ABSTRACT
Gibbon ape leukemia viruses (GALVs) are part of a larger group of pathogenic gammaretroviruses present across phylogenetically diverse host species of Australasian mammals. Despite the biomedical utility of GALVs as viral vectors and in cancer gene therapy, full genome sequences have not been determined for all of the five identified GALV strains, nor has a comprehensive evolutionary analysis been performed. We therefore generated complete genomic sequences for each GALV strain using hybridization capture and high-throughput sequencing. The four strains of GALV isolated from gibbons formed a monophyletic clade that was closely related to the woolly monkey virus (WMV), which is a GALV strain that likely originated in a gibbon host. The GALV-WMV clade in turn formed a sister group to the koala retroviruses (KoRVs). Genomic signatures of episodic diversifying selection were detected among the gammaretroviruses with concentration in the env gene across the GALV strains that were particularly oncogenic and KoRV strains that were potentially exogenous, likely reflecting their adaptation to the host immune system. In vitro studies involving vectors chimeric between GALV and KoRV-B established that variable regions A and B of the surface unit of the envelope determine which receptor is used by a viral strain to enter host cells.
IMPORTANCE The gibbon ape leukemia viruses (GALVs) are among the most medically relevant retroviruses due to their use as viral vectors for gene transfer and in cancer gene therapy. Despite their importance, full genome sequences have not been determined for the majority of primate isolates, nor has comprehensive evolutionary analysis been performed, despite evidence that the viruses are facing complex selective pressures associated with cross-species transmission. Using hybridization capture and high-throughput sequencing, we report here the full genome sequences of all the GALV strains and demonstrate that diversifying selection is acting on them, particularly in the envelope gene in functionally important domains, suggesting that host immune pressure is shaping GALV evolution.
INTRODUCTION
Gibbon ape leukemia virus (GALV) is an exogenous gammaretrovirus associated with hematopoietic neoplasms in captive colonies of white-handed gibbon (Hylobates lar). Five strains of GALV have been isolated from gibbons. The first was isolated from an animal with lymphocytic leukemia in a colony at the San Francisco Medical Center (strain SF) (1, 2). GALV was later isolated from gibbons displaying malignant tumors, notably an individual gibbon with granulocytic leukemia, at the Southeast Asia Treaty Organization Medical Research Laboratory in Bangkok, Thailand (strain SEATO) (3, 4), and another gibbon with lymphocytic leukemia from a colony on Hall's Island, near Bermuda (strain GALV-H) (5, 6). The Brain strain was isolated from two healthy gibbons injected with brain extracts from human patients with kuru and from an uninoculated cage mate (7). The SEATO strain has been shown to cause chronic myelogenous leukemia when injected into juvenile gibbons (8).
A closely related retrovirus isolated from a 3-year-old male woolly monkey (Lagothrix lagotricha) with multiple fibrosarcomas was originally designated SSAV (for simian sarcoma-associated virus) and now renamed woolly monkey virus (WMV). WMV is considered a member of the GALV lineage (9). WMV isolated from the woolly monkey exists as a mixture of a replication-defective acute transforming virus and its associated replication-competent helper virus (10). Replication-competent WMV is related to GALV as supported by immunological (11) and serological tests (9), antigenic similarities in some gene products (7, 12, 13), and high RNA sequence homology (5, 7). Since the woolly monkey from which WMV was isolated was reported to have been in contact with a gibbon for the 3 months before its death, WMV is likely the product of a single horizontal transmission of GALV from a gibbon to a woolly monkey.
The GALV genomes deposited in GenBank are not representative of any one of the five GALV strains. Rather, the GALV-SEATO genome deposited by Delassus et al. (14) (M26927) represents a GALV-SEATO/SF chimeric genome that contains an envelope open reading frame (ORF) encoding a truncated form of the envelope protein lacking an R peptide (14). The R peptide in the cytoplasmic terminus of the gammaretroviral envelope protein prevents membrane fusion before budding. Transfection of this truncated form of GALV-SEATO envelope into human cells resulted in the expression of a hyperfusogenic GALV envelope protein with strong cytotoxic effects (15, 16). The second GALV genome sequence available in GenBank (U60065) is from a GALV discovered as a contaminant of an HIV-infected human cell line originally referred to as retrovirus X (17) and subsequently designated the GALV-X strain (18). The provenance of GALV-X remains unknown.
Only envelope sequences of the remaining GALV strains—GALV-Brain, Hall's Island, and SF—have been determined (19). Phylogenetic analysis of the two full-genome GenBank sequences and related retroviruses has revealed that GALV is most closely related to the koala retrovirus (KoRV) among viruses sequenced to date (20). KoRV and GALV occur in taxonomically distant mammalian hosts from different continents, suggesting that these viruses may be the products of a recent cross-species transmission, most likely originating in a common intermediate vector to both species (20, 21). In a recent study attempting to identify such an intermediate host, the Melomys burtoni retrovirus (MbRV) was isolated from the grassland mosaic-tailed rat, an Australian murid rodent, and showed a high nucleotide identity (93%) and close phylogenetic relatedness to GALV-SEATO (M26927) (21). Nevertheless, because of the different geographic distribution of M. burtoni and gibbons, MbRV cannot be considered the source of GALV, and therefore the origins of GALV are still unclear.
To better characterize GALV phylogenetic relationships and functional domains in viral control regions and structural genes besides env, we applied two methods to determine the complete genomic sequence of all known GALV strains. A PCR-based approach on DNA extracted from GALV-infected cell lines using primers designed on the limited GALV sequences available in GenBank was applied, but it did not recover the full genome sequences of all the strains because of the unsuccessful amplification of certain portions of the genomes. Therefore, hybridization capture and high-throughput sequencing were performed to determine the full-length GALV genomes (22, 23). We report the complete nucleotide sequence of all GALV strains, their genomic structure, the phylogenetic relationships within the GALVs, their relationship to other gammaretroviruses, and the selection pressures driving evolution within this retroviral clade.
MATERIALS AND METHODS
Cell lines and viruses.
GALV wild-type viruses were obtained from the following productively infected cell lines: SEATO-88, GALV-SEATO-infected bat lung fibroblasts; GALV-4-88, GALV-Brain-infected bat lung fibroblasts; 71-AP-1, WMV-infected marmoset fibroblasts; MLA-144, GALV-SF-infected primate T cells; 6G1-PB, GALV-Hall's Island-infected lymphocytes; and HOS (ATCC CRL-1543) GALV-SF-infected human osteosarcoma cells. GALV-SF was represented by two different cultures, one from the MLA-144 cell line and another cultured in HOS cells.
DNA extraction.
Genomic DNA extraction from the cell lines was performed using the Wizard Genomic DNA purification kit (Promega) according to the manufacturer's protocol. The DNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies).
PCR.
Two primer pairs were designed, based on the alignment of the GALV sequences available in GenBank (SEATO, M26927; GALV-X, U60065), to target two regions, each ca. 4 kb in length, which together cover the GALV genome. Primers U5 (5′-CCCGTGTGTCCAATAAAACCTCT-3′) and PolR1 (5′-CTAGCCCATACCGTCCGC-3′) were used to amplify the first 4 kb of the GALV genome (the 5′ long terminal repeat [5′ LTR], gag, and part of the pol gene) and primers PolF1 (5′-TGGTATACAGACGGTAGCAGT-3′) and U3 (5′-AGCGAGAGGCAAGGTAAT-3′) for the second 4 kb (part of the pol gene, gag, and the 3′ LTR). The PCRs were performed in a final volume of 23 μl using 100 ng of DNA extract, a 0.6 μM final concentration of each primer, 12.5 μl of 2× MyFi Mix (Bioline), and sterile-distilled water. The thermal cycling conditions were as follows: 95°C for 4 min; 40 cycles at 95°C for 30 s, 53 to 57°C (based on the best PCR product yield per strain determined empirically) for 30 s, and 72°C for 6 min; and finally 72°C for 10 min. An aliquot of each PCR product was visualized on 1.5% (wt/vol) agarose gels stained with GelRed (Biotium). In cases of positive amplification, the PCR products were purified using the MSB Spin PCRapace kit (Stratec Molecular GmbH), quantified using a NanoDrop ND-1000 spectrophotometer, and Sanger sequenced by primer walking. The primers that yielded high-quality Sanger sequences are listed in Table 1.
TABLE 1.
Primers that yielded PCR products and high-quality Sanger sequences
| Primer | Sequence (5′-3′) | GALV strain(s)a |
|---|---|---|
| SSAVF | CAAGAACTCCCACATGACCG | WMV |
| SSAVR | GAACACGTCTGCTCGCTAC | WMV |
| U5 | CCCGTGTGTCCAATAAAACCTCT | SF, SEATO, WMV |
| PolF1 | TGGTATACAGACGGTAGCAGT | SEATO, Br, WMV |
| EnvR1 | CACAAYYCCATTCTTTACAGTAT | SF, SEATO, H, Br, WMV |
| EnvR2 | GGAGGTCAGCATCTATGGCGATC | SF, SEATO, H, Br, WMV |
| U3 | AGCGAGAGGCAAGGTAAT | H, WMV |
| PolR2 | GCAAACCCAGGGATCCAGAGTCTACA | SF, H, Br |
| PolR1 | CTAGCCCATACCGTCCGC | SF, SEATO |
| GagF1 | CCCCTATCTCCCTCACTCT | SEATO, H, Br |
| GagF2 | GACCTCGCTCAGAGTCCCCCACCATG | SEATO |
| F2 | GCCTTCCCCCTCAATCGACCTC | SEATO |
| F3 | ACTAGACAAAGACCAGTGCGCATAC | SEATO |
| F4 | TGGCTCCAGCTTTTCCCCACTG | SEATO |
| EnvF | ACCTCCKGAYTCAGACTATAC | SEATO |
| HallsR | CACGTCTGTTCGCTACTCAC | H |
| HallsF | CTTCTCGCTTCTGTACCCG | H |
Abbreviations: SF, San Francisco; H, Hall’s Island; Br, Brain.
Illumina library preparation.
The extracted DNA from each cell line was sheared using a Covaris M220 (Covaris) to an average size of 250 bp. Aliquots from each fragmented DNA extract were used to generate Illumina libraries as described by Meyer and Kircher (24) with the modifications described in Alfano et al. (25). Each library contained a unique index adapter to allow for subsequent discrimination among samples after the sequencing of pooled libraries. A negative-control extraction library was also prepared and indexed separately to monitor for experimental cross-contamination. Each library was amplified in three replicate reactions to minimize amplification bias in individual PCRs. The amplifications of the libraries were performed using Herculase II Fusion DNA polymerase (Agilent Technologies) in 50-μl volume reactions, with the cycling conditions of 95°C for 5 min, followed by five cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 40 s and then finally 72°C for 7 min. After pooling the three replicate PCR products for each sample, amplified libraries were purified using the QIAquick PCR purification kit (Qiagen) and quantified using the 2200 TapeStation (Agilent Technologies) on D1K ScreenTapes. Three additional amplification cycles were performed for SEATO and SF-HOS libraries using Herculase II Fusion DNA polymerase with P5 and P7 Illumina library outer primers with the same cycling conditions to balance library concentrations.
