ABSTRACT
Gag intracellular assembly and export are very important processes for lentiviruses replication. Previous studies have demonstrated that equine infectious anemia virus (EIAV) matrix (MA) possesses distinct phosphoinositide affinity compared with HIV-1 MA and that phosphoinositide-mediated targeting to peripheral and internal membranes is a critical factor in EIAV assembly and release. In this study, we compared the cellular assembly sites of EIAV and HIV-1. We observed that the assembly of EIAV particles occurred on interior cellular membranes, while HIV-1 was targeted to the plasma membrane (PM) for assembly. Then, we determined that W7 and K9 in the EIAV MA N terminus were essential for Gag assembly and release but did not affect the cellular distribution of Gag. The replacement of EIAV MA with HIV-1 MA directed chimeric Gag to the PM but severely impaired Gag release. MA structural analysis indicated that the EIAV and HIV-1 MAs had similar spatial structures but that helix 1 of the EIAV MA was closer to loop 2. Further investigation indicated that EIAV Gag accumulated in the trans-Golgi network (TGN) but not the early and late endosomes. The 9 N-terminal amino acids of EIAV MA harbored the signal that directed Gag to the TGN membrane system. Additionally, we demonstrated that EIAV particles were transported to the extracellular space by the cellular vesicle system. This type of EIAV export was not associated with multivesicular bodies or microtubule depolymerization but could be inhibited by the actin-depolymerizing drug cytochalasin D, suggesting that dynamic actin depolymerization may be associated with EIAV production.
IMPORTANCE In previous studies, EIAV Gag was reported to localize to both the cell interior and the plasma membrane. Here, we demonstrate that EIAV likely uses the TGN as the assembly site in contrast to HIV-1, which is targeted to the PM for assembly. These distinct assembly features are determined by the MA domain. We also identified two sites in the N terminus of EIAV MA that were important for Gag assembly and release. Furthermore, the observation of EIAV transport by cellular vesicles but not by multivesicular bodies sheds light on the mechanisms underlying EIAV cellular replication.
INTRODUCTION
Equine infectious anemia virus (EIAV) is a lentivirus that causes a lifelong persistent infection in equids. The EIAV genome is the simplest among the lentivirus family and only encodes three accessory proteins (Tat, Rev, and S2) (1). Similar to all retroviruses, EIAV Gag is the major viral structural protein, and its expression in appropriate cells is sufficient to generate extracellular virus-like particles (VLPs). The polyprotein of EIAV Gag comprises four major domains: matrix (MA), capsid (CA), nucleocapsid (NC), and p9. These domains are cleaved by a viral protease during the final stages of Gag assembly to form infectious particles (2, 3). The four proteins that compose the Gag polyprotein are critical for retrovirus assembly and budding: the MA domain targets Gag to the site of viral assembly and facilitates Gag-membrane binding, the central CA domain has been shown to mediate the Gag-Gag interaction and homo-oligomerization in an ordered manner during viral assembly and determines the particle morphology, and NC contains an RNA-binding domain and enables the packaging of the viral genome and Gag multimerization (4). The EIAV p9 contains an YPDL L domain that connects to the ESCRT pathway via AIP1/Alix and is critical for viral release during the late stage of virus budding (5–7). Similarly, HIV Gag contains both PTAP and YPLA L domains that bind directly to TSG101/ESCRT-I and ALIX, respectively, to promote viral budding (4, 8, 9).
It is widely accepted that HIV-1 Gag assembly and budding occur predominantly on the plasma membrane (PM) and are mediated by the Gag N-terminal myristoylated MA domain. The myristyl group is essential for facilitating membrane anchoring and Gag assembly. Disrupting the conserved basic residues of the myristyl group leads to inefficient Gag targeting to the PM, which results in reduced virus production (10, 11). Additionally, the association of HIV-1 MA with phosphatidylinositol 4,5-biphosphate [PI(4,5)P2], a phosphoinositide present on the inner leaflet of the plasma membrane, is essential for Gag targeting to the PM (12). Gag multimerization and pH are regarded as “internal modulators” that regulate the exposure of the myristyl group. Similar to the HIV-1 MA, the MA domains of other retroviruses also interact with phosphoinositides. However, the type of phosphoinositide and the degree of phosphoinositide binding may vary among different retroviruses. The crystal structure of EIAV MA has been reported. Despite the lack of apparent sequence similarity with the MAs of primate lentiviruses, the EIAV MA shows striking overall structural similarity to the MAs of HIV-1 and SIV (13). However, the EIAV MA differs from the HIV-1 MA in that it lacks a myristoylation signal and binds to phosphatidylinositol 3-phosphate [PI(3)P] with a higher affinity than PI(4,5)P2 (14, 15). Moreover, EIAV virus-like particle (VLP) release is inhibited by YM201636 [a kinase inhibitor that blocks the production of PI(4,5)P2 from PI(3)P] and induces Gag colocalization with aberrant compartments (15). EIAV VLP release is not significantly inhibited by the coexpression of 5ptase IV, which can lead to the accumulation of HIV-1 Gag at multivesicular body (MVB) membranes, thereby inhibiting HIV-1 assembly and viral production (15). Recently, the HIV-1 MA was reported to bind almost exclusively to specific tRNAs in the cytosol, thereby regulating Gag binding to cellular membranes (16).
