stimulated gene 56 (ISG56) Fwd, 5′-GAAGCAGGCAATCACAGAAA-3′, and ISG56 Rev, 5′-TGAAACCGACCATAGTGGAA-3′; GAPDH Fwd 5′-GAAGGTGAAGGTCGGAGTC-3′, and GAPDH Rev 5′-GAAGATGGTGATGGGATTTC-3′.
Transfection of herring DNA.
Tzm-bl CON and Tzm-bl STING cells were seeded in 24-well plates (7 × 104 cells/well). The following day, the cells were treated with 2 μl Lipofectamine 2000 transfection reagent (Life Technologies) alone or with 1 μg DNA sodium salt from herring testes (Sigma) per well. The medium was replaced 4 h posttransfection, and 24 h posttransfection, the cells were lysed for RNA extraction or prepared for 7-AAD labeling.
Western blotting.
Cells were lysed in 2× SDS lysis buffer and boiled. Cleared lysates equivalent to 5 × 104 cells were separated on 12% SDS-PAGE and blotted onto nitrocellulose membranes. Blocked membranes were probed with the following primary antibodies: mouse anti-IFI16 monoclonal antibody, mouse anti-DDX41 monoclonal antibody (Santa Cruz), rabbit anti-TMEM173 (STING) polyclonal antibody, rabbit anti-MB21D1 (cGAS) polyclonal antibody, rabbit anti-PQBP1 polyclonal antibody (Sigma), and mouse anti-GAPDH monoclonal antibody (Santa Cruz). Secondary antibodies conjugated to horseradish peroxidase were used for enhanced chemiluminescence (ECL)-based detection.
Statistical analysis.
Statistical analysis of data sets was carried out using Prism6 software (GraphPad, Inc.). Statistical significance was analyzed by Student's t test.
RESULTS
Tzm-bl cells lack detectable expression of STING.
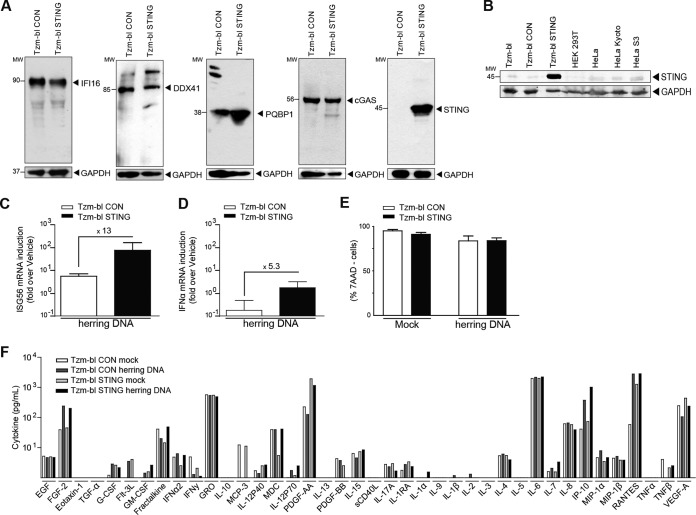
The goal of this study was to establish an accessible cell culture model competent for cell-autonomous sensing of HIV-1 infection and suitable for high-throughput analyses. Due to their high permissiveness to HIV infection, their ability to quantify productive infection events via an integrated long terminal repeat (LTR)-luciferase reporter, and their suitability for high-throughput screening, we focused on Tzm-bl cells (34–36). The reported ability of Tzm-bl cells or other derivatives of the parental HeLa cells for sensing HIV infection, as evidenced by the induction of cytokine mRNA, range from a mild induction upon HIV infection to a complete lack thereof (44, 45). Since mRNA levels of ISG56 or IFN-β, two genes often used as markers for effective immune recognition (46–48), were not increased upon infection with HIV in our Tzm-bl cells (data not shown), we first tested whether factors that have been implicated in the cell-autonomous recognition of HIV infection were expressed. Western blotting revealed robust expression of the cytoplasmic DNA sensors reported to recognize HIV reverse transcription products, IFI16, DDX41 (49), and cGAS (Fig. 1A, Tzm-bl CON), as well as PQBP1, which was recently identified as an adaptor between HIV-1 DNA and cGAS (50). In contrast, expression of STING, an essential adaptor for innate signaling downstream of cGAS (27, 51, 52), was weak or undetectable on the protein level (Fig. 1A) in parental Tzm-bl cells, various other HeLa derivatives, or HEK 293T cells (Fig. 1B). These low levels of STING expression in a range of cell lines were surprising considering that HeLa and HEK 293T cells have been described as expressing robust levels of the signaling adaptor (53). Since basal levels of STING expression in HEK 293T cells were reported as insufficient for many aspects of DNA sensing and this defect could be compensated by stable expression of ectopic STING (24), we generated lentiviral vectors encoding STING or an empty vector control and established stable bulk cell lines by transduction and selection. Tzm-bl CON and Tzm-bl STING cells maintained robust levels of IFI16, DDX41, cGAS, and PQBP1, and significant amounts of STING protein were detected in Tzm-bl STING cells (Fig. 1A). To test whether expression of STING resulted in sensing of cytoplasmic DNA, cells were transfected with herring DNA and induction of ISG56 (Fig. 1C) or IFN-α (Fig. 1D) mRNA was assessed. While at best mild mRNA induction was observed in Tzm-bl CON cells, ISG56 and IFN-α mRNA induction levels were increased by 13- and 5-fold in Tzm-bl STING cells, respectively, without compromising cell viability (Fig. 1E). Quantification of 42 cytokines/chemokines in the supernatant of Tzm-bl CON or Tzm-bl STING cells 24 h after mock transfection or transfection with herring DNA, however, revealed that cytoplasmic DNA sensing does not trigger enhanced cytokine release (Fig. 1F).
FIG 1.
