ABSTRACT
Mucosal surfaces are vulnerable to human immunodeficiency virus (HIV)/simian immunodeficiency virus (SIV) infection and thus are key sites for eliciting vaccine-mediated protection. Vaccine protocols carried out at the Yerkes Primate Research Center utilized SIVmac239-based immunization strategies with intrarectal and intravaginal SIVsmE660 challenge of rhesus macaques. We investigated whether there were genetic signatures associated with SIVsmE660 intrarectal and intravaginal transmissions in vaccinated and unvaccinated monkeys. When transmitted/founder (T/F) envelope (Env) sequences from 49 vaccinated and 15 unvaccinated macaques were compared to each other, we were unable to identify any vaccine breakthrough signatures. In contrast, when the vaccinated and control T/F Envs were combined and compared to the challenge stock, residues at gp120 positions 23, 45, 47, and 70 (Ile-Ala-Lys-Asn [I-A-K-N]) emerged as signatures of mucosal transmission. However, T/F Envs derived from intrarectal and intravaginal infections were not different. Our data suggest that the vaginal and rectal mucosal environments both imposed a strong selection bias for SIVsmE660 variants carrying I-A-K-N that was not further enhanced by immunization. These findings, combined with the strong conservation of A-K-N in most HIV-2/SIVsmm isolates and the analogous residues in HIV-1/SIVcpz isolates, suggest that these residues confer increased transmission fitness to SIVsmE660.
IMPORTANCE Most HIV-1 infections occur across a mucosal barrier, and it is therefore important to understand why these sites are vulnerable and how to protect them with a vaccine. To gain insight into these questions, we studied rhesus macaques that were vaccinated with SIVmac239 and unvaccinated controls to determine whether the SIVsmE660 viral variants that infected these two groups were different. We did not find differences between viral variants in the absence versus presence of vaccination-induced immunity, but we did find that the SIVsmE660 viral variants that infected the monkeys, regardless of vaccination, were different from the dominant population found in the viral challenge inoculum. Our data suggest that the mucosal environments of the vagina and rectum both impose a strong selection for the SIVsmE660 variants in the challenge inoculum that are most like SIV and HIVs that circulate in nature.
INTRODUCTION
The majority of human immunodeficiency virus type 1 (HIV-1) transmission events occur across the genital or rectal mucosa and involve a reduction in the diversity of the infecting viral quasispecies (1). This genetic bottleneck of HIV-1 transmission has been attributed to various features of the HIV-1 envelope (Env) glycoproteins (2–8), resistance to interferon (7, 9), and a selection bias toward consensus residues that confer increased in vivo fitness (10). These studies notwithstanding, it has been difficult to pinpoint the attributes that directly facilitate transmission of a particular HIV-1 variant from a genetically diverse quasispecies. A primary focus of nonhuman primate simian immunodeficiency virus (SIV) challenge models is therefore to recapitulate the critical events that occur during HIV-1 transmission so that any observed effect, including protection, is accurately reflected by the model. An approach in which rhesus macaques are inoculated intravaginally or intrarectally at regular intervals with a relatively low dose of SIV has been shown to recapitulate the hallmark reduction in viral diversity characteristic of HIV-1 transmission (11–14). As a result, the repeated, low-dose challenge method has become widely accepted and commonly utilized to model protection against mucosal SIV infection (14–32). Despite these numerous studies, the characteristics of the virus that influence intravaginal and intrarectal transmission and protection are incompletely understood.
We previously demonstrated that an SIVmac239-based regimen consisting of two DNA primes followed by two modified vaccinia virus Ankara (MVA) boosts provided significant protection against repeated, low-dose SIVsmE660 intrarectal challenge. Including granulocyte-macrophage colony-stimulating factor (GM-CSF) or CD40 ligand (CD40L) as an adjuvant with the DNA primes further enhanced this protection (16–18, 33). We followed up on these results with a trial testing the protective capacity of DNA/MVA vaccination regimens and a novel protein-based prime boost regimen against intravaginal SIVsmE660 challenge. These studies were conducted at a single primate center using standard methodology, parallel immunization schedules, and Env immunogens all derived from SIVmac239. In addition, the challenge stock utilized for the follow-up trials (NM2010) was derived from the same SIVsmE660 seed stock as the challenge stock used in the initial trials (VH2000) and was expanded in vitro using the same protocol. The samples collected from these trials therefore provided a unique opportunity to explore genetic bottlenecks in SIVsmE660 Env that could be associated with SIVmac239-based vaccine protection and/or mucosal transmission.
MATERIALS AND METHODS
Ethics statement.
The Emory University Institutional Animal Care and Use Committee (AWA no. A3180-01) approved these studies of nonhuman primates under protocol YER-2000936-061014GA. This study was conducted in strict accordance with U.S. Department of Agriculture regulations and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (34). SIV-infected animals were housed in standard nonhuman primate cages, received standard primate feed as well as fresh fruit and enrichment daily, and had continual access to water. Animals had continuous access to enrichment resources, including objects for perching and other manipulanda. Animal welfare was monitored daily. Appropriate procedures were performed to ensure that potential distress, pain, or discomfort was alleviated. The sedative ketamine (10 mg/kg of body weight) or telazol (4 mg/kg) was used for blood draws. Euthanasia using pentobarbital (100 mg/kg) under anesthesia was performed only when deemed clinically necessary by veterinary medical staff and according to IACUC endpoint guidelines.
Rhesus macaque vaccine trials.