The L domain describes motifs whose mutation induces the characteristic defective assembly phenotype, whereby virions fail to separate from the cell membranes. The L domain also serves as a docking site for the recruitment of cellular factors, thereby providing functions that are essential for virion assembly and budding. To date, three types of L domains have been identified: PTAP, PPXY, and YPXL (8). The originally identified motif is the HIV-1 PTAP motif, which acts by recruiting Tsg101, an essential component of the ESCRT I complex that is responsible for the formation of MVBs (17, 18). PPXY is a consensus sequence for interaction with the WW domain of proteins; this domain is present in multiple copies in HECT ubiquitin ligases such as Nedd4 (19). Interestingly, the EIAV YPDL motif specifically interacts with AIP1/Alix, which acts as a bridging factor between ESCRT-I and ESCRT-III (5). Moreover, the μ subunit of the AP-2 adaptor protein complex was reported to interact with the EIAV YPDL motif, thereby facilitating virion production (6). Although these L domains function by interacting with different cellular proteins, interchangeability is allowed. For example, both the PTAP and PPPY motifs can substitute for the YPDL domain to recover EIAV replication. However, the function of PTAP cannot be replaced by the YPDL motif (20, 21). These observations suggest that retroviruses and other enveloped viruses have evolved different L domains to recruit distinct cellular factors and achieve virus budding and release by specific cellular pathways.
The recruitment and interaction of retroviral Gag proteins and cellular factors determine the viral assembly pathway. Two distinct assembly pathways have been described for orthoretroviruses. For gammaretroviruses such as murine leukemia virus (MLV) and HIV, viral Gag proteins are targeted to the plasma membrane, where viral particles are assembled and subsequently released (22, 23). However, MLV and HIV can also bud into late endosomes or MVBs in some cell types (24–26). EIAV Gag has been shown to localize at both the interior cellular membrane and the plasma membrane (6, 15). The interaction of the EIAV L domain with AP-2 and AIP-1 suggests that EIAV assembly may be associated with the formation of both early endosomes and late MVBs (6). However, the colocalization of EIAV Gag with both early endosome antigen 1 (EEA1) and the limiting membrane of LE/MVBs occurs at a low level (15). Therefore, where EIAV Gag assembly occurs and which region guides Gag trafficking to the assembly site remain to be determined. In the present study, we demonstrate that the interior cellular compartment is the main assembly site of EIAV Gag. This assembly feature is determined by the MA domain of EIAV Gag, especially 9 amino acids at the N terminus of MA. We also found that W7 and K9 in helix 1 of the EIAV MA are important for Gag assembly. The mutations of these two sites completely inhibited Gag release but did not affect Gag intracellular localization. Additionally, we found that this assembly site is not associated with early endosomes and MVBs and that the intracellular trafficking of EIAV Gag multimers does not depend on the movement of microtubules and endosomes. However, the accumulation of EIAV Gag in the trans-Golgi network (TGN) was detected for the first time. The results of this study demonstrate a previously unrecognized involvement of the TGN machinery in EIAV Gag assembly.
MATERIALS AND METHODS
Cells and plasmids.
Human embryonic kidney (HEK) 293T cells, equine dermal (ED) cells and HeLa cells were cultured in Dulbecco high-glucose modified Eagle medium (HyClone, USA) supplemented with 10% fetal bovine serum, penicillin (100 U/ml; HyClone), and streptomycin (100 μg/ml; HyClone).
Plasmids expressing HIV-1 and EIAV Gag-green fluorescent protein (GFP)/Gag-DsRed proteins were constructed by amplifying codon-optimized HIV-1 and EIAV Gag sequences. These fragments were inserted into the pEGFP-N1 or pDsRed2-N1 expression vector. Truncated versions of HIV-1 and EIAV Gag lacking the MA globular head were generated using PCR and were inserted between the HindIII-NotI sites of TOPOpcDNA-GSTV5. The MA mutants of EIAV Gag were generated using PCR and were inserted between the HindIII-SacII sites of pEGFP-N1. To generate the HMA-EGag and EMA-HGag plasmids, HIV-1 MA, EIAV MA, HIV-1 CA-NC-P6, and EIAV CA-NC-P9 fragments were separately amplified; then, the HIV-1 MA and EIAV MA fragments were fused with EIAV CA-NC-P9 and HIV-1 CA-NC-P6, respectively, through overlapping extension PCR. The fused fragments were cloned into pEGFP-N1 using the HindIII and SacII restriction sites. H9-EGag-red fluorescent protein (RFP) and E9-HGag-RFP were constructed by the same method except that the fragments were inserted into the pDsRed2-N1 expression vector. Rab5 and Rab7 were amplified from 293T cells; these fragments were inserted into pEGFP-C1 using the KpnI and BamHI restriction sites. Rab5 Q79L and Rab7 Q67L were generated using the FasMut system mutagenesis kit (TransGen Biotech, China). The plasmids TGN38-yellow fluorescent protein (YFP), Lamp1-YFP, and CD63-YFP were kindly provided by Walther Mothes (Yale University).
Western blotting.
All Western blotting assays were performed using the Odyssey infrared imaging system (LI-COR Biosciences, USA). All EIAV and HIV-1 Gag proteins were detected using a mouse monoclonal anti-V5 antibody (Sigma, USA), followed by a secondary goat anti-mouse IRD800-conjugated monoclonal antibody (Sigma). The anti-actin polyclonal antibody was obtained from Sigma. EIAV p26 proteins were detected using EIAV-positive serum, followed by a secondary goat anti-horse IRD800-conjugated antibody (Sigma). HIV-1 p24 proteins were detected using a mouse monoclonal anti-p24 antibody, followed by a secondary goat anti-mouse IRD800-conjugated antibody (Sigma). Rab5 and Rab7 proteins were detected using an anti-GFP polyclonal antibody (Beyotime, China). The signal intensities of Gag-associated bands were analyzed by using software WCIF-ImageJ. The relative virus release efficiency was calculated as the amount of VLP-associated Gag as a fraction of total Gag present in cell and VLP lysates and normalized to the virus release efficiency in wild-type or non-drug-treatment groups. All experiments were performed at least in triplicate.