Expression of STING reconstitutes sensing of cytoplasmic DNA in Tzm-bl cells. (A) Tzm-bl cells were transduced with CON- or STING-expressing lentiviral vectors and selected as bulk cultures. Expression of IFI16, DDX41, PQBP1, cGAS, and STING was assessed in Tzm-bl CON and Tzm-bl STING cells by Western blotting. GAPDH serves as a loading control. (B) The expression of STING was assessed in Tzm-bl, Tzm-bl CON, Tzm-bl STING, HEK 293T, HeLa, HeLa Kyoto, and HeLa S3 cells by Western blotting. GAPDH serves as a loading control. (C, D, and E) At 24 h posttransfection with herring DNA, ISG56 mRNA (C), IFN-α mRNA (D), and cell viability (E) were quantified. ISG56 and IFN-α mRNA levels were quantified by qPCR and normalized to GAPDH mRNA. Data are expressed as fold increases relative to results for mock-transfected cells. Cell viability was measured by flow cytometry following 7-AAD labeling. Shown is the percentage of 7-AAD-negative cells. Values are averages ± SD of the results of three experiments. (F) TZM-bl CON and STING cells were transfected with herring DNA, and 24 h posttransfection, the levels of 42 cytokines/chemokines in the cell culture supernatant were determined. Results are expressed in picograms per milliliter. Molecular weight (MW) markers are indicated in thousands.
Expression of STING increases induction of ISG56 mRNA in Tzm-bl cells in response to HIV-1 infection.
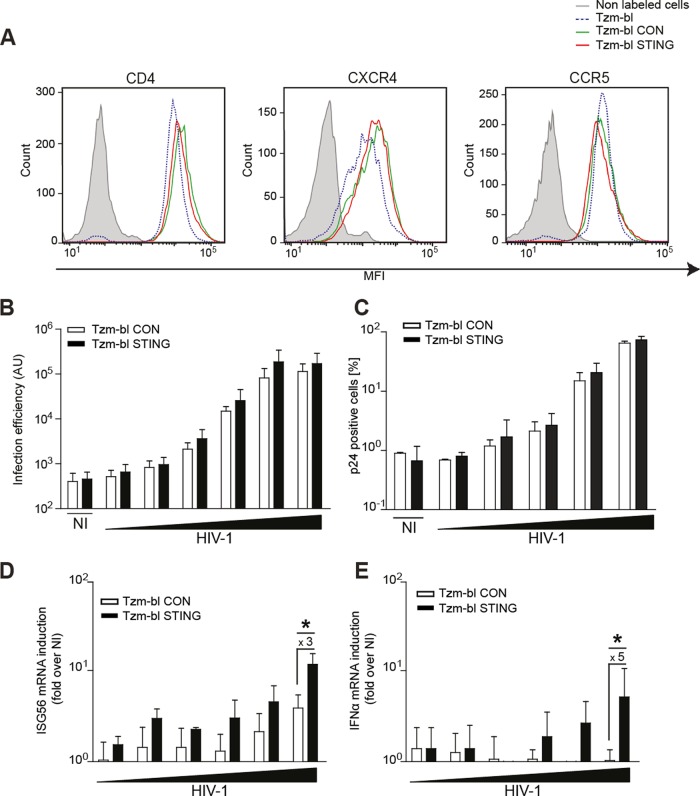
We next tested whether the expression of STING enabled Tzm-bl cells to sense HIV-1 infection. Cell surface levels of the viral entry receptor CD4 and the coreceptors CXCR4 and CCR5 were comparable between Tzm-bl CON and STING cells (Fig. 2A). Consistently, both cell lines were equally permissive to infection with HIV-1 NL4-3, resulting in a dose-dependent increase of luciferase expressed from the LTR reporter present in Tzm-bl cells (Fig. 2B) or in the amount of productively infected cells, as judged by intracellular p24 staining (Fig. 2C). Of note, luciferase activity plateaued at approximately 105 arbitrary units (AU). Intracellular p24 staining in turn revealed that infection with the two highest doses of virus input resulted in 20% ± 9% and 73% ± 11% productively infected Tzm-bl STING cells, respectively, and similar infection rates were obtained in Tzm-bl CON cells. Parallel quantification of ISG56 mRNA levels revealed that at high doses of virus input with which over 50% of cells were productively infected, the levels of ISG56 mRNA induced in Tzm-bl STING cells were approximately 3-fold higher than the mild induction in Tzm-bl CON cells (Fig. 2D). Similarly, IFN-α mRNA was induced approximately 5-fold in Tzm-bl STING cells upon HIV-1 infection (Fig. 2E). Expression of STING in Tzm-bl cells thus facilitates the induction of innate immunity genes in response to HIV-1 infection.
FIG 2.
STING expression in Tzm-bl cells elevates ISG56 mRNA induction by HIV-1 but does not affect infection rates. (A) Cell surface expression levels of HIV-1 entry receptors. Cells were labeled with fluorophore-conjugated antibodies against CD4, CCR5, and CXCR4 and subjected to FACS analysis. Results are representative of three experiments. (B, C, D, and E) Cells were infected with a logarithmic serial dilution (1 to 10−4 or 1 to 10−5) of HIV-1 WT (1.9 × 108 pU/ml), and 48 hpi, the infection efficiency was determined by luminescence (B) and by flow cytometry following intracellular p24 labeling (C). In parallel, ISG56 mRNA (D) and IFN-α mRNA (E) levels were quantified. Data are expressed as fold increases relative to results for noninfected (NI) cells. Values are averages ± SD of the results of three independent experiments. *, P < 0.05.
HIV-1 ΔVpr triggers more potent antiviral gene expression than HIV-1 WT in Tzm-bl STING cells.