Hybridization capture baits.
PCR products used as baits for capturing GALV sequences from the Illumina libraries were generated from the SEATO and SF-MLA strains. A preliminary phylogenetic analysis of the envelope nucleotide sequences of SEATO, Hall's Island, Brain, SF, and WMV strains deposited in GenBank by Ting et al. (19) (AF055060 to AF055064) suggested that baits from these two strains would cover sufficient genetic diversity to allow for capture of unknown and divergent GALV sequences, since SEATO and SF represent each of the two main branches in which the GALV strains are clustered and thus cover much GALV diversity (data not shown). The phylogenetic analysis was carried out in Seaview v4 (26) using the neighbor-joining method (27) and the HKY model (28). Node robustness was estimated with 100 bootstrap replicates. KoRV (AF151794) was used as outgroup. Primer pairs U5-PolR1 and PolF1-U3 were used to amplify the genome of SEATO and SF-MLA, with the same reaction setup and thermal profile described in the PCR methods. PCR products were purified using the MSB Spin PCRapace kit, quantified using a NanoDrop ND-1000 and Sanger sequenced to verify that the target region had been amplified. After sequence verification, the PCR products were then pooled to equimolar concentrations to produce a mixed SEATO/SF-MLA bait and fragmented using a Covaris M220 to generate 250-bp fragments. The GALV fragments were then blunt ended using the Quick Blunting kit (New England BioLabs), ligated to a biotin adaptor using the Quick Ligation kit (New England BioLabs), and immobilized in separated individual tubes on streptavidin-coated magnetic beads as described previously (22).
Hybridization capture.
Each amplified Illumina library was mixed with blocking oligonucleotides (200 μM) that help prevent cross-linking of Illumina library adapters, Agilent 2× hybridization buffer, and Agilent 10× blocking agent and heated at 95°C for 3 min to separate the DNA strands (22). Each Illumina library hybridization mixture was then combined in separate tubes with the biotinylated baits bound to the streptavidin beads. Samples were incubated in a mini-rotating incubator (Labnet) for 48 h at 65°C, during which the hybridization took place. After 48 h, the beads were washed to remove off-target DNA as described previously (22), and the hybridized libraries were eluted by incubation at 95°C for 3 min. The DNA concentration for each eluted sample was measured using the 2200 TapeStation on D1K ScreenTapes and further amplified accordingly using P5 and P7 Illumina outer primers (24). The enriched amplified libraries were then pooled in equimolar amounts to a final library concentration of 8 nM for paired-end sequencing (2 × 250) on an Illumina MiSeq platform with the v2 reagents kit at the Danish National High-Throughput DNA Sequencing Centre in Copenhagen, Denmark. As a control, a 1% PhiX genome library spike-in was used.
Genome sequence assembly and annotation.
A total of 12,949,200 paired-end sequence reads 250-bp long were generated (average = 2,158,200 paired-end reads per sample, standard deviation [SD] = 451,197.4) and then sorted by index sequences. Adaptor sequences were trimmed from the reads using Cutadapt v1.2.1 (29), and low-quality reads were removed using Trimmomatic v0.27 (30), with a quality cutoff set at 20. Reads that were shorter than 20 bp were excluded from further analyses. After adaptor and quality trimming, 97.6% of the sequences were retained. Reads were then mapped to the GALV-X full genome reference sequence (U60065) using BWA v0.7.10 with default parameters (BWA-MEM algorithm) (31). Reads from the SEATO strain were also mapped to the SEATO full genome reference sequence (M26927), and the results of the two alignments were compared. Samtools v1.2 (32) was used to convert, sort, and index the aligned data files, while potential duplicates were removed using Picard (http://broadinstitute.github.io/picard). Variant call analysis was performed using GATK v1.6-11 (33), setting the minimum variant frequency to 0.2, the depth of coverage to 20, quality to 30, and the quality by depth to 5. To get better variant calling results, paired-end reads were first merged into single reads using FLASH with default parameters (34). The alignments were then visualized and manually curated using Geneious v7.1.7 (Biomatters, Inc.). Consensus sequences were generated as the majority character state at every position in an alignment of sequences. Regions that mapped poorly, likely corresponding to regions diverging from the reference sequence, were resolved by comparison with previously generated Sanger sequences. Nucleotide positions that could not be resolved by variant calling or Sanger sequencing due to the presence of multiple nucleotides at a given position were identified as polymorphisms and assigned IUPAC ambiguity codes. Exact counts for homopolymer stretches must be considered tentative due to the limitations of the Illumina platform in distinguishing their lengths. Homopolymer lengths were defined by assigning the number of nucleotides detected in the most abundant reads. In order to identify protein domains and regulatory motifs, the nucleotide sequence of each strain was compared to the annotated genome sequences available in GenBank for GALV-X (U60065), SEATO (M26927), and KoRV (AF151794) and also analyzed using the NCBI Conserved Domains Database (CDD; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). The consensus sequence and annotations of each GALV strain genome were deposited in GenBank. Illumina reads mapping to GALV-X for each captured GALV strain were deposited in the NCBI Sequence Read Archive.
Cell lines used in the GALV/KoRV-B chimeric envelope experiment.
293T human embryonic kidney cells (ATCC CCL 11268) and murine Mus dunni tail fibroblast MDTF cells (35) were maintained in Dulbecco modified Eagle medium with high glucose, supplied with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. MDTF cells expressing human PiT1 and human THTR1 individually were described previously (36).
Construction of GALV/KoRV-B chimeric envelope.
Both chimeric envelope proteins were generated using overlap extension PCR cloning as described previously (37), and DNA sequencing analysis confirmed the sequence of each chimeric envelope. The PCR fragment of KoRV-B VRA was used to replace the corresponding VRA of GALV SEATO envelope protein (residues 46 to 100 of GALV) to construct GALV-VRAKoRV-B. The VRA region of KoRV-B, corresponding to envelope residues 49 to 107, was PCR amplified using the following primer pairs flanking the VRA regions of the KoRV-B envelope gene: sense (5′-GTCCTGGGAACTGGAAAAGACTGATCATCCTCTTAAG-3′) and antisense (5′-CTTCTGAAAGGGTCCGGCCATCCCGGGG-3′). GALV-VRAKoRV-B was used as a template to replace the VRB of GALV SEATO (residues 46 to 100) with that of KoRV-B (residues 193 to 204) for the generation of GALV-VRA/VRBKoRV-B. To generate GALV-VRA/VRBKoRV-B, a modified overlap extension PCR cloning was used, where a primer pair containing KoRV-B VRB sequences was used instead of a PCR fragment. The sense primer of the primer pair contain GALV sequences upstream of the VRA region (underlined) sense primer, 5′-GTGTTCGCATGTCCCCGTAGGGTGGCCCAGGCCTACAGTTATGAGGTCTTTTGAGGATTTAGATAGCCA-3′, and the antisense primer contains GALV sequences downstream of the VRA region (underlined): 5′-GTAGGCCTGGGCCACCCTACGGGGACATGCGAACACACCGGCTGGTGTAACCCCCTTAAAATAGATTTC-3′.
V5 epitope tagging of GALV and KoRV-B envelope proteins.
Using the modified overlap extension PCR as mentioned above, the DNA sequence encoding the V5 epitope tag (GKPIPNPLLGLDST) was engineered into the primer pair to be used as the oversized primer for overlap extension PCR cloning to construct tagged KoRV-B and GALV SEATO envelope protein with a V5 epitope inserted downstream of signal peptide at the N-terminal of envelope sequences.
Retroviral vector production and transduction.
A ProFection mammalian transfection system-calcium phosphate kit (Promega) was used for transfection of 293T cells 10-cm plates. For binding assay, 20 μg of expression plasmid encoding individual V5-epitope tagged-envelope protein was transfected into 293T cells. For assessment of envelope function of the different chimeras, pCI-neo plasmid encoding individual envelope protein was cotransfected with an MLV gag-pol, and a retroviral genome encoding β-galactosidase (lacZ gene) as an indicator of transduction. At 48 to 72 h posttransfection, viral supernatants was collected, filtered through a 0.45-μm-pore-size syringe and stored at −80°C. For transduction, target cells were seeded at a density of 4 × 104 per well of a 24-well plate and exposed 24 h later to retroviral particles bearing one of the GALV, GALV-VRAKoRV-B, GALV-VRA/VRBKoRV-B, or KoRV-B envelopes in the presence of 10 μg of Polybrene/ml. At 48 h postexposure, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining was performed, and β-galactosidase expression was evaluated by counting blue colonies to calculate the titers of the viral vectors. The titers of the viral vectors were averaged from at least three independent experiments and are expressed as mean numbers of β-galactosidase-expressing cells ± the SD of the mean.
Envelope binding analysis.
V5 epitope-tagged envelope proteins were transfected into 293T cells and, after 48 to 72 h, the supernatant was filtered and used for binding assays. MDTFPiT1 or MDTFTHTR1 cells were trypsinized from a tissue culture flask, and 106 cells were resuspended with supernatant containing each of the V5-tagged envelope proteins, followed by incubation at 37°C for 45 min with shaking. To detect the presence of V5-tagged envelope on the surface of the target mouse cell, anti-V5 monoclonal antibody (Bio-Rad) was used as the first antibody, followed by a secondary antibody, a goat anti-mouse antibody conjugated to phycoerythrin (Invitrogen). The cells were then subjected to flow cytometric analysis using a FACSCalibur (BD Biosciences), and data were analyzed using CellQuest software (BD Biosciences).
Evolutionary analyses.
To characterize the phylogenetic relationships among the GALV strains and other gammaretroviruses, we inferred phylogenetic trees using the translated amino acid sequences. The sequences of Env, Gag, and Pol proteins of each gammaretrovirus were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/GenBank) (Table 2). Individual gene sequences for env, gag, and pol were aligned by preserving the protein-coding frame in TranslatorX (38) using MAFFT (39). Sequences presenting premature stop codons were excluded from the analyses. For this reason, OOEV and MbRV were removed from the alignment of the pol gene. Phylogenetic analysis was carried out using maximum likelihood as an optimality criterion and the general time-reversible substitution model (40) for nucleotide sequences and the rtREV model (41) for amino acid sequences with among-site rate heterogeneity modeled by the Γ distribution and four rate categories (42), as implemented in the POSIX-threads build of RAxML v8 (43). Node robustness was assessed with rapid bootstrap pseudoreplicates (44). The bootstopping criterion (45) as implemented in RAxML showed that more than 100 (for amino acid sequences) and 500 (for nucleotide sequences) rapid bootstrap pseudoreplicates were unlikely to alter node support. Gene alignments were checked for recombination using the Φw test statistic (otherwise referred to as the pairwise homoplasy index) (46). The signature of natural selection was examined using the mixed effects model evolution (MEME) that allows the ratio ω of the rate of nonsynonymous substitution (dN) to the rate of synonymous substitution (dS) to vary along the tree branches and across codons (47), Fast Unconstrained Bayesian AppRoximation (FUBAR) that estimates codon-wise trends of negative or positive selection (48), and the branch-site random effects likelihood (BSREL) method that is able to detect the branches on which a proportion of codons evolve with ω > 1 (49). The protein-coding sequences of env, gag, and pol were concatenated and analyzed in a partitioned framework, where each partition was allowed to evolve under its own substitution model.