Drug treatment.
A total of 106 293T cells were plated per well of a six-well plate and transfected 12 h later with 4 μg of DNA using 10 μl of Lipofectamine 2000 (Invitrogen, USA). After 8 to 10 h, the media were replaced with cell culture media containing nocodazole (Sigma), U18666A (Biomol, USA), GW4869 (Sigma), or cytochalasin D (Sigma). All of the aforementioned drugs were dissolved in dimethyl sulfoxide (DMSO; Sigma) and diluted in medium immediately prior to use. Control cultures were treated with an equal amount of DMSO. Drugs were left on the cells until 48 h after transfection. Cells and culture supernatants were collected for Western blotting assays.
Immunofluorescence microscopy and analysis.
293T, ED or HeLa cells grown on polystyrene coverslips (NEST Biotechnology, China) were transfected with 1 μg of EIAV Gag-GFP or Gag-DsRed and one of the following plasmids using Lipofectamine 2000 (Invitrogen): 1 μg of Rab5-GFP, 1 μg of Rab7-GFP, 1 μg of CD63-YFP, 1 μg of Lamp1-YFP, or 1 μg of TGN38-YFP. At 2 days posttransfection, the cells were fixed in 4% paraformaldehyde in PBS for 30 min and permeabilized in 0.1% Triton X-100 in PBS for 15 min. Nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI; Beyotime) for 5 min. Images were captured using a Leica DM-IRE2 confocal microscope (Leica, Germany). The degree of colocalization or lack of colocalization for two kinds of different fluorescence in cells was quantified by using software WCIF-ImageJ. Briefly, in ImageJ software, red and green images were opened and converted into 8-bit style. Colocalization finder plugin was used to analyze colocalization degree. The colocalization part was shown in white color. Then, the value for colocalization degree was calculated. For each sample, at least three cells were used for colocalization quantification analysis.
A high-content screen (HCS) assay was used to evaluate the presence frequency of the cells with colocalization of green and red fluorescence in all of the cells observed. All the cells transfected by different plasmids were cultured in 96-well plates, and the fluorescence signals were observed by operetta (Perkin-Elmer, USA) equipped with a 460- to 490-nm/500- to 530-nm enhanced GFP (EGFP) (EYFP) excitation/emission filter, a 520- to 550-nm/560- to 630-nm DsRed excitation/emission filter, and a 360- to 400-nm/410- to 480-nm DAPI excitation/emission filter. Images for each well were acquired in the three different channels for EGFP (EYFP), DsRed, and DAPI using a 20× dry objective. The plates were exposed for 200 ms to acquire EGFP (EYFP), DsRed, and DAPI images. The colocalization signals of green and red fluorescence were identified by “Finding spots block” in harmony analysis software (Perkin-Elmer). The frequency was determined by dividing the number of green and red fluorescence-colocalized single cells by the number of single cells stained by DAPI in each well. This procedure was repeated for each well. All experiments were performed in triplicate.
Transmission electron microscopy.
First, the cells infected by viruses or transfected with EIAV and HIV-1 Gag-Pol plasmids were harvested and prepared for thin sectioning. In brief, the harvested specimen was fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate-buffered saline (PBS; pH 7.4) for 2 h, rinsed with three changes of PBS, and postfixed with 1% (vol/vol) OsO4 in PBS for 2 h. After being washed, the specimen was dehydrated in a graded series of ethanol and embedded in epoxy (agar low-viscosity) resin according to the standard protocol. Ultrathin sections were collected on carbon-coated 200-mesh copper grids and stained with 1% uranyl acetate and 1% lead citrate. In addition, clarified stool suspension was used for direct electron microscopy (EM) examination of the sample after negative staining with 2% phosphotungstic acid (pH 4.5). EM grids were screened at 80 kV in a Hitachi-7650 transmission electron microscope (Hitachi, Japan).
Statistical analysis.
The experiments in this study have been replicated for at least three times and the data are calculated by software SAS8.1 and presented as mean values of the standard errors. For statistical analysis, the data were calculated by using a Student t test and charted with the software GraphPad Prism v5.01. A difference was considered statistically significant when the P value was <0.05.
RESULTS
EIAV Gag accumulates in internal cellular compartments.