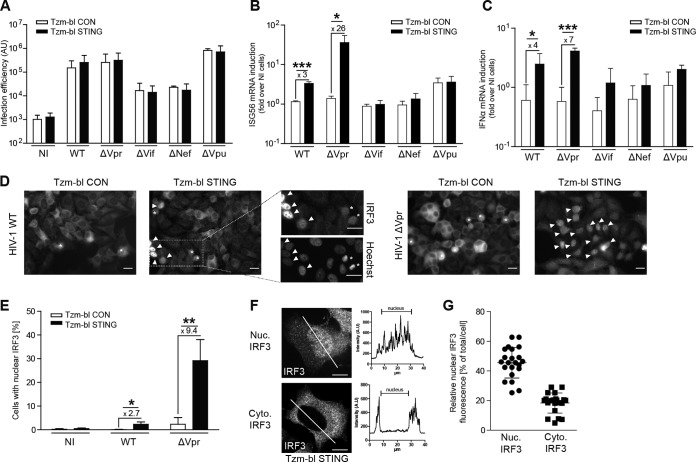
Although HIV-1 WT infection triggered ISG56 mRNA production in Tzm-bl STING cells, the magnitude of this effect was limited. To examine whether expression of a viral antagonist of cell-autonomous sensing restricted this response, we tested a panel of isogenic HIV-1 variants, each lacking the expression of one of the accessory genes, vpr, vif, nef, or vpu. Expectedly, the lack of functional vpr or vpu genes did not impact the efficiency of HIV-1 infection. As expected from the ability of the Nef and Vif proteins to enhance virion infectivity (54, 55), disruption of vif or nef substantially reduced infection rates in Tzm-bl CON and Tzm-bl STING cells (Fig. 3A). The absence of Vif, Nef, or Vpu did not significantly alter the expression of ISG56 (Fig. 3B) or IFN-α (Fig. 3C) mRNA in response to infection in the two cell lines. In contrast, infection of Tzm-bl STING cells with HIV-1 ΔVpr triggered a marked upregulation of both mRNAs in comparison to infection of Tzm-bl CON cells (26 ± 9-fold for ISG56, 7 ± 1-fold for IFN-α). This enhanced recognition of HIV-1 ΔVpr in Tzm-bl STING cells was not associated with an increase in cell toxicity (data not shown). Similar results were obtained when using nuclear translocation of IRF3, a central transcription factor driving expression of cytokine genes downstream of STING (56, 57) as the readout for cell-autonomous sensing (Fig. 3D to G). Parallel Hoechst staining allowed discrimination of cells with nuclear translocation of IRF3 from mitotic cells (Fig. 3D). While nuclear translocation of IRF3 was absent in Tzm-bl CON cells and HIV-1 WT targeted IRF3 to the nucleus in only a small fraction of Tzm-bl STING cells, up to 30% of Tzm-bl STING cells displayed pronounced nuclear localization of IRF3 upon infection with HIV-1 ΔVpr (Fig. 3E). In cells with nuclear IRF3, over 45% ± 10% of the total IRF3 signal was nuclear, while only around 18% ± 7% of the total IRF3 signal per cell was found in the nucleus of cells without apparent IRF3 translocation (Fig. 3F and G).
FIG 3.
Vpr reduces sensing of HIV-1 infection in Tzm-bl STING cells. (A to C) Tzm-bl CON or Tzm-bl STING cells were infected with 1.9 × 108 pU/ml of the indicated viruses and analyzed for infection efficiency (A) and induction of ISG56 (B) or IFN-α (C) mRNA 48 hpi. Values are averages ± SD of the results of three independent experiments. (D to F) Subcellular localization of IRF3 in Tzm-bl CON or Tzm-bl STING cells. (D) Representative wide-field micrographs of the indicated cells after immunofluorescence staining for endogenous IRF3. Arrowheads indicate cells with prominent nuclear translocation of IRF3, and asterisks denote mitotic cells in which IRF3 appears nuclear due to breakdown of the nuclear envelope, as identified by parallel Hoechst staining (see zoom magnifications of the indicated box on the right). (E) Percentage of cells with nuclear IRF3 as judged by immunofluorescence as shown in panel D in noninfected cells (NI) or 48 hpi with HIV-1 WT (WT) or HIV-1 ΔVpr (ΔVpr). Cells were considered positive for nuclear IRF3 when a nuclear signal could clearly be detected. Values are averages ± SD of the results of four independent experiments with at least 150 cells analyzed each per condition. (F) Nuclear localization of IRF3 in Tzm-bl STING cells after HIV-1 ΔVpr infection. Shown are representative single-plane confocal micrographs of IRF3 in cells classified as with (Nuc. IRF3) or without (Cyt. IRF3) pronounced nuclear IRF3 localization. Panels on the right show profile plot representations of the IRF3 signal intensity along the cross section of the cell, which is indicated by a white line. (G) Quantification of the nuclear fluorescence intensity relative to the total IRF3 fluorescence intensity in individual cells with and without apparent nuclear IRF3 staining (left and right columns, respectively). At least 20 cells per condition from three independent experiments are represented, with the mean ± SD indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001 Scale bars = 20 μm.
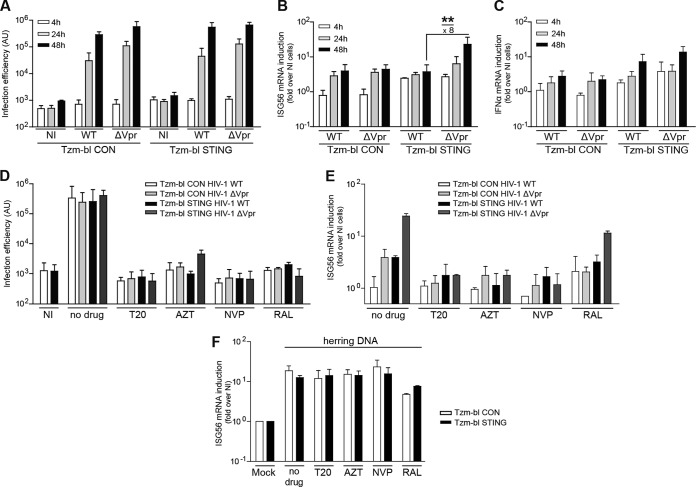
We next assessed the kinetics of infection and cell-autonomous sensing for HIV-1 WT and HIV-1 ΔVpr on Tzm-bl CON and STING cells. Robust infection was detected 24 hpi with both viruses on both target cells, and these levels of infection were slightly increased an additional 48 hpi (Fig. 4A). At the 24 hpi time point, ISG56 mRNA levels were induced 3- to 5-fold over those in noninfected cells independently of the virus and target cell used. In contrast, at 48 hpi, an additional significant induction of ISG56 mRNA was observed in Tzm-bl STING cells with HIV-1 lacking Vpr (Fig. 4B; HIV-1 ΔVpr versus HIV-1 WT at 48 hpi, P = 0.0027). Similarly, IFN-α mRNA was induced 48 hpi, but the requirement for deletion of Vpr was less pronounced than for ISG56 (Fig. 4C). This late induction of mRNA induction was consistent with that of IRF3 nuclear translocation induced by HIV-1 ΔVpr, which was first apparent 34 hpi and most pronounced 48 hpi (data not shown). These results suggest that the sensing of HIV-1 infection that leads to nuclear translocation of IRF3 and the expression of ISG56 mRNA in Tzm-bl cells rely on STING for downstream signaling and are more efficient in the absence of Vpr.