TABLE 2.
Gammaretrovirus sequences used for phylogenetic analyses in this study
| Strain (accession no.) | Full name | Host | gag | pol | env | Reference or GenBank accession no. |
|---|---|---|---|---|---|---|
| GALV SF | Gibbon ape leukemia virus strain San Francisco | Gibbon | ✓ | ✓ | ✓ | This study |
| GALV Brain | Gibbon ape leukemia virus strain Brain | Gibbon | ✓ | ✓ | ✓ | This study |
| GALV Hall's Island | Gibbon ape leukemia virus strain Hall’s Island | Gibbon | ✓ | ✓ | ✓ | This study |
| GALV SEATO | Gibbon ape leukemia virus strain SEATO | Gibbon | ✓ | ✓ | ✓ | This study |
| WMV | Woolly monkey virus | Gibbon | ✓ | ✓ | ✓ | This study |
| GALV SEATO (M26927) | Gibbon ape leukemia virus strain SEATO | Gibbon | ✓ | ✓ | ✓ | M26927 |
| GALV-X | Gibbon ape leukemia virus strain X | Gibbon | ✓ | ✓ | ✓ | U60065 |
| GALV SF (AF055063) | Gibbon ape leukemia virus strain San Francisco | Gibbon | ✓ | AF055063 | ||
| GALV SEATO (AF055060) | Gibbon ape leukemia virus strain SEATO | Gibbon | ✓ | AF055060 | ||
| WMV (AF055064) | Woolly monkey virus | Gibbon | ✓ | AF055064 | ||
| GALV Brain (AF055062) | Gibbon ape leukemia virus strain Brain | Gibbon | ✓ | AF055062 | ||
| GALV Hall’s Island (AF055061) | Gibbon ape leukemia virus strain Hall’s Island | Gibbon | ✓ | AF055061 | ||
| KoRV-A (KF786280) | Koala retrovirus, variant A | Koala | ✓ | ✓ | ✓ | KF786280 |
| KoRV-A (KF786284) | Koala retrovirus, variant A | Koala | ✓ | ✓ | ✓ | KF786284 |
| KoRV-A (AF151794) | Koala retrovirus, variant A (strain “Cindy”) | Koala | ✓ | ✓ | ✓ | AF151794 |
| KoRV-A (AB721500) | Koala retrovirus, variant A (strain “Aki”) | Koala | ✓ | ✓ | ✓ | AB721500 |
| KoRV-B | Koala retrovirus, variant B (strain Br2-1CETTG) | Koala | ✓ | ✓ | ✓ | KC779547 |
| KoRV-A (AB823238) | Koala retrovirus, variant A (strain OJ-4) | Koala | ✓ | AB823238 | ||
| KoRV-C | Koala retrovirus, variant C (strain OJ-4) | Koala | ✓ | AB828005 | ||
| KoRV-D | Koala retrovirus, variant D (strain OJ-4) | Koala | ✓ | AB828004 | ||
| KoRV-J | Koala retrovirus, variant J (strain OJ-4) | Koala | ✓ | AB822553 | ||
| MDEV | Mus dunni endogenous virus | Mouse | ✓ | ✓ | ✓ | AF053745 |
| McERV | Mus caroli endogenous virus | Mouse | ✓ | ✓ | ✓ | KC460271 |
| MmERV | Mus musculus retrovirus | Mouse | ✓ | ✓ | ✓ | AC005743 |
| MbRV | Melomys burtoni retrovirus | Mouse | ✓ | ✓ | KF572483 to KF572486 | |
| PERV-A 1 | Porcine endogenous retrovirus A | Pig | ✓ | ✓ | ✓ | AJ293656 |
| PERV-A 2 | Porcine endogenous retrovirus A | Pig | ✓ | ✓ | ✓ | HQ540592 |
| PERV-B 1 | Porcine endogenous retrovirus B | Pig | ✓ | ✓ | ✓ | HQ540593 |
| PERV-B 2 | Porcine endogenous retrovirus B | Pig | ✓ | ✓ | ✓ | AY099324 |
| PERV-C 1 | Porcine endogenous retrovirus C | Pig | ✓ | ✓ | ✓ | HQ536013 |
| PERV-C 2 | Porcine endogenous retrovirus C | Pig | ✓ | ✓ | ✓ | AM229311 |
| PERV-C MSL | Porcine endogenous retrovirus MSL | Pig | ✓ | ✓ | ✓ | AF038600 |
| RlRV | Rousettus leschenaultii retrovirus | Bat | ✓ | ✓ | JQ951957 to JQ951958 | |
| MlRV | Megaderma lyra retrovirus | Bat | ✓ | ✓ | JQ951955 to JQ951956 | |
| RfRV | Rhinolophus ferrumequinum retrovirus | Bat | ✓ | ✓ | ✓ | JQ303225 |
| CrERV | Odocoileus hemionus endogenous virus | Mule deer | ✓ | ✓ | ✓ | JN592050 |
| OOEV | Orcinus orca endogenous retrovirus | Killer whale | ✓ | ✓ | ✓ | GQ222416 |
| BaEV | Baboon endogenous virus | Baboon | ✓ | ✓ | ✓ | D10032 |
| RD114 | Feline RD114 retrovirus | Cat | ✓ | ✓ | ✓ | EU030001 |
| REV | Reticuloendotheliosis virus | Bird | ✓ | ✓ | ✓ | AY842951 |
| PreXMRV-1 | Prexenotropic MuLV-related virus 1 | Mouse | ✓ | ✓ | ✓ | FR871849 |
| M-CRV | Murine type C retrovirus | Mouse | ✓ | ✓ | ✓ | X94150 |
| M-MuLV | Moloney murine leukemia virus | Mouse | ✓ | ✓ | ✓ | AF033811 |
| F-MuLV | Friend murine leukemia virus | Mouse | ✓ | ✓ | ✓ | Z11128 |
| R-MuLV | Rauscher murine leukemia virus | Mouse | ✓ | ✓ | ✓ | U94692 |
| FeLV | Feline leukemia virus | Cat | ✓ | ✓ | ✓ | AF052723 |
Accession numbers.
The consensus sequence and annotations of each GALV strain genome were deposited in GenBank under accession numbers KT724047 to KT724051. Illumina reads mapping to GALV-X for each captured GALVstrain were deposited in the NCBI Sequence Read Archive as BioProject PRJNA306599.
RESULTS
PCR and Sanger sequencing of GALV strains.
DNA was extracted from six cell lines, each infected with a different strain of GALV. Two primer sets (U5-PolR1 and PolF1-U3) based on the full genome sequences of GALV-X (U60065) and SEATO (M26927) were designed to generate two overlapping PCR products, each 4 kb long, in order to cover the whole GALV genome from each cell line. However, full sequences of the GALV strains were not recovered by PCR, since one of the two primer pairs generally failed to yield an amplification product or readable Sanger sequence, presumably due to the coamplification of different products. Furthermore, the PCR approach has the disadvantage of omitting sequences at the genome ends covered by the primers. The primers that yielded products and high-quality Sanger sequences are listed in Table 1. The Sanger sequences, however, were subsequently used to confirm the proper assembly of high-throughput sequences obtained by hybridization capture.
Hybridization capture and high-throughput sequencing of GALV strains.
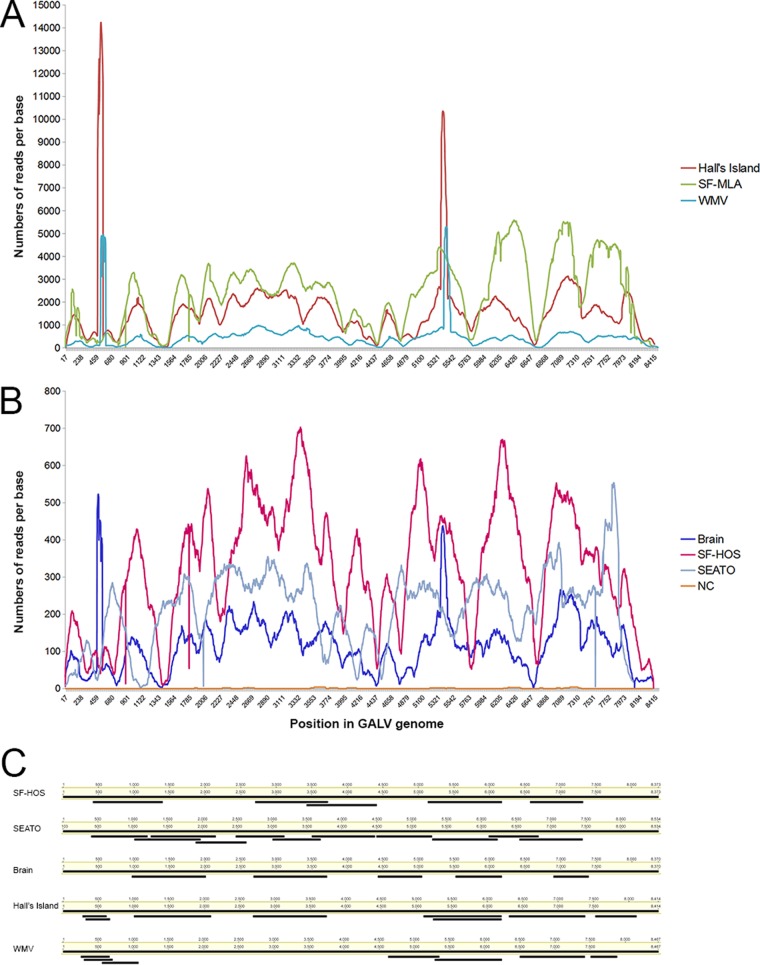
Illumina libraries were prepared from each cell line DNA extract and indexed to allow all samples to be processed in a single Illumina sequencing experiment. Two amplicons 4 kb in length, together covering the entire GALV genome from SF-MLA and SEATO strains, were generated as hybridization capture baits (23). Equimolar amounts of indexed libraries were hybridized to the GALV baits and the enriched GALV libraries sequenced on an Illumina MiSeq platform. The enrichment (proportion of on-target reads mapping to GALV), which ranged from 0.6% (Brain) to 15% (Hall's Island), was comparable to previous reports (22), although the rates for the Brain, SEATO, and SF-HOS strains were relatively low (0.6 to 0.9%). This might be in part due to low sequence identity between baits used and some of the strains targeted. Nonetheless, full coverage of the GALV genome was obtained from each of the cell lines included in the study. The capture enrichment yielded very high per-base coverage, with average values ranging from 2,362× for SF-MLA to 116× for Brain (Fig. 1A and B). Although the per-base coverage differed among strains, the coverage profiles were similar among the GALV strains (Fig. 1A and B). The negative control generated few sequence reads, which only sporadically mapped to GALV (33 of 560 total reads) (Fig. 1A and B). This low frequency of target-mapping reads was well within the known misindex error reading rate on the Illumina platform (0.3%) (50) and is consistent with the rate reported by previous studies (23).
FIG 1.
Hybridization capture sequence and Sanger sequence coverage across the proviral genome for the GALV strains. The sequence coverage is shown for each nucleotide position, numbered as in the corresponding strain consensus sequence. Mapping results for a negative control (NC) are also shown. Each sample is color coded. Panel A shows a coverage profile of the strains that reached very high values (up to 14,000 reads per base), while panel B shows the coverage profile of the strains with lower coverage (up to 700 reads per base). Panel C shows the position of each Sanger sequence generated by PCR in comparison to the full genome consensus sequences of the GALV strain from which it was generated. The Sanger sequences presented here were all of high quality and were used to confirm the bioinformatics assembly of sequences obtained by hybridization capture.
GALV consensus sequence determination.
A nucleotide consensus sequence was generated for each GALV strain, with the exception of SF-MLA, in which the presence of multiple distinct viral sequences prevented assembly. Therefore, the genome of GALV-SF was derived from an infected HOS cell line (SF-HOS), which lacks the defective GALV-SF variants (M. V. Eiden, unpublished data).
The consensus sequences were confirmed by the previously generated Sanger sequences covering parts of the GALVs genomes (Fig. 1C). There was concordance between the hybridization capture and PCR-derived sequences. Polymorphisms detected among sequences in the hybridization capture data were confirmed as double peak signals in the Sanger electropherograms. By comparison of the GALV consensus sequences with the primer sequences, we found that the failures in the PCRs or Sanger sequencing were due to indels and polymorphisms that presumably prevented the primers from binding to the templates.
GALV strain genome structures and regulatory motifs.
All GALV strains had comparable genome sizes ranging from 8,370 bp (Brain) to 8,534 bp (SEATO) (Table 3 and Fig. 2A). In an attempt to precisely localize the coding regions and the regulatory motifs within the genome of each strain, the nucleotide sequence of each strain was compared to the annotated genomes available in GenBank of GALV-X (U60065) and SEATO (M26927) and of the closely related KoRV (AF151794). Each strain was characterized by the common genetic structure of simple type C mammalian retroviruses with a 5′ LTR-gag-pol-env-3′ LTR organization. Furthermore, the following regulatory motifs were readily identified in each strain: a tRNAPro primer binding site, a CAAT box, a TATA box, a Cys-His box, a polypurine tract, and a polyadenylation [poly(A)] signal. No differences in these motifs were detected among GALV strains with the exception of four polymorphisms in the Cys-His box, three of which were mutations unique to WMV (positions 2518, 2536, and 2539), along with a G-to-A (position 2530) transition and a C-to-G (position 2536) transversion, both found in GALV-X and SF (data not shown).
TABLE 3.
Length and coordinates of the genomic regions of the GALV strainsa
| Strain | Total length (nt) | 5′ LTR |
gag |
pol |
env |
3′ LTR |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (nt) | Coordinates (nt) | Length (nt) | Coordinates |
Length (nt) | Coordinates |
Length (nt) | Coordinates |
Length (nt) | Coordinates (nt) | |||||
| nt | aa | nt | aa | (nt) | aa | |||||||||
| SF-HOS | 8,373 | 463 | 1–463 | 1,566 | 910–2475 | 1–521 | 3,384 | 2590–5973 | 1–1127 | 2,013 | 5855–7867 | 1–670 | 463 | 7911–8373 |
| SEATO | 8,534 | 463 | 1–463 | 1,563 | 954–2516 | 1–520 | 3,384 | 2631–6014 | 1–1127 | 2,058 | 5875–7932 | 1–685 | 559 | 7976–8534 |
| Brain | 8,370 | 453 | 1–453 | 1,572 | 899–2470 | 1–523 | 3,375 | 2585–5959 | 1–1124 | 2,046 | 5829–7874 | 1–681 | 453 | 7918–8370 |
| Hall’s Island | 8,414 | 469 | 1–469 | 1,572 | 915–2486 | 1–523 | 3,384 | 2601–5984 | 1–1127 | 2,058 | 5845–7902 | 1–685 | 469 | 7946–8414 |
| WMV | 8,467 | 507 | 1–507 | 1,566 | 963–2528 | 1–521 | 3,384 | 2643–6026 | 1–1127 | 2,010 | 5908–7917 | 1–669 | 507 | 7961–8467 |
aa, amino acids; nt, nucleotides.
FIG 2.
Genomic structure of the GALV strains. Alignment of the newly generated nucleotide sequences of the GALV strains with GALV GenBank reference sequences (SEATO, M26927; GALV-X, U60065). Panel A shows the full genomes of each GALV strain, with the positions of proviral genes, proteins, and regulatory motifs indicated. Panels B and C show the differences among the GALV strains in the 5′ and 3′ LTRs, respectively. Nucleotide positions identical among the strains are indicated in light gray, while mismatches are shown in black. Gaps are shown as dashes. The green bar above the alignment indicates the percent identity among the sequences (green, highest identity; red, lowest identity). The following structural regions are shown: the 5′ and 3′ LTRs with the typical U3-R-U5 structure (in light blue), the CAAT box and TATA box (in red), the polyadenylation [poly(A)] signal (in dark blue), the primer binding site (PBS) (in green), the Cys-His box (in orange), and the polypurine tract (PPT) (in gray). The ORFs of gag, pol, and env genes are shown in yellow, while protein domains are in sky blue. Protein domain abbreviations: MA, matrix; CA, capsid; NC, nucleocapsid; Pro, protease; RT, reverse transcriptase; IN, integrase; SU, surface unit; TM, transmembrane subunit.
The 5′ and 3′ LTRs of the GALV strains were 463 to 559 bp long (Table 3) with a retrovirus-typical U3-R-U5 region structure (Fig. 2B and C). The 5′ and 3′ LTRs were compared for each strain and were found to be identical, further validating the sequencing and assembly methods used. The overall average nucleotide identity of LTRs across the GALV strains was 82.2%, lower than that calculated for the open reading frames (ORFs). However, between GALV-X and SF-HOS, the LTRs were 100% identical, and the LTRs of Brain and Hall's Island were similar (93.2% sequence identity) (Table 4). The differences among GALV strain LTRs were concentrated in the U3 region, which was the most variable (average identity, 75.8%). In addition to small insertions, deletions, and point mutations, there were three notable differences among the strains: (i) a 16-bp deletion at the 5′ end of the U3 region of the Brain strain (compared to GALV-X, positions 9 to 25); (ii) two fragments, 21 and 22 bp in length, present only in WMV (positions 52 to 72 and positions 101 to 122, respectively); and (iii) a 48-bp perfect tandem direct repeat present only in SEATO (positions 136 to 183), as previously reported (51) (Fig. 2B and C). The 48-bp motif is found in two copies in the 3′ LTR in the SEATO sequence from Delassus et al. (14) and Trainor et al. (51). However, in the current study different variants with two to four copies were observed among the Illumina sequences (three copies are reported in the 3′ LTR of the consensus sequence). The GenBank entry for SEATO (M26927) (14) does not include the first 320 bp of the 5′ LTR, and the data presented here fill in the genome sequence.
TABLE 4.
Similarities among the GALV strains in the LTRs and in the full genomes sequencesa
| Strain | LTRs (% identity) |
Full genome (% identity) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GALV-X | SEATO* | SF-HOS | SEATO | Brain | Hall’s Island | WMV | GALV-X | SEATO* | SF-HOS | SEATO | Brain | Hall’s Island | WMV | |
| GALV-X | 84 | 100 | 83.5 | 80 | 82.9 | 80.8 | 87.6 | 99 | 87.1 | 88.2 | 88.3 | 90 | ||
| SEATO* | 75.2 | 84 | 100 | 87.7 | 87 | 90.3 | 87.6 | 87.9 | 98.7 | 91.4 | 91.5 | 89.6 | ||
| SF-HOS | 100 | 75.2 | 83.5 | 80 | 82.9 | 80.8 | 99 | 87.9 | 87.4 | 88.4 | 88.5 | 90.5 | ||
| SEATO | 69.6 | 89.5 | 69.6 | 84 | 88 | 81.9 | 87.1 | 98.7 | 87.4 | 90.9 | 91.1 | 89 | ||
| Brain | 79.5 | 74.5 | 79.5 | 69.9 | 93.2 | 79.4 | 88.2 | 91.4 | 88.4 | 90.9 | 97.7 | 89.9 | ||
| Hall’s Island | 82.5 | 78.9 | 82.5 | 73.1 | 93.2 | 81.6 | 88.3 | 91.5 | 88.5 | 91.1 | 97.7 | 89.9 | ||
| WMV | 80.8 | 72.4 | 80.8 | 68.9 | 79.4 | 81.5 | 90 | 89.6 | 90.5 | 89 | 89.9 | 89.9 | ||
The similarities are reported as percent nucleotide identities between nucleotide sequences. For the LTRs, the values above the diagonal represent the percent nucleotide identities among the 5′ LTR sequences of GALV strains, whereas the values below the diagonal represent the percent identities among the 3′ LTR sequences. GALV reference sequences from GenBank (SEATO, M26927, indicated by SEATO*; GALV-X, U60065) are included in the comparison.
An imperfect 7-bp inverted repeat (e.g., TGAAAGA/TCTCTCA in SF-HOS), which is known to mark the boundaries of the LTR ends (18, 51), was identified in each strain with minor differences. An AAAAATAC motif, which was found to correlate with leukemogenicity in several MuLVs (52), was identified in SEATO, Brain, and Hall's Island GALVs. The insertions and deletions previously reported by Trainor et al. (51) in the U5 region of GALV strains, including a deletion affecting the poly(A) signal in SEATO, were not detected in the current study. In fact, among the GALV strains the U5 region was overall more conserved (85.2% sequence identity) than the U3 region (75.8%).
When the full nucleotide sequences were compared, all of the GALV strains demonstrated a high degree of similarity overall, with an average nucleotide identity of 90.6% (Table 4). Specifically, as expected, the SEATO sequence generated here was almost identical to the GenBank SEATO (98.7% identity), while SF-HOS shared 99% identity with GALV-X. The Brain and Hall's Island strains were very closely related (97.7% nucleotide identity) and together more similar to GenBank SEATO (average nucleotide identity, 91.4%) than to GALV-X (88.2%). WMV did not show strong affinity with any specific GALV strain, although identity with the other GALVs was high (89 to 90.5%, Table 4).
Three ORFs corresponding to the gag, pol, and env genes were identified in the genome of each GALV strain. The ORF average length was 1,568 bp (1,563 to 1,572 bp) for gag, 3,382 bp (3,375 to 3,384 bp) for pol, and 2,037 bp (2,010 to 2,058 bp) for env, indicating low ORF size variability among the GALV strains. All ORFs were undisrupted. The gag and pol ORFs were in the same reading frame, while env was in a different frame, with the end of pol and the beginning of env ORFs overlapping, as found in many retroviruses. The GALV strains displayed a 93.3% average amino acid similarity for gag, 96.2% for pol, and 87.6% for env (Table 5).
TABLE 5.
Amino acid similarity among the GALV strains from this study for the Gag, Pol, and Env proteins
| Strain | Similarity (% identity)a |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag (avg, 93.3%) |
Pol (avg, 96.2%) |
Env (avg, 87.6%) |
|||||||||||||
| SF-HOS | SEATO | Brain | Hall’s Island | WMV | SF-HOS | SEATO | Brain | Hall’s Island | WMV | SF-HOS | SEATO | Brain | Hall’s Island | WMV | |
| SF-HOS | 91.4 | 90.8 | 90.6 | 92.1 | 95.7 | 95.5 | 94.9 | 96.5 | 85.3 | 85 | 86.1 | 85.6 | |||
| SEATO | 91.4 | 96.4 | 96.6 | 92.5 | 95.7 | 96 | 95.5 | 96.5 | 85.3 | 92.8 | 94 | 83.1 | |||
| Brain | 90.8 | 96.4 | 97.7 | 92.7 | 95.5 | 96 | 99.2 | 96.4 | 85 | 92.8 | 97.8 | 82.8 | |||
| Hall’s Island | 90.6 | 96.6 | 97.7 | 92.4 | 94.9 | 95.5 | 99.2 | 96 | 86.1 | 94 | 97.8 | 83.7 | |||
| WMV | 92.1 | 92.5 | 92.7 | 92.4 | 96.5 | 96.5 | 96.4 | 96 | 85.6 | 83.1 | 82.8 | 83.7 | |||
The similarities are reported as percent identities between amino acid sequences. The average amino acid similarity among strains for each of the protein is indicated in parentheses in the column heading. The average amino acid similarities among strains for each of the protein domains were as follows: (i) within Gag, p15 MA (89.14%), p12 (88.81), p30 CA (98.53), and p10 NC (85.6%); (ii) within Pol, Pro (97.31%), RT (96.44%), and IN (95.66%); and (iii) within Env, gp70 SU (84.83%) and p15e TM (94.8%). Abbreviations: MA, matrix; CA, capsid; NC, nucleocapsid; Pro, protease; RT, reverse transcriptase; IN, integrase; SU, surface unit; TM, transmembrane subunit.
For each GALV strain, we identified the matrix p15 (MA), p12, capsid p30 (CA), and nucleocapsid p10 (NC) proteins within Gag; the protease (PR), reverse transcriptase (RT), and integrase (IN) proteins within Pol, and the surface unit gp70 (SU) and transmembrane subunit p15E (TM) within Env (Fig. 2A). Furin sites with the motif R-X-K-R for the cleavage of the Env precursor into SU and TM subunits were identified in each GALV strain at the C terminus of the SU. Also, the CWLC motif, which is thought to play a role in the assembly and function of the Env complex (53), was conserved across all GALV strains (positions 355 to 358 of the Env protein). Among Gag protein domains, the capsid was by far the most conserved among GALV strains with 98.5% amino acid identity, while the nucleocapsid was the most variable (85.6% among strain similarity). All Pol protein domains were highly conserved, while within Env the surface unit was much more variable than the transmembrane subunit (84.8 and 94.8% identity, respectively) (Table 5 and Fig. 2A). On average, 34% of the polymorphisms identified in Gag, Pol, and Env were mutations unique to SF-HOS (17.5, 22.2, and 16.3%, respectively) and WMV (12.5, 16.6, and 18.2%, respectively). These unique polymorphisms were concentrated in the p12 domain in Gag, in the integrase domain in Pol, and in the surface unit in Env.
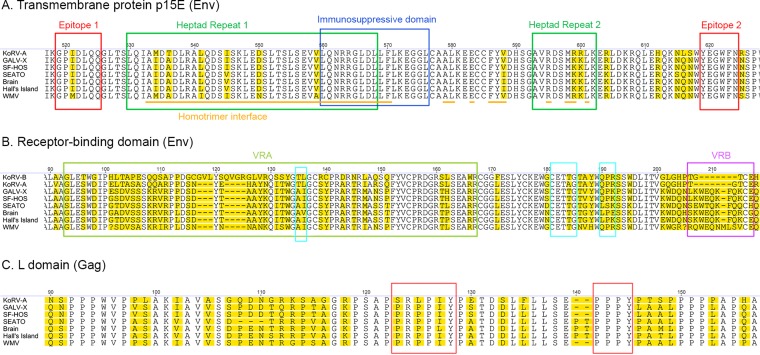
The transmembrane protein p15E of the envelope is known to contain several motifs that are highly conserved among gammaretroviruses (54). The epitopes E1 (residues 519 to 525) and E2 (residues 619 to 624), the immunosuppressive domain (residues 560 to 576), the homotrimer interface (interspersed residues 533 to 601), and the heptad repeats 1 and 2 (residues 530 to 568 and residues 593 to 602, respectively) were conserved across all GALV strains (Fig. 3A). These domains are mainly involved in viral fusion and are highly conserved among GALVs, KoRVs, and PERVs (54). Nevertheless, one polymorphism each within the E1 and heptad repeat 2 and five polymorphisms in the overlapping region between heptad repeat 1 and the homotrimer interface were observed among GALVs. Six of the seven detected polymorphisms were identified in WMV. Of these six polymorphisms identified in WMV, two were shared with KoRV (Fig. 3A).
FIG 3.
Differences among GALV strains and KoRV in the Env and Gag domains regulating viral fusion, infectivity, and host range. Alignment of Env and Gag amino acid sequences of GALV strains with relevant GenBank reference sequences (GALV-X, U60065; KoRV, AF151794) for the domains affecting viral fusion (epitopes 1 and 2, heptad repeats 1 and 2, homotrimer interface, and immunosuppressive domain of the transmembrane protein p15E of Env) (A), receptor specificity (variable regions A and B of Env) (B), and viral infectivity (receptor-binding domain of Env and L domain of Gag) (B and C, respectively). The three motifs influencing infectivity within the receptor-binding domain are marked by turquoise squares (B), while the PRPPIY and PPPY motifs are marked by brown squares within the L domain (C). Positions where amino acids vary are highlighted in yellow. Since KoRV-B was used to investigate the functional differences between GALV and KoRV in the VRA and VRB regions, KoRV-B has been included in panel B.
Differences in the variable regions A and B (VRA and VRB) of the receptor-binding domain (RBD) of the envelope protein are responsible for variation in receptor specificity for WMV and the other GALV strains (19). Sixteen polymorphisms in the VRA, and eight polymorphisms in the VRB were observed, as well as an insertion of one amino acid in the VRB of WMV compared to other GALVs (Fig. 3B). WMV, which is the only GALV strain to show a difference from other strains in the host range (it cannot infect E36 hamster cells), exhibited a high degree of diversification in these two regions, with an average of 13 amino acid residue differences in the VRA and of 8.5 amino acid residue differences in VRB sequences relative to other GALVs (Fig. 3B). Similarly to WMV, KoRV-A also fails to infect E-36 cells (Eiden, unpublished). Thus, the ability to infect hamster E36 cells is a distinguishing feature of the GALVs, with the exception of WMV. It has been previously shown that glycosylation does not account for the inability of WMV to use the E36 GALV receptors, and it has been postulated that cellular factors, such as the expression of inhibiting factors or the lack of accessory proteins, may be involved (19).
We also confirmed the high variability detected by Oliveira et al. (55) among GALV strains in the motifs of the RBD of the envelope protein, which are known to influence the differential infectivity of GALV and KoRV (55). All GALV strains presented the AI residues at positions 135 to 136 of the envelope surface unit, with the exception of Brain, which had AV at these positions. WMV was the only strain to show at residues 190 to 192 the same QPR residues displayed by KoRV (55) (Fig. 3B). Oliveira et al. (55) showed that when these five residues of the GALV envelope are replaced by the corresponding residues of KoRV, the resulting mutant vectors exhibit substantially reduced titers similar to those observed with KoRV vectors. In contrast, no polymorphisms among GALV strain envelopes were observed in the CETTG motif (residues 181 to 185 of the surface unit) (Fig. 3B), which is highly conserved among infectious gammaretroviruses, including KoRV-B, although is mutated in KoRV-A (55). It has been hypothesized that these mutations played a key role in the endogenization process of KoRV-A into the koala genome (55).
Few differences were observed among GALV strains in the PRPPIY and PPPY motifs of the L domain of the Gag protein (residues 123 to 128 and residues 142 to 145, respectively, of the matrix protein) (Fig. 3C), which are known to play a key role in the release of viral particles from the plasma membrane after viral budding. Replacement of GALV PRPPIY with KoRV SRLPIY motif causes a substantial reduction in viral titer (55), while the disruption of the PPPY motif has been reported to be involved in the reduction of KoRV viral budding (56, 57). The only difference observed in the PRPPIY motif was an I-to-L residue replacement in GALV-Brain at the fifth position of the motif, while the PPPY motif was identical across all GALV strains (Fig. 3C). A high level of conservation was observed in the major homology region, which is the most conserved region among retroviruses of the Gag CA protein and whose residues are necessary for the proper assembly of mature capsids (58). Only one polymorphism (an A-to-T change in Brain) was found at the sixth position of the motif (VLQGPAEPPSVFLERLMEAY, positions 348 to 367 of the Gag protein).
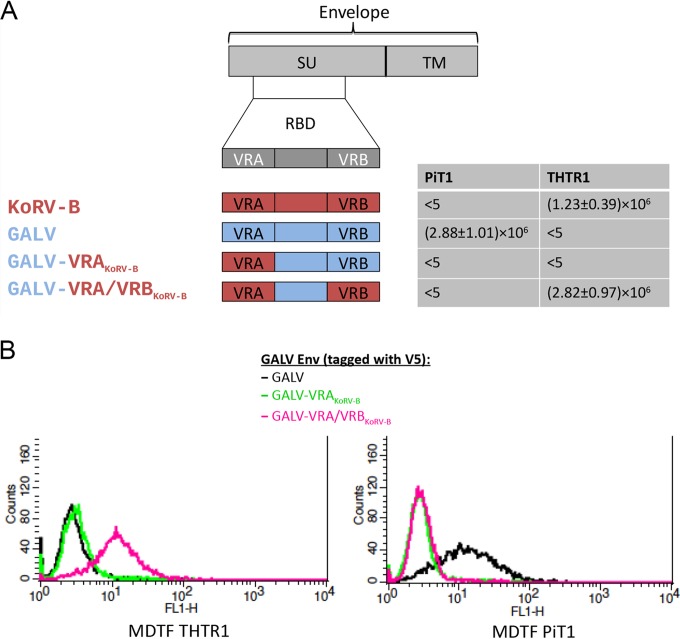
Functional differences between GALV and KoRV VRA and VRB regions.
The GALV polymorphisms identified within the VRA and VRB regions may have functional consequences for receptor binding. Within the KoRV/GALV group, KoRV-A and all GALVs use the sodium-dependent phosphate transporter 1 (PiT1) as a receptor (59), whereas KoRV-B and -J infect cells via the thiamine transporter 1 (THTR1) (36). In order to understand which part of the envelope of KoRV and GALV influences receptor specificity, we constructed vectors endowed with GALV-SEATO chimeric envelopes in which regions of the RBD were replaced by the corresponding region of KoRV-B (Fig. 4). These vectors were used to infect Mus dunni tail fibroblast (MDTF) cells. Murine MDTF cells are resistant to all KoRVs and GALVs, but the expression of PiT1 renders them susceptible to KoRV-A and GALVs but not KoRV-B, whereas the expression of THTR1 renders them susceptible to KoRV-B but not GALVs or KoRV-A (36). Chimeric vectors with a GALV envelope in which the GALV VRA was replaced by the VRA from KoRV-B failed to infect MDTF cells expressing PiT1 or THTR1 (Fig. 4A). However, when the GALV vector had both VRA and VRB replaced by the corresponding regions from KoRV-B, MDTF cells expressing THTR1 were successfully infected, and the vector titer was similar to that of vectors bearing the full-length KoRV-B envelope (Fig. 4A). Therefore, although KoRV-B VRA was by itself insufficient to confer infectivity, the combination of VRA and VRB was sufficient to confer the infectivity properties of KoRV-B to GALV. Binding studies involving MDTF cells expressing either PiT1 or THTR1 were conducted (Fig. 4B). These studies demonstrated that the reason why the vector bearing both KoRV-B VRA and VRB does not infect MDTF cells expressing PiT1 (Fig. 4A) is that this vector does not bind PiT1 (Fig. 4B). Therefore, the block to infection is not mediated at a postbinding stage of entry. Similarly, the inability of vector bearing only KoRV-B VRA to infect MDTF cells expressing THTR1 is due to the failure to bind THTR1 (Fig. 4B).
FIG 4.
Exposition of murine MDTF cells expressing the receptor for GALV (PiT1) or the receptor for KoRV-B (THTR1) to vectors bearing different GALV/KoRV-B envelopes. The structure of a gammaretroviral envelope protein with the surface unit (SU) and the transmembrane subunit (TM) is schematically depicted at the top of the figure underneath which is a depiction of the receptor-binding domain (RBD) located within the surface unit gene. In the schematic representation of the chimeric envelopes, sequences from KoRV-B envelope are in red, and those from GALV-SEATO are in blue. The GALV chimeric envelope within which the VRA of GALV-SEATO was replaced by the corresponding region of KoRV-B is designated GALV-VRAKoRV-B, whereas the GALV-SEATO chimeric envelope containing both KoRV-B VRA and VRB is designated GALV-VRA/VRBKoRV-B. Murine MDTF cells expressing PiT1 or THTR1 were exposed to vectors bearing GALV, KoRV-B, GALV-VRAKoRV-B, or GALV-VRA/VRBKoRV-B envelopes and assessed for susceptibility to these vectors using a conventional β-galactosidase assay. The titers of the viral vectors were averaged from at least three independent experiments and are expressed as mean numbers of β-galactosidase-expressing cells ± the SD of the mean. Panel B demonstrates the ability of GALV (black line), GALV-VRAKoRV-B (green line) and GALV-VRA/VRBKoRV-B (pink line) envelopes, each with a V5 epitope tag, to bind to MDTF cells expressing either PiT1 or THTR1. The binding ability of the vectors was assessed using flow cytometry.
Phylogenetic and selection analysis of GALV strains.
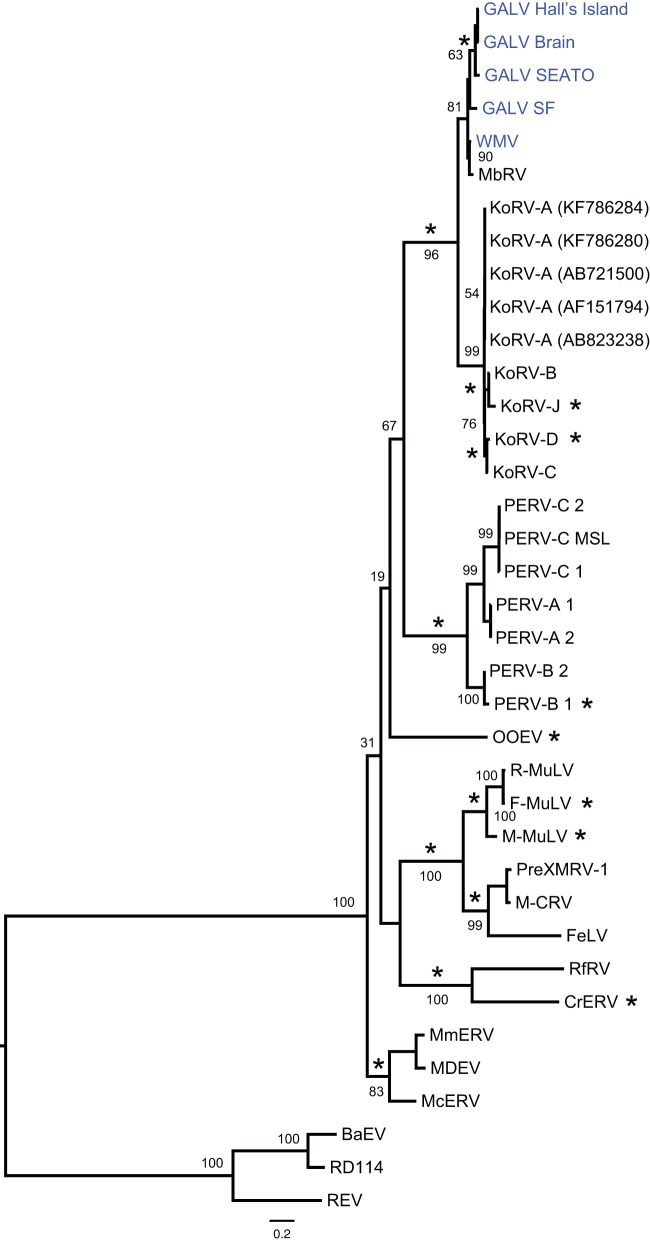
Nucleotide mismatches were observed between the sequences from GenBank and those generated in this study for the same GALV strain, many of the differences representing nonsynonymous substitutions. This was pronounced in env for which sequences of each GALV strain are available in GenBank. For example, we detected 24 nucleotide differences in the GALV Hall's Island env, 8 of which were nonsynonymous substitutions. All GALV GenBank sequences were generated more than 15 years ago (14, 18, 19) by Sanger sequencing, while the sequences reported here were confirmed both by hybridization capture and bidirectional Sanger sequencing with an updated BigDye chemistry kit (v3.1). In order to account for the potential of errors in the GenBank sequences, the selection analysis was run with and without the GenBank sequences. While the results of the selection analysis for gag and pol did not change, in env three GenBank-derived GALV sequences (Hall's Island AF055061, SEATO M26927, and SF AF055063) were found to have undergone episodic diversifying selection, whereas all other GALV tree terminal branches were not. Even though the GALV env GenBank sequences grouped with their strain counterparts from our sequences (data not shown), the evidence of episodic diversifying selection on the GenBank sequences is likely an artifact of either mistakes in the GenBank sequences or mutations that have occurred over time in cell culture. Therefore, the results of the evolutionary analyses on the env are presented without GALV GenBank sequences (Fig. 5).
FIG 5.
Maximum-likelihood phylogenetic tree of gammaretroviruses inferred using complete env nucleotide sequences, excluding GALV GenBank sequences. GALV GenBank sequences were excluded to avoid any influence of possible errors in these sequences on the analysis of selection. Node robustness was assessed with 500 rapid bootstrap pseudoreplicates. Numbers above or below the internode branches indicate bootstrap support. The GALV strain sequences generated in this study are highlighted in blue. Branches with significant (P < 0.05) evidence of episodic diversifying selection as indicated by the BSREL method are marked with an asterisk. GenBank accession codes are shown in brackets. The scale bar indicates 0.2 nucleotide substitutions per site. The tree is midpoint-rooted for purposes of clarity. All abbreviations can be found in Table 2.
All GALV strains formed a monophyletic clade sister to WMV, with the clade of the GALVs and WMV forming a sister group to the KoRVs, both at the nucleotide (Fig. 5 and Fig. 6) and amino acid level (data not shown). The highest level of internode branch support was observed when the protein sequences of the three protein-coding genes were concatenated and analyzed in a partitioned maximum likelihood framework (data not shown). The evolutionary relationships among GALV strains were robust regardless of the data type analyzed. Both concatenated, partitioned protein sequences (data not shown) and concatenated, partitioned nucleotide sequences (data not shown) (that included noncoding LTRs and spacers) grouped the two SEATO isolates, sister to the Brain and Hall's Island strains with the SF and X strains (98 to 100% bootstrap support).
FIG 6.
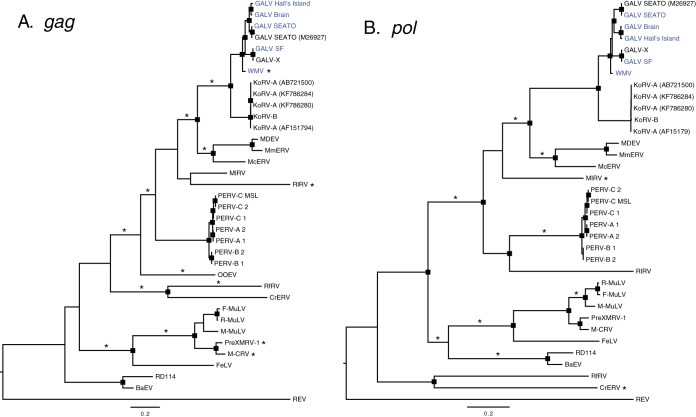
Maximum-likelihood phylogenetic trees of gammaretroviruses inferred using complete pol (A) and gag (B) nucleotide sequences. Node robustness was assessed with 500 rapid bootstrap pseudoreplicates. The rectangles on the nodes indicate a bootstrap support of >80%. The GALV strain sequences generated in this study are highlighted in blue. Branches with significant (P < 0.05) evidence of episodic diversifying selection, as indicated by the BSREL method, are marked with an asterisk. GenBank accession codes are shown in brackets. The scale bar indicates 0.2 nucleotide substitutions per site. The trees are midpoint rooted for purposes of clarity. All abbreviations can be found in Table 2.
Recombination was not detected in any of the protein-coding loci. Signs of positive diversifying selection were detected using the consensus results of MEME and FUBAR methods: only codons found to be under positive selection by both methods were considered. FUBAR detected only codons 98 and 360 under positive selection in the env gene, with a posterior probability (PP) > 0.97 and an empirical Bayes factor (EBF) of >180. By relaxing the PP threshold to 0.7 (EBF > 12), 11 more codons were found under episodic diversifying selection. MEME analysis identified many more codons (data not shown). The consensus consisted only in codon 98 of the env gene or with the relaxed threshold in codons 89, 96, 98, 211, 212, 282, 345, and 396. These codons correspond to residues 14, 21, 23, 118, 119, 154, 202, and 227, respectively, of the surface unit gp70 (SU) of the Env protein. Residues 118, 119, 154, and 202 represent four of the polymorphisms that we detected among GALV strains in the variable regions A and B (VRA/VRB) of the N-terminal region of the envelope and which are thought to influence the receptor specificity of these viruses. Although identified by both FUBAR and MEME, the codons identified by FUBAR only at a lower threshold should be treated with caution. We uncovered signs of episodic diversifying selection along the branches of the gag, pol, and env gene trees using the BSREL method (Fig. 5 and 6). A fraction of the codons of gag were found to deviate from purifying selection and neutrality on the branch unifying the GALV and KoRV clades and on the WMV terminal branch (Fig. 6). In the env gene, the branches connecting GALV-Hall's Island, Brain, and SEATO, and KoRV-B/KoRV-J strains were found to be under episodic diversifying selection (Fig. 5).
DISCUSSION
Because of its broad host range, GALV-based retroviral vectors have been developed for use in gene transfer (60). GALV has also been used in cancer gene therapy. GALV envelope fusogenic membrane glycoprotein (a C-terminal truncated form of GALV envelope glycoprotein, GALV.fus), which has strong cytotoxic effects, can be transduced into a range of human tumor cells to efficiently kill the cells through a process of syncytial formation (61). The use of this system in the treatment of lung cancer has already given encouraging results (62). In addition to its utility as a clinical tool, GALV is an epizootic agent. Therefore, it is surprising that, with the exception of two strains, SEATO and GALV-X, most GALV laboratory strains have not been fully sequenced.
Hybridization capture advantages for viral genomics.
Part of the difficulty in characterizing the GALV strains by PCR was the high failure rate of primer combinations given that the underlying diversity was unknown. Hybridization capture outperformed PCR amplification and Sanger sequencing in determining the uncharacterized genomic regions of the five GALV strains. PCR is subject to primer target mismatches and is sensitive to GC content. Hybridization capture, in contrast, can tolerate bait and target mismatches well over 15% (63). Multiplexing can be performed and yields high per-base coverage across the genome while allowing for discrimination of polymorphism or viral variant cooccurrence. The result was full coverage of all GALV strain genomes (Fig. 1A and B) with an average per-base fold coverage of 848.6. Where the Sanger sequencing and hybridization capture results overlapped, the sequences were identical. The consistency of results between Sanger sequencing and hybridization capture suggests that the capture results can be relied upon to yield the correct sequences. The GALV-SEATO and SF derived baits were suitable for examining viruses with up to 12.9% divergence and will likely be applicable to viruses with greater divergence, as observed by whole-genome cross hybridization experiments (64). Therefore, hybridization capture will likely be a valuable tool for viral discovery among closely and distantly related gammaretroviruses, which could be generally applied to retroviral discovery. However, when multiple similar viral strains are present in a sample, genome assembly can be hindered due to their sequence similarity. In our case it was not possible to recover the genome sequence of SF-MLA because of the presence of multiple distinct viral sequences. MLA-144 is a T-lymphoid cell line established from tumor cells of a gibbon with lymphoid leukemia (1). In contrast to other GALV cell lines, MLA-144 is thought to harbor several different defective recombinant GALV-SF proviruses, which may contain cell-derived, nonviral sequences (65). Furthermore, it was found that the MLA-144 cell line contains two GALV insertions in the IL-2 gene, which allow the cell line to produce interleukin 2 constitutively (66). Together, these anomalies of the MLA-144 cell line hindered the capture experiment and complicated the assembly of the sequencing reads of SF-MLA.
Significance of genomic structural differences of GALV.
As with other gammaretroviruses, malignancies induced by GALV or KoRV involve both viral and cellular determinants. The viral determinants include the transcription elements contained within the long terminal repeats (LTRs) and the envelope protein that affects cell tropism, in vivo spread and cytopathicity. Cellular determinants of infectivity and pathogenesis include viral receptors and cellular oncogenes activated by the adjacent integration of a transcriptionally active LTR. The only GALV sequences previously available in GenBank were the SEATO and GALV-X genome sequences (14, 18) and the envelope sequences of each GALV strain (19). Therefore, the sequences of the LTRs and the gag and pol genes were missing for most of the strains. Furthermore, the GenBank entry for SEATO (14) is chimeric, with part of the pol gene of SF strain incorporated into the SEATO genome, and also excludes the first 320 bp of the 5′ LTR. We have determined that the env gene of the GenBank SEATO is wrongly annotated since it does not include the sequence corresponding to the R peptide. Thus, the data presented in this study fill in these gaps in the SEATO genome completing its sequence. GALV-X was found to be almost identical to GALV-SF, suggesting that they could represent the same virus.
The five GALV strains showed high degree of similarity at the genome level with an average nucleotide identity above 90% (Table 4). However, we found high variability among the GALV strains in the LTRs (Table 4), especially in the U3 region. Notably, the insertions in the LTRs of WMV compared to the other strains and the 48-bp perfect tandem direct repeat present only in SEATO (Fig. 2B and C) are located in an area likely to contain transcriptional enhancers and could be relevant to the leukemogenic potential of these two strains, as already suggested (51). Of note, an AAAAATAC motif, reported by Villemur et al. (52) to be present specifically in the U3 of leukemogenic strains of MuLV, was identified in the LTRs of the SEATO, Brain, and Hall's Island strains.
At the amino acid level, the GALV strains demonstrated high degree of conservation in the pol and gag genes, with an average amino acid identity above 93% (Table 5). However, multiple distinct mutations could be identified in SF-HOS and WMV in both proteins, particularly in the p12 domain of Gag and in the integrase domain of Pol. The env gene was more variable, particularly in the surface unit (average amino acid identity 84.8%), which is known to contain motifs influencing viral infectivity (e.g., RBD) and receptor specificity (e.g., VRA/VRB). A high percentage of the polymorphisms were attributable to mutations found in SF-HOS and WMV in this domain. Functional analysis of differences, particularly between these two strains and the other GALVs may reveal further insights into the different biological properties of these viruses.
Until now only the env gene sequences were available for all the GALV strains, thus most functional analyses have been confined to domains within this gene. The only two determinants of infectivity identified in gag—the PRPPIY and PPPY motifs of the L domain, which are known to influence the release of viral particles from the plasma membrane after viral budding (56, 57)—were highly conserved across the GALV strains (Fig. 3C). The only exception was one amino acid difference found in Brain.
Our study confirmed the high degree of conservation in env, already highlighted among gammaretroviruses and specifically between KoRV and GALV (54), in the amino acid sequences of the domains and epitopes of the transmembrane envelope protein p15E that are important for viral fusion (Fig. 3A). The exception was WMV, which was variable in most motifs in comparison with other GALVs and shared some polymorphisms with KoRV. Similarly, WMV demonstrated unique amino acid changes relative to the other GALVs in the variable regions A and B (VRA and VRB) within the RBD of the envelope (Fig. 3B). These two regions are involved in receptor utilization and variation has been demonstrated to be responsible for the difference in host range between WMV and the other GALVs (19). Although both WMV and GALVs use PiT1 (SLC20A1) to infect human cells, WMV cannot infect hamster E36 cells that are susceptible to all other GALVs (19). The difference in host range is due to residues in the RBD of WMV (19). When GALV-SEATO RBD residues were substituted for the corresponding residues in WMV, the block to E36 infection was circumvented (19). A similar host range restriction extends to KoRV-A with respect to its inability to infect hamster E36 cells. The high degree of residue variation detected in the RBD region between WMV and KoRV-A and the other GALVs (Fig. 3B) supports the role of VRA and VRB in modulating receptor specificity (19).
Despite their genetic similarity, KoRV-B, unlike KoRV-A and the GALVs, does not use PiT1 as a receptor. THTR1 serves instead as KoRV-B receptor (36). Using chimeric envelopes derived from KoRV-B and GALV, we determined that both VRA and VRB comprising the RBD are required for GALV to switch to KoRV-B receptor usage (Fig. 4). Thus, we provided a second example among the KoRVs, WMV, and GALVs of the importance of RBD in receptor utilization.
Evolutionary analyses.
Episodic diversifying selection is associated with selection pressure at the host-pathogen interface. Less pathogenic or endogenous retroviruses may be expected to elicit a less severe immune or antiretroviral response and exhibit reduced evidence of selection. Episodic diversifying selection was found to be acting on most of the gammaretroviral clades examined (Fig. 5 and 6). However, each gene exhibited a different pattern of selection. Selection on gag was observed on most clades except for the BaEV/RD114 clade (Fig. 6). There was also no evidence for specific selection on the GALV/KoRV lineages, even though the general clade to which GALV and KoRV belong is under selection. This was also true for the pol gene. In contrast, for the env gene there was evidence for selection on the GALV/KoRV clade, and specifically on the GALV Hall's Island/Brain/SEATO, KoRV-B/KoRV-J, and KoRV-C/D subclades (Fig. 5). In the case of the GALV strains under episodic diversifying selection, they represent some of the strains associated with leukemias in captive gibbons, GALV-SEATO and Hall's Island strains. The codon-oriented FUBAR and MEME analyses indicated that positive selection in these gammaretroviruses was concentrated on eight amino acids within the SU of the envelope, the most accessible portion of the virus to the immune system, supporting the potential involvement of host-pathogen interactions.
The only KoRVs exhibiting episodic diversifying selection are those associated with greater pathogenicity and which have switched receptor usage from Pit-1 to THTR1 (36, 67). In both cases it has been posited that these variants of KoRV are recently evolved strains that are exogenous (23, 36, 67). The concentration of selection in the env gene is consistent with analysis of historical koala KoRV-A derived sequences that suggest that the env gene is one of the few genes under longer-term selection, although weak (23, 68). The results are also consistent with our functional analysis of the importance of the VRA and VRB domains to receptor specificity in KoRV and GALV. The concentration of polymorphisms in the VRA and VRB regions among GALVs and the selective forces acting on the SU region of the env gene suggest that selection is strongly influencing GALV and KoRV interactions with host cells. The lack of observable positive selection on the KoRV-A clade is consistent with the endogenization of KoRV-A viruses in the koala genome (54).
Conclusions.
Although most GALV strains are highly similar at the nucleotide and amino acid sequence level, WMV is the most divergent GALV, and it shares some traits with KoRV, i.e., host range and infectivity motifs in the env gene, which could explain the biological differences observed between WMV and other GALV strains. Episodic diversifying selection is concentrated on the Env protein likely as a consequence of adaptation to host immune responses. Among the GALVs and KoRVs, episodic diversifying selection acts most prominently on GALVs associated with leukemia in captive gibbons and KoRVs thought to be exogenous. Because viruses with affinity to GALVs are regularly being discovered in wildlife species such as rodents and bats (21, 69), our findings and the methods applied provide a comparative framework for analyzing GALV-like retroviruses as they are discovered. The full GALV strain genomes reported here provide a resource to functionally explore and augment or improve existing retroviral vector biology.
Funding Statement
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Kawakami TG, Huff SD, Buckley PM, Dungworth DL, Synder SP, Gilden RV. 1972. C-type virus associated with gibbon lymphosarcoma. Nat New Biol 235:170–171. doi: 10.1038/newbio235170a0. [DOI] [PubMed] [Google Scholar]
- 2.Snyder SP, Dungworth DL, Kawakami TG, Callaway E, Lau DT. 1973. Lymphosarcomas in two gibbons (Hylobates lar) with associated C-type virus. J Natl Cancer Inst 51:89–94. [DOI] [PubMed] [Google Scholar]
- 3.DePaoli A, Johnsen DO, Noll MD. 1973. Granulocytic leukemia in white handed gibbons. J Am Vet Med Assoc 163:624–628. [PubMed] [Google Scholar]
- 4.Kawakami TG, Buckley PM. 1974. Antigenic studies on gibbon type-C viruses. Transplant Proc 6:193–196. [PubMed] [Google Scholar]
- 5.Gallo RC, Gallagher RE, Wong-Staal F, Aoki T, Markham PD, Schetters H, Ruscetti F, Valerio M, Walling MJ, O'Keeffe RT, Saxinger WC, Smith RG, Gillespie DH, Reitz MS Jr. 1978. Isolation and tissue distribution of type-C virus and viral components from a gibbon ape (Hylobates lar) with lymphocytic leukemia. Virology 84:359–373. doi: 10.1016/0042-6822(78)90255-6. [DOI] [PubMed] [Google Scholar]
- 6.Reitz MS Jr, Wong-Staal J-F, Haseltine WA, Kleid DG, Trainor CD, Gallagher RE, Gallo RC. 1979. Gibbon ape leukemia virus-Hall's Island: new strain of gibbon ape leukemia virus. J Virol 29:395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todaro GJ, Lieber MM, Benveniste RE, Sherr CJ. 1975. Infectious primate type C viruses: three isolates belonging to a new subgroup from the brains of normal gibbons. Virology 67:335–343. doi: 10.1016/0042-6822(75)90435-3. [DOI] [PubMed] [Google Scholar]
- 8.Kawakami TG, Kollias GV Jr, Holmberg C. 1980. Oncogenicity of gibbon type-C myelogenous leukemia virus. Int J Cancer 25:641–646. doi: 10.1002/ijc.2910250514. [DOI] [PubMed] [Google Scholar]
- 9.Theilen GH, Gould D, Fowler M, Dungworth DL. 1971. C-type virus in tumor tissue of a woolly monkey (Lagothrix spp.) with fibrosarcoma. J Natl Cancer Inst 47:881–889. [PubMed] [Google Scholar]
- 10.Wolfe LG, Smith RK, Deinhardt F. 1972. Simian sarcoma virus, type 1 (Lagothrix): focus assay and demonstration of nontransforming associated virus. J Natl Cancer Inst 48:1905–1908. [PubMed] [Google Scholar]
- 11.Hino S, Stephenson JR, Aaronson SA. 1975. Antigenic determinants of the 70,000 molecular weight glycoprotein of woolly monkey type C RNA virus. J Immunol 115:922–927. [PubMed] [Google Scholar]
- 12.Rangan SR. 1974. Antigenic relatedness of simian C-type viruses. Int J Cancer 13:64–70. doi: 10.1002/ijc.2910130108. [DOI] [PubMed] [Google Scholar]
- 13.Reitz MS Jr, Luczak JC, Gallo RC. 1979. Mapping of related and nonrelated sequences of RNA from woolly monkey virus and gibbon ape leukemia virus. Virology 93:48–56. doi: 10.1016/0042-6822(79)90274-5. [DOI] [PubMed] [Google Scholar]
- 14.Delassus S, Sonigo P, Wain-Hobson S. 1989. Genetic organization of gibbon ape leukemia virus. Virology 173:205–213. doi: 10.1016/0042-6822(89)90236-5. [DOI] [PubMed] [Google Scholar]
- 15.Fielding AK, Chapel-Fernandes S, Chadwick MP, Bullough FJ, Cosset FL, Russell SJ. 2000. A hyperfusogenic gibbon ape leukemia envelope glycoprotein: targeting of a cytotoxic gene by ligand display. Hum Gene Ther 11:817–826. doi: 10.1089/10430340050015437. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A, Bullough F, Murphy S, Emiliusen L, Lavillette D, Cosset FL, Cattaneo R, Russell SJ, Vile RG. 2000. Fusogenic membrane glycoproteins as a novel class of genes for the local and immune-mediated control of tumor growth. Cancer Res 60:1492–1497. [PubMed] [Google Scholar]
- 17.Burtonboy G, Delferriere N, Mousset B, Heusterspreute M. 1993. Isolation of a C-type retrovirus from an HIV infected cell line. Arch Virol 130:289–300. doi: 10.1007/BF01309661. [DOI] [PubMed] [Google Scholar]
- 18.Parent I, Qin Y, Vandenbroucke AT, Walon C, Delferriere N, Godfroid E, Burtonboy G. 1998. Characterization of a C-type retrovirus isolated from an HIV infected cell line: complete nucleotide sequence. Arch Virol 143:1077–1092. doi: 10.1007/s007050050357. [DOI] [PubMed] [Google Scholar]
- 19.Ting YT, Wilson CA, Farrell KB, Chaudry GJ, Eiden MV. 1998. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J Virol 72:9453–9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J Virol 74:4264–4272. doi: 10.1128/JVI.74.9.4264-4272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simmons G, Clarke D, McKee J, Young P, Meers J. 2014. Discovery of a novel retrovirus sequence in an Australian native rodent (Melomys burtoni): a putative link between gibbon ape leukemia virus and koala retrovirus. PLoS One 9:e106954. doi: 10.1371/journal.pone.0106954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maricic T, Whitten M, Paabo S. 2010. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One 5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsangaras K, Siracusa MC, Nikolaidis N, Ishida Y, Cui P, Vielgrader H, Helgen KM, Roca AL, Greenwood AD. 2014. Hybridization capture reveals evolution and conservation across the entire koala retrovirus genome. PLoS One 9:e95633. doi: 10.1371/journal.pone.0095633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protoc pdb.prot5448 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [DOI] [PubMed] [Google Scholar]
- 25.Alfano N, Courtiol A, Vielgrader H, Timms P, Roca AL, Greenwood AD. 2015. Variation in koala microbiomes within and between individuals: effect of body region and captivity status. Sci Rep 5:10189. doi: 10.1038/srep10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 29.Martin M. 2012. Cutadapt removes adapter sequences from high-throughput sequencing reads. Bioinformatics Action 17:10–12. [Google Scholar]
- 30.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303:3997. [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lander MR, Chattopadhyay SK. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol 52:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Stadler CK, Gorman K, Jensen N, Kim D, Zheng H, Tang S, Switzer WM, Pye GW, Eiden MV. 2013. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc Natl Acad Sci U S A 110:11547–11552. doi: 10.1073/pnas.1304704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryksin AV, Matsumura I. 2010. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanave C, Preparata G, Saccone C, Serio G. 1984. A new method for calculating evolutionary substitution rates. J Mol Evol 20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- 41.Dimmic MW, Rest JS, Mindell DP, Goldstein RA. 2002. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J Mol Evol 55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol 39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 45.Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A. 2010. How many bootstrap replicates are necessary? J Comput Biol 17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- 46.Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SD, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol 28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trainor CD, Scott ML, Josephs SF, Fry KE, Reitz MS Jr. 1984. Nucleotide sequence of the large terminal repeat of two different strains of gibbon ape leukemia virus. Virology 137:201–205. doi: 10.1016/0042-6822(84)90025-4. [DOI] [PubMed] [Google Scholar]
- 52.Villemur R, Rassart E, DesGroseillers L, Jolicoeur P. 1983. Molecular cloning of viral DNA from leukemogenic Gross passage A murine leukemia virus and nucleotide sequence of its long terminal repeat. J Virol 45:539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinter A, Kopelman R, Li Z, Kayman SC, Sanders DA. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol 71:8073–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishida Y, McCallister C, Nikolaidis N, Tsangaras K, Helgen KM, Greenwood AD, Roca AL. 2015. Sequence variation of koala retrovirus transmembrane protein p15E among koalas from different geographic regions. Virology 475:28–36. doi: 10.1016/j.virol.2014.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. 2007. Changes in viral protein function that accompany retroviral endogenization. Proc Natl Acad Sci U S A 104:17506–17511. doi: 10.1073/pnas.0704313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Demirov DG, Freed EO. 2004. Retrovirus budding. Virus Res 106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Shojima T, Hoshino S, Abe M, Yasuda J, Shogen H, Kobayashi T, Miyazawa T. 2013. Construction and characterization of an infectious molecular clone of koala retrovirus. J Virol 87:5081–5088. doi: 10.1128/JVI.01584-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purdy JG, Flanagan JM, Ropson IJ, Rennoll-Bankert KE, Craven RC. 2008. Critical role of conserved hydrophobic residues within the major homology region in mature retroviral capsid assembly. J Virol 82:5951–5961. doi: 10.1128/JVI.00214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oliveira NM, Farrell KB, Eiden MV. 2006. In vitro characterization of a koala retrovirus. J Virol 80:3104–3107. doi: 10.1128/JVI.80.6.3104-3107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller AD, Garcia JV, von Suhr N, Lynch CM, Wilson C, Eiden MV. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol 65:2220–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higuchi H, Bronk SF, Bateman A, Harrington K, Vile RG, Gores GJ. 2000. Viral fusogenic membrane glycoprotein expression causes syncytium formation with bioenergetic cell death: implications for gene therapy. Cancer Res 60:6396–6402. [PubMed] [Google Scholar]
- 62.Zhu B, Yang JR, Jiang YQ, Chen SF, Fu XP. 2014. Gene therapy of lung adenocarcinoma using herpes virus expressing a fusogenic membrane glycoprotein. Cell Biochem Biophys 69:583–587. doi: 10.1007/s12013-014-9836-4. [DOI] [PubMed] [Google Scholar]
- 63.Mason VC, Li G, Helgen KM, Murphy WJ. 2011. Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens. Genome Res 21:1695–1704. doi: 10.1101/gr.120196.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Enk JM, Devault AM, Kuch M, Murgha YE, Rouillard JM, Poinar HN. 2014. Ancient whole genome enrichment using baits built from modern DNA. Mol Biol Evol 31:1292–1294. doi: 10.1093/molbev/msu074. [DOI] [PubMed] [Google Scholar]
- 65.Eiden M, Trainor CD, Reitz MS. 1986. Gibbon ape leukaemia virus RNA in leukaemic T-lymphoid cell lines: expression of a novel RNA transcript. J Gen Virol 67(Pt 7):1455–1460. [DOI] [PubMed] [Google Scholar]
- 66.Chen SJ, Holbrook NJ, Mitchell KF, Vallone CA, Greengard JS, Crabtree GR, Lin Y. 1985. A viral long terminal repeat in the interleukin 2 gene of a cell line that constitutively produces interleukin 2. Proc Natl Acad Sci U S A 82:7284–7288. doi: 10.1073/pnas.82.21.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shojima T, Yoshikawa R, Hoshino S, Shimode S, Nakagawa S, Ohata T, Nakaoka R, Miyazawa T. 2013. Identification of a novel subgroup of koala retrovirus from koalas in Japanese zoos. J Virol 87:9943–9948. doi: 10.1128/JVI.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Avila-Arcos MC, Ho SY, Ishida Y, Nikolaidis N, Tsangaras K, Honig K, Medina R, Rasmussen M, Fordyce SL, Calvignac-Spencer S, Willerslev E, Gilbert MT, Helgen KM, Roca AL, Greenwood AD. 2013. One hundred twenty years of koala retrovirus evolution determined from museum skins. Mol Biol Evol 30:299–304. doi: 10.1093/molbev/mss223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cui J, Tachedjian G, Tachedjian M, Holmes EC, Zhang S, Wang LF. 2012. Identification of diverse groups of endogenous gammaretroviruses in mega- and microbats. J Gen Virol 93:2037–2045. doi: 10.1099/vir.0.043760-0. [DOI] [PMC free article] [PubMed] [Google Scholar]