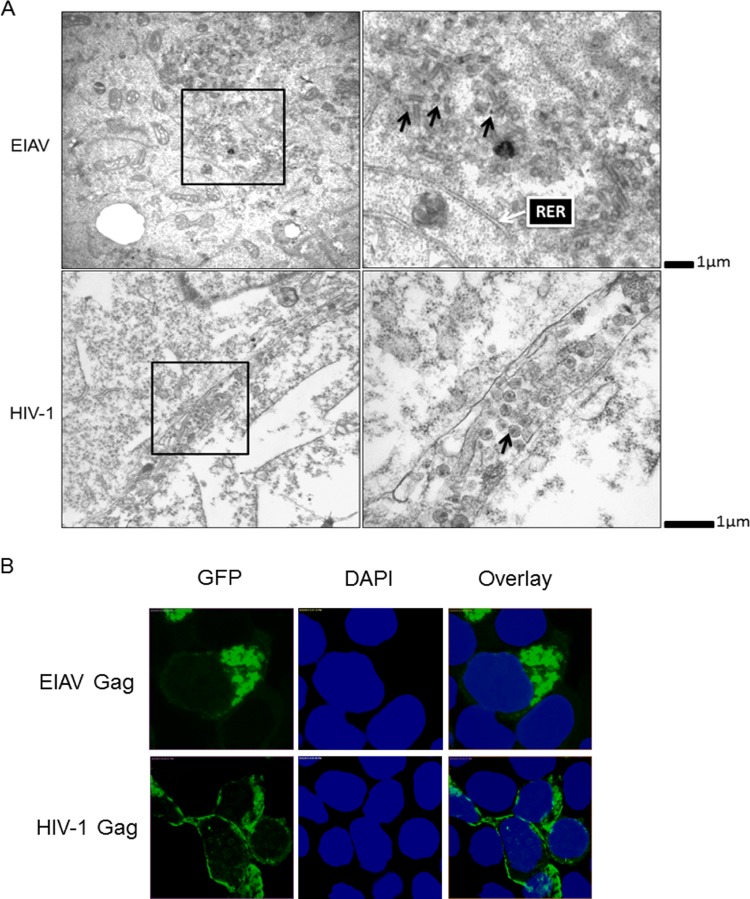
In contrast to HIV-1 Gag, which assembles on the plasma membrane, EIAV Gag mainly localizes to the interior cellular membrane (15). EIAV Gag budding is also unique. The process appears to be ubiquitin independent, and its unique YPDL L domain recruits AIP1 and AP-2 as its budding partners (5, 6, 27). However, little is known about the trafficking pathway and assembly sites of EIAV Gag. To better understand the trafficking and assembly of EIAV, we analyzed the subcellular distribution of EIAV viral particles using transmission electron microscopy (EM). We consistently found large amounts of unclosed, closed and tubular particles in the area adjacent to the rough endoplasmic reticulum (RER) (Fig. 1A). With EM, RER often has a tubular appearance, whereas SER is more dilated and convoluted. The same RER structure was observed and described by Voeltz et al. (28). These particles were immature and did not contain viral cores. In contrast, HIV-1 particles were mainly found on the plasma membrane (PM), and these particles contained typical viral cores (Fig. 1A). These results suggest that EIAV assembly is initiated and completed at the interior cellular membrane, while HIV-1 is selectively targeted to the plasma membrane to induce efficient particle release. Gag proteins are central to the assembly of all retroviruses. Their expression in appropriate cells is sufficient to generate extracellular VLPs (with the exception of foamy virus, the assembly and budding of which rely on the envelope glycoproteins) (4, 29). To determine the targeting sites of the EIAV and HIV-1 Gag proteins, 293T cells were transfected with EIAV or HIV-1 Gag-GFP plasmids. After 24 h, the cells were harvested for fluorescence analysis. The results showed that EIAV Gag accumulated mainly in the cell interior, with only a small number of EIAV Gag proteins detected at the plasma membrane (Fig. 1B). In contrast, HIV-1 Gag was mainly detected at the plasma membrane (Fig. 1B). Therefore, we concluded that the EIAV and HIV-1 Gag proteins are key factors that determine the viral assembly sites; however, EIAV Gag assembly occurs mainly in the cell interior, while HIV-1 Gag is targeted to the cell surface for assembly.
FIG 1.
Comparison of cellular localization of EIAV and HIV-1 viral particles. (A) EIAV VLPs were mainly localized at interior cellular membranes, while HIV-1 VLPs were detected at the plasma membrane. 293T cells were transfected with EIAV and HIV-1 Gag-Pol plasmids. After 48 h, the supernatants were discarded, and the cells were washed three times with PBS prior to harvesting for transmission electron microscopy. The intracellular VLPs are indicated by black arrows, and a typical RER that had a tubular appearance is indicated by a white arrow (28). (B) Cellular localization of EIAV and HIV-1 Gag were distinct. EIAV and HIV-1 Gag-GFP were transfected into 293T cells. After 48 h, the cells were fixed using formalin and stained with DAPI (blue). Gag-GFP expression and localization were analyzed by confocal microscopy.
The EIAV MA globular head is essential for Gag assembly.
