ABSTRACT
Porcine epidemic diarrhea virus (PEDV) is an enteropathogenic coronavirus causing lethal watery diarrhea in piglets. Since 2010, a PEDV variant has spread rapidly in China, and it emerged in the United States in 2013, posing significant economic and public health concerns. The ability to circumvent the interferon (IFN) antiviral response, as suggested for PEDV, promotes viral survival and regulates pathogenesis of PEDV infections, but the underlying mechanisms remain obscure. Here, we show that PEDV-encoded 3C-like protease, nsp5, is an IFN antagonist that proteolytically cleaves the nuclear transcription factor kappa B (NF-κB) essential modulator (NEMO), an essential adaptor bridging interferon-regulatory factor and NF-κB activation. NEMO is cleaved at glutamine 231 (Q231) by PEDV, and this cleavage impaired the ability of NEMO to activate downstream IFN production and to act as a signaling adaptor of the RIG-I/MDA5 pathway. Mutations specifically disrupting the cysteine protease activity of PEDV nsp5 abrogated NEMO cleavage and the inhibition of IFN induction. Structural analysis suggests that several key residues outside the catalytic sites of PEDV nsp5 probably impact NEMO cleavage by modulating potential interactions of nsp5 with their substrates. These data show that PEDV nsp5 disrupts type I IFN signaling by cleaving NEMO. Previously, we and others demonstrated that NEMO is also cleaved by 3C or 3C-like proteinases of picornavirus and artertivirus. Thus, NEMO probably represents a prime target for 3C or 3C-like proteinases of different viruses.
IMPORTANCE The continued emergence and reemergence of porcine epidemic diarrhea virus (PEDV) underscore the importance of studying how this virus manipulates the immune responses of its hosts. During coevolution with its hosts, PEDV has acquired mechanisms to subvert host innate immune responses for its survival advantage. At least two proteins encoded by PEDV have been identified as interferon (IFN) antagonists, papain-like protease (PLP) and N protein. Here, we report that the PEDV nsp5 gene, which encodes the 3C-like protease of PEDV, is another IFN antagonist. Mechanistically, the cysteine protease activity of PEDV nsp5 mediates proteolysis of NEMO, the key adaptor for IFN synthesis, and NEMO is cleaved at glutamine 231 (Q231). The new molecular details and determinants impacting NEMO scission by PEDV nsp5 delineated in this study are fundamental to our understanding of critical virus-host interactions that determine PEDV pathogenesis.
INTRODUCTION
Coronaviruses (CoVs) have the largest RNA viral genomes of viruses belonging to the order Nidovirales, which includes the families Coronaviridae, Arteriviridae, Mesoniviridae, and Roniviridae. Based on genotypic and serological characterizations, the CoV family is divided into four genera: Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV), and Deltacoronavirus (δ-CoV) (1, 2). CoVs are enveloped viruses with a single-stranded, positive-sense RNA genome ranging from approximately 27 to 32 kb. Their genomes contain 7 to 14 open reading frames (ORFs), and ORF1a and ORF1b encode two polyproteins. Upon cleavage by two virus-encoded proteinases, a papain-like proteinase encoded by the gene for nonstructural protein 3 (nsp3) and a 3C-like protease (3CLpro) encoded by the gene for nsp5, the polyproteins are processed into intermediate precursors and mature, individual nonstructural proteins that execute distinct functions in the viral life cycle (3).
CoVs have drawn extensive attention since the outbreaks of severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 (4). CoVs are prone to variation, resulting in new variants and reemergence. For example, porcine epidemic diarrhea virus (PEDV), an enteropathogenic CoV causing lethal watery diarrhea in piglets, was first reported in the early 1970s in Europe (5). In 2010, a large-scale outbreak of porcine epidemic diarrhea (PED) caused by a PEDV variant occurred in China (6). This virus emerged in the United States in 2013 and spread rapidly (7, 8). Furthermore, this PEDV variant possesses the potential to infect humans, posing significant economic and public health concerns (9). The continued emergence and reemergence of PEDV highlight the importance of studying how viruses manipulate the immune responses of their hosts. Understanding this process is critical to enabling successful control of PEDV infections with antiviral therapeutics or prophylaxis through vaccination.
Innate immune responses are activated through host pattern recognition receptors (PRRs), which recognize pathogen-associated molecular patterns (10). CoVs can be sensed by retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), primarily melanoma differentiation-associated protein 5 (MDA5) (11–13). Upon engaging cytoplasmic viral double-strand RNAs (dsRNAs), the RLRs recruit mitochondrial antiviral signaling protein (MAVS, also known as IPS-1/VISA/Cardiff), transmitting signals to an essential bridging adaptor, the NF-κB essential modulator (NEMO; also called IKKγ). Subsequently, classical IKK and IKK-related kinases lead to the phosphorylation of NF-κB and interferon (IFN)-regulatory factor 3 (IRF3). These transcription factors directly activate promoters of type I IFNs such as IFN-β (10, 14). During coevolution with their hosts, many viruses have acquired mechanisms to circumvent these host innate immune responses. We and others have previously found that the 3C protease (3Cpro) of picornaviruses, including hepatitis A virus (HAV) (15–17), foot-and-mouth disease virus (FMDV) (18), coxsackievirus B3 (CVB3) (19), and enterovirus 71 (EV71) (20, 21), target a signaling component(s) of the IFN-inducing pathway(s) for proteolysis, thereby antagonizing innate immune signaling pathways. CoV polyprotein precursors, pp1a and pp1b, are mainly cleaved by a 3CLpro, which displays similarity to the picornavirus 3Cpro in its chymotrypsin fold and cleavage specificity. However, it remains unclear whether the 3CLpro of PEDV antagonizes the induction of type I IFNs by targeting or cleaving a component(s) of the IFN signaling pathway.
Herein, we provide evidence that the reemerging CoV PEDV 3CLpro nsp5 disrupts type I IFN signaling by cleaving a critical signaling protein, NEMO, which is also a substrate shared by 3Cpro or 3CLpro of FMDV, HAV, and porcine reproductive and respiratory syndrome virus (PRRSV) (22). Furthermore, we also dissected the critical residues in PEDV nsp5 that govern NEMO cleavage. These data uncover a new mechanism acquired by PEDV to disarm host innate immune responses.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney cells (HEK-293T), African green monkey kidney epithelial cells (Vero), and porcine kidney cells (PK-15) were obtained from the China Center for Type Culture Collection and maintained in Dulbecco's modified Eagle medium (DMEM, Invitrogen, USA) supplemented with 10% heat-inactivated fetal calf serum (FCS) at 37°C with 5% CO2 in a humidified incubator. PEDV strain AJ1102 (GenBank accession number JX188454), which was isolated from a suckling piglet with acute diarrhea in China in 2011, was used in this study (23). PEDV strain AJ1102 was propagated in Vero cells, which are widely used for PEDV isolation, propagation, vaccine production, and basic research. Sendai virus (SEV) was obtained from the Centre of Virus Resource and Information, Wuhan Institute of Virology, Chinese Academy of Sciences.
Plasmids.