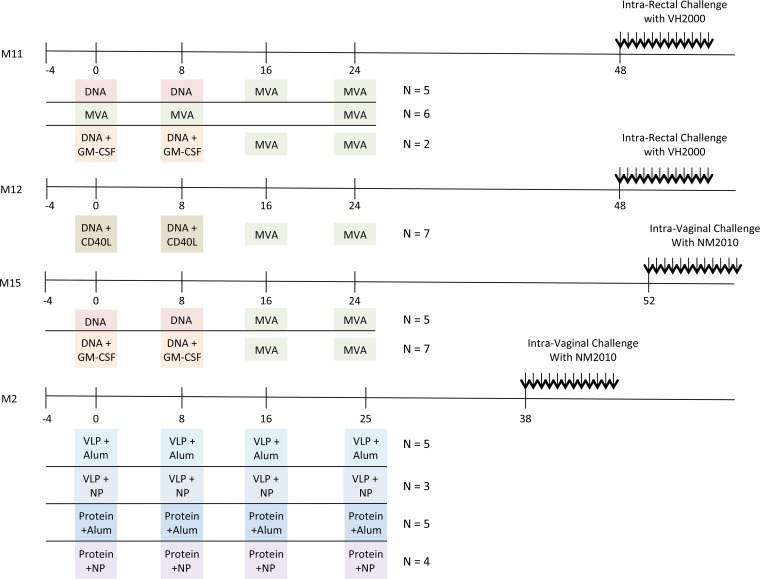
Plasma samples were obtained from vaccinated or unvaccinated rhesus macaques following SIVsmE660 challenge during four SIV vaccine trials carried out previously at Yerkes National Primate Research Center (designated M2, M11, M12, and M15). All trials utilized SIVmac239 immunogens and a repeated, low-dose SIVsmE660 mucosal challenge using equivalent animal infectious doses in rhesus macaques. Immunized and control animals in the M11 and M12 trials were challenged 12 times intrarectally, while animals in the M2 and M15 trials were challenged 12 times intravaginally. The M11 trial was conducted by Rama Amara and consisted of three vaccine groups: DNA prime/MVA boost (DDMM), DNA prime with GM-CSF adjuvant/MVA boost (DgDgMM), and three MVA immunizations (MMM). An additional arm was later included, designated M12, which consisted of DNA prime with CD40 ligand adjuvant followed by MVA boost (D40LD40LMM). The M2 trial was led by Bali Pulendran and consisted of three gp140 protein or virus-like particle (VLP) immunizations delivered with alum (protein+alum or VLP+alum) or nanoparticles (NP) (monophosphoryl lipid A and resiquimod) (protein+NP or VLP+NP) as adjuvants. The M15 trial was conducted by Rama Amara and was similar to M11 in that the vaccination arms were DNA prime, with or without the GM-CSF adjuvant, with two MVA boosts.
SGA and sequencing of SIVsmE660 env genes.
Plasma collected at the second positive viral load test result during weekly challenges was used for 384-well single genome amplification (SGA) of SIVsmE660 env genes. Viral RNA was extracted from plasma using a QIAmp viral RNA kit according to the manufacturer's instructions (Qiagen). Reverse transcription of viral RNA into cDNA was performed using a SuperScript III kit according to the manufacturer's instructions (Invitrogen), with reverse primer SIVsm/macEnvR1 (5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′). cDNA was diluted to result in <30% positive wells for SGA. First-round PCR was performed in a 15-μl volume using Phusion Hotstart II high-fidelity DNA polymerase (Thermo Scientific) with SIVsm/macEnvF1 (5′-CCTCCCCCTCCAGGACTAGC-3′) and SIVsm/macEnvR1 (5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′). Cycling conditions were 98°C for 2 min; 10 cycles of 95°C for 15 s, 54°C for 60 s, and 68°C for 4 min; 25 cycles of 95°C for 15 s, 54°C for 60 s, and 68°C for 4 min, adding 5 s to the extension per cycle; 72°C for 30 min; and 4°C hold. Second-round PCR was performed with the same enzyme in a 10-μl volume with 1 μl from the first round of PCR and SIVsmEnvF2 (5′-CACCTATGATAGACATGGAGACACCCTTGAAGGAGC-3′) and SIVsmEnvR2 (5′-ATGAGACATRTCTATTGCCAATTTGA-3′). Cycling conditions were 95°C for 2 min; 30 cycles of 95°C for 15 s, 54°C for 60 s, and 72°C for 2.5 min; 72°C for 10 min; and 4°C hold. PCR amplicons were purified using a Qiagen PCR clean-up kit following the manufacturer's instructions (Qiagen). Amplicons were sequenced with Beckman Coulter Genomics. The following primers were used: F1 (5′-ATCCATTTCAGAAGTGGATGTGCCCACTCC-3′), F2 (5′-AGGGTTAAAAAGGGACAAAAGGATAGAATA-3′), F3 (5′-TTGTAGAGGAGAATTCTTATACTGCAAAAT-3′), and F4N (5′-AAACATTGGTGCCTAATTGGAGCAATATGA-3′) and R1 (5′-GCAAAGCATAACCTGGCGGTGCACAATATC-3′), R2 (5′-CTCCTTCCCTAGGAGGCAAATATACATTTT-3′), R3N (5′-TTGCAATTCATACATATTCTTTTCTTGCTG-3′), and R4 (5′-TCATCTTCATCATCCACATCATCCATGTTT-3′).
Sequence analyses.
Sequencher v5 was used to generate nucleotide sequence contigs, and sequences with evidence of mixed peaks were omitted from the analysis. Geneious v6.1.7 was used to translate nucleotide sequences, create alignments, and calculate pairwise diversity. Nucleotide alignments were exported from Geneious in FASTA format and used to generate PhyML (http://www.hiv.lanl.gov/content/sequence/PHYML/interface.html) and Rainbow (http://www.hiv.lanl.gov/content/sequence/RAINBOWTREE/rainbowtree.html) trees. GenBank accession numbers for the VH2000 challenge stock sequences are FJ579014 to FJ579055, and those for the transmitted/founder (T/F) Env and NM2010 challenge stock sequences are KP734847 to KP735110 and KT288416 to KT288894.