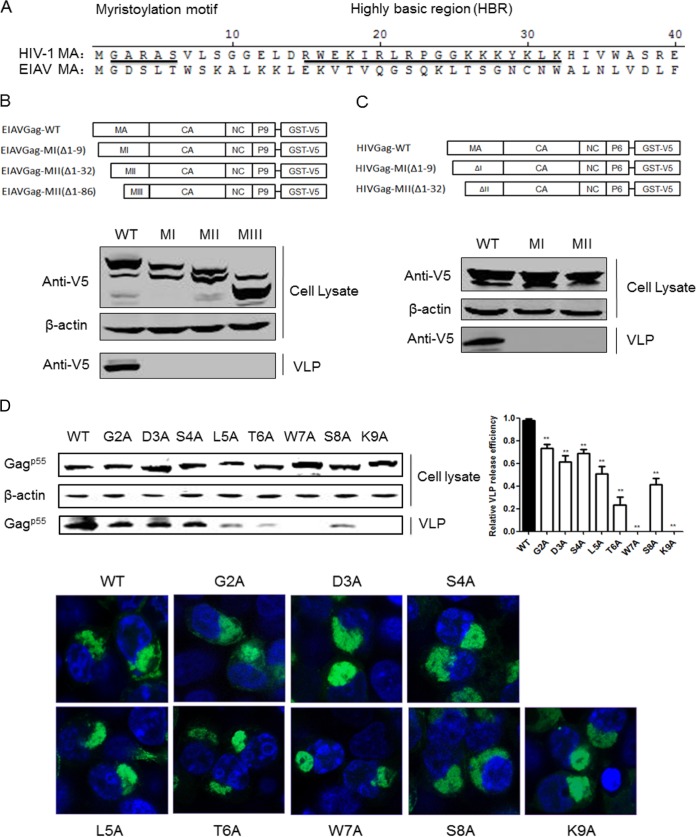
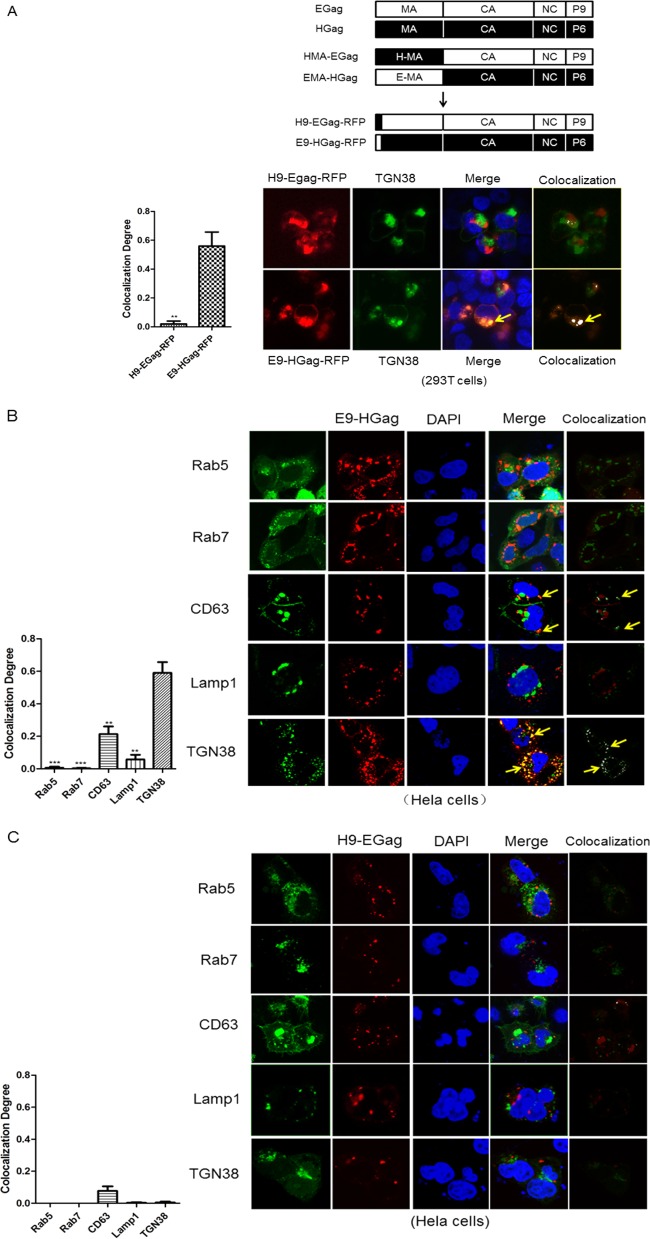
HIV-1 assembly is a complex process that occurs at the plasma membrane; thus, binding and targeting of Gag to the plasma membrane are the first steps in this assembly process. The N-terminal 14 amino acids of HIV-1 MA are myristylated, and this myristylation is required for efficient membrane association and virion formation (30). Additionally, a highly basic region (HBR) spanning residues 14 to 31 is essential for efficient membrane binding and proper targeting of Gag to the PM (31). However, the EIAV MA lacks both the myristylation domain and the HBR that compose the HIV-1 membrane-binding motif (Fig. 2A). Several residues (S100 and K49) in the EIAV MA have been reported to control PI binding; these residues are important determinants of Gag targeting and assembly (14, 15). To determine whether the N-terminal residues of EIAV MA are essential for Gag assembly and release, we truncated the EIAV MA and analyzed the release efficiency of these truncated Gag proteins. As revealed by Western blotting, the MI, MII, and MIII truncations of EIAV Gag severely inhibited VLP release, whereas wild-type (Fig. 2, WT) Gag formed normal VLPs (Fig. 2B). Similar results were observed in the HIV-1 MA truncation experiments, where HIV Gag-MI and MII abrogated VLP formation (Fig. 2C). Deletion of the 9 N-terminal amino acids from EIAV MA completely inhibited Gag release. This result suggests that, even though these amino acids are not myristylated, they harbor an undefined function that promotes Gag assembly. To identify the residues that were determinants of efficient Gag assembly, we separately mutated these amino acids to alanine and analyzed their effect on Gag assembly. The results showed that all of the mutants had identical p55 expression levels in the cell lysate but exhibited variable influences on secretion of the Gag protein into the supernatant (Fig. 2D). G2A, D3A, and S4A resulted in a slight inhibition of Gag release, L5A, T6A, and S8A inhibited Gag release at medium levels, and W7A and K9A severely reduced the amount of Gag p55 protein in the supernatant (Fig. 2D). Two mutations (K29E and K31E) in HIV-1 MA led to viral particle assembly in a multivesicular body compartment and defective release of cell-free particles in HeLa and 293T cells (32). To determine whether W7A and K9A changed the localization of Gag from the interior cellular membrane, Gag localization of these mutants was confirmed by confocal microscopy. However, no distinct localization was observed. W7A and K9A were mainly detected in the interior cellular membrane adjacent to the nucleus, which was identical to the results obtained with wild-type Gag (Fig. 2D). Consequently, we concluded that the N-terminal residues (1–9) of EIAV and HIV-1 MA are essential for the release of EIAV and HIV Gag VLPs, and W7 and K9 in the EIAV MA regulate Gag assembly but do not affect Gag localization.