FIG 4.
Vpr reduces the sensitivity of HIV-1 for innate immune recognition in Tzm-bl STING cells. (A to C) Tzm-bl CON or Tzm-bl STING cells were infected with HIV-1 WT or HIV-1 ΔVpr and analyzed for infection efficiency (A) and induction of ISG56 mRNA (B) or IFN-α mRNA (C) at 4, 24, and 48 hpi. (D and E) Tzm-bl CON or Tzm-bl STING cells were infected with HIV-1 WT or HIV-1 ΔVpr in the absence of drugs (no drug) or in the presence of T20 (50 μM), AZT (5 μM), nevirapine (NVP) (5 μM), or raltegravir (RAL) (500 μM). Infection efficiency (D) and ISG56 mRNA induction (E) were analyzed 48 hpi. (F) Tzm-bl CON or Tzm-bl STING cells were transfected with herring DNA before incubation in the absence of drugs (no drug) or in the presence of T20 (50 μM), AZT (5 μM), NVP (5 μM), or RAL (500 μM). At 24 h posttransfection, ISG56 mRNA was quantified and normalized to GAPDH mRNA. Data are expressed as fold increases relative to results for mock-transfected cells. Values are averages ± SD of the results of three independent experiments. **, P < 0.01.
Sensing of HIV-1 ΔVpr in Tzm-bl STING cells depends on virus entry and reverse transcription but not on genome integration.
We next sought to determine where in the viral life cycle Vpr-sensitive, STING-dependent recognition occurs and examined the effect of anti-HIV drugs that arrest virus replication at defined steps. With same variability in efficiency, all drugs markedly reduced infection rates close to background levels at the concentrations used (Fig. 4D). Inhibition of virus entry (by the fusion inhibitor T-20) or reverse transcription (by the reverse transcriptase inhibitors azidothymidine [AZT] or nevirapine [NVP]) also abrogated the upregulation of ISG56 mRNA in Tzm-bl STING cells by HIV-1 ΔVpr (Fig. 4E). In contrast, induction of ISG56 mRNA by HIV-1 ΔVpr in Tzm-bl STING cells was preserved, albeit at slightly reduced levels, when the integration of viral DNA into the host genome was prevented by the integrase inhibitor raltegravir (RAL). T20, AZT, and NVP had no effect on ISG56 mRNA induction following transfection of herring DNA, while RAL had a slight inhibitory effect (Fig. 4F). These results place Vpr-sensitive, STING-dependent recognition of HIV infection between reverse transcription and integration in the HIV life cycle, a result that is consistent with recognition of cytoplasmic viral DNA. RAL seems to exert moderate inhibitory effects on such cytoplasmic DNA sensing events.
STING and Vpr affect the magnitude and specificity of the cytokine mRNA profile triggered by cell-autonomous sensing of HIV-1.
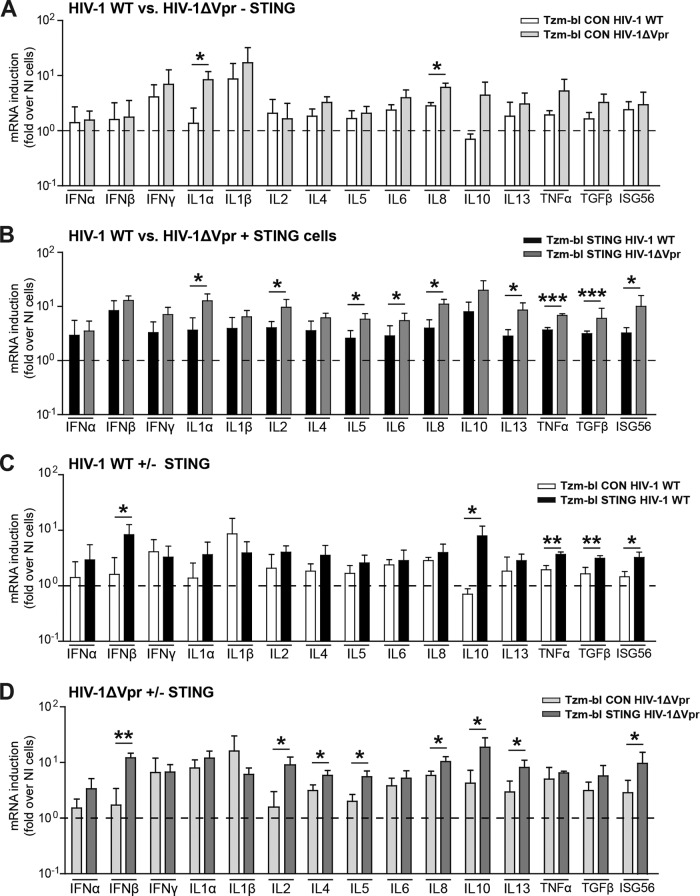
To characterize the scope of effectors triggered by sensing of HIV-1 in our cell models, we next quantified the mRNA levels of a set of cytokines in Tzm-bl CON or STING cells 48 h following infection with HIV-1 WT or HIV-1 ΔVpr (Fig. 5). This analysis revealed that only a few cytokine mRNAs (IFN-γ, IL-1β) were induced up to 10-fold by HIV-1 WT infection in Tzm-bl CON cells, and this effect was only slightly more pronounced with HIV-1 ΔVpr (Fig. 5A). Presumably due to differences in levels in activity and/or expression of host cell sensing machinery between the HeLa-derived cell lines used, we did not observe the slight induction of IFN-α and IFN-β mRNA described recently (44). For other cytokine mRNAs, significant induction was observed only upon infection with HIV-1 ΔVpr, not with HIV-1 WT (statistically significant for IL-1α and IL-8, tendency for IL-10 and TNF-α). In Tzm-bl STING cells (Fig. 5B), cytokine mRNA induction in response to infection with HIV-1 WT was broader and more pronounced than in Tzm-bl CON cells, and this responsiveness was significantly boosted upon infection with HIV-1 ΔVpr. Surprisingly included (among IL-1α, IL-2, IL-5, IL-6, IL-8, IL-13, TNF-α, TGF-β, and ISG56) a number of cytokines typically associated with adaptive T lymphocyte immune functions (e.g., IL-2, IL-13). Pairwise plotting of these results for HIV-1 WT in Tzm-bl CON versus Tzm-bl STING cells (Fig. 5C) illustrated that the presence of STING emphasized the induction of IFN-β, IL-10, TNF-α, and TGF-β. The same analysis for HIV-1 ΔVpr revealed a similar STING-dependent induction of IFN-β and IL-10 mRNA but also of various other cytokine mRNAs (IL-2, IL-4, IL-5 IL-8, IL-13, and ISG56) (Fig. 5D). Together, these results demonstrate that HIV-1 infection triggers innate immune signaling in Tzm-bl cells and that expression of STING increases the breadth and magnitude of this response. Vpr appears to limit cytokine mRNA induction in both the presence and absence of STING in the cells, but only for selected, not all, cytokines.
FIG 5.
STING and Vpr modulate the magnitude and specificity of the cytokine mRNA expression profile induced by HIV-1 infection. Tzm-bl CON or Tzm-bl STING cells were infected with HIV-1 WT or HIV-1 ΔVpr, and cytokine mRNA levels were determined 48 h later. Four histograms are presented for comparison of two conditions each. (A) HIV-1 WT versus HIV-1 ΔVpr in Tzm-bl CON cells; (B) HIV-1 WT versus HIV-1 ΔVpr in Tzm-bl STING cells; (C) Tzm-bl CON versus STING cells infected by HIV-1 WT; (D) Tzm-bl CON versus STING cells infected by HIV-1 ΔVpr. Data are expressed as fold increases relative to results for noninfected cells. Values are averages ± SD of the results of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
A Tzm-bl STING variant expressing IRF3.GFP.
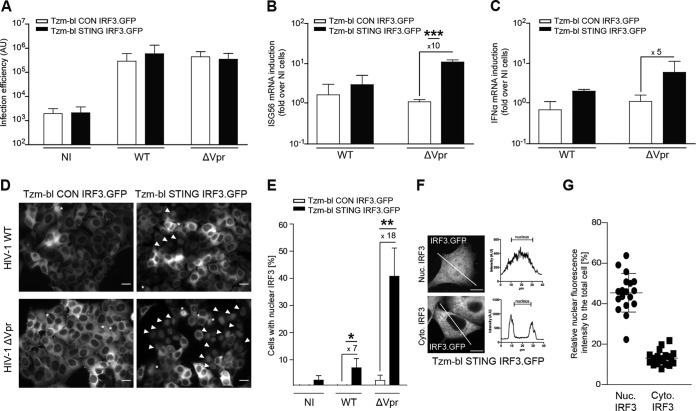
While the above results established Tzm-bl STING cells as a model system in which innate immune recognition of HIV-1 infection and the efficiency of infection can easily be quantified, single-cell visualization of innate immune recognition was restricted to immunostaining of fixed cells. We therefore generated derivatives of Tzm-bl CON as well as Tzm-bl STING cells that stably expressed IRF.GFP (Tzm-bl CON IRF.GFP and Tzm-bl STING IRF.GFP). Expression of IRF.GFP did not affect infection rates of HIV-1 WT or HIV-1 ΔVpr (Fig. 6A), and HIV-1 ΔVpr induced ISG56 and IFN-α mRNA in Tzm-bl STING IRF3.GFP cells to an extent similar to that observed before in the absence of IRF.GFP (Fig. 6B and C). Translocation of IRF3.GFP to the cell nucleus was observed in few cells upon infection with HIV-1 WT (7% ± 4%) but was much more pronounced when using HIV-1 ΔVpr for infection (41% ± 10%) (Fig. 6D and E), and the magnitude of IRF3.GFP redistribution in cells with translocation to the nucleus was comparable to that of endogenous IRF3 (Fig. 6F and G). These results establish Tzm-bl STING IRF3.GFP cells as a convenient reporter system for the parallel quantification of HIV infection and innate immune recognition thereof.
FIG 6.
Innate immune recognition of HIV-1 ΔVpr infection in Tzm-bl STING IRF3.GFP cells. (A to C) Tzm-bl CON IRF3.GFP or Tzm-bl STING IRF3.GFP cells were infected with 1.9 × 108 pU/ml of the indicated viruses and analyzed for infection efficiency (A) and induction of ISG56 mRNA (B) or IFN-α mRNA (C) 48 hpi. Values are averages ± SD of the results of three independent experiments. (D to F) Nuclear localization of IRF3 in Tzm-bl CON IRF3.GFP or Tzm-bl STING IRF3.GFP cells. (D) Representative wide-field micrographs of the indicated cells. Arrowheads indicate cells with prominent nuclear translocation of IRF3.GFP, and asterisks denote mitotic cells in which IRF3.GFP appears nuclear due to the breakdown of the nuclear envelope. (E) Percentage of cells with nuclear IRF3.GFP in noninfected cells (NI) or 48 hpi with HIV-1 WT (WT) or HIV-1 ΔVpr (ΔVpr). Values are averages ± SD of the results of three independent experiments with at least 150 cells analyzed each per condition. (F) Nuclear localization of IRF3.GFP in Tzm-bl STING IRF3.GFP cells after HIV-1 ΔVpr infection. Shown are representative single-plane confocal micrographs of IRF3.GFP in cells with (Nuc. IRF3) or without (Cyt. IRF3) pronounced nuclear IRF3.GFP localization. Panels on the right show profile plot representations of IRF3 signal intensity along the cross section of the cell, which is indicated by a white line. (G) Quantification of the nuclear fluorescence intensity relative to the total IRF3.GFP fluorescence intensity in individual cells with and without apparent nuclear IRF3 staining (left and right columns, respectively). At least 20 cells per condition from three independent experiments are represented, with the mean ± SD indicated. Scale bars = 20 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
We describe here that reconstituted expression of the innate signaling adaptor STING enhances the ability of Tzm-bl cells to sense HIV infection and to convert this information into expression of cytokine mRNA. STING-dependent sensing of HIV-1 was particularly efficient for an HIV-1 variant lacking expression of the accessory protein Vpr, suggesting an important role of the viral protein in circumventing such recognition. Induced cytokine expression depended on virus entry and reverse transcription but not on genome integration, indicating that reverse transcription DNA products trigger this innate immune response. Finally, Vpr as well as STING had significant impacts on the magnitude and breadth of the cytokine mRNA expression profile induced upon HIV-1 infection.
Together these studies establish Tzm-bl STING cells as a convenient cell culture model for studies of innate immune recognition of HIV-1 in which important aspects of sensing but also viral evasion thereof are recapitulated. Induction of cytokine mRNAs in response to infection was significantly enhanced by reconstituted expression of STING and use of HIV-1 ΔVpr for infection and was in magnitude almost comparable to that observed upon infection of primary HIV target cells (45) (58). However, the Tzm-bl STING model clearly still has important limitations in its present form. While cytoplasmic DNA sensing triggered the expression of cytokine mRNA, this did not result in enhanced release of these cytokines to the cell culture supernatant. In addition, the induction of downstream targets of IRF3 such as IFN-α and -β is moderate despite the efficient translocation of the transcription factor in cells infected with HIV-1 ΔVpr. Similar to the parental HeLa cells (59), the Tzm-bl-based cell systems described here thus bear important defects in the signaling events downstream of innate sensing that lead to increased production and/or secretion of cytokines. The molecular nature of these defects remains to be elucidated. This lack of production of potentially antiviral cytokines likely explains why innate immune recognition did not limit HIV infection in our experiments. Even though we did not formally exclude effects of innate immune sensing on uninfected bystander cells, the lack of cytokine secretion suggests that the mRNA induction observed is a consequence of cell-autonomous innate recognition events. Tzm-bl STING cells thus provide a convenient tool for the analysis of early events in innate immune recognition of HIV infection but clearly do not yet mimic any of the scenarios HIV encounters in relevant primary target cells. Future efforts will be targeted at further reconstituting Tzm-bl STING cells to not only sense HIV-1 infection but also monitor the potentially protective effects of this response. Such attempts to render Tzm-bl-derived cells more similar to natural target cells with respect to HIV sensing and antiviral response will likely prove useful for the identification of essential components of the involved host cell machinery. Currently, these cells will facilitate high-throughput as well as biochemistry-based and visualization approaches to further identify and dissect the molecular regulation and dynamics of innate sensing of HIV infection and viral counteraction. In addition, they may serve as a valuable tool for the identification of relevant components of the effector machinery triggered by these recognition events.
While we did not specifically address the molecular mechanisms underlying sensing in this system, the requirement for STING, the lack of cell death associated with sensing, and the induction of IRF3 translocation to the nucleus, as well as the mapping of the recognition event to a step between reverse transcription and integration, are consistent with sensing via the cGAS-cGAMP-STING module. However, low levels of cytokine mRNA induction were already observed in Tzm-bl CON cells in response to HIV-1 infection, which may reflect the activity of undetectable background levels of STING expression and/or the involvement of STING-independent pathways. Similarly, STING and Vpr impacted the mRNA profile induced in infected cells, indicating the presence of multiple recognition pathways or of differential thresholds for triggering select effectors downstream of the same recognition event. Based on the currently limited antiviral potency of the innate response triggered by HIV-1 infection of these cells, it would be premature to speculate about the potential biological consequences of these different cytokine mRNA signatures, but the current model may facilitate the assessment of interconnections between individual recognition events.
Another aspect of this study relates to the ability of Vpr to limit innate sensing of HIV-1 infection in the Tzm-bl STING cell model. Vpr, a 14-kDa multifunctional viral protein that is efficiently packaged into virus particles, has been implicated in the regulation of apoptosis of HIV-infected cells, the efficiency and accuracy of the reverse transcription process, and the nuclear import of viral genomes (60, 61). Vpr has been most intensively studied for its ability to arrest the cell cycle of HIV target cells in G2 (62–68). Consistent with our results, several laboratories have previously reported that infection of several cell types with Vpr-negative viruses provokes more potent cytokine responses than infection with HIV WT (69–72). Together, these results suggest Vpr as an antagonist of innate sensing of HIV-1 infection. The recent landmark study by Laguette and colleagues suggested a first molecular explanation for this activity by unraveling the interaction of Vpr with SLX4, a nuclease scaffold with important roles in DNA damage and repair (44, 73). Via this interaction, Vpr prematurely activates SLX4 complexes, which results in cell cycle arrest but also reduces the proinflammatory cytokine response to HIV infection (44, 59, 74). As speculated by these authors, the effect of Vpr on cytokine induction may reflect a reduction of viral genomes available for sensing that results from triggered endonuclease activity of the SLX4 complex. Following virus entry, these genomes remain packaged together with Vpr within subviral cores that uncoat and deliver viral genomes to the nucleus via poorly defined mechanisms (60, 75, 76). An alternative and not mutually exclusive explanation for this shaping of innate immune recognition of HIV infection is that Vpr regulates the accessibility of viral genomes to cytoplasmic DNA sensors. This may reflect the effects of Vpr on core stability, on which transport pathway is used by incoming subviral cores, or on the regulation of the abundance of host cell sensing machinery, activities that likely involve Vpr's ability to target cargo molecules to proteasomal degradation (44, 77–81). Irrespective of the underlying molecular mechanisms, it is conceivable that an increase in the amount and/or accessibility of viral DNA enhances recognition by cytoplasmic sensors such as cGAS, which would explain the involvement of STING in the reduction of innate recognition by Vpr.
Together, we here describe Tzm-bl STING and Tzm-BL IRF3.GFP cells as an informative and accessible model system to quantify and mechanistically dissect innate recognition of HIV-1 that will also be useful for studies on the molecular mechanisms by which the viral protein Vpr evades such recognition.
ACKNOWLEDGMENTS
We are grateful to Valerie Bosch and Oliver T. Keppler for the kind gift of reagents, Sakshi Arora for comments on the manuscript, the FACS sorting facility at the ZMBH Heidelberg for cell sorting, Vibor Laketa (Infectious Diseases Imaging Platform at the Department of Infectious Diseases, University Hospital Heidelberg) for expert microscopy support, and the NIH AIDS Reagent Program for reagents.
This project was supported by the BMBF consortium ImmunoQuant (Teilprojekt Q 0316170C) and by the German Center for Infection Research (project 8.2 TTU HIV). O.T.F. is a member of the cluster of excellence Cellnetworks (EXC81).
REFERENCES
- 1.Randow F, MacMicking JD, James LC. 2013. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science 340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacMicking JD. 2012. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol 12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz PD. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol 5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 5.Yan N, Chen ZJ. 2012. Intrinsic antiviral immunity. Nat Immunol 13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen MS, Shaw GM, McMichael AJ, Haynes BF. 2011. Acute HIV-1 infection. N Engl J Med 364:1943–1954. doi: 10.1056/NEJMra1011874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang JJ, Altfeld M. 2010. Innate immune activation in primary HIV-1 infection. J Infect Dis 202(Suppl 2):S297–S301. doi: 10.1086/655657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C. 2013. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, Hue S, Barclay WS, Schulz R, Malim MH. 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, del Prete GQ, Hill BJ, Timmer JK, Reiss E, Yarden G, Darko S, Contijoch E, Todd JP, Silvestri G, Nason M, Norgren RB Jr, Keele BF, Rao S, Langer JA, Lifson JD, Schreiber G, Douek DC. 2014. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds MR, Weiler AM, Piaskowski SM, Kolar HL, Hessell AJ, Weiker M, Weisgrau KL, Leon EJ, Rogers WE, Makowsky R, McDermott AB, Boyle R, Wilson NA, Allison DB, Burton DR, Koff WC, Watkins DI. 2010. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J Virol 84:9190–9199. doi: 10.1128/JVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh WW, Rao SS, Lim SY, Zhang J, Hraber PT, Brassard LM, Luedemann C, Todd JP, Dodson A, Shen L, Buzby AP, Whitney JB, Korber BT, Nabel GJ, Mascola JR, Letvin NL. 2011. The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J Virol 85:10389–10398. doi: 10.1128/JVI.00854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, Shaw GM, Hahn BH, Ochsenbauer C, Kappes JC, Borrow P. 2013. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Towers GJ, Noursadeghi M. 2014. Interactions between HIV-1 and the cell-autonomous innate immune system. Cell Host Microbe 16:10–18. doi: 10.1016/j.chom.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki A. 2012. Innate immune recognition of HIV-1. Immunity 37:389–398. doi: 10.1016/j.immuni.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lepelley A, Louis S, Sourisseau M, Law HK, Pothlichet J, Schilte C, Chaperot L, Plumas J, Randall RE, Si-Tahar M, Mammano F, Albert ML, Schwartz O. 2011. Innate sensing of HIV-infected cells. PLoS Pathog 7:e1001284. doi: 10.1371/journal.ppat.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N. 2005. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest 115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chattergoon MA, Latanich R, Quinn J, Winter ME, Buckheit RW III, Blankson JN, Pardoll D, Cox AL. 2014. HIV and HCV activate the inflammasome in monocytes and macrophages via endosomal Toll-like receptors without induction of type 1 interferon. PLoS Pathog 10:e1004082. doi: 10.1371/journal.ppat.1004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier A, Chang JJ, Chan ES, Pollard RB, Sidhu HK, Kulkarni S, Wen TF, Lindsay RJ, Orellana L, Mildvan D, Bazner S, Streeck H, Alter G, Lifson JD, Carrington M, Bosch RJ, Robbins GK, Altfeld M. 2009. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med 15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, El Marjou A, Lacabaratz C, Lelievre JD, Manel N. 2013. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Tengchuan J, Laustsen A, Hansen K, Ostergaard L, Fitzgerald KA, Xiao TS, Mikkelsen JG, Mogensen TH, Paludan SR. 2013. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci U S A 110:E4571-E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. 2014. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galao RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. 2012. Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell Host Microbe 12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hotter D, Sauter D, Kirchhoff F. 2013. Emerging role of the host restriction factor tetherin in viral immune sensing. J Mol Biol 425:4956–4964. doi: 10.1016/j.jmb.2013.09.029. [DOI] [PubMed] [Google Scholar]
- 33.Cocka LJ, Bates P. 2012. Identification of alternatively translated Tetherin isoforms with differing antiviral and signaling activities. PLoS Pathog 8:e1002931. doi: 10.1371/journal.ppat.1002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/JVI.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puck TT, Fisher HW. 1956. Genetics of somatic mammalian cells. I. Demonstration of the existence of mutants with different growth requirements in a human cancer cell strain (HeLa). J Exp Med 104:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stutz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, Gagneur J, Korbel JO, Huber W, Steinmetz LM. 2013. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3:1213–1224. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Binder M, Kochs G, Bartenschlager R, Lohmann V. 2007. Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology 46:1365–1374. doi: 10.1002/hep.21829. [DOI] [PubMed] [Google Scholar]
- 40.Geist MM, Pan X, Bender S, Bartenschlager R, Nickel W, Fackler OT. 2014. Heterologous Src homology 4 domains support membrane anchoring and biological activity of HIV-1 Nef. J Biol Chem 289:14030–14044. doi: 10.1074/jbc.M114.563528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fackler OT, Moris A, Tibroni N, Giese SI, Glass B, Schwartz O, Krausslich HG. 2006. Functional characterization of HIV-1 Nef mutants in the context of viral infection. Virology 351:322–339. doi: 10.1016/j.virol.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 42.Pizzato M, Erlwein O, Bonsall D, Kaye S, Muir D, McClure MO. 2009. A one-step SYBR green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J Virol Methods 156:1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 43.Fritz JV, Tibroni N, Keppler OT, Fackler OT. 2012. HIV-1 Vpu's lipid raft association is dispensable for counteraction of the particle release restriction imposed by CD317/Tetherin. Virology 424:33–44. doi: 10.1016/j.virol.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Laguette N, Bregnard C, Hue P, Basbous J, Yatim A, Larroque M, Kirchhoff F, Constantinou A, Sobhian B, Benkirane M. 2014. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156:134–145. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Rasaiyaah J, Tan CP, Fletcher AJ, Price AJ, Blondeau C, Hilditch L, Jacques DA, Selwood DL, James LC, Noursadeghi M, Towers GJ. 2013. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy D, Larner A, Chaudhuri A, Babiss LE, andDarnell JE Jr. 1986. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci U S A 83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo J, Peters KL, Sen GC. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- 48.Levy DE, Marie I, Prakash A. 2003. Ringing the interferon alarm: differential regulation of gene expression at the interface between innate and adaptive immunity. Curr Opin Immunol 15:52–58. doi: 10.1016/S0952-7915(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 49.Stavrou S, Blouch K, Kotla S, Bass A, Ross SR. 2015. Nucleic acid recognition orchestrates the anti-viral response to retroviruses. Cell Host Microbe 17:478–488. doi: 10.1016/j.chom.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoh SM, Schneider M, Seifried J, Soonthornvacharin S, Akleh RE, Olivieri KC, De Jesus PD, Ruan C, de Castro E, Ruiz PA, Germanaud D, des Portes V, Garcia-Sastre A, Konig R, Chanda SK. 2015. PQBP1 is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell 161:1293–1305. doi: 10.1016/j.cell.2015.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shu C, Yi G, Watts T, Kao CC, Li P. 2012. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol 19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalamvoki M, Roizman B. 2014. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci U S A 111:E611-E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basmaciogullari S, Pizzato M. 2014. The activity of Nef on HIV-1 infectivity. Front Microbiol 5:232. doi: 10.3389/fmicb.2014.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goila-Gaur R, Strebel K. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 58.Li G, Cheng M, Nunoya J, Cheng L, Guo H, Yu H, Liu YJ, Su L, Zhang L. 2014. Plasmacytoid dendritic cells suppress HIV-1 replication but contribute to HIV-1 induced immunopathogenesis in humanized mice. PLoS Pathog 10:e1004291. doi: 10.1371/journal.ppat.1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blondot ML, Dragin L, Lahouassa H, Margottin-Goguet F. 2014. How SLX4 cuts through the mystery of HIV-1 Vpr-mediated cell cycle arrest. Retrovirology 11:117. doi: 10.1186/s12977-014-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guenzel CA, Herate C, Benichou S. 2014. HIV-1 Vpr—a still “enigmatic multitasker”. Front Microbiol 5:127. doi: 10.3389/fmicb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kogan M, Rappaport J. 2011. HIV-1 accessory protein Vpr: relevance in the pathogenesis of HIV and potential for therapeutic intervention. Retrovirology 8:25. doi: 10.1186/1742-4690-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Re F, Luban J. 1997. HIV-1 Vpr: G2 cell cycle arrest, macrophages and nuclear transport. Prog Cell Cycle Res 3:21–27. [DOI] [PubMed] [Google Scholar]
- 64.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol 69:6304–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Planelles V, Bachelerie F, Jowett JB, Haislip A, Xie Y, Banooni P, Masuda T, Chen IS. 1995. Fate of the human immunodeficiency virus type 1 provirus in infected cells: a role for vpr. J Virol 69:5883–5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Romani B, Cohen EA. 2012. Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase. Curr Opin Virol 2:755–763. doi: 10.1016/j.coviro.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bregnard C, Benkirane M, Laguette N. 2014. DNA damage repair machinery and HIV escape from innate immune sensing. Front Microbiol 5:176. doi: 10.3389/fmicb.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med 4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 69.Harman AN, Lai J, Turville S, Samarajiwa S, Gray L, Marsden V, Mercier SK, Jones K, Nasr N, Rustagi A, Cumming H, Donaghy H, Mak J, Gale M Jr, Churchill M, Hertzog P, Cunningham AL. 2011. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood 118:298–308. doi: 10.1182/blood-2010-07-297721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Majumder B, Venkatachari NJ, O'Leary S, Ayyavoo V. 2008. Infection with Vpr-positive human immunodeficiency virus type 1 impairs NK cell function indirectly through cytokine dysregulation of infected target cells. J Virol 82:7189–7200. doi: 10.1128/JVI.01979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okumura A, Alce T, Lubyova B, Ezelle H, Strebel K, Pitha PM. 2008. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology 373:85–97. doi: 10.1016/j.virol.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muthumani K, Choo AY, Premkumar A, Hwang DS, Thieu KP, Desai BM, Weiner DB. 2005. Human immunodeficiency virus type 1 (HIV-1) Vpr-regulated cell death: insights into mechanism. Cell Death Differ 12(Suppl 1):S962–S970. [DOI] [PubMed] [Google Scholar]
- 73.Cybulski KE, Howlett NG. 2011. FANCP/SLX4: a Swiss army knife of DNA interstrand crosslink repair. Cell Cycle 10:1757–1763. doi: 10.4161/cc.10.11.15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berger G, Lawrence M, Hue S, Neil SJ. 2015. G2/M cell cycle arrest correlates with primate lentiviral Vpr interaction with the SLX4 complex. J Virol 89:230–240. doi: 10.1128/JVI.02307-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Noronha CM, Sherman MP, Lin HW, Cavrois MV, Moir RD, Goldman RD, Greene WC. 2001. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science 294:1105–1108. doi: 10.1126/science.1063957. [DOI] [PubMed] [Google Scholar]
- 76.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. 2002. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belzile JP, Duisit G, Rougeau N, Mercier J, Finzi A, Cohen EA. 2007. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog 3:e85. doi: 10.1371/journal.ppat.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeHart JL, Zimmerman ES, Ardon O, Monteiro-Filho CM, Arganaraz ER, Planelles V. 2007. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol J 4:57. doi: 10.1186/1743-422X-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, Margottin-Goguet F. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- 80.Schrofelbauer B, Hakata Y, Landau NR. 2007. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc Natl Acad Sci U S A 104:4130–4135. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan L, Ehrlich E, Yu XF. 2007. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J Virol 81:10822–10830. doi: 10.1128/JVI.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]