Statistical analyses.
Amino acid alignments were exported from Geneious in FASTA format for Sequence Harmony comparisons (http://www.ibi.vu.nl/programs/shmrwww/) and for generation of probability Weblogos (http://weblogo.threeplusone.com/create.cgi). Fisher's exact tests were calculated via Prism 6 for Mac OS X.
Nucleotide sequence accession numbers.
The NM2010 challenge stock sequences, M2 and M15 T/F Env sequences, and additional M11 and M12 T/F Env sequences are available in GenBank under accession numbers KT288416 to KT288894.
RESULTS
SIVsmE660 challenge stocks used for intrarectal and intravaginal challenges were genetically indistinguishable.
The M11, M12, M15, and M2 trials were preclinical SIVmac239-based immunization studies carried out at the Yerkes National Primate Research Center in rhesus macaques (16–18; unpublished data). M11, M12, and M15 vaccine regimens delivered immunogens via two DNA primes followed by two heterologous MVA boosts (DDMM) at 8-week intervals, followed by SIVsmE660 challenge at 48 to 52 weeks (Fig. 1). These trials also tested GM-CSF (DNA+GM-CSF) or CD40L (DNA+CD40L) adjuvanted DNA primes or a homologous prime boost using only MVA. The M2 trial delivered SIVmac239 Gag p55 and Env gp140 (protein) or virus-like particle (VLP) immunizations with alum or nanoparticles (NP) containing Toll-like receptor ligands (Fig. 1), followed by challenge at 38 weeks. Two SIVsmE660 challenge stocks were used; VH2000 was provided by Vanessa Hirsch and used for intrarectal challenges in M11 and M12, and NM2010 was provided by Nancy Miller and used for intravaginal challenges in M15 and M2. Ranajit Pal at ABL prepared NM2010 using the same seed stock (E660/PT71) and culture procedures (growth in pigtail macaque peripheral blood mononuclear cells [PBMC]) used to derive VH2000. To determine the genetic similarity of the two SIVsmE660 challenge stocks, we analyzed 37 SGA Env sequences (provided by Brandon Keele) from the VH2000 challenge stock and 59 SGA Env sequences from the NM2010 challenge stock. The sequences from the two stocks comingled on a PhyML phylogenetic tree (Fig. 2), and the maximum pairwise nucleotide difference was 3%. At the amino acid level, there were no signatures detected by Sequence Harmony analysis that could differentiate the two stocks using a Z-score of −3 as the threshold for significance (data not shown) (35). Based on the common lineage of the two stocks and their high genetic similarity, we combined the sequences to investigate viral determinants of vaccine breakthrough or transmission.
FIG 1.
Timelines of nonhuman primate vaccine trials. The immunization schedule for each vaccine trial (M11, M12, M15, and M2) is plotted along a timeline, in weeks. The agents used for priming and boosting for each trial are indicated in colored boxes, highlighting similarities and differences between the trials. The number of animals that were analyzed from each group is indicated to the right of each vaccination regimen. Fifteen unvaccinated control animals were also included. Twelve low-dose, repeated challenges with either the VH2000 or the NM2010 SIVsmE660 stock were initiated at week 38, 48, or 52, either intrarectally or intravaginally, as indicated. NP, nanoparticles; VLP, virus-like particles; Protein, soluble envelope gp140. Challenge results for M11 and M12 have been published previously (16–18). Challenge results for M2 and M15 are unpublished.
FIG 2.
Phylogenetic tree of SGA-derived env nucleotide sequences from the VH2000 and NM2010 challenge stocks. A Geneious v6 nucleotide alignment containing all VH2000 and NM2010 env sequences was uploaded to the LANL PhyML tool to generate a neighbor-joining phylogenetic tree, which was annotated using the LANL Rainbow Tree tool. The horizontal bar shows the scale of genetic distance.
No vaccination breakthrough signature was identified in Env.
To investigate potential sequence signatures in vaccination breakthrough, we analyzed an average of 10 ± 4 SGA T/F Env sequences from 49 immunized monkeys and 15 unvaccinated control monkeys (Fig. 1). The T/F Env sequences did not cluster according to vaccination group or status or with particular challenge stock sequences when aligned and analyzed phylogenetically (see Fig. S1 in the supplemental material), which is consistent with our previous analysis (14). The sequence analysis also revealed that a small minority of the animals (3 of 64) was clearly infected with more than one variant; however, we did not perform exhaustive sequencing to identify minor variants, as this was not the focus of our analysis. To explore the possibility of a vaccine sieving effect, the T/F Env sequences from the 49 vaccinated animals were compared against those from the 15 unvaccinated controls using Sequence Harmony (35). This analysis did not reveal significant differences (Z-score < −3) at any amino acid position that could be associated with vaccine breakthrough (data not shown).
Signature residues in gp120 C1 are associated with mucosal transmission.
In the absence of a vaccine breakthrough signature, we compared the combined challenge stock sequences (n = 96) against all T/F Env sequences regardless of vaccination status (n = 67) to search for signatures that might favor mucosal transmission of SIVsmE660. Four amino acid positions (23, 45, 47, and 70) in the gp120 C1 region (based on SIVmac239 numbering and consistent with numbering in references 15, 36, and 37) achieved a Z-score of less than −3 in Sequence Harmony (Fig. 3A and B), indicating that these residues could be associated with a selective advantage during mucosal transmission of SIVsmE660. The four signature positions were dominated by Val-Thr-Arg-Ser (V-T-R-S) in the SIVsmE660 challenge stocks (Fig. 3A). Using an independent Fisher exact test for each of the four positions, we found significantly higher frequencies of A-K-N at these positions in the T/F Envs than in the challenge stocks (Fig. 3B; also see Table S1 in the supplemental material) (P ≤ 0.002), while enrichment of I at position 23 was less significant (P = 0.017). This result is consistent with a recent meta-analysis performed by Gonzalez et al. (37).
FIG 3.
Weblogo and Sequence Harmony comparison of challenge stocks to T/F Env sequences. Sequence Harmony analysis comparing challenge stock to T/F Envs identified four amino acid positions with Z-scores below −3: positions 23, 45, 47, and 70 in gp120. Differences in amino acid composition at each of these locations are illustrated by Weblogos for the challenge stock (A) and all T/F Env sequences combined (B). The height of the amino acid within the stack at each position indicates its relative frequency. For illustrative purposes, I-A-K-N residues are highlighted in red and V-T-R-S residues in green. P values for Fischer's exact tests at each position were ≤0.01 (see Table S1 in the supplemental material).
As described above, Sequence Harmony analysis did not identify any signature residues associated with vaccination breakthrough; however, another study had reported that I-A-K-N was favored over V-T-R-S in breakthrough infections of SIVmac239 Env-vaccinated rhesus macaques (15). We therefore compared the frequencies of amino acids at these positions in the 51 vaccinated and 16 control T/F Envs using a Fisher exact test. There was no significant difference between the two (see Table S1 in the supplemental material), as illustrated in Fig. 4A and B, and thus no evidence for enhanced selection of I-A-K-N variants during vaccination breakthrough in the experiments analyzed here.
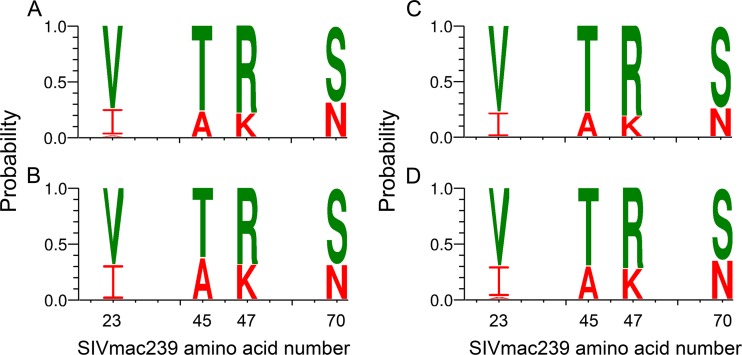
FIG 4.
Weblogo comparison of vaccinated versus control T/F Envs and intrarectal versus intravaginal T/F Envs. Weblogos were generated to illustrate similarity in amino acid composition at positions 23, 45, 47, and 70 between T/F Envs from vaccinated (A) and control (B) animals or intrarectally challenged (C) and intravaginally challenged (D) animals. P values for Fischer's exact tests comparing results shown in panels A versus B and panels C versus D at each position were not significant (see Table S1 in the supplemental material). The height of the amino acid within the stack at each position indicates its relative frequency. For illustrative purposes, I-A-K-N residues are highlighted in red and V-T-R-S residues in green.
A Fisher exact test was also used to compare amino acid frequencies at positions 23, 45, 47, and 70 between animals infected intrarectally and intravaginally. There were no statistically significant differences at any of the signature positions between T/F Envs from these two groups (Fig. 4C and D; also see Table S1 in the supplemental material). A similar approach was used to investigate whether an association of amino acids at positions 23, 45, 47, and 70 with the number of challenges required to transmit infection (low, 1 to 6 doses; high, 7 to 12 doses) or TRIM5α encoded host susceptibility to infection. Neither of these was statistically significantly different in terms of the frequencies of I-A-K-N (see Table S1).
I-A-K-N restores transmission fitness to SIVsmE660.
Overall, the data suggest that while V-T-R-S variants transmit across the mucosa, they do so at a lower frequency than expected by their dominance in the challenge stocks. Gonzalez et al. recently hypothesized that I-A-K-N variants of SIVsmE660 have higher transmission fitness than those carrying V-T-R-S (37). To explore this hypothesis with our data, we used challenge stock and T/F Env sequences to calculate the relative transmission fitness (W) of the V-T-R-S and I-A-K-N genotypes and all other genotypes combined (38). WIAKN (TransmittedIAKN/StockIAKN, 2.5) was approximately 5-fold greater than WVTRS (TransmittedVTRS/StockVTRS, 0.48) and 2-fold greater than WOthers (TransmittedOthers/StockOthers, 1.2) by these calculations. We used these values to extrapolate population compositions after an additional nine transmission cycles (Fig. 5A). By transmission cycle 3, I-A-K-N variants are predicted to overtake V-T-R-S and other variants as the majority genotype (60%). By transmission cycle 10, I-A-K-N virtually extinguished V-T-R-S and the other genotypes, making up >99% of the population. Thus, according to our extrapolations from a single infection cycle, I-A-K-N provides a strong advantage for in vivo SIVsmE660 mucosal transmission.
FIG 5.
Relative fitness model showing I-A-K-N displacement of V-T-R-S variants. (A) The frequencies in the challenge stock and T/F Envs of V-T-R-S, I-A-K-N, and all other permutations combined were used to calculate the relative transmission fitness value for each group. These values were then used to model the number of transmission events needed for I-A-K-N to overtake the V-T-R-S sequences predominant in the SIVsmE660 challenge stock. (B) Weblogo illustrating the amino acid compositions of 112 HIV-2/SIVsmm reference strains (excluding SIVsmE660) downloaded from the LANL HIV sequence database. The height of the amino acid within the stack at each position indicates its relative frequency. For illustrative purposes, I-A-K-N residues are highlighted in red and V-T-R-S residues in green.
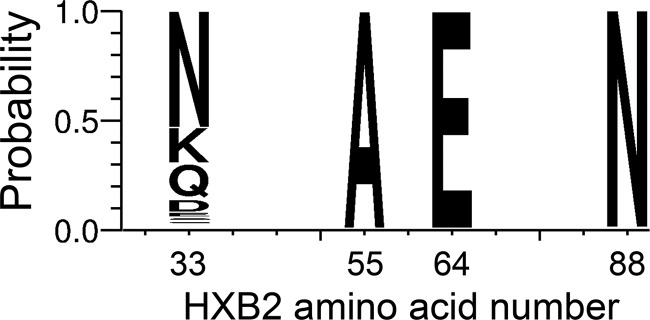
To gain further insight into the advantageous nature of the I-A-K-N genotype and determine whether it is also more fit for aspects of viral replication other than transmission, we investigated the amino acid compositions at positions 23, 45, 47, and 70 in the HIV-2/SIVsmm lineage, excluding SIVsmE660-derived sequences. One hundred twelve HIV-2/SIVsmm reference Env sequences were downloaded from the LANL HIV sequence database (http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html) and examined alongside the challenge stock and T/F Env sequences. Interestingly, V and I, the only residues found at position 23 in SIVsmE660, occurred in only a small minority of the HIV-2/SIVsmm reference Env sequences, which displayed high entropy at this position (Fig. 5B). For the other three positions, A-K-N were highly conserved in the HIV-2/SIVsmm reference Env sequences, reflecting their increased frequency in the T/F Envs. An analysis of the analogous residues in more than 5,000 Env sequences from the HIV-1/SIVcpz lineage downloaded from the LANL database followed a pattern similar to that of the HIV-2/SIVsmm sequences (Fig. 6), in this case, with A-E-N dominating. Thus, diversifying selection at position 23 and high conservation at positions 45, 47, and 70 occur in the great majority of natural SIV and HIV variants.
FIG 6.
Weblogo of HIV-1/SIVcpz reference strains. The amino acid frequencies at positions 33, 55, 64, and 88 (analogous to positions 23, 45, 47, and 70 in the HIV-2/SIVsmm lineage) of 5,132 HIV-1/SIVcpz reference strains are illustrated via Weblogo. The height of the amino acid within the stack at each position indicates its relative frequency.
DISCUSSION
This study and two other recent publications have identified amino acids 23, 45, 47, and 70 in the C1 region of gp120 as important residues during mucosal SIVsmE660 challenge of rhesus macaques (15, 37). Roederer et al. attributed an enrichment of I-A-K-N to neutralization resistance (emphasizing A-K) that facilitated vaccine breakthrough infection, while a retrospective analysis of multiple independent trials carried out by Gonzalez et al. hypothesized that the enrichment of I-A-K-N was the result of increased transmission fitness and was independent of preexisting immunity. Due to the purposeful design of the M11, M12, M2, and M15 vaccine trials, we recognized a unique opportunity to explore both of these hypotheses.
Regarding the association between positions 45 and 47 and resistance to neutralizing antibody, in-depth analysis of prechallenge antibody responses against the autologous breakthrough T/F Env variants in the M11 and M12 trials yielded results that were not consistent with those of Roederer et al. in that both A-K and T-R variants were susceptible to heterologous and autologous neutralization (14). In the M11/M12 study, neither T-R nor A-K predicted susceptibility to heterologous or vaccine-induced neutralizing antibody responses, and breakthrough variants from vaccinated animals were not enriched for A-K over the T/F Envs from unvaccinated control animals. The Sequence Harmony signature analysis presented here, performed on a larger number of sequences from the M2, M11, M12, and M15 trials, also failed to identify any amino acid signatures in T/F Envs from vaccinated versus control animals. Furthermore, results from direct Fisher's exact tests of amino acids at positions 23, 45, 47, and 70 between these two groups were not significant. However, the results of our current study are in agreement with those of Gonzalez et al. and suggest that I-A-K-N is advantageous for mucosal transmission regardless of whether vaccine-induced antibodies are present or absent (37). It is important to note that the trials analyzed here used entirely different vaccination strategies and challenge stocks from the Roederer et al. study, which could explain the inconsistencies between our observations. Nevertheless, our findings do argue against positions 45 and 47 being a universal marker for neutralization resistance (14, 36, 37, 39, 40).
Even if amino acid changes at positions 45 and 47 alter SIVsmE660 neutralization sensitivity, the ability to evade host antibody responses cannot be the dominant selective pressure on transmission. Roederer et al. suggested that stepwise mutations from T-R to T-K, A-R, and A-K at positions 45 to 47 resulted in increased neutralization resistance (T-R < T-K < A-R < A-K) (15). However, we did not isolate any sequences with T-K at positions 45 and 47 in the 96 challenge stock sequences or among the 67 T/F Envs. This could be due to the fact that SGA, not deep sequencing, was used here. Even so, a search of all SIVsmm Env sequences available in the LANL HIV sequence database for those that contain the T-K motif CTTKN (amino acid positions 44 to 48) identified only 22 sequences, 21 of which were from a single SIVsmE660-infected monkey (15). Thus, even if the T-K intermediate confers an increase in neutralization resistance, there appears to be selective pressure against this amino acid combination during in vitro propagation of viral stocks, in animal transmission models, and in the natural animal host environment. The reasons underlying selection for or against certain residues in the SIVsmE660 gp120 C1 region could shed light upon this region's potential contributions to viral fitness.
The question remains as to why I-A-K-N variants of SIVsmE660 are favored during mucosal transmission and what they can teach us about HIV-1 transmission and protection. These variants could be more efficient than others at any number of steps, including crossing the mucosal epithelium, infecting a susceptible CD4+ target cell, evading innate immunity, expanding locally to infect other CD4+ target cells, or disseminating out of the mucosa (41). However, residues A-K-N are so highly conserved in HIV-2/SIVsmm isolates that only the atypical amino acid composition of the SIVsmE660 challenge stock Env quasispecies allowed this selection event to be observed. It might then be tempting to alter the genetic composition of SIV or simian-human immunodeficiency virus (SHIV) challenge stocks beyond what is found in nature to enhance our ability to detect informative sites and learn more about the transmission process. However, this approach runs the risk of generating a viral quasispecies that is no longer representative of circulating HIV-1 variants. If these populations deviate too far from nature, the dominant selective pressure could be adaptation back to the animal host, obfuscating the vaccine-mediated protection that these models are set up to test. Indeed, V-T-R-S, despite being the dominant genotype in the SIVsmE660 quasispecies, is disadvantageous not only for transmission but also during natural SIV infection. In considering the nearly invariant nature of the analogous residues in HIV-1 (A-E-N) (Fig. 6), it becomes clear that the chance of a vaccinated individual encountering an HIV-1 variant as atypical as SIVsmE660 V-T-R-S during a transmission event in the human host is extremely low. It is also improbable that this signature would be easily identified in acute versus chronic or donor versus recipient comparisons of HIV-1 Env sequences, due to the invariant nature of the residues. Our results, combined with those of Gonzalez et al. (37), should prompt a careful consideration of the authenticity of nonhuman primate challenge models, wherein protection may be influenced by the effects of reinstating in vivo fitness, a phenomenon which does not occur in natural HIV-1 infection.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Brandon Keele for providing SGA sequences from the SIVsmE660 VH200 challenge stock, Lori Spicer and Samantha Burton for technical assistance, and Harriet Robinson for helpful discussions.
Funding Statement
The work was also funded by grants NIH P51-RR000165 and P51-OD011132 to the Yerkes National Primate Research Center. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02711-15.
REFERENCES
- 1.Shaw GM, Hunter E. 2012. HIV transmission, p 1–23. In Bushman FD, Nabel GJ, Swanstrom R (ed), Cold Spring Harbor perspectives in medicine, vol 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 3.Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol 79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, Lapedes AS, Shen T, Gaschen B, Krishnamoorthy M, Li H, Decker JM, Salazar-Gonzalez JF, Wang S, Jiang C, Gao F, Swanstrom R, Anderson JA, Ping LH, Cohen MS, Markowitz M, Goepfert PA, Saag MS, Eron JJ, Hicks CB, Blattner WA, Tomaras GD, Asmal M, Letvin NL, Gilbert PB, Decamp AC, Magaret CA, Schief WR, Ban YE, Zhang M, Soderberg KA, Sodroski JG, Haynes BF, Shaw GM, Hahn BH, Korber B. 2011. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog 7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sagar M, Laeyendecker O, Lee S, Gamiel J, Wawer MJ, Gray RH, Serwadda D, Sewankambo NK, Shepherd JC, Toma J, Huang W, Quinn TC. 2009. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis 199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker ZF, Iyer SS, Wilen CB, Parrish NF, Chikere KC, Lee FH, Didigu CA, Berro R, Klasse PJ, Lee B, Moore JP, Shaw GM, Hahn BH, Doms RW. 2013. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J Virol 87:2401–2411. doi: 10.1128/JVI.02964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP Jr, Bjorkman PJ, Wilen CB, Doms RW, O'Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson SI, Gray ES, Mkhize NN, Sheward DJ, Lambson BE, Wibmer CK, Masson L, Werner L, Garrett N, Passmore JA, Karim QA, Karim SS, Williamson C, Moore PL, Morris L. 2015. South African HIV-1 subtype C transmitted variants with a specific V2 motif show higher dependence on alpha4beta7 for replication. Retrovirology 12:54. doi: 10.1186/s12977-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, Shaw GM, Hahn BH, Ochsenbauer C, Kappes JC, Borrow P. 2013. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin TH, Peng J, Seese AM, Shapiro R, Frater J, Ndung'u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJ, Allen TM, Allen S, Hunter E. 2014. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345:1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, Mascola JR, Nabel GJ, Haynes BF, Bhattacharya T, Perelson AS, Korber BT, Hahn BH, Shaw GM. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med 206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Keele BF, Li H, Keating S, Norris PJ, Carville A, Mansfield KG, Tomaras GD, Haynes BF, Kolodkin-Gal D, Letvin NL, Hahn BH, Shaw GM, Barouch DH. 2010. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J Virol 84:10406–10412. doi: 10.1128/JVI.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone M, Keele BF, Ma ZM, Bailes E, Dutra J, Hahn BH, Shaw GM, Miller CJ. 2010. A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J Virol 84:7083–7095. doi: 10.1128/JVI.00481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton SL, Kilgore KM, Smith SA, Reddy S, Hunter E, Robinson HL, Silvestri G, Amara RR, Derdeyn CA. 2015. Breakthrough of SIV strain smE660 challenge in SIV strain mac239-vaccinated rhesus macaques despite potent autologous neutralizing antibody responses. Proc Natl Acad Sci U S A 112:10780–10785. doi: 10.1073/pnas.1509731112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roederer M, Keele BF, Schmidt SD, Mason RD, Welles HC, Fischer W, Labranche C, Foulds KE, Louder MK, Yang ZY, Todd JP, Buzby AP, Mach LV, Shen L, Seaton KE, Ward BM, Bailer RT, Gottardo R, Gu W, Ferrari G, Alam SM, Denny TN, Montefiori DC, Tomaras GD, Korber BT, Nason MC, Seder RA, Koup RA, Letvin NL, Rao SS, Nabel GJ, Mascola JR. 2013. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505:502–508. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwa S, Lai L, Gangadhara S, Siddiqui M, Pillai VB, Labranche C, Yu T, Moss B, Montefiori DC, Robinson HL, Kozlowski PA, Amara RR. 2014. CD40L-adjuvanted DNA/modified vaccinia virus Ankara simian immunodeficiency virus SIV239 vaccine enhances SIV-specific humoral and cellular immunity and improves protection against a heterologous SIVE660 mucosal challenge. J Virol 88:9579–9589. doi: 10.1128/JVI.00975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai L, Kwa S, Kozlowski PA, Montefiori DC, Ferrari G, Johnson WE, Hirsch V, Villinger F, Chennareddi L, Earl PL, Moss B, Amara RR, Robinson HL. 2011. Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine. J Infect Dis 204:164–173. doi: 10.1093/infdis/jir199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai L, Kwa SF, Kozlowski PA, Montefiori DC, Nolen TL, Hudgens MG, Johnson WE, Ferrari G, Hirsch VM, Felber BK, Pavlakis GN, Earl PL, Moss B, Amara RR, Robinson HL. 2012. SIVmac239 MVA vaccine with and without a DNA prime, similar prevention of infection by a repeated dose SIVsmE660 challenge despite different immune responses. Vaccine 30:1737–1745. doi: 10.1016/j.vaccine.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouts TR, Bagley K, Prado IJ, Bobb KL, Schwartz JA, Xu R, Zagursky RJ, Egan MA, Eldridge JH, LaBranche CC, Montefiori DC, Le Buanec H, Zagury D, Pal R, Pavlakis GN, Felber BK, Franchini G, Gordon S, Vaccari M, Lewis GK, DeVico AL, Gallo RC. 2015. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc Natl Acad Sci U S A 112:E992–E999. doi: 10.1073/pnas.1423669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrareddy SN, Kallam B, Arthos J, Cicala C, Nawaz F, Hiatt J, Kersh EN, McNicholl JM, Hanson D, Reimann KA, Brameier M, Walter L, Rogers K, Mayne AE, Dunbar P, Villinger T, Little D, Parslow TG, Santangelo PJ, Villinger F, Fauci AS, Ansari AA. 2014. Targeting alpha4beta7 integrin reduces mucosal transmission of simian immunodeficiency virus and protects gut-associated lymphoid tissue from infection. Nat Med 20:1397–1400. doi: 10.1038/nm.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon SN, Doster MN, Kines RC, Keele BF, Brocca-Cofano E, Guan Y, Pegu P, Liyanage NP, Vaccari M, Cuburu N, Buck CB, Ferrari G, Montefiori D, Piatak M Jr, Lifson JD, Xenophontos AM, Venzon D, Robert-Guroff M, Graham BS, Lowy DR, Schiller JT, Franchini G. 2014. Antibody to the gp120 V1/V2 loops and CD4+ and CD8+ T cell responses in protection from SIVmac251 vaginal acquisition and persistent viremia. J Immunol 193:6172–6183. doi: 10.4049/jimmunol.1401504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selinger C, Strbo N, Gonzalez L, Aicher L, Weiss JM, Law GL, Palermo RE, Vaccari M, Franchini G, Podack ER, Katze MG. 2014. Multiple low-dose challenges in a rhesus macaque AIDS vaccine trial result in an evolving host response that affects protective outcome. Clin Vaccine Immunol 21:1650–1660. doi: 10.1128/CVI.00455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrareddy SN, Ayash-Rashkovsky M, Kramer VG, Lee SJ, Correll M, Novembre FJ, Villinger F, Johnson WE, von Gegerfelt A, Felber BK, Ruprecht RM. 2013. Live attenuated Rev-independent Nef SIV enhances acquisition of heterologous SIVsmE660 in acutely vaccinated rhesus macaques. PLoS One 8:e75556. doi: 10.1371/journal.pone.0075556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulkarni V, Rosati M, Valentin A, Jalah R, Alicea C, Yu L, Guan Y, Shen X, Tomaras GD, LaBranche C, Montefiori DC, Irene C, Prattipati R, Pinter A, Sullivan SM, Pavlakis GN, Felber BK. 2013. Vaccination with Vaxfectin((R)) adjuvanted SIV DNA induces long-lasting humoral immune responses able to reduce SIVmac251 viremia. Hum Vaccin Immunother 9:2069–2080. doi: 10.4161/hv.25442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cervasi B, Carnathan DG, Sheehan KM, Micci L, Paiardini M, Kurupati R, Tuyishime S, Zhou XY, Else JG, Ratcliffe SJ, Ertl HC, Silvestri G. 2013. Immunological and virological analyses of rhesus macaques immunized with chimpanzee adenoviruses expressing the simian immunodeficiency virus Gag/Tat fusion protein and challenged intrarectally with repeated low doses of SIVmac. J Virol 87:9420–9430. doi: 10.1128/JVI.01456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurupati R, Tuyishime S, Kossenkov AV, Sazanovich M, Haut LH, Lasaro MO, Ratcliffe SJ, Bosinger SE, Carnathan DG, Lewis M, Showe LC, Silvestri G, Ertl HC. 2013. Correlates of relative resistance against low-dose rectal simian immunodeficiency virus challenges in peripheral blood mononuclear cells of vaccinated rhesus macaques. J Leukoc Biol 93:437–448. doi: 10.1189/jlb.0612287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenbusch M, Ignatius R, Temchura V, Nabi G, Tippler B, Stewart-Jones G, Salazar AM, Sauermann U, Stahl-Hennig C, Uberla K. 2012. Risk of immunodeficiency virus infection may increase with vaccine-induced immune response. J Virol 86:10533–10539. doi: 10.1128/JVI.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao P, Patterson LJ, Kuate S, Brocca-Cofano E, Thomas MA, Venzon D, Zhao J, DiPasquale J, Fenizia C, Lee EM, Kalisz I, Kalyanaraman VS, Pal R, Montefiori D, Keele BF, Robert-Guroff M. 2012. Replicating adenovirus-simian immunodeficiency virus (SIV) recombinant priming and envelope protein boosting elicits localized, mucosal IgA immunity in rhesus macaques correlated with delayed acquisition following a repeated low-dose rectal SIV(mac251) challenge. J Virol 86:4644–4657. doi: 10.1128/JVI.06812-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winstone N, Wilson AJ, Morrow G, Boggiano C, Chiuchiolo MJ, Lopez M, Kemelman M, Ginsberg AA, Mullen K, Coleman JW, Wu CD, Narpala S, Ouellette I, Dean HJ, Lin F, Sardesai NY, Cassamasa H, McBride D, Felber BK, Pavlakis GN, Schultz A, Hudgens MG, King CR, Zamb TJ, Parks CL, McDermott AB. 2011. Enhanced control of pathogenic simian immunodeficiency virus SIVmac239 replication in macaques immunized with an interleukin-12 plasmid and a DNA prime-viral vector boost vaccine regimen. J Virol 85:9578–9587. doi: 10.1128/JVI.05060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alpert MD, Rahmberg AR, Neidermyer W, Ng SK, Carville A, Camp JV, Wilson RL, Piatak M Jr, Mansfield KG, Li W, Miller CJ, Lifson JD, Kozlowski PA, Evans DT. 2010. Envelope-modified single-cycle simian immunodeficiency virus selectively enhances antibody responses and partially protects against repeated, low-dose vaginal challenge. J Virol 84:10748–10764. doi: 10.1128/JVI.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds MR, Weiler AM, Piaskowski SM, Kolar HL, Hessell AJ, Weiker M, Weisgrau KL, Leon EJ, Rogers WE, Makowsky R, McDermott AB, Boyle R, Wilson NA, Allison DB, Burton DR, Koff WC, Watkins DI. 2010. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J Virol 84:9190–9199. doi: 10.1128/JVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, Yeh WW, Asmal M, Gelman RS, Shen L, Whitney JB, Seoighe C, Lacerda M, Keating S, Norris PJ, Hudgens MG, Gilbert PB, Buzby AP, Mach LV, Zhang J, Balachandran H, Shaw GM, Schmidt SD, Todd JP, Dodson A, Mascola JR, Nabel GJ. 2011. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci Transl Med 3:81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer SS, Gangadhara S, Victor B, Gomez R, Basu R, Hong JJ, Labranche C, Montefiori DC, Villinger F, Moss B, Amara RR. 2015. Codelivery of envelope protein in alum with MVA vaccine induces CXCR3-biased CXCR5+ and CXCR5- CD4 T cell responses in rhesus macaques. J Immunol 195:994–1005. doi: 10.4049/jimmunol.1500083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 35.Brandt BW, Feenstra KA, Heringa J. 2010. Multi-Harmony: detecting functional specificity from sequence alignment. Nucleic Acids Res 38:W35–W40. doi: 10.1093/nar/gkq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilgore KM, Murphy MK, Burton SL, Wetzel KS, Smith SA, Xiao P, Reddy S, Francella N, Sodora DL, Silvestri G, Cole KS, Villinger F, Robinson JE, Pulendran B, Hunter E, Collman RG, Amara RR, Derdeyn CA. 2015. Characterization and implementation of a diverse simian immunodeficiency virus SIVsm envelope panel in the assessment of neutralizing antibody breadth elicited in rhesus macaques by multimodal vaccines expressing the SIVmac239 envelope. J Virol 89:8130–8151. doi: 10.1128/JVI.01221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez MW, DeVico AL, Lewis GK, Spouge JL. 2015. Conserved molecular signatures in gp120 are associated with the genetic bottleneck during simian immunodeficiency virus (SIV), SIV-human immunodeficiency virus (SHIV), and HIV type 1 (HIV-1) transmission. J Virol 89:3619–3629. doi: 10.1128/JVI.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holland JJ, de la Torre JC, Clarke DK, Duarte E. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol 65:2960–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopker M, Easlick J, Sterrett S, Decker JM, Barbian H, Learn G, Keele BF, Robinson JE, Li H, Hahn BH, Shaw GM, Bar KJ. 2013. Heterogeneity in neutralization sensitivities of viruses comprising the simian immunodeficiency virus SIVsmE660 isolate and vaccine challenge stock. J Virol 87:5477–5492. doi: 10.1128/JVI.03419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee F-H, Mason R, Welles H, Learn GH, Keele BF, Roederer M, Bar KJ. 2015. Breakthrough virus neutralization resistance as a correlate of protection in a nonhuman primate heterologous simian immunodeficiency virus vaccine challenge study. J Virol 89:12388–12400. doi: 10.1128/JVI.01531-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keele BF, Estes JD. 2011. Barriers to mucosal transmission of immunodeficiency viruses. Blood 118:839–846. doi: 10.1182/blood-2010-12-325860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.