FIG 2.
The N terminus of EIAV and HIV-1 MA was essential for Gag assembly. (A) Comparison of N-terminal amino acid sequences in MAs from EIAV and HIV-1. The myristoylation motif and the highly basic region (HBR) are underlined and indicated in HIV-1 MA sequences. (B and C) 293T cells were transfected with the truncated EIAV and HIV-1 Gag constructs. After 48 h, the cells were lysed, and Gag VLPs in the supernatant were purified by centrifugation at 20,000 × g for 2 h at 4°C. Gag proteins in the cell lysate and supernatant were detected using anti-V5 antibodies. Cell lysates were also analyzed for equal amounts of total proteins using the anti-β-actin monoclonal antibody. (D) All EIAV Gag mutants were fused with GFP in the C terminus and transfected into 293T cells. Gag expression and release were detected with anti-GFP antibodies. The relative virus release efficiency was calculated as the amount of VLP-associated Gag as a fraction of the total Gag present in the cell and VLP lysates and normalized to the virus release efficiency in wild-type group. Additionally, the cellular distribution of the mutants was analyzed by confocal microscope. **, P < 0.005.
EIAV MA determines Gag targeting to the interior cellular membrane.
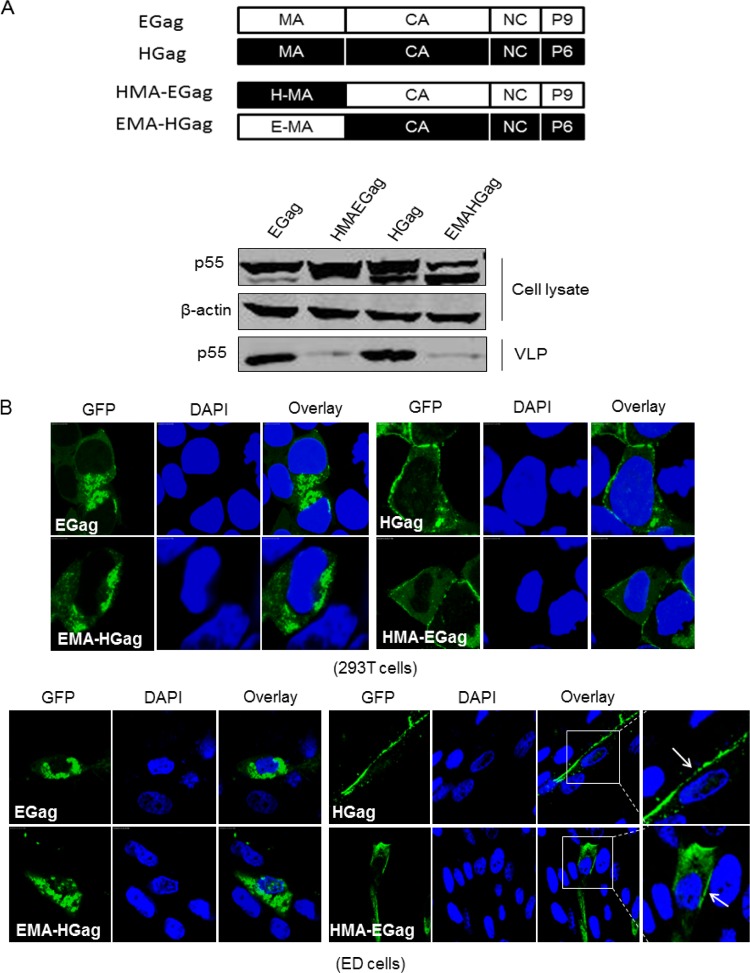
The functional interchangeability of retrovirus L domains was previously reported. For instance, the PTAP motif of HIV-1 can replace the YPDL domain of EIAV to support EIAV replication, while the PTAP function for HIV-1 assembly can be substituted by PPPY but not YPDL (20). We then addressed whether the EIAV and HIV-1 MAs could be mutually exchanged to support the normal assembly of Gag. The MA motifs of the EIAV and HIV-1 Gag proteins were swapped (Fig. 3A), and the assembly rates of the chimeric Gag proteins were analyzed. The results showed that the wild-type EIAV and HIV-1 Gag proteins were expressed well and could effectively release Gagp55 VLPs into the cell culture supernatants. Gag proteins were expressed by HIVMA-Egag at levels that were similar to those of wild-type EIAV Gag. However, VLP release was severely inhibited by this substitution (Fig. 3A). VLP release was also inhibited when HIV-1 MA was replaced by EIAV MA; this replacement also simultaneously decreased the quantity of Gag proteins in the cell lysate (Fig. 3A).
FIG 3.
EIAV MA determines Gag targeting to internal cellular membranes. (A) The MA swap between EIAV and HIV-1 impaired the release of chimeric Gag. Schematic structures of chimeric Gags were shown. Wild-type Gag and HMA-EGag and EMA-HGag fused with green fluorescent protein (GFP) in the C terminus were transfected into 293T cells. Expression and release efficiency were detected by Western blotting as in Fig. 2B. (B) 293T and ED cells were transfected with wild-type and chimeric Gag. The cellular localization of Gag was analyzed by confocal microscopy.
