ABSTRACT
Hepatitis B virus (HBV) has been implicated as a potential trigger of hepatic steatosis although molecular mechanisms involved in the pathogenesis of HBV-associated hepatic steatosis still remain elusive. Our prior work has revealed that the expression level of liver fatty acid binding protein 1 (FABP1), a key regulator of hepatic lipid metabolism, was elevated in HBV-producing hepatoma cells. In this study, the effects of HBV X protein (HBx) mediated FABP1 regulation on hepatic steatosis and the underlying mechanism were determined. mRNA and protein levels of FABP1 were measured by quantitative RT-PCR (qPCR) and Western blotting. HBx-mediated FABP1 regulation was evaluated by luciferase assay, coimmunoprecipitation, and chromatin immunoprecipitation. Hepatic lipid accumulation was measured by using Oil-Red-O staining and the triglyceride level. It was found that expression of FABP1 was increased in HBV-producing hepatoma cells, the sera of HBV-infected patients, and the sera and liver tissues of HBV-transgenic mice. Ectopic overexpression of HBx resulted in upregulation of FABP1 in HBx-expressing hepatoma cells, whereas HBx abolishment reduced FABP1 expression. Mechanistically, HBx activated the FABP1 promoter in an HNF3β-, C/EBPα-, and PPARα-dependent manner, in which HBx increased the gene expression of HNF3β and physically interacted with C/EBPα and PPARα. On the other hand, knockdown of FABP1 remarkably blocked lipid accumulation both in long-chain free fatty acids treated HBx-expressing HepG2 cells and in a high-fat diet-fed HBx-transgenic mice. Therefore, FABP1 is a key driver gene in HBx-induced hepatic lipid accumulation via regulation of HNF3β, C/EBPα, and PPARα. FABP1 may represent a novel target for treatment of HBV-associated hepatic steatosis.
IMPORTANCE Accumulating evidence from epidemiological and experimental studies has indicated that chronic HBV infection is associated with hepatic steatosis. However, the molecular mechanism underlying HBV-induced pathogenesis of hepatic steatosis still remains to be elucidated. In this study, we found that expression of liver fatty acid binding protein (FABP1) was dramatically increased in the sera of HBV-infected patients and in both sera and liver tissues of HBV-transgenic mice. Forced expression of HBx led to FABP1 upregulation, whereas knockdown of FABP1 remarkably diminished lipid accumulation in both in vitro and in vivo models. It is possible that HBx promotes hepatic lipid accumulation through upregulating FABP1 in the development of HBV-induced nonalcoholic fatty liver disease. Therefore, inhibition of FABP1 might have therapeutic value in steatosis-associated chronic HBV infection.
INTRODUCTION
Hepatitis B virus (HBV) infection is a serious health problem worldwide, causing acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma (1). Emerging evidence from epidemiological and experimental studies suggests that chronic HBV infection, as well as HCV infection, is associated with hepatic steatosis (2, 3). The frequency of hepatic steatosis in subjects with a chronic HBV infection ranges from 27 to 51% (4). Furthermore, HBV X protein (HBx) is known to cause hepatic lipid deposition by inhibiting the secretion of apolipoprotein B (5). A previous report showed that the increased HBx expression can cause lipid accumulation in hepatocytes, likely mediated by sterol regulatory element binding protein 1 and peroxisome proliferator-activated receptor γ (PPARγ) (4). The molecular mechanism by which HBV induces the pathogenesis of hepatic steatosis remains elusive. Using fluorescent two-dimensional difference gel electrophoresis and matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis, we found that the protein level of liver fatty acid binding protein (L-FABP or FABP1) in HBV-producing cells was significantly increased compared to that in control cells (6).
Liver fatty acid binding protein belongs to a multigene FABP family of 14- to 15-kDa cytoplasmic proteins involved in the uptake, transport, and metabolism of cellular long-chain fatty acids and other lipid ligands (7). The human FABP1 gene is localized in the centromeric p12-q11 region of chromosome 2 (8). FABP1 is found abundantly in the cytosol of liver parenchymal cells (9, 10) and represents ca. 0.2% of the total cytosolic proteins in the human hepatoblastoma cell line HepG2 (11). Similar to its family members, FABP1 plays a central role in intracellular fatty acid transport and utilization (12) and is also involved in the modulation of mitosis (13), cell growth, and differentiation (14). Studies in HepG2 cells suggested that the overexpression of FABP1 markedly increased the rate of fatty acid uptake. In contrast, fatty acid uptake significantly decreased following FABP1 antisense RNA expression (15). FABP1 gain-of-function experiments also revealed an increase in fatty acid uptake and lipoprotein secretion from rat hepatoma cells (16). Moreover, the Thr94Ala mutation in the FABP1 gene, which was supposed to abolish the binding of fatty acid, led to decreased triglycerides compared to the wild-type cells when incubated with extracellular fatty acids (17). In a transgenic mouse model, ablation of FABP1 gene impaired the ability of the liver to efficiently import and transfer fatty acids into glycerolipid biosynthesis resulting in a reduction of hepatic triglyceride (TG) accumulation (18). These changes were not due to an inability to upregulate fatty acid oxidation or TG synthesis but rather resulted from decreased rates of hepatic fatty acid uptake and trafficking in the face of short-term (48-h) mobilization of adipose TG stores and increased fatty acid availability (18). The germ line FABP1−/− mice exhibited decreased hepatic triglyceride content with altered fatty acid uptake kinetics (18). Interestingly, FABP1−/− mice fed a high-saturated-fat, high-cholesterol “Western” diet were protected against diet-induced obesity and hepatic steatosis, reflecting an alteration in the kinetics of saturated fatty acid utilization (19, 20). Proteomic screens showed FABP1 to be overexpressed in obese subjects with simple steatosis, along with paradoxically decreased expression in the progressive versus mild forms of nonalcoholic steatohepatitis (20).
Given the key role of FABP1 in lipid metabolism and its potential influence on metabolic disorders and the fact that subjects with chronic HBV infection frequently develop hepatic steatosis, we sought to determine whether FABP1 was involved in HBV-associated hepatic steatosis and to explore the molecular mechanism responsible for the regulation of FABP1 by HBx.
MATERIALS AND METHODS
Cell lines and patients.
HepG2 (HB-8065; American Type Culture Collection [ATCC], Manassas, VA), Huh7 (JCRB0403; Japan), and Hep3B (HB-8064; ATCC) cells were maintained in Dulbecco modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Invitrogen). HepG2.2.15 cells (CRL-11997; ATCC) harboring four copies of HBV-DNA were cultured in modified Eagle medium (Invitrogen) with G418 (Invitrogen) at 380 μg/ml. HepG2-HBV3 cell line harboring 1.2 unit lengths of the HBV genome and control HepG2-RepSal1 cell line (6) were maintained in DMEM supplemented with 10% FBS and hygromycin B (Roche Diagnostics, GmbH, Mannheim, Germany) at 250 μg/ml.
A total of 150 patients with chronic hepatitis B were collected from the Fuzhou Hospital for Infectious Diseases for FABP1 detection in serum by enzyme-linked immunosorbent assay (ELISA). These patients were positive for HBsAg, HBeAg, anti-HBc, and HBV-specific DNA in serum. The HBV viral load was quantified by using an HBV diagnostic kit (Abbott Laboratories, Chicago, IL) according to the manufacturer's instructions. No antiviral drugs were administered before the blood samples were drawn. A total of 30 normal non-HBV-infected sera were obtained as controls. The clinical section of the research was reviewed and ethically approved by the Institutional Ethics Committee of Fujian Medical University (Fuzhou, China). All patients had given informed and written consent.
Plasmids, oligonucleotide siRNA transfection, and generation of recombinant adenoviruses and lentiviruses.
pGL3B-255, pGL3B-255HNF3βmut, and pGL3B-255C/EBPαmut were constructed as previously described (21). pGL3B-255PPARαmut and pGL3B-255mut (which included triple binding site mutations for HNF3β, C/EBPα, and PPARα) were synthesized chemically by the Sangon Biotech (Shanghai, China). HNF3β and C/EBPα small interfering RNAs (siRNAs) were described in our earlier study (21). PPARα siRNA mix and FABP1 siRNA mix were obtained from Santa Cruz Biotechnology, and a nontargeting siRNA was used as a negative control (NC). pRep-HBV (harboring 1.2 unit lengths of the HBV genome) and control plasmid pRep-Sal were described previously (6). The pRep-HBV-X(–) mutation plasmid, which contains an stop codon at HBx codon 8 (CAA to UAA), was generated by site-specific mutagenesis as described previously (22). DNA transfection was carried out using Lipofectamine 2000 (Invitrogen), and siRNA transfections were performed by use of Lipofectamine RNAiMAX (Invitrogen).
HBx-overexpressing adenovirus (Ad-HBx) and empty control (Ad-GFP) were generated using the AdEasy XL system (Stratagene, CA) as we previously described (23). HepG2, Huh7, and Hep3B cells were seeded in six-well plates at a density of 5 × 105 per well and infected with recombinant adenoviruses at a multiplicity of infection (MOI) of 10, 100, or 200 or the indicated doses, respectively. Cell culture supernatants were collected, and cells were harvested at 72 h postinfection. Recombinant lentivirus LV-FABP1-shRNA containing short hairpin RNA (shRNA) against mouse FABP1 was constructed by GeneChem Co., Ltd. (Shanghai, China), the target sequence was 5′-GTCAGAAATCGTGCATGAA-3′. The negative-control lentiviruses LV-GFP were provided by GeneChem.
Animal experiments.
Male, age-matched (6 to 8 weeks old) HBV (HBV-Tg) or HBx+/– transgenic mice (HBx-Tg) and their wild-type (WT) littermates (24) were used in this study. Mice were housed in humidity- and temperature-controlled rooms, with free access to food and water. High-fat-diet (HFD)-induced obese mice were given an HFD (Diet Research, USA) for 8 weeks, while the control mice were fed a normal diet (ND). Control lentivirus (LV-GFP) or LV-FABP1-shRNA recombinant lentivirus (5 × 107 infectious lentivirus particles per mouse) were delivered by hydrodynamic tail vein injection to mice (25). Blood samples were obtained by heart puncture at time of sacrifice. The hepatic index was calculated as liver wet weight/body weight. Liver tissues were extracted, immediately snap-frozen using liquid nitrogen, and stored at −80°C until analyzed. The animal study was approved by the Fujian Medical University Institutional Animal Care and Use Committee (IACUC). The IACUC protocol number for the study is M00186.
In vitro model of cellular steatosis.
Sodium palmitate (catalog no. P9767) and sodium oleate (catalog no. O7501) were obtained from Sigma-Aldrich (St. Louis, MO). Fat-overloading induction of cells was performed mainly according to previously established methods (26), where HepG2 cells at 80% confluence were exposed to saturated (palmitate) and unsaturated (oleate) long-chain free fatty acid (FFA) mixtures at a ratio of 2:1. Fatty acid-free bovine serum albumin (BSA) was used as a control. To assess the influence of FABP1 knockdown on cellular steatosis, cells were treated with FFA mixture after 48 h of siRNA-FABP1 transfection. After incubation with the FFA mixture, cellular lipid accumulation was measured using Oil-Red-O staining and the triglyceride level.
Determination of cellular fat content and triglyceride level.
Oil-Red-O staining was performed on liver sections and HepG2 cells using standard procedures as previously described (27). The triglyceride content was determined using a commercially available kit (Sigma-Aldrich) according to the manufacturer's instructions and normalized to the protein content by BCA assay (Pierce, Rockford, IL).
Liver morphology.
Liver tissues were fixed in 4% formalin and embedded in paraffin according to standard methods. Paraffin-embedded tissues were sectioned (5 μm) and stained with hematoxylin and eosin (H&E) for morphological analysis.
qPCR and Western blot analysis.
Total RNA and protein were extracted from HepG2 cell lysates or mouse liver tissues for gene expression analysis.
qPCR was performed with the ABI StepOne real-time PCR system (Applied Biosystems, Foster City, CA) and a SYBR Premix Ex Taq kit (TaKaRa) in accordance with the manufacturer's instructions. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as an internal control. The forward and reverse primers used were 5′-CAAGTTCACCATCACCGCTG-3′ and 5′-ATTATGTCGCCGTTGAGTTCG-3′ for FABP1 and 5′-TGCACCACCAACTGCTTAGC-3′ and 5′-AGCTCAGGGATGACCTTGCC-3′ for GAPDH.
Western blot analysis was performed using anti-FABP1 (1:1,000 dilution, HPA028275; Sigma), anti-Flag M2 (1:1,000 dilution; Sigma), anti-HNF3β (sc-6554; 1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), anti-C/EBPα (sc-61, 1:500 dilution; Santa Cruz Biotechnology), anti-PPARα (sc-9000; Santa Cruz Biotechnology), or anti-β-actin (1:4,000 dilution; Sigma) and then probed with alkaline phosphatase (AP)-conjugated appropriate secondary antibody and chemiluminescence detection. Immunoreactive protein bands were visualized by adding CDP STAR reagents (Roche). Intensities of band signals were quantified using the densitometric software Quantity One (Bio-Rad, Hercules, CA).
Luciferase assay.
HepG2 cells were plated in a 12-well culture plate and transiently cotransfected with 0.2 μg of promoter reporter plasmids or pGL3-Basic (Promega, Madison, WI)/well and 0.1 μg of the pRL-TK plasmid (Promega) containing the Renilla luciferase gene used for internal normalization/well. After 48 h, the cells were harvested, and a total of 20 μg of cell lysate was used for the detection of intracellular luciferase activity according to the recommendations of the Promega Corporation. Luminescence measurement was carried out on a luminometer (Orion II microplate luminometer; Berthold Detection Systems, Germany). All assays were performed at least in triplicate.
Coimmunoprecipitation assay.
Lysates from HepG2 cells were immunoprecipitated with anti-FLAG M2 agarose (Sigma) or HNF3β, C/EBPα, and PPARα antibodies and protein A/G agarose (Santa Cruz Biotechnology) at 4°C overnight. The immune complexes were eluted and subjected to Western blot analysis with the respective antibodies.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assay was performed according to Abcam's X-ChIP protocol. For immunoprecipitation, sonicated cell lysates were incubated with anti-HNF3β, anti-C/EBPα, anti-PPARα, or anti-Flag M2 (the same amount of normal IgG as control) antibodies for protein-DNA binding detection. Bound target DNA fractions were analyzed by PCR with the paired primers 5′-CATTTCGTGGGGTGCGGAGA-3 (sense) and 5′-AAGAACAGGATGGGCACAGC-3′ (antisense), which amplified the region from nucleotides −136 to −251 (region 1 [R1]) containing HNF3β and C/EBPα binding sites, and the paired primers 5′-GCTGTGCCCATCCTGTTCTT-3′ (sense) and 5′-GGGGCTCCCTTCCCTTCGAA-3′ (antisense), which amplified the region from nucleotides −36 to −155 (region 2 [R2]) containing PPARα binding sites. A distant upstream 5′ untranslated region (−4723 to −4460, region 3 [R3]) of the FABP1 gene was amplified as a negative control with the primers 5′-TCTCACCCTCACCCTCAACT-3′ (sense) and 5′-AATGCCCACCAACAGTAGACT-3′ (antisense).
ELISA.
The amounts of FABP1 present in HBV-infected serum or in non-HBV-infected serum were determined using an anti-human FABP1 ELISA kit (GWB-HFABP1; GeneWay Biotech, Inc., San Diego, CA) or a specific anti-mouse FABP1 ELISA kit (RFBP10; R&D Systems) according to the manufacturer's instructions.
Statistical analysis.
The differences between two groups were analyzed by using a Student t test. All values are expressed as means ± standard deviations (SD) from triplicate experiments. A P value of <0.05 was considered statistically significant.
RESULTS
HBV infection upregulates FABP1 expression.
The effect of HBV on FABP1 expression was first evaluated by qPCR and Western blot analysis in the stable HBV-producing hepatoma cell lines HepG2-HBV3 and HepG2.2.15 that had been proven to constitutively produce infectious HBV particles (6, 28). Both FABP1 mRNA (Fig. 1A) and protein (Fig. 1B) levels were significantly elevated in HepG2-HBV3 and HepG2.2.15 cells compared to their respective controls, HepG2-RepSal and HepG2. A similar result was obtained with Hep3B, Huh7, and HepG2 cells transiently transfected with 1.2 unit lengths of the HBV genome (pRep-HBV), showing the increased FABP1 mRNA and protein levels in the HBV-DNA transfected cells (Fig. 1C and D). We also analyzed the expression of FABP1 in the sera of HBV-infected patients and HBV-Tg mice by ELISA. As expected, serum FABP1 levels were significantly higher in HBV-positive patients, with the HBV titer exceeding 107 copies/ml (Fig. 1E), and mice than in their control groups (Fig. 1F). Similarly, the expression of FABP1 was markedly upregulated in the liver tissues of HBV-Tg mice, relative to that of wild type (WT) mice (Fig. 1G and H). These data suggest that FABP1 is elevated under the condition of HBV infection and may play an important role in HBV infection-induced hepatic diseases.
FIG 1.
Effect of HBV on FABP1 mRNA and protein level. (A and B) mRNA and protein levels of FABP1 in HBV-producing cell lines. HepG2.2.15 and HepG2-HBV3 cells were HBV-producing cell lines. (C and D) mRNA and protein levels of FABP1 in HBV DNA-transfected cells. HepG2, Huh7, or Hep3B cells were transfected with pRep-HBV and control plasmid pRep-Sal. (E) Serum levels of FABP1 protein in HBV-infected patients with HBV copies ranging from 101 to >109 (n = 30/group). (F) Serum levels of FABP1 protein in HBV-Tg mice versus wild-type (WT) mice (n = 10). (G and H) mRNA and protein levels of FABP1 in liver tissues from HBV-Tg mice versus WT mice. β-Actin served as a loading control for Western blotting. GAPDH served as an internal control for qPCR. *, P < 0.05 versus control.
HBx induces FABP1 gene expression.
HBx appears to have the most pathogenic potential. To evaluate the effect of HBx on FABP1 gene expression, recombinant adenoviruses expressing HBx protein (Ad-HBx) or green fluorescent protein (GFP) control (Ad-GFP) were generated and used to infect cells, as we previously described (23). The result showed that the expression of HBx significantly increased the FABP1 mRNA and protein levels in HepG2, Huh7, and Hep3B cells (Fig. 2A and B). Interestingly, transfection of the HBx mutant pRep-1.2HBV-X(–) into HepG2 cells reduced FABP1 expression compared to cells transfected with pRep-HBV (Fig. 2C and D). The fact that HBx constitutively upregulates FABP1 gene expression raised the question as to whether HBx would behave differently in the presence of fatty acid (FFA), a functional FABP1 inducer (10, 29). Expectedly, the induction of FABP1 mRNA and protein expression was further increased in the FFA-treated HBx-expressing HepG2 cells (Fig. 2E and F). In addition, HBx-Tg mice also had a significant increase of FABP1 expression in sera and liver tissues compared to WT mice at the same age (Fig. 3). Taken together, these data suggest that HBx is a critical regulator for FABP1 expression during HBV infection.
FIG 2.
FABP1 is upregulated in HBx-expressing cells. (A and B) mRNA and protein levels of FABP1 in HepG2, Huh7, and Hep3B cells infected with Ad-HBx or Ad-GFP (control). *, P < 0.05 versus Ad-GFP. (C and D) Impaired HBx reduces FABP1 expression. pRep-HBV or HBx stop mutant pRep-HBV-X(–) was transfected into HepG2 cells, respectively. The mRNA (C) and protein (D) levels of FABP1 were measured. *, P < 0.05 versus pRep-HBV. (E and F) Effect of HBx on FFA induction of FABP1 at mRNA (E) or protein (F) levels in HepG2 cells. HepG2 cells were infected with Ad-HBx or Ad-GFP in the presence of a 0.5 mM FFA mixture (oleate/palmitate) at the final ratio of 2:1 for 24 h. Fatty acid-free BSA uses as a control. *, P < 0.05 versus Ad-GFP+BSA.
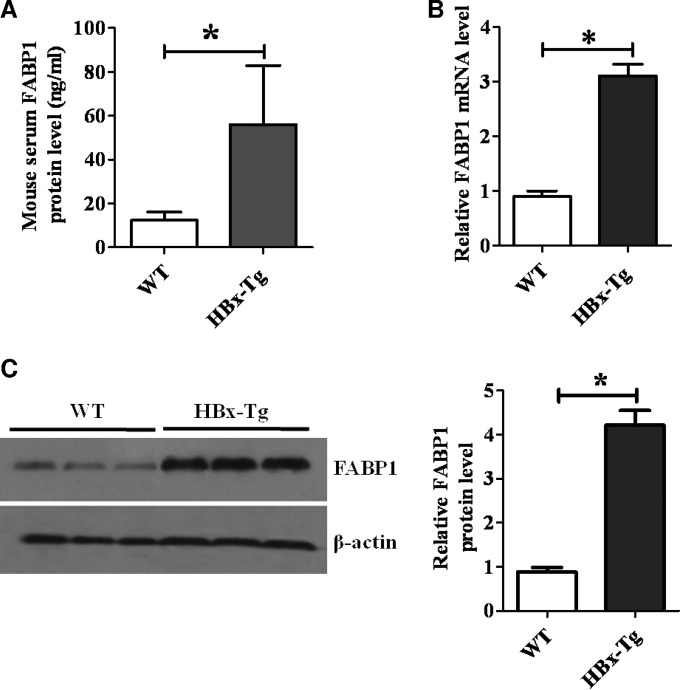
FIG 3.
FABP1 is upregulated in HBx-Tg mice. (A) Serum levels of FABP1 protein in HBx-Tg mice. *, P < 0.05 versus WT mice. (B and C) mRNA (B) and protein (C) levels of FABP1 in the livers of HBx-Tg mice. n = 10 for both HBx-Tg and WT groups; *, P < 0.05 versus WT mice.
HBx facilitates HNF3β, C/EBPα, and PPARα binding to FABP1 promoter.
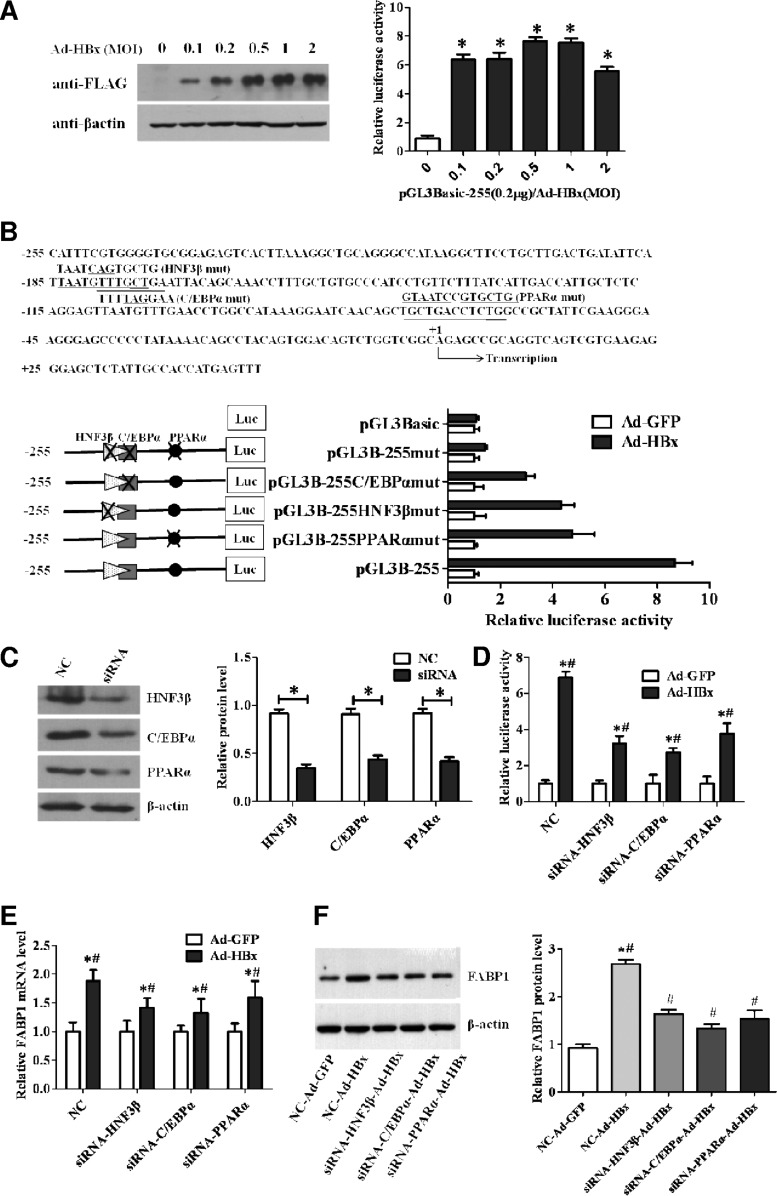
To determine whether HBx induces FABP1 transcription, a reporter plasmid pGL3B-255 containing the human FABP1 promoter sequence from positions −255 to +50, a region known to possess maximum FABP1 promoter activity (21), was cotransfected with Ad-HBx or Ad-GFP control into HepG2 cells. The luciferase assay revealed that the FABP1 promoter activity was markedly enhanced by the HBx protein (Fig. 4A). Previous studies from our laboratory have demonstrated that FABP1 gene was regulated predominantly by liver-enriched transcription factors HNF3β and C/EBPα (21), and a functional PPARα-responsive element was recently reported to play an important role in FABP1 gene regulation (30). The putative binding sites of HNF3β, C/EBPα, and PPARα on the FABP1 promoter were shown in Fig. 4B (upper panel). To determine which elements were involved in HBx-related FABP1 gene transcription, we introduced mutations in the binding sites for HNF3β, C/EBPα, and PPARα. We found that mutations at either HNF3β, C/EBPα, or PPARα binding site significantly decreased the HBx-induced promoter activity (Fig. 4B, lower panel). Notably, mutations at all these three binding sites abolished its responsiveness to HBx-mediated FABP1 transcription. To further confirm that HBx-induced FABP1 expression was mediated through HNF3β, C/EBPα, or PPARα, we knocked down the expression of these transcription factors in Ad-HBx- or Ad-GFP-transduced HepG2 cells (Fig. 4C) and measured the FABP1 promoter activity. As shown in Fig. 4D, knockdown of either HNF3β, C/EBPα, or PPARα reduced the HBx-mediated luciferase activities. Meanwhile, the expression of FABP1 at both mRNA (Fig. 4E) and protein (Fig. 4F) levels was also reduced significantly. Taken together, these results suggest that HNF3β, C/EBPα, and PPARα, through binding to their corresponding sites at the FABP1 promoter region, are essential for HBx-mediated FABP1 promoter activation and FABP1 transcription.
FIG 4.
HBx activates the FABP1 promoter in an HNF3β-, C/EBPα-, and PPARα-dependent manner. (A) HepG2 cells were transfected with reporter construct pGL3B-255 (0.2 μg/well) and infected with increasing amounts of Ad-HBx at a ratio (μg of pGL3B-255/MOI of Ad-HBX) of 0, 0.1, 0.2, 0.5, 1, or 2. Cells were harvested 48 h after transfection. The increased protein levels of FLAG-tagged HBx were determined by Western blotting with anti-FLAG antibody (left panel), and the relative luciferase activity was determined (right panel). (B) Effects of mutations at transcription factor binding sites on promoter activities. HNF3β, C/EBPα, and PPARα binding sites and mutated sequences are underlined (upper panel). Mutated constructs of FABP1 promoter are depicted schematically in the lower panel (left); 0.2 μg of various mutated constructs or pGL3-Basic were transfected into Ad-HBx-infected-HepG2 cells. At 48 h after transfection, the relative luciferase activity was determined (lower panel, right). (C) Expression of HNF3β, C/EBPα, and PPARα were measured by Western blotting in HepG2 cells transfected with siRNA targeting HNF3β, C/EBPα, or PPARα (100 nM) or control siRNA (NC, 100 nM). (D) Luciferase activities were measured in Ad-HBx-infected-HepG2 cells cotransfected with pGL3B-255 (0.2 μg/well) and siRNA (100 nM) or control siRNA (NC; 100 nM), respectively. (E and F) Expression of FABP1 was detected by qPCR (E) and Western blotting (F) in Ad-HBx-infected-HepG2 cells transfected with control siRNA (NC; 100 nM) or siRNA (100 nM), respectively. Data are shown as means ± the SD of three independent experiments after being normalized to the empty vector control Ad-GFP. *, P < 0.05 versus Ad-GFP.
HBx increases HNF3β expression and interacts with C/EBPα and PPARα.
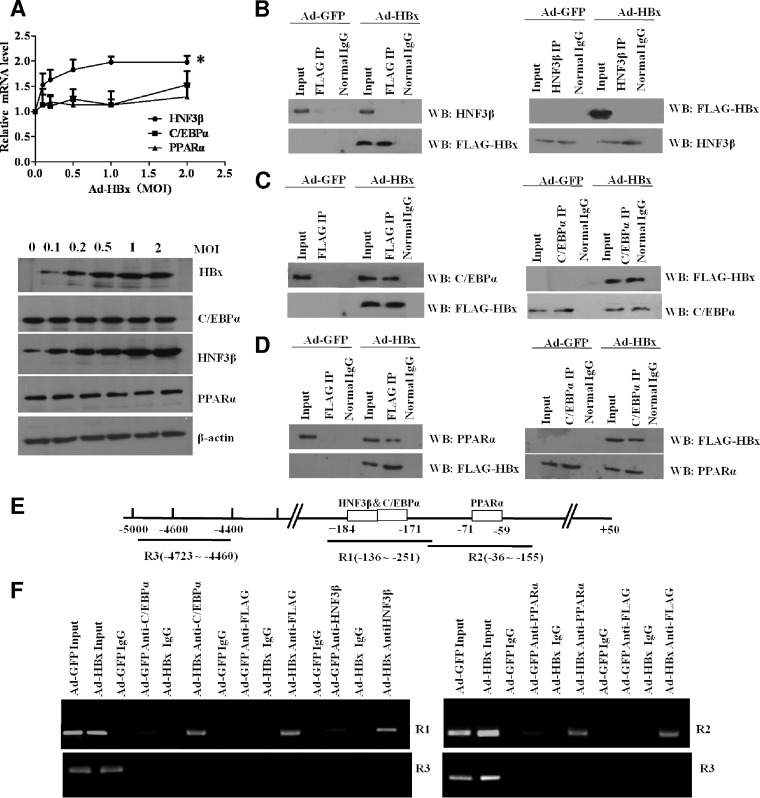
To further investigate the mechanism underlying HBx-enhanced FABP1 expression, we examined whether HBx could directly affect the expression of HNF3β, C/EBPα, and PPARα. We found that ectopic expression of HBx in HepG2 cells increased HNF3β mRNA and protein levels but caused no significant changes in C/EBPα or PPARα expression (Fig. 5A). To assess whether HBx could physically interact with endogenous HNF3β, C/EBPα, or PPARα, we performed a coimmunoprecipitation study with HepG2 cells infected with recombinant virus encoding FLAG-tagged HBx. The results showed that FLAG-tagged HBx was immunoprecipitated with C/EBPα and PPARα but not HNF3β, and these interactions were further confirmed by reverse immunoprecipitation using HNF3β, C/EBPα, and PPARα antibodies (Fig. 5B to D). To test whether HBx binds FABP1 gene promoter, we performed a ChIP assay in Ad-HBx-infected HepG2 cells with specific antibodies against HNF3β, C/EBPα, PPARα, and FLAG-tagged HBx and PCR using the primers to amplify the fragments within the FABP1 promoter region corresponding to the C/EBPα and HNF3β binding sites (approximately −136 to −251) and the PPARα binding site (approximately −36 to −155). The ChIP data indicated that the antibodies to HNF3β, C/EBPα, and PPARα or the antibody to FLAG for capture of the FLAG-tagged HBx efficiently immunoprecipitated the corresponding FABP1 promoter region (Fig. 5E and F). These results suggests that HBx upregulated FABP1 transcription via enhancing the expression of HNF3β and interacting with C/EBPα and PPARα.
FIG 5.
Effect of HBx on HNF3β, C/EBPα, and PPARα. (A) The effect of HBx on the expression of HNF3β, C/EBPα, and PPARα was detected by qPCR (upper panel) and Western blotting (lower panel) in HepG2 cells infected with increasing amounts of Ad-HBx (i.e., a 0-, 0.1-, 0.2-, 0.5-, 1-, or 2-fold excess in MOI), respectively. The increased protein levels of FLAG-tagged HBx were determined with anti-FLAG antibody. *, P < 0.05 versus control. (B to D) Interaction between HBx and HNF3β, C/EBPα, and PPARα as determined by coimmunoprecipitation assay. FLAG-tagged HBx was immunoprecipitated with anti-FLAG antibody, and HNF3β, C/EBPα, or PPARα was detected by Western blotting with specific antibodies (left panel; B, C, and D, respectively). Reciprocal coimmunoprecipitation was carried out using HNF3β, C/EBPα, and PPARα antibodies, and HBx was detected by Western bloting with anti-FLAG antibody (right panel; B, C, and D, respectively). Normal IgG served as a control. (E) Schematic representation of PCR-amplified fragments of FABP1 promoter region. (F) ChIP assays were performed to confirm the interaction of HBx with the promoter region of FABP1. Lysates of HepG2 cells infected with Ad-HBx underwent ChIP using antibodies against FLAG-tagged HBx, HNF3β, C/EBPα, or PPARα. PCR was conducted with immunoprecipitated DNA. Amplicon R1 covers the HNF3β and C/EBPα binding site, amplicon R2 contains the PPARα site, and upstream regulatory region R3 was used as a negative control. An empty vector containing GFP and an isotype control IgG served as external and internal controls, respectively.
HBx-induced FABP1 gene expression causes lipid accumulation in HepG2 and HBx-Tg mice.
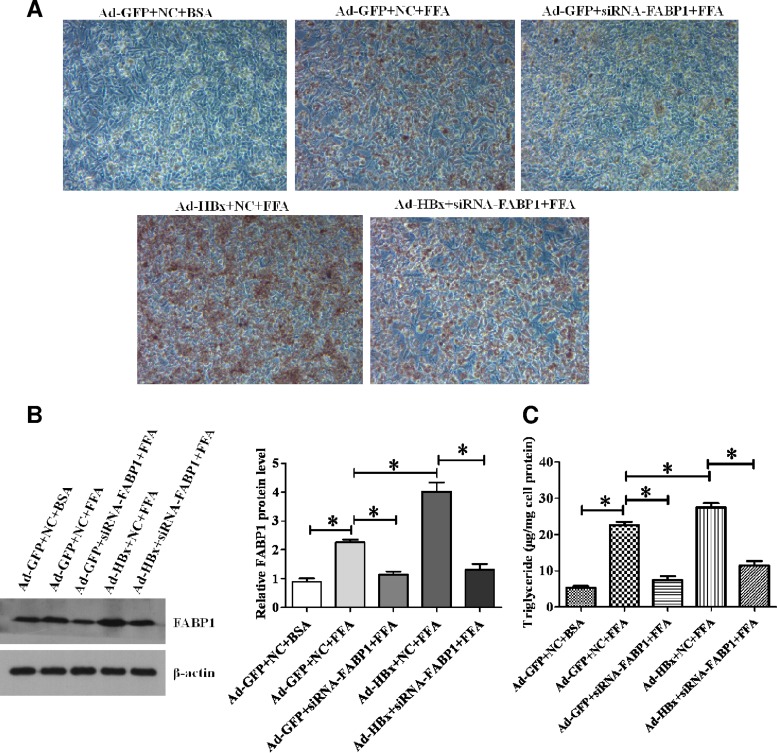
To investigate the influence of HBx-mediated FABP1 gene activation on hepatic lipid metabolism at the cellular level, we treated HepG2 cells with a 0.5 mM FFA mixture (oleate/palmitate [2:1]) to induce an in vitro cellular steatosis. As revealed by Oil-Red-O staining (Fig. 6A), FFA induced substantial lipid accumulation in HepG2 cells, and the percentage of positive cells among HBx expressing Ad-HBx cells was much higher than in the control Ad-GFP cells. In contrast, knockdown of FABP1 significantly prevented lipid accumulation in the cells. To evaluate whether the lipid accumulation in HepG2 cells resulted from differences in FABP1 expression, the protein levels of FABP1 were determined by Western blot analysis after the indicated treatments (Fig. 6B). Consistently, the cellular TG contents were significantly higher in Ad-HBx cells than in Ad-GFP cells (Fig. 6C), and siRNA-FABP1 treated cells markedly reduced TG accumulation. The results indicate that FABP1 expression level is correlated with the extent of hepatic steatosis.
FIG 6.
HBx increases lipid accumulation in HepG2 cells through regulation of FABP1. (A) Inhibition of FABP1 expression in HBx-expressing HepG2 cells reduced FFA-induced cellular lipid accumulation, as revealed by Oil-Red-O staining. Representative images of Oil-Red-O staining of HepG2 cells treated with indicated treatments are presented. Red staining indicates neutral lipid. (B) Western blot analysis of the effect of indicated treatments on FABP1 protein levels. The ratio of FABP1 protein levels in the treated cells relative to that in the control Ad-GFP+NC+BSA cells was calculated densitometrically after normalization to the level of β-actin. Values are means ± the SD (n = 3). (C) The triglyceride content was measured in HepG2 cell lysates with the indicated treatments using a commercially assay kit, as described in Materials and Methods. Triglyceride levels were normalized to the protein concentration (μg/mg) and is given as means ± the SD of three separate experiments. *, P < 0.05.
To further confirm our findings under physiologic conditions, we investigated the hepatic lipid content in the liver tissues of HBx-Tg mice under HFD-induced hepatic steatosis. To overcome drawbacks of short-time action and lack of regenerating ability of siRNA and to permit robust inducible RNAi mediated FABP1 silencing, lentiviral LV-FABP1-shRNA was generated. The efficiencies of infection were observed under fluorescence microscope (Fig. 7A), and gene silencing in mouse liver were determined in Fig. 7B. After 8 weeks of HFD induction, HBx mice developed a more prominent fatty liver phenotype featuring an enlarged and light-colored liver caused by extensive fat accumulation compared to WT mice at the same age (Fig. 7C). In contrast, knockdown of the mouse FABP1 gene significantly prevented lipid accumulation in the liver (Fig. 7C). Consistently, liver weights and liver TG contents were significantly higher in HBx-Tg mice than in WT mice and were reduced by LV-FABP1-shRNA treatment (Fig. 7D and E). We then examined the histologic changes of the hepatic sections. Similar to our findings in HepG2 cells, H&E and Oil-Red-O staining of liver sections revealed the presence of diffuse intracellular lipid droplets in HBx-Tg mice compared to the WT mice, and treatment of the LV-FABP1-shRNA markedly reduced lipid accumulation (Fig. 7F).
FIG 7.
FABP1 mediates HBx-induced hepatic steatosis in vivo. (A) Representative fluorescence images of liver infected with LV-FABP1-shRNA expressing FABP1-GFP and control lentivirus LV-GFP. (B) FABP1 protein level was determined by Western blotting. (C) Representative liver images from mice with the indicated treatments. (D and E) Liver weight and hepatic triglyceride levels. (F) Representative images of liver sections from the treated mice stained using H&E and Oil-Red-O. The droplets indicate neutral lipid accumulation in H&E staining. Red staining indicates neutral lipid in Oil-Red-O staining. Values represent means ± the SD (n = 5 for each group). *, P < 0.05.
DISCUSSION
Nonalcoholic fatty liver disease (NAFLD) has attracted considerable attention due to its high morbidity. This disease features an excessive triglyceride accumulation in hepatocytes (31). Prolonged NAFLD is associated with steatohepatitis, a condition that can result in end-stage liver diseases (32, 33). Increasing evidence has indicated that chronic HBV infection, as well as HCV infection, is associated with hepatic steatosis through alterations in lipid metabolism (2, 3). Among the four HBV-encoding proteins, the HBV X protein (HBx) apparently has the most pathogenic potential involving in HBV infection, replication, apoptosis, cell proliferation, DNA damage, and even hepatocarcinogenesis (34–36). Thus, we hypothesized that HBx may be involved in HBV-induced hepatic steatosis.
To address this hypothesis, we first investigated the effect of HBV on FABP1 expression in HBV-producing hepatoma cell lines and in the sera of HBV-infected patients and HBV-Tg mice. Our results showed that FABP1 was upregulated by HBV. We further observed that FABP1 was activated markedly in HBx-expressing hepatoma cell lines and HBx-Tg mice. Thus, we conclude that HBx is able to upregulate FABP1 in hepatoma cells, which may contribute to the development of HBV-related hepatic steatosis.
Numerous studies have shown that HBx does not bind DNA directly, but it can transactivate multiple cellular promoters by interacting with nuclear transcription factors (34), such as TFIIB (37), RXR (38), AP1 (39), and NF-κB (40). In our previous report, we identified that the −255/+50 fragment contained the core region of the FABP1 promoter (21). Results from this study showed that HBx was indeed able to activate the −255/+50 region in a dose-dependent manner. Intriguingly, we also found that liver-enriched transcription factors HNF3β and C/EBPα elements were present in the −255/+50 FABP1 promoter region (21).
HNF3β and C/EBPα belong to the liver-enriched transcription factors families. HNF3β belongs to the HNF-3 gene family, which includes three proteins (HNF3α, HNF3β, and HNF3γ) that mediate hepatocyte-enriched transcription of numerous genes necessary for organ function (41). C/EBPα belongs to the C/EBP family, which is composed of six related proteins that belong to the larger family of basic region leucine zipper (bZIP) transcription factors (42). High expression levels of human C/EBPα have been found in liver and other organs (43). C/EBPα is involved in diverse physiological functions in liver biology that include energy metabolism (44), liver regeneration (45), cellular differentiation (46), and cell cycle control (42). PPARα, a lipid-sensor nuclear factor, is highly expressed in liver tissue (47) and plays a key role in hepatic lipid metabolism. PPARα was reported to lower plasma TG by decreasing production of very-low-density lipoproteins, enhancing fatty acid oxidation and stimulating clearance of TG-rich lipoproteins (48). Mice deficient in PPARα are reported to be prone to massive hepatic lipid accumulation, hypercholesterolemia, and hypertriglyceridemia (49). Thus, we sought to determine whether HNF3β, C/EBPα, and PPARα were involved in HBx-enhanced FABP1 expression. Our data demonstrated that HBx failed to activate the FABP1 promoter when either the HNF3β, C/EBPα, or PPARα binding site in the promoter was mutated and that the promoter activity of FABP1 was reduced when either HNF3β, C/EBPα, or PPARα was knocked down by the respective siRNA. Meanwhile, the expression levels of FABP1 induced by HBx were also downregulated. Moreover, HBx turned out to induce FABP1 gene expression by upregulating the HNF3β expression level, but not those of C/EBPα or PPARα. Furthermore, HBx interacted with C/EBPα or PPARα, but not HNF3β, leading to the enhanced transcriptional activity of C/EBPα and PPARα. Accordingly, we validated by ChIP assay that HBx was able to indirectly bind to the promoter of FABP1 by HNF3β, C/EBPα, and PPARα. Therefore, it is conceivable that HBx activates the FABP1 promoter through HNF3β, C/EBPα, and PPARα.
In functional studies, we elucidated the important role of FABP1 in governing hepatic lipid metabolism using a HepG2 in vitro cellular steatosis model and an in vivo dietary-fat-overloading mouse model. The observation that FFA-induced lipid accumulation was dramatically reduced in HepG2 cells and that fat accumulation was diminished in HFD-treated mice supports a protective role of FABP1 deficiency in HBx-induced hepatic steatosis.
In summary, we demonstrate that HBx promotes hepatic lipid accumulation through upregulating FABP1 that involves HNF3β, C/EBPα, and PPARα in the development of HBV-induced NAFLD. An interaction between HBx and FABP1 may be a key central driver for pathogenesis of hepatic steatosis. The present data may provide a potential therapeutic option to protect HBV-infected livers from HBx-induced lipid deposition and deterioration of hepatic functions by inhibition of FABP1 expression.
ACKNOWLEDGMENTS
This study was supported by the grants from the National Natural Science Foundation of China (grants 81271822, 81473047, and 81401657), the State Key Project Specialized for Infectious Diseases (2012ZX10002002-004-006 and 2013ZX10002002-005-002), the Training Program Foundation for Middle-Aged and Young Talents from Sanitation System of Fujian province (2014-ZQN-ZD-23), and Foundation of Fujian Educational Committee (JK2013021).
REFERENCES
- 1.El-Serag HB, Rudolph KL. 2007. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Gordon A, McLean CA, Pedersen JS, Bailey MJ, Roberts SK. 2005. Hepatic steatosis in chronic hepatitis B and C: predictors, distribution, and effect on fibrosis. J Hepatol 43:38–44. doi: 10.1016/j.jhep.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 3.Yoon EJ, Hu KQ. 2006. Hepatitis C virus (HCV) infection and hepatic steatosis. Int J Med Sci 3:53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim KH, Shin HJ, Kim K, Choi HM, Rhee SH, Moon HB, Kim HH, Yang US, Yu DY, Cheong J. 2007. Hepatitis B virus X protein induces hepatic steatosis via transcriptional activation of SREBP1 and PPARγ. Gastroenterology 132:1955–1967. doi: 10.1053/j.gastro.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 5.Kang SK, Chung TW, Lee JY, Lee YC, Morton RE, Kim CH. 2004. The hepatitis B virus X protein inhibits secretion of apolipoprotein B by enhancing the expression of N-acetylglucosaminyltransferase III. J Biol Chem 279:28106–28112. doi: 10.1074/jbc.M403176200. [DOI] [PubMed] [Google Scholar]
- 6.Huang Q, Wang L, Bai S, Lin W, Chen W, Lin J, Lin X. 2009. Global proteome analysis of hepatitis B virus expressing human hepatoblastoma cell line HepG2. J Med Virol 81:1539–1550. doi: 10.1002/jmv.21593. [DOI] [PubMed] [Google Scholar]
- 7.Hertzel AV, Bernlohr DA. 2000. The mammalian fatty acid-binding protein multigene family: molecular and genetic insights into function. Trends Endocrinol Metab 11:175–180. doi: 10.1016/S1043-2760(00)00257-5. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman AW, Veerkamp JH. 2002. New insights into the structure and function of fatty acid-binding proteins. Cell Mol Life Sci 59:1096–1116. doi: 10.1007/s00018-002-8490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Storch J, Corsico B. 2008. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr 28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 10.Wolfrum C, Ellinghaus P, Fobker M, Seedorf U, Assmann G, Borchers T, Spener F. 1999. Phytanic acid is ligand and transcriptional activator of murine liver fatty acid binding protein. J Lipid Res 40:708–714. [PubMed] [Google Scholar]
- 11.Gibbons GF. 1994. A comparison of in-vitro models to study hepatic lipid and lipoprotein metabolism. Curr Opin Lipidol 5:191–199. doi: 10.1097/00041433-199405030-00006. [DOI] [PubMed] [Google Scholar]
- 12.Schroeder F, Jolly CA, Cho TH, Frolov A. 1998. Fatty acid binding protein isoforms: structure and function. Chem Phys Lipids 92:1–25. doi: 10.1016/S0009-3084(98)00003-6. [DOI] [PubMed] [Google Scholar]
- 13.Sorof S. 1994. Modulation of mitogenesis by liver fatty acid binding protein. Cancer Metastasis Rev 13:317–336. doi: 10.1007/BF00666102. [DOI] [PubMed] [Google Scholar]
- 14.Schroeder F, Atshaves BP, Starodub O, Boedeker AL, Smith RR 3rd, Roths JB, Foxworth WB, Kier AB. 2001. Expression of liver fatty acid binding protein alters growth and differentiation of embryonic stem cells. Mol Cell Biochem 219:127–138. doi: 10.1023/A:1010851130136. [DOI] [PubMed] [Google Scholar]
- 15.Wolfrum C, Buhlmann C, Rolf B, Borchers T, Spener F. 1999. Variation of liver-type fatty acid binding protein content in the human hepatoma cell line HepG2 by peroxisome proliferators and antisense RNA affects the rate of fatty acid uptake. Biochim Biophys Acta 1437:194–201. doi: 10.1016/S1388-1981(99)00008-6. [DOI] [PubMed] [Google Scholar]
- 16.Linden D, Lindberg K, Oscarsson J, Claesson C, Asp L, Li L, Gustafsson M, Boren J, Olofsson SO. 2002. Influence of peroxisome proliferator-activated receptor alpha agonists on the intracellular turnover and secretion of apolipoprotein (Apo) B-100 and ApoB-48. J Biol Chem 277:23044–23053. doi: 10.1074/jbc.M110416200. [DOI] [PubMed] [Google Scholar]
- 17.Gao N, Qu X, Yan J, Huang Q, Yuan HY, Ouyang DS. 2010. L-FABP T94A decreased fatty acid uptake and altered hepatic triglyceride and cholesterol accumulation in Chang liver cells stably transfected with L-FABP. Mol Cell Biochem 345:207–214. doi: 10.1007/s11010-010-0574-7. [DOI] [PubMed] [Google Scholar]
- 18.Newberry EP, Xie Y, Kennedy S, Han X, Buhman KK, Luo J, Gross RW, Davidson NO. 2003. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J Biol Chem 278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- 19.Newberry EP, Xie Y, Kennedy SM, Luo J, Davidson NO. 2006. Protection against Western diet-induced obesity and hepatic steatosis in liver fatty acid-binding protein knockout mice. Hepatology 44:1191–1205. doi: 10.1002/hep.21369. [DOI] [PubMed] [Google Scholar]
- 20.Newberry EP, Kennedy SM, Xie Y, Luo J, Davidson NO. 2009. Diet-induced alterations in intestinal and extrahepatic lipid metabolism in liver fatty acid binding protein knockout mice. Mol Cell Biochem 326:79–86. doi: 10.1007/s11010-008-0002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu YL, Peng XE, Wang D, Chen WN, Lin X. 2012. Human liver fatty acid binding protein (hFABP1) gene is regulated by liver-enriched transcription factors HNF3β and C/EBPα. Biochimie 94:384–392. doi: 10.1016/j.biochi.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Lin YJ, Huang LR, Yang HC, Tzeng HT, Hsu PN, Wu HL, Chen PJ, Chen DS. 2010. Hepatitis B virus core antigen determines viral persistence in a C57BL/6 mouse model. Proc Natl Acad Sci U S A 107:9340–9345. doi: 10.1073/pnas.1004762107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YL, Wang D, Peng XE, Chen YL, Zheng DL, Chen WN, Lin X. 2013. Epigenetic silencing of NAD(P)H:quinone oxidoreductase 1 by hepatitis B virus X protein increases mitochondrial injury and cellular susceptibility to oxidative stress in hepatoma cells. Free Radic Biol Med 65:632–644. doi: 10.1016/j.freeradbiomed.2013.07.037. [DOI] [PubMed] [Google Scholar]
- 24.Wu BK, Li CC, Chen HJ, Chang JL, Jeng KS, Chou CK, Hsu MT, Tsai TF. 2006. Blocking of G1/S transition and cell death in the regenerating liver of hepatitis B virus X protein transgenic mice. Biochem Biophys Res Commun 340:916–928. doi: 10.1016/j.bbrc.2005.12.089. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Calvisi DF. 2014. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol 184:912–923. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Lechon MJ, Donato MT, Martinez-Romero A, Jimenez N, Castell JV, O'Connor JE. 2007. A human hepatocellular in vitro model to investigate steatosis. Chem Biol Interact 165:106–116. doi: 10.1016/j.cbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Joshi-Barve S, Barve SS, Amancherla K, Gobejishvili L, Hill D, Cave M, Hote P, McClain CJ. 2007. Palmitic acid induces production of proinflammatory cytokine interleukin-8 from hepatocytes. Hepatology 46:823–830. doi: 10.1002/hep.21752. [DOI] [PubMed] [Google Scholar]
- 28.Sells MA, Chen ML, Acs G. 1987. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newberry EP. 2009. Liver fatty acid binding protein (L-FABP) as a target for the prevention of high-fat diet induced obesity and hepatic steatosis immunity. Endocrinol Metab Agents Med Chem 9:30–37. [Google Scholar]
- 30.Guzman C, Benet M, Pisonero-Vaquero S, Moya M, Garcia-Mediavilla MV, Martinez-Chantar ML, Gonzalez-Gallego J, Castell JV, Sanchez-Campos S, Jover R. 2013. The human liver fatty acid binding protein (FABP1) gene is activated by FOXA1 and PPARα and repressed by C/EBPα: implications in FABP1 down-regulation in nonalcoholic fatty liver disease. Biochim Biophys Acta 1831:803–818. doi: 10.1016/j.bbalip.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Adams LA, Lindor KD. 2007. Nonalcoholic fatty liver disease. Ann Epidemiol 17:863–869. doi: 10.1016/j.annepidem.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Powell EE, Jonsson JR, Clouston AD. 2005. Steatosis: co-factor in other liver diseases. Hepatology 42:5–13. doi: 10.1016/j.jhep.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Browning JD, Horton JD. 2004. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest 114:147–152. doi: 10.1172/JCI200422422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchard MJ, Schneider RJ. 2004. The enigmatic X gene of hepatitis B virus. J Virol 78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouchard MJ, Wang L, Schneider RJ. 2006. Activation of focal adhesion kinase by hepatitis B virus HBx protein: multiple functions in viral replication. J Virol 80:4406–4414. doi: 10.1128/JVI.80.9.4406-4414.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang H, Oishi N, Kaneko S, Murakami S. 2006. Molecular functions and biological roles of hepatitis B virus X protein. Cancer Sci 97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rui E, Moura PR, Goncalves KA, Rooney RJ, Kobarg J. 2006. Interaction of the hepatitis B virus protein HBx with the human transcription regulatory protein p120E4F in vitro. Virus Res 115:31–42. doi: 10.1016/j.virusres.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Barnabas S, Andrisani OM. 2000. Different regions of hepatitis B virus X protein are required for enhancement of bZip-mediated transactivation versus transrepression. J Virol 74:83–90. doi: 10.1128/JVI.74.1.83-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benn J, Su F, Doria M, Schneider RJ. 1996. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol 70:4978–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su F, Schneider RJ. 1996. Hepatitis B virus HBx protein activates transcription factor NF-κB by acting on multiple cytoplasmic inhibitors of rel-related proteins. J Virol 70:4558–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samadani U, Costa RH. 1996. The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression. Mol Cell Biol 16:6273–6284. doi: 10.1128/MCB.16.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrem H, Klempnauer J, Borlak J. 2004. Liver-enriched transcription factors in liver function and development. II. The C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev 56:291–330. [DOI] [PubMed] [Google Scholar]
- 43.Antonson P, Xanthopoulos KG. 1995. Molecular cloning, sequence, and expression patterns of the human gene encoding CCAAT/enhancer binding protein alpha (C/EBPα). Biochem Biophys Res Commun 215:106–113. doi: 10.1006/bbrc.1995.2439. [DOI] [PubMed] [Google Scholar]
- 44.Crosson SM, Davies GF, Roesler WJ. 1997. Hepatic expression of CCAAT/enhancer binding protein alpha: hormonal and metabolic regulation in rats. Diabetologia 40:1117–1124. doi: 10.1007/s001250050796. [DOI] [PubMed] [Google Scholar]
- 45.Diehl AM, Yang SQ. 1994. Regenerative changes in C/EBP alpha and C/EBP beta expression modulate binding to the C/EBP site in the c-fos promoter. Hepatology 19:447–456. doi: 10.1002/hep.1840190225. [DOI] [PubMed] [Google Scholar]
- 46.Diehl AM, Michaelson P, Yang SQ. 1994. Selective induction of CCAAT/enhancer binding protein isoforms occurs during rat liver development. Gastroenterology 106:1625–1637. [DOI] [PubMed] [Google Scholar]
- 47.Storch J, McDermott L. 2009. Structural and functional analysis of fatty acid-binding proteins. J Lipid Res 50(Suppl):S126–S131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barter PJ, Rye KA. 2006. Cardioprotective properties of fibrates: which fibrate, which patients, what mechanism? Circulation 113:1553–1555. doi: 10.1161/circulationaha.105.620450. [DOI] [PubMed] [Google Scholar]
- 49.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. 1999. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest 103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]