The luciferase reporter plasmids IFN-β-Luc, IRF3-Luc, and NF-κB-Luc have been described previously (17, 24). The luciferase reporter plasmids, IRF-3-Luc and NF-κB-Luc, each contained four copies of the IRF- or NF-κB-binding motif of the porcine IFN-β promoter upstream of the firefly luciferase reporter gene. cDNA expression constructs encoding human MAVS, MDA5, RIG-I, NEMO, a constitutively activated form of NEMO (NEMO-K277A), and porcine NEMO have been described previously (17, 18). The cDNA expression constructs encoding PEDV nsp5 and nsp4-5 were PCR amplified and cloned into pCAGGS-HAC or pCAGGS-FlagC, which encode a C-terminal hemagglutinin (HA) tag and a Flag tag. Mutagenesis of NEMO and PEDV nsp5 and nsp4-5 constructs was carried out by overlap extension PCR using specific mutagenic primers (available upon request). All constructs were validated by DNA sequencing.
RNA extraction and quantitative real-time RT-PCR.
Briefly, total RNA was extracted from the cells using TRIzol reagent (Invitrogen), and RNA (1 μg) was reverse transcribed to cDNA using avian myeloblastosis virus (AMV) reverse transcriptase (TaKaRa, Japan). The cDNA (1 μl of 25 μl cDNA) was subsequently used in a SYBR green PCR assay (Applied Biosystems). The abundance of individual mRNA transcript in each sample was assayed three times and normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The primers are as follows: human IFN-β (GenBank accession number NM_002176; 5′-TCTTTCCATGAGCTACAACTTGCT-3′ and 5′-GCAGTATTCAAGCCTCCCATTC-3′); human GAPDH (NM_002046; 5′-TCATGACCACAGTCCATGCC-3′ and 5′-GGATGACCTTGCCCACAGCC-3′); monkey NEMO (XM_007993157; 5′-GTAGAGCACCTGAAGAGATG-3′ and 5′-AGAGTCTGGCATTCCTTAGT-3′); monkey GAPDH (XM_007967342; 5′-AAGGTGAAGGTCGGAGTC-3′ and 5′-GGTGGAATCATACTGGAACA-3′).
Luciferase reporter gene assays.
HEK-293T cells or PK-15 cells grown in 48-well plates were cotransfected with a reporter plasmid, pRL-TK (Promega) (used as an internal control for normalization of transfection efficiency), and various expression plasmids or an empty control plasmid. Where indicated, cells were further infected with SEV 24 h after the initial cotransfection. Cells were lysed 16 h later and firefly luciferase and Renilla luciferase activities were determined using the dual-luciferase reporter assay system (Promega, USA) according to the manufacturer's protocol. The data represent relative firefly luciferase activity normalized to Renilla luciferase activity and are representative of three independently conducted experiments. Data are presented as means and standard deviations (SD). Values of P of <0.05 were considered statistically significant, and P values of <0.01 were considered highly statistically significant.
Western blot analyses.
Briefly, HEK-293T cells or PK-15 cells cultured in 60-mm dishes were transfected with various plasmids. After 30 h, the cells were harvested by the addition of lysis buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 20 nM phenylmethylsulfonyl fluoride [PMSF]), and protein concentrations were measured in whole cellular extracts. Equal amounts of the samples were then subjected to SDS-PAGE and analyzed for their expression of RIG-I, MDA5, MAVS, or NEMO proteins by Western blotting using an anti-Flag antibody (Macgene, China). To confirm the expression levels of HA-tagged wild-type (WT) and mutant PEDV nsp5, an anti-HA antibody (MBL, Japan) was used for immunoblotting. Expression of β-actin was detected with an anti-β-actin mouse monoclonal antibody (Beyotime, China) to confirm loading of equal protein amounts.
RESULTS
Identification of PEDV nsp5 as an IFN antagonist.
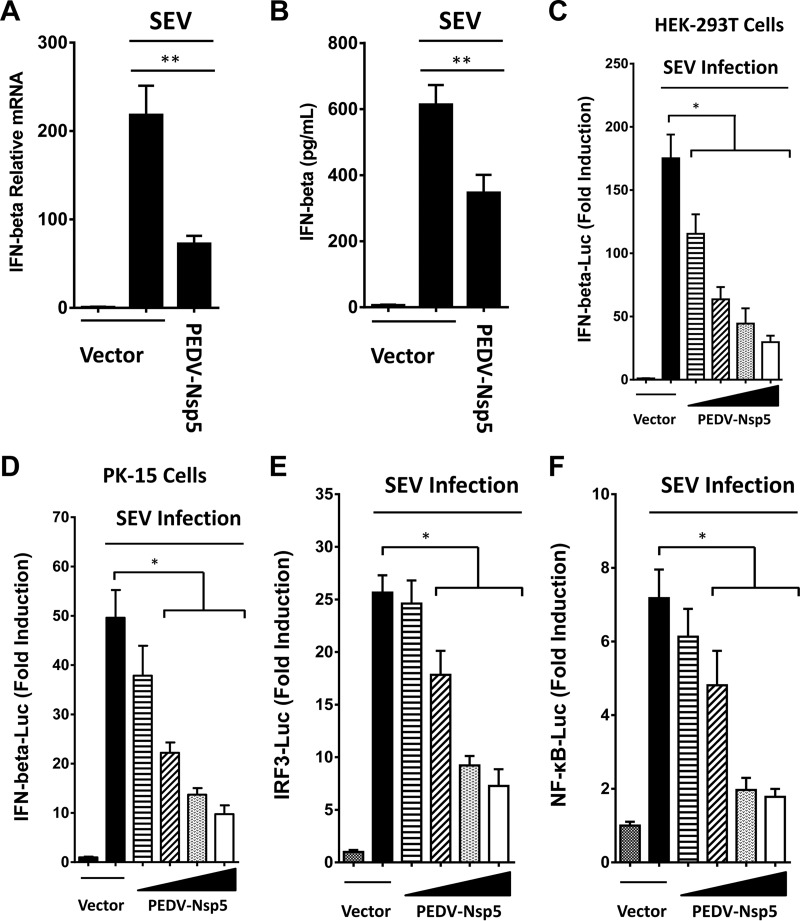
To determine if PEDV nsp5 has the ability to inhibit IFN-β expression, we constructed an expression vector encoding the nsp5 of PEDV and determined the impact of its ectopic expression on Sendai virus (SEV)-induced IFN-β synthesis in transiently transfected HEK-293T cells. As shown in Fig. 1A, B, and C, PEDV nsp5 significantly inhibited SEV-induced IFN-β synthesis, as revealed by real-time reverse transcription-PCR (RT-PCR), enzyme-linked immunosorbent assay (ELISA), and promoter-based luciferase reporter assay. We also found that PEDV nsp5 inhibited SEV-induced activation of IFN-β promoter in PK-15 cells (Fig. 1D). These results demonstrate that PEDV nsp5 is a new CoV-encoded IFN antagonist. Furthermore, activation of both IRF3-dependent and NF-κB-dependent promoters was dose-dependently impaired by PEDV nsp5 in HEK-293T cells (Fig. 1E and F), suggesting that PEDV nsp5 likely targets a step in the IFN-inducing pathway prior to the bifurcation of IRFs and NF-κB.
FIG 1.
PEDV nsp5 inhibits IFN-β promoter activation. (A and B) HEK-293T cells cultured in 48-well plates were transfected with PEDV nsp5 expression plasmid or an empty vector (0.5 μg). Twenty-four hours after the initial transfection, the cells were further infected or mock infected with SEV. The cells and supernatants were collected at 16 h postinfection (hpi) and analyzed for IFN-β levels by real-time RT-PCR (A) and ELISA (B). (C and D) HEK-293T cells or PK-15 cells cultured in 48-well plates were transfected with IFN-β-Luc plasmid (0.1 μg) along with pRL-TK plasmid (0.01 μg, for normalization of transfection efficiency) and increasing quantities of plasmids encoding the indicated PEDV nsp5 (0, 0.05, 0.1, 0.2, or 0.4 μg). Twenty-four hours after the initial transfection, the cells were further infected or mock infected with SEV. Luciferase assays were performed at 16 hpi. (E and F) The experiments were performed similarly to those whose results are shown in panel C, except that the IRF3-Luc (E) or NF-κB-Luc (F) promoter reporter plasmid was used. *, P < 0.05; **, P < 0.01.
Protease activity governs the ability of PEDV nsp5 to suppress IFN induction.
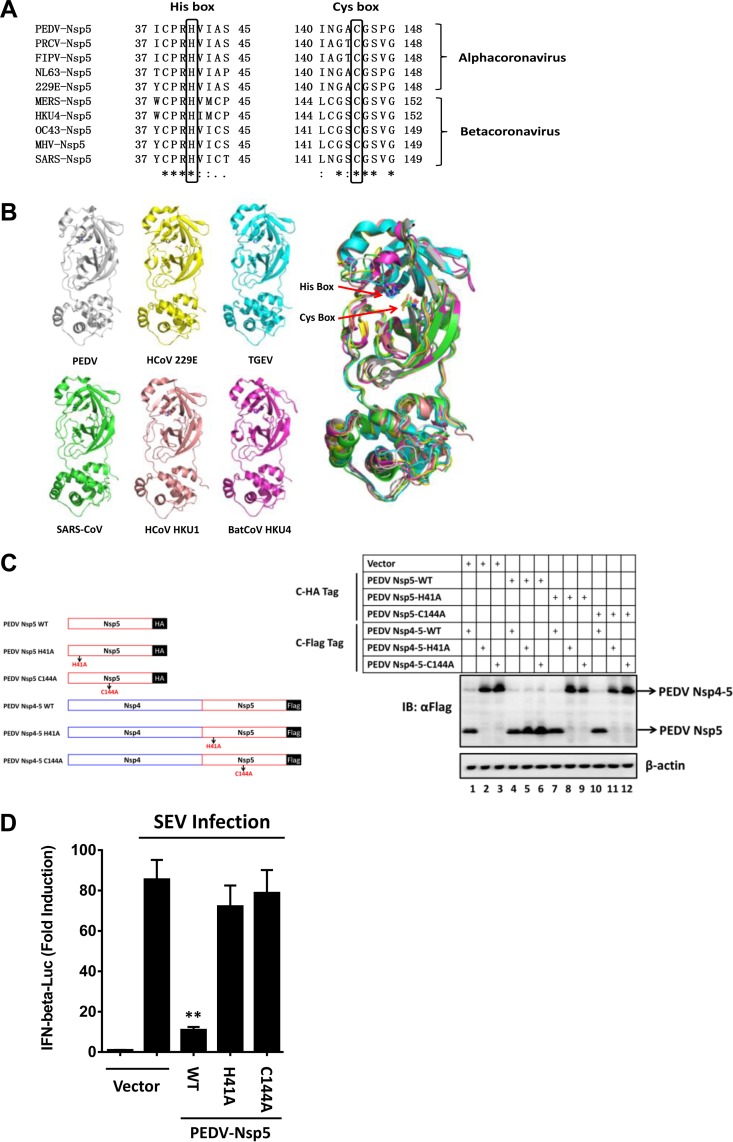
CoV nsp5 is a cysteine proteinase that is responsible for most cleavages within the viral polyprotein (3). The structures of their catalytic domains exhibit a high degree of homology among all α- and β-CoV nsp5s, mainly in two regions known as cysteine (Cys [C]) and histidine (His [H]) boxes that surround the catalytic Cys and His residues (25–28). Sequence alignments showed that His41 and Cys144 (numbering based on PEDV nsp5) are highly conserved among both α-CoVs and β-CoVs (Fig. 2A). A comparison of all available α-CoV and β-CoV nsp5 three-dimensional (3D) structures revealed that the locations of the catalytic residues are also well conserved (Fig. 2B). As further evidence supporting this idea, a mutation at either residue (Ala for His41 [H41A] or Ala for Cys144 [C144A]) eliminated the capacity of PEDV nsp5 to catalytically cleave a precursor of PEDV nsp5, nsp4-5 (Fig. 2C), suggesting that residues His41 and Cys144 of PEDV nsp5 form a catalytic dyad. In contrast to wild-type (WT) PEDV nsp5, PEDV nsp5-H41A and nsp5-C144A were incapable of suppressing a SEV-induced IFN-β promoter (Fig. 2D). Thus, PEDV nsp5 depends on its protease activity to antagonize IFN-β production.
FIG 2.
The protease activity governs the ability of PEDV nsp5 to suppress virus-induced IFN response. (A) Amino acid alignment of the conserved region surrounding Cys145 and His41 (numbering is based on PEDV nsp5) in CoV nsp5s. Black boxes indicate conserved enzymatic proteolysis residues. The sequences were derived from GenBank entries with the following accession numbers: PEDV, AFQ37597.1; porcine respiratory coronavirus (PRCV), ABG89316.1; feline infectious peritonitis virus (FIPV), AGZ84525.1; human coronaviruses NL63 (HCoV NL63), AFV53147.1; human coronaviruses 229E (HCoV 229E), AGW80947.1; Middle East respiratory syndrome coronavirus (MERS-CoV), AGV08401.1; bat coronavirus HKU4 (BatCoV HKU4), YP_001039952.1; human coronaviruses OC43 (HCoV OC43), AAR01012.1; murine hepatitis virus (MHV), NP_068668.2; SARS-CoV, NP_828850.1. (B) Structural alignment of the conserved residues Cys145 and His41 in CoV nsp5s. Red arrows indicate conserved enzymatic proteolysis residues. The 3D structures were derived from the Protein Data Bank with the following accession numbers: PEDV, 4XFQ; HCoV 229E, 2ZU2; porcine transmissible gastroenteritis virus (TGEV), 2AMP; SARS-CoV, 3IWM; human coronavirus HKU1 (HCoV HKU1), 3D23; BatCoV HKU4, 2YNA. (C) Identification of conserved enzymatic proteolysis residues in PEDV nsp5. (Left) Schematic representation of PEDV nsp5 and its precursors. (Right) HEK-293T cells were transfected with the indicated HA-tagged PEDV nsp5 and Flag-tagged PEDV nsp5 precursors (nsp4-5). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (D) HEK-293T cells were cotransfected with IFN-β-Luc plasmid, pRL-TK plasmid, and PEDV nsp5 expression plasmid (0.4 μg). Twenty-four hours after the initial transfection, the cells were further infected or mock infected with SEV. Luciferase assays were performed 16 h after infection. **, P < 0.01.
PEDV nsp5 disrupts RIG-I/MDA5 signaling and cleaves NEMO.
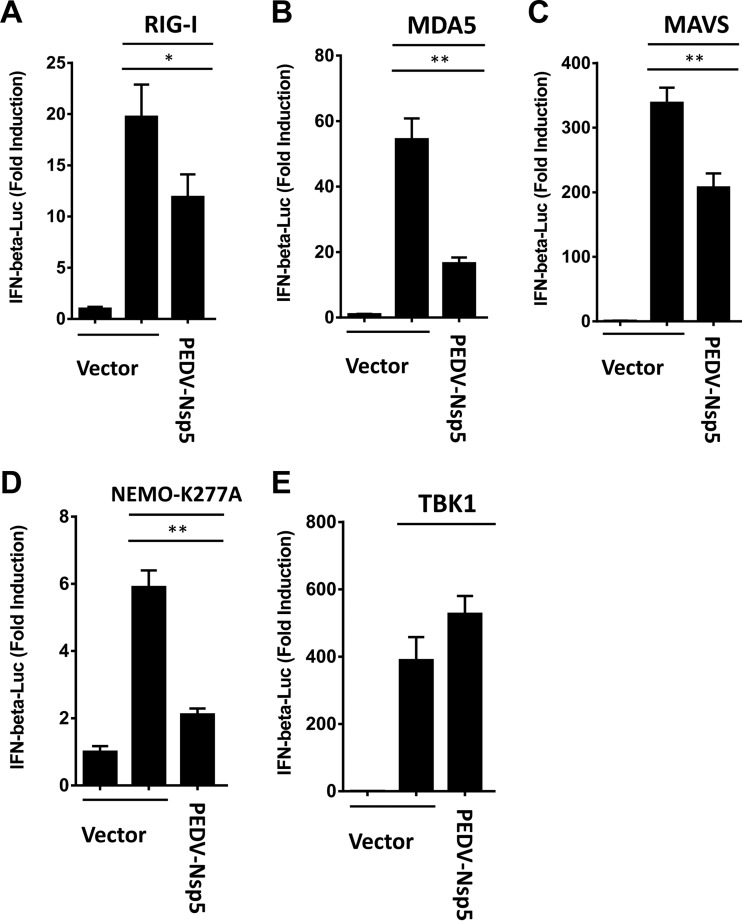
Given the pivotal role of RIG-I/MDA5 in mediating the viral induction of IFN-β (10, 14), we further investigated if PEDV nsp5 inhibits RIG-I/MDA5-mediated IFN signaling. Overexpression of RIG-I, MDA5, MAVS, a constitutively activated NEMO mutant (NEMO-K277A), or TBK1 significantly activated the IFN-β promoter, compared with the empty plasmid control (Fig. 3). However, activation of the IFN-β promoter driven by NEMO-K277A or its upstream molecules (RIG-I, MDA-5, and MAVS) was inhibited by PEDV nsp5 (Fig. 3A to D). In contrast, TBK-1-induced IFN-β promoter activation was not affected by PEDV nsp5 (Fig. 3E), suggesting that PEDV nsp5 inhibits RIG-I/MDA5 signaling upstream of TBK1.
FIG 3.
PEDV nsp5 disrupts RIG-I/MDA5 signaling. HEK-293T cells were cotransfected with IFN-β-Luc plasmid, pRL-TK plasmid, and plasmid encoding PEDV nsp5 (0.2 μg), together with the human RIG-I (A), MDA5 (B), MAVS (C), NEMO-K277A (D), or TBK1 (E) expression vector (0.2 μg). Luciferase assays were performed 36 h after transfection. *, P < 0.05; **, P < 0.01.
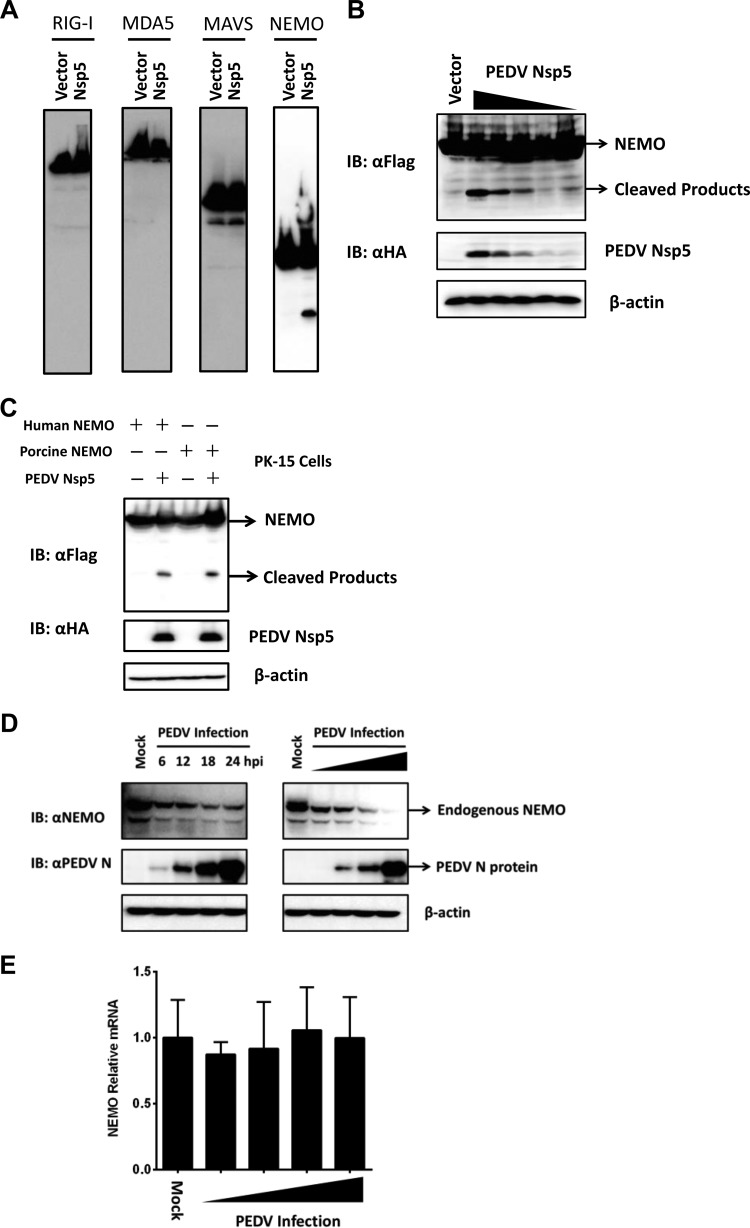
Because the protease activity was critical for PEDV nsp5-mediated inhibition of the IFN response (Fig. 2D), we speculated that nsp5 may target a signaling component(s) of the IFN-inducing pathway(s) for cleavage. Considering that PEDV nsp5 inhibits RIG-I/MDA5 signaling upstream of TBK1 (Fig. 3), we thus investigated if RIG-I, MAVS, NEMO, and MDA5, the upstream signaling molecules of TBK1, serve as substrates for PEDV nsp5. As shown in Fig. 4A, coexpression of Flag-NEMO and PEDV nsp5 resulted in the appearance of faster-migrating bands that were detected by an anti-Flag antibody and had molecular masses of approximately 24 kDa. Presumably, this band is the PEDV nsp5 cleavage product. In contrast, no cleavage products were observed for MAVS, RIG-I, or MDA5. Notably, the degree of NEMO cleavage was positively correlated with the expression levels of PEDV nsp5 (Fig. 4B). Because of species differences between human and porcine NEMO, we analyzed whether PEDV nsp5 also cleaves porcine NEMO. To this end, we transfected PK-15 cells with the human or porcine Flag-NEMO-encoding vector along with an empty vector or plasmid encoding PEDV nsp5. As shown in Fig. 4C, both porcine NEMO and human NEMO were cleaved in PEDV nsp5-transfected PK-15 cells, concomitant with the same PEDV nsp5 cleavage products.
FIG 4.
PEDV nsp5 cleaves NEMO. (A) HEK-293T cells cultured in 60-mm dishes were transfected with a Flag-tagged human RIG-I, MDA5, MAVS, or NEMO expression plasmid (4 μg) along with an empty vector or plasmid encoding PEDV nsp5 (2 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (B) HEK-293T cells were transfected with a Flag-tagged human NEMO expression plasmid (4 μg), along with increasing quantities of plasmids encoding HA-tagged PEDV nsp5 (0, 0.125, 0.25, 0.5, 1, and 2 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (C) PK-15 cells were transfected with a Flag-tagged human NEMO or porcine NEMO expression plasmid (6 μg) along with an empty vector or plasmid encoding PEDV nsp5 (3 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (D) (Left) HEK-293T cells were infected with PEDV AJ1102 strain (multiplicity of infection [MOI] of 0.2), lysed at different postinfection times, and analyzed by Western blotting. (Right) Vero cells were infected with different doses (MOI of 0.0002, 0.002, 0.02, or 0.2) of PEDV, lysed 24 h postinfection, and analyzed for NEMO protein levels by Western blotting. (E) Vero cells were infected with different doses (MOI of 0.0002, 0.002, 0.02, or 0.2) of PEDV, lysed 24 h postinfection, and analyzed for NEMO mRNA levels by real-time RT-PCR.
NEMO is reduced in PEDV-infected cells.
To exclude the possibility that the observed PEDV nsp5-mediated NEMO cleavage was an artificial effect of plasmid overexpression in cell culture, we analyzed NEMO cleavage in the context of PEDV infection. Compared with uninfected cells, NEMO protein abundance was substantially reduced in PEDV-infected Vero cells. In these experiments, the degree of NEMO reduction correlated with the PEDV infection dose, and substantial NEMO degradation coincided with the level of PEDV replication at 6 h postinfection and beyond (Fig. 4D). Furthermore, the NEMO mRNA content was comparable between mock-infected and PEDV-infected cells (Fig. 4E).
PEDV nsp5 cleaves NEMO at glutamine 231.
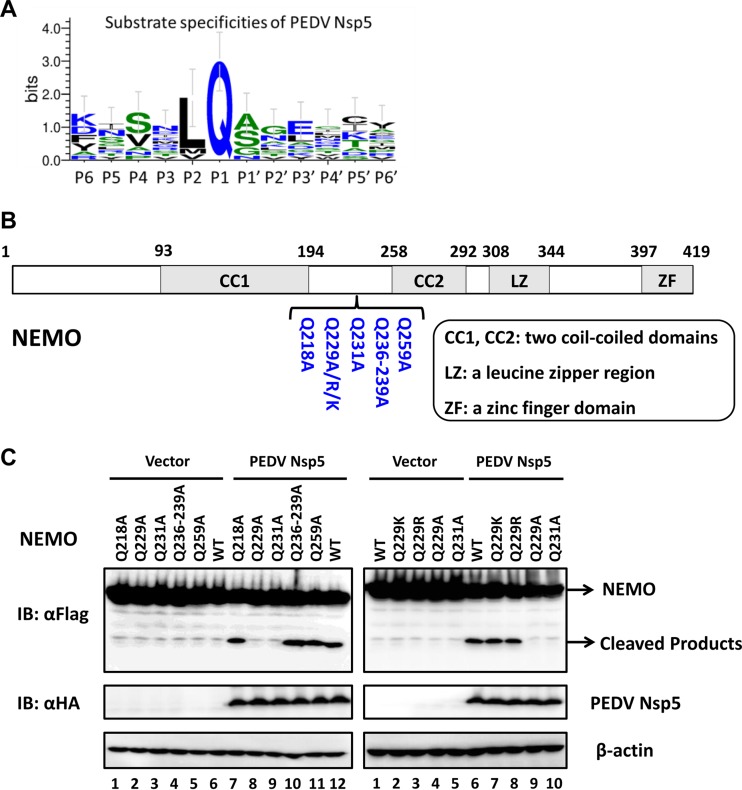
We next examined the sequence of NEMO for potential PEDV nsp5 cleavage sites. Previous studies on CoV nsp5 substrate specificity documented a preference for glutamine (Gln [Q]) at the P1 position (29, 30). As NEMO cleavage produced an approximately 24-kDa product by PEDV nsp5, we focused on the sequence of NEMO for potential PEDV nsp5 cleavage sites that could be capable of generating fragments of the appropriate size following nsp5 scission. We constructed a series of NEMO mutants in which the invariant Gln at each potential P1 position was replaced with alanine (Ala [A]) and examined their cleavage by PEDV nsp5 (Fig. 5A and B). PEDV nsp5-mediated NEMO cleavage was not affected by Q218A, Q236A/Q239A, or Q259A mutations. In contrast, either Q229A or Q231A substitution (Leu-Ala-Gln229-Leu-Gln231-Val-Ala-Tyr) blocked the appearance of the cleavage product as well as the reduction in abundance of full-length NEMO (Fig. 5C, left, lanes 8 and 9). Escarmís et al. showed that a mutation of the FMDV capsid precursor, which is not at the cleavage site, may also affect 3Cpro-mediated capsid precursor cleavage (31). Since the CoV nsp5 substrate specificity showed preferences for leucine (Leu [L]) at the P2 position and lysine (Lys [K])/arginine (Arg [R]) at the P3 position (30), we speculated that a Q229A substitution may affect 3Cpro-mediated NEMO cleavage at Q231. We next constructed two NEMO mutants in which Q229 at the potential P3 position was substituted with lysine or arginine. Remarkably, PEDV nsp5-mediated NEMO cleavage was not affected by Q229K or Q229R mutations (Fig. 5C, right, lanes 7 and 8), suggesting that PEDV nsp5 cleaves NEMO at Gln231 but not at Gln229.
FIG 5.
Glutamine 231 is the site of PEDV nsp5-mediated cleavage of NEMO. (A) Sequence logos of the polyprotein junctions cleaved by PEDV nsp5. An amino acid sequence logo of the substrate was generated by WebLogo 3 (http://weblogo.threeplusone.com/). (B) Schematic representation of WT NEMO and its derivatives. (C) HEK-293T cells were transfected with Flag-tagged WT NEMO or NEMO mutants as indicated, along with PEDV nsp5. Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting.
PEDV nsp5-induced NEMO cleavage fragments lose the ability to induce type I IFN.
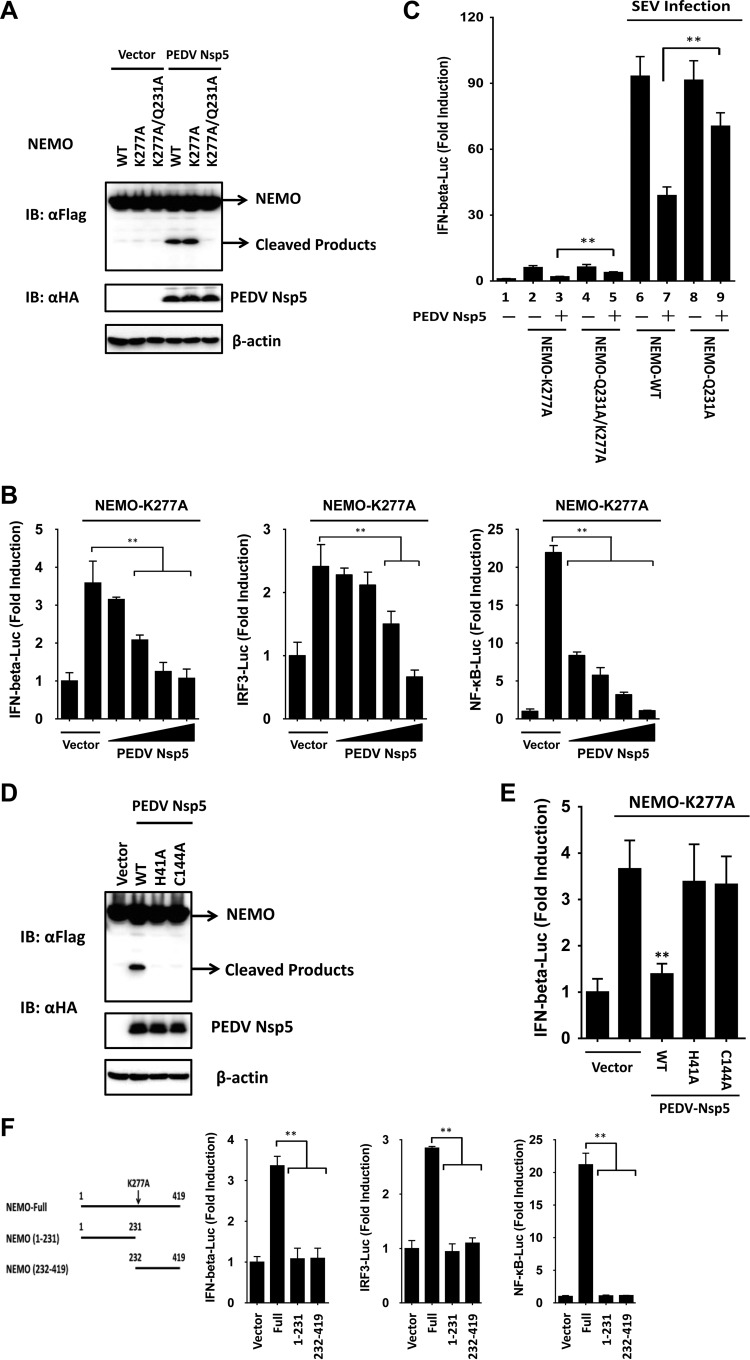
Because WT NEMO did not significantly activate IRFs and NF-κB to induce IFN-β, we used a constitutively active NEMO mutant, NEMO-K277A (17, 32), to examine the effect of PEDV nsp5-mediated NEMO cleavage on IFN-β induction. The K277A substitution did not alter the susceptibility of NEMO to PEDV nsp5 (Fig. 6A), whereas the Q231A mutations rendered NEMO-K277A resistant to cleavage by PEDV nsp5 (Fig. 6A). When it was transfected into HEK-293T cells, NEMO-K277A resulted in the constitutive activation of the IFN-β promoter. However, this activation was significantly inhibited in the presence of PEDV nsp5 in a dose-dependent manner (Fig. 6B). Similar results were obtained when the cells were transfected with IRF3-Luc or NF-κB-Luc in place of IFN-β-Luc (Fig. 6B). Taken together, these data show that PEDV nsp5-mediated cleavage of NEMO-K277A likely impairs the ability of the NEMO-K277A to induce type I IFN expression by disrupting activation of IRFs and NF-κB. We also found that the Q231A mutant significantly restored the ability of NEMO-K277A and NEMO to activate the IFN-β promoter and to potentiate SEV-induced IFN-β promoter activity, respectively, in the presence of PEDV nsp5 overexpression (Fig. 6C, compare columns 3, 5, 7, and 9). The incomplete restoration of IFN activation by the Q231A mutant NEMO could be attributed to the ability of PEDV nsp5 to associate with NEMO (and NEMO-Q231A), which disrupts NEMO-containing signaling complex formation, in addition to NEMO cleavage. This effect was observed in a previous study (18) with 3Cpro of FMDV, which also cleaves NEMO.
FIG 6.
PEDV nsp5-mediated NEMO cleavage is involved in the inhibition of type I IFN induction. (A) HEK-293T cells were transfected with a constitutively activated form of NEMO (NEMO-K277A) or its mutants as indicated (4 μg), along with PEDV nsp5 (2 μg). Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (B) HEK-293T cells were transfected with the indicated reporter plasmids, pRL-TK plasmid, and Flag-tagged NEMO-K277A expression plasmid (0.2 μg), along with increasing quantities of the plasmid encoding PEDV nsp5 (0.025, 0.05, 0.1, and 0.2 μg). Luciferase assays were performed at 36 h after the transfection. (C) HEK-293T cells were cotransfected with IFN-β-Luc, pRL-TK plasmid, and a plasmid encoding PEDV nsp5 (0.15 μg) together with the empty vector or the NEMO-WT, NEMO-Q231A, NEMO-K277A, or NEMO-Q231A/K277A expression vector (0.25 μg). Twenty-four hours after the initial transfection, the cells were further infected or mock infected with SEV. Luciferase assays were performed at 16 h after infection. (D) HEK-293T cells were transfected with the Flag-tagged NEMO expression plasmid, along with the indicated PEDV nsp5 expression plasmids. Cell lysates were prepared 30 h posttransfection and analyzed by Western blotting. (E) HEK-293T cells were cotransfected with IFN-β-Luc plasmid, pRL-TK plasmid, and Flag-tagged NEMO-K277A expression plasmid along with the designated PEDV nsp5 expression plasmids. Luciferase assays were performed at 36 h after the transfection. (F) HEK-293T cells were cotransfected with the indicated reporter plasmids, pRL-TK plasmid, and either a plasmid encoding Flag-fused NEMO-K277A (Full), a plasmid encoding putative PEDV nsp5-induced cleavage fragments of NEMO-K277A, or an empty vector (0.4 μg). Luciferase assays were performed at 36 h after the transfection. **, P < 0.01.
We next determined if PEDV nsp5 cleaves NEMO by means of its protease activity. To this end, the abilities of WT and various catalytically inactive mutants of PEDV nsp5 (PEDV nsp5-H41A and PEDV nsp5-C144A) to cleave ectopically coexpressed Flag-NEMO-K277A were compared (Fig. 6D). Whereas PEDV WT nsp5 induced NEMO cleavage, none of the nsp5 mutants did so. Thus, NEMO is a proteolytic substrate for PEDV nsp5. Importantly, none of the PEDV nsp5 catalytically inactive mutants inhibited activation of the IFN-β promoter by ectopically expressed NEMO-K277A (Fig. 6E). This result is consistent with the inability of these nsp5 mutants to cleave NEMO (Fig. 6D), which may explain our earlier observation that the catalytically inactive nsp5 mutants lost their capacity to inhibit the SEV-induced IFN response (Fig. 2D).
To further determine if either of the PEDV nsp5-induced cleavage fragments of NEMO retains the ability to activate the IFN response, we ectopically expressed the constitutively active NEMO-K277A, the N-terminal fragment of NEMO-K277A (encompassing residues 1 to 231), or the C-terminal NEMO fragment (encompassing residues 232 to 419) that would result from PEDV nsp5 cleavage and examined their abilities to activate IFN-β-, IRF3-, and NF-κB-dependent promoters. In contrast to full-length NEMO-K277A, which was competent in inducing all three promoters, none of the N-terminal or C-terminal fragments had such effects (Fig. 6F), suggesting that PEDV nsp5-mediated cleavage of NEMO efficiently inactivates NEMO-mediated downstream signaling.
DISCUSSION
During coevolution with their hosts, CoVs have acquired mechanisms to subvert host innate immune responses. At least eight proteins encoded by SARS-CoV, the most studied CoV, have been identified as IFN antagonists: nsp1, papain-like protease (PLP), nsp7, nsp15, N, M, ORF3b, and ORF6 (33–40). Additionally, the ORF4a- and ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus (MERS-CoV) have been identified as IFN antagonists (41, 42). In this study, we present evidence that PEDV nsp5 is another CoV-encoded protein capable of antagonizing type I IFN signaling, highlighting the multifaceted control of host innate immunity by CoVs. Furthermore, we demonstrate that PEDV nsp5 adopts a new mechanism from other known CoV-encoded IFN antagonists.
Of the several known viral evasion strategies, the cleavage of innate immune adaptor molecules is a particularly powerful means of inactivating antiviral responses, because this strategy efficiently suppresses common downstream targets of key innate immune signaling pathways. For example, CVB3, HAV, and EV71 cleave both the RIG-I/MDA5 adaptor protein MAVS and the Toll-like receptor 3 (TLR3) adaptor, Toll-IL-1 receptor domain-containing adaptor inducing interferon beta (TRIF) (15, 16, 19, 20, 43). NEMO, a crucial adaptor at the converging point of multiple signaling pathways leading to type I IFN expression has been reported to be cleaved by FMDV, HAV, and PRRSV (17, 18, 22). In this study, we found for the first time that NEMO is also a substrate shared by PEDV nsp5, a 3CLpro. Based on data presented here and those of others mentioned above, we speculate that NEMO represents a prime target for 3C or 3C-like proteinases of different viruses and that NEMO proteolysis facilitates evasion of innate immunity by and survival of 3C- or 3C-like protease-encoding viruses.
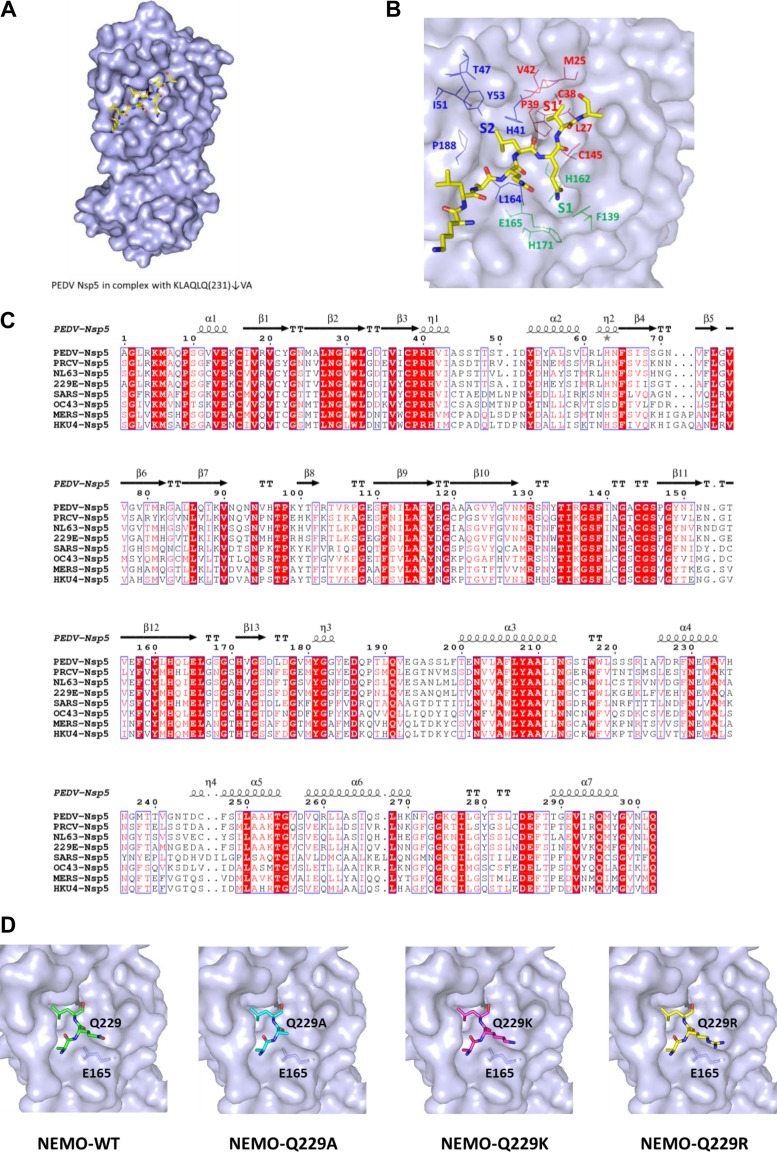
Because the structure of PEDV nsp5 in complex with its peptide substrate was unavailable, and the structures of all CoV nsp5s are very similar (44), we constructed a homology model for PEDV in complex with its peptide substrate of NEMO based on the structure of SARS-CoV nsp5 (PDB ID 2Q6G). The residues KLAQLQ231↓VA (P6 to P2′; numbering is based on NEMO) occupy, and thereby define, the subsites S6 to S2′ of PEDV (Fig. 7A). The side chains of PEDV nsp5 Phe139, His162, Glu165, and His171 are involved in constituting the S1 subsite, which has an absolute requirement for Gln at the P1 position via two hydrogen bonds. In fact, all of these residues are conserved in all known coronavirus 3CLpro (Fig. 7B and C) (25–28). The deep and wide hydrophobic S2 subsite of PEDV nsp5 is formed by the side chains of Leu164, Ile51, Thr47, His41, Pro188, and Tyr53 and is able to accommodate the relatively large side chain of conserved Leu or Met (Fig. 7B). At the substrate binding site of CoV nsp5, the orientation of the long side chain P3 residues (Arg, Lys, or Asp) is in accordance with that of Glu165, which would stabilize the substrate (or inhibitor) binding at this position via a van der Waals interaction between these two residues (28). The interaction suggests that a mutation of the P3 position with a relatively short side chain, such as Ala, is potentially harmful for nsp5-substrate binding (Fig. 7D). This idea is strongly supported by the observation that the ability of PEDV nsp5 to cleave the NEMO mutant Q229A, which has a shorter side chain than the Q229, Q229K, and Q229R mutants at the P3 position, is significantly impaired.
FIG 7.
The modeled structure of PEDV nsp5 in complex with a peptide substrate. (A) Homology model for PEDV and MERS-CoV nsp5 in complex with their peptide substrates based on the structure of SARS-CoV nsp5 (PDB ID 2Q6G). The residues KLAQLQ231↓VA (P6 to P2′, numbering based on NEMO) occupy, and thereby define, the subsites S6 to S2′ of PEDV nsp5. (B) The positions of P6 to P2′, S2 to S1′, and the potential residues forming the S2 (blue), S1 (green), and S1′ (red) subsites are labeled. (C) Structure-based sequence alignment of CoV nsp5 from two genera. The α-CoVs were PEDV, PRCV, HCoV 229E, and HCoV NL63; the β-CoVs were MERS-CoV, SARS-CoV, HCoV OC43, and BatCoV HKU4. Secondary structures of PEDV nsp5 are indicated above the sequence. (D) Schematic diagram of the potential van der Waals interactions between P3 residues (NEMO-Q229, NEMO-Q229A, NEMO-Q229K, or NEMO-Q229R) and Glu165 in PEDV nsp5.
Although all tested 3C and 3C-like proteases can cleave NEMO, distinct sites in NEMO are targeted by different viruses. For example, FMDV and HAV 3Cpro cleave NEMO at Q383 and Q304, respectively, whereas PRRSV nsp4 specifically targets NEMO at E349 (17, 18, 22). Results from the current study also reveal that, NEMO is cleaved at Gln231 by PEDV nsp5. However, to our knowledge, the structural basis for distinct NEMO cleavage sites had not been examined to date. Although the protease activity was critical for PEDV nsp5-mediated NEMO cleavage and inhibition of the IFN response, we cannot exclude the possibility that other nonactive site residues of PEDV nsp5 are also involved. Future investigation to identify other nonactive site residues but do not affect the protease activity will be required.
In summary, our data identify PEDV nsp5 as a new IFN antagonist encoded by PEDV and show that the nsp5-mediated proteolytic cleavage of NEMO is directly involved in inhibition of IFN-β transcription. In revealing these, our study describes a new mechanism by which PEDV counteracts host innate antiviral responses.
ACKNOWLEDGMENTS
This work was supported by the National Natural Sciences Foundation of China (31225027), the Key Technology R&D Programme of China (2015BAD12B02), the National Basic Research Program (973) of China (2014CB542700), the Natural Science Foundation of Hubei Province (2014CFA009), and the Fundamental Research Funds for the Central Universities (2013PY043).
We thank Qingye Zhang at Huazhong Agricultural University for fruitful discussions.
REFERENCES
- 1.Graham RL, Donaldson EF, Baric RS. 2013. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris JS, Yuen KY, Osterhaus AD, Stohr K. 2003. The severe acute respiratory syndrome. N Engl J Med 349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 5.Wood EN. 1977. An apparently new syndrome of porcine epidemic diarrhoea. Vet Rec 100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- 6.Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. 2012. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis 18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang YW, Dickerman AW, Pineyro P, Li L, Fang L, Kiehne R, Opriessnig T, Meng XJ. 2013. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4:e00737-13. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Li G, Stasko J, Thomas JT, Stensland WR, Pillatzki AE, Gauger PC, Schwartz KJ, Madson D, Yoon KJ, Stevenson GW, Burrough ER, Harmon KM, Main RG, Zhang J. 2014. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J Clin Microbiol 52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Tang J, Ma Y, Liang X, Yang Y, Peng G, Qi Q, Jiang S, Li J, Du L, Li F. 2015. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol 89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat Immunol 7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 11.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Liu Y, Zhang X. 2010. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J Virol 84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneyama M, Fujita T. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev 18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, Lemon SM. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A 104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu L, Feng Z, Yamane D, Liang Y, Lanford RE, Li K, Lemon SM. 2011. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog 7:e1002169. doi: 10.1371/journal.ppat.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Fang L, Wei D, Zhang H, Luo R, Chen H, Li K, Xiao S. 2014. Hepatitis A virus 3C protease cleaves NEMO to impair induction of beta interferon. J Virol 88:10252–10258. doi: 10.1128/JVI.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Fang L, Li K, Zhong H, Fan J, Ouyang C, Zhang H, Duan E, Luo R, Zhang Z, Liu X, Chen H, Xiao S. 2012. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J Virol 86:9311–9322. doi: 10.1128/JVI.00722-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. 2011. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei X, Sun Z, Liu X, Jin Q, He B, Wang J. 2011. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by Toll-like receptor 3. J Virol 85:8811–8818. doi: 10.1128/JVI.00447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei X, Han N, Xiao X, Jin Q, He B, Wang J. 2014. Enterovirus 71 3C inhibits cytokine expression through cleavage of the TAK1/TAB1/TAB2/TAB3 complex. J Virol 88:9830–9841. doi: 10.1128/JVI.01425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C, Zhang Q, Guo XK, Yu ZB, Xu AT, Tang J, Feng WH. 2014. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 antagonizes beta interferon expression by targeting the NF-kappaB essential modulator. J Virol 88:10934–10945. doi: 10.1128/JVI.01396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi J, Zeng S, Xiao S, Chen H, Fang L. 2012. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J Virol 86:10910–10911. doi: 10.1128/JVI.01919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Fang L, Li P, Sun L, Fan J, Zhang Q, Luo R, Liu X, Li K, Chen H, Chen Z, Xiao S. 2011. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol 85:3758–3766. doi: 10.1128/JVI.02589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 26.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci U S A 100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H, Xie W, Xue X, Yang K, Ma J, Liang W, Zhao Q, Zhou Z, Pei D, Ziebuhr J, Hilgenfeld R, Yuen KY, Wong L, Gao G, Chen S, Chen Z, Ma D, Bartlam M, Rao Z. 2005. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol 3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, Li J, Zhou Z, Ding Y, Zhao Q, Zhang XC, Liao M, Bartlam M, Rao Z. 2008. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J Virol 82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuck CP, Chong LT, Chen C, Chow HF, Wan DC, Wong KB. 2010. Profiling of substrate specificity of SARS-CoV 3CL. PLoS One 5:e13197. doi: 10.1371/journal.pone.0013197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chuck CP, Chow HF, Wan DC, Wong KB. 2011. Profiling of substrate specificities of 3C-like proteases from group 1, 2a, 2b, and 3 coronaviruses. PLoS One 6:e27228. doi: 10.1371/journal.pone.0027228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escarmís C, Perales C, Domingo E. 2009. Biological effect of Muller's ratchet: distant capsid site can affect picornavirus protein processing. J Virol 83:6748–6756. doi: 10.1128/JVI.00538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloor S, Ryzhakov G, Wagner S, Butler PJ, Smith DL, Krumbach R, Dikic I, Randow F. 2008. Signal processing by its coil zipper domain activates IKK gamma. Proc Natl Acad Sci U S A 105:1279–1284. doi: 10.1073/pnas.0706552105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol 81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu X, Pan J, Tao J, Guo D. 2011. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ, Jin DY. 2009. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem 284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, Lui PY, Chan CP, Tse H, Woo PC, Yuen KY, Jin DY. 2014. Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol 88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. 2014. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol 95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, Jin Q, Zhao Z. 2013. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog 9:e1003231. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Q, Li S, Xue F, Zou Y, Chen C, Bartlam M, Rao Z. 2008. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J Virol 82:8647–8655. doi: 10.1128/JVI.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]