The functional domains of MA from EIAV and HIV-1 determine Gag protein targeting and assembly. To confirm the location of these chimeric Gag proteins, 293T cells were transfected with EIAVgag-GFP, HIVMA-Egag-GFP, HIVgag-GFP, and EMA-HIVgag-GFP. After 24 h, the cells were harvested and stained by DAPI, and fluorescence was observed using confocal microscopy. The results showed that EIAV Gag targeted to the interior cell membrane, while HIV-1 Gag targeted to the plasma membrane, similar to the results described above (Fig. 3B). HIV-1 MA could induce EIAV Gag accumulation near the plasma membrane of the cells. In contrast, EIAV MA induced HIV-1 Gag multimerization in the cell interior (Fig. 3B). The same experiments were also performed in ED cells. The result was identical to that from 293T cells, suggesting that this finding was not cell type specific (Fig. 3B). Taking our results together, we concluded that EIAV MA determines Gag targeting to the internal cell membrane to complete assembly, while HIV-1 MA guides EIAV Gag to the plasma membrane but does not support Gag assembly.
EIAV Gag proteins accumulate in the trans-Golgi network (TGN).
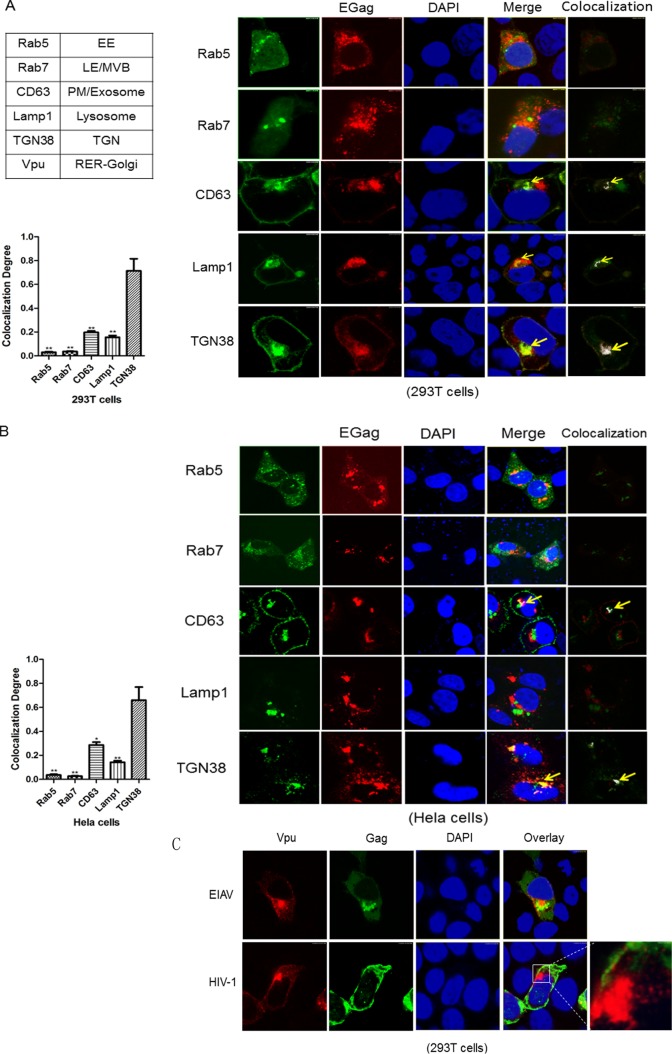
To identify the nature of the membranous structures where EIAV Gag assembly occurs, subcellular membrane markers and EIAV Gag-DsRed were used to detect costaining of Gag and intracellular membranes. EIAV Gag-DsRed accumulated into a mass at the periphery of the cell nucleus, which was consistent with the Gag-GFP results described above. Costaining of Gag with early and late endosome markers (i.e., Rab5 and Rab7) was detected, but no colocalization was observed (Fig. 4A). The lack of Gag in early and late endosomes was consistent with previous results that demonstrated a low level of colocalization of EIAV Gag with early endosome antigen 1 (EEA1) and the limiting membrane of LE/MVBs (15). The lysosomal membrane was labeled by Lamp1-YFP, but only slight accumulation of EIAV Gag was observed in intracellular and cell surface Lamp1-positive vesicles (Fig. 4A). Thus, these results suggested that EIAV Gag does not target MVBs to complete viral assembly.
FIG 4.
The subcellular localization of EIAV Gag proteins. (A and B) EIAV Gag-RFP was coexpressed in 293T cells or in HeLa cells with the indicated cellular membrane markers tagged with YFP (CD63, Lamp1, and TGN38) or GFP (Rab5 and Rab7). The cells were fixed 48 h posttransfection, stained with DAPI (blue), and imaged by confocal microscopy. Images from three different views were used for quantitative colocalization analysis. The colocalization is indicated in white. The statistics were calculated and charted by GraphPad PrismV5.1. **, P < 0.01; *, P < 0.05. (C) EIAV Gag-GFP or HIV-1 Gag-GFP was cotransfected with Vpu-HA into 293T cells. After 48 h, the cell nuclei were stained with DAPI (blue), and the cells were stained with anti-hemagglutinin antibodies (red) to observe Vpu.
A fraction of EIAV Gag was observed with intracellular pools of CD63(the value of colocalization degree is 0.18 ± 0.017), which are proteins of the tetraspannin family that localize to the plasma membrane and to intracellular exosomes (Fig. 4A). However, most EIAV Gag proteins colocalized with TGN38(the value of colocalization degree is 0.66 ± 0.144), which is predominantly in the TGN (Fig. 4A). Furthermore, HCS analysis showed that the percentage of the cells in which the colocalization of CD63 or TGN38 with Gag among all the cells observed was 21.1% ± 7.4% or 24.6% ± 3.3%. Costaining of EIAV Gag and subcellular membrane markers was also detected in HeLa cells. EIAV Gag primarily colocalized with TGN38 (the value of colocalization degree is 0.69 ± 0.069) but not with other cellular markers, which was consistent with the results in 293T cells (Fig. 4B). Previous studies have shown that the HIV-1 Vpu protein is transported predominantly to RER/Golgi complex compartments (33). Thus, we cotransfected Vpu-HA with EIAV Gag-GFP or HIV-1 Gag-GFP into 293T cells and evaluated their cellular distribution. The results showed that HIV-1 Gag was mainly detected on plasma membranes and did not colocalize with Vpu, whereas EIAV Gag localized to an interior cellular membrane surrounding the Vpu compartments (Fig. 4C). These results suggested that EIAV Gag assembly is associated with the TGN.
In Fig. 2, we demonstrated that the N-terminal 9 amino acids of MA (especially W7 and K9) regulate EIAV Gag assembly. Therefore, we investigated whether these amino acids were important for guiding Gag to the TGN. We separately substituted this region in EIAV and HIV-1 Gag and analyzed their colocalization with TGN in 293T cells. The results revealed that H9-EGag-RFP was still detected in the cell interior, but it only showed slight colocalization with the TGN (Fig. 5A). In contrast, E9-HGag-RFP was not only detected at the interior cellular membrane but was also highly colocalized with the TGN (the value of colocalization degree is 0.57 ± 0.066) (Fig. 5A). Finally, we evaluated the colocalization of H9-EGag and E9-HGag with other cellular markers in HeLa cells. E9-HGag exhibited an identical cellular distribution with wild-type EIAV Gag, which mainly colocalized with TGN38 and less partial colocalized with CD63 but not with other markers. The values of colocalization degree were 0.20 ± 0.066 and 0.70 ± 0.094, respectively (Fig. 5B). The percentages of the colocalization cells among all the cells observed were 29.5% ± 6.7% and 19.2% ± 6.8%. However, no colocalization was observed between H9-EGag and the tested cellular makers (Fig. 5C). These results suggest that the 9 N-terminal amino acids of EIAV MA are determinants that can direct Gag to the appropriate location of viral assembly.
FIG 5.
The N-terminal 9 amino acids of EIAV MA are determinants for directing Gag to the TGN. (A) Schematic structures depicting the substitutions of the 9 N-terminal amino acids between EIAV and HIV-1 Gag were designated H9-EGag-RFP and E9-HGag-RFP. These two constructs were cotransfected with TGN38 tagged with YFP into 293T cells. After 48 h, the cells were stained with DAPI and imaged using confocal microscopy. (B and C) E9-HGag-RFP and H9-EGag-RFP were cotransfected with other cellular markers into HeLa cells. Images were captured using the same method. Three independent images of each sample were used for quantitative colocalization analysis. The colocalization was analyzed by software WCIF-ImageJ and is shown in white. The statistics were calculated and charted by GraphPad Prism v5.1. **, P < 0.01; *, P < 0.05.
Three-dimensional (3D) structure analysis of EIAV and HIV-1 MA.
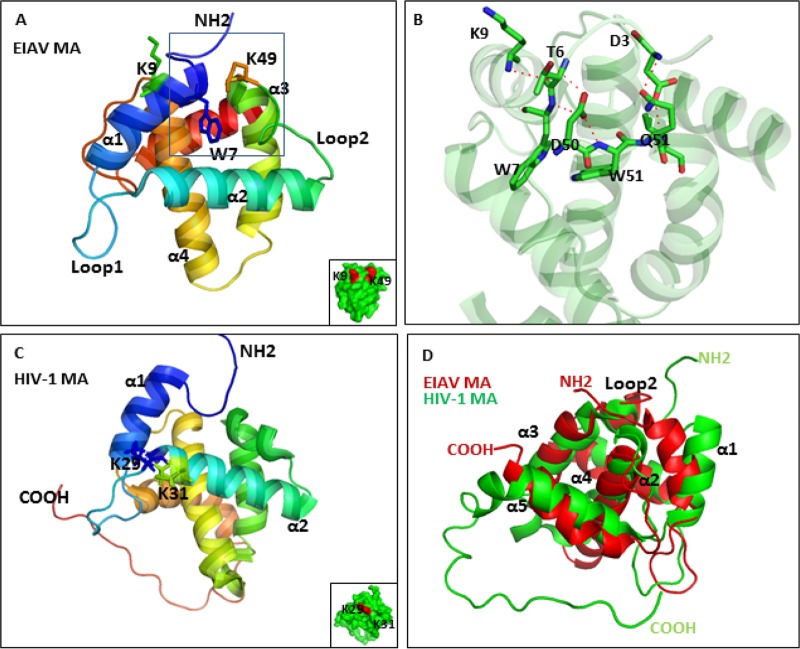
The EIAV MA structure has been reported (13). Here, we compared the structure of the EIAV and HIV-1 MAs. As shown in Fig. 6A, the EIAV MA contains 5 helices; the K9, W7 and K49 residues that were important for Gag assembly were labeled. Loop 2 between helix 2 and helix 3 was also indicated; this loop was reported to be the main region involved in binding to phosphoinositides. K9 and K49 were on the surface of MA, while the hydrophobic W7 residue was hidden inside the molecule. Above, we found that the 9 N-terminal amino acids of MA could direct the Gag protein to the assembly site. In the structure, we found that these amino acids were located in helix 1 and had an internal interaction with loop 2. The hydrogen bonds between W51, D50, and W7 are represented (Fig. 6B). Additionally, D50 also formed hydrogen bonds with T6, and an interaction between D3 and Q51 was observed (Fig. 6B). The crystal structure of the EIAV MA is strikingly similar to the crystal structures of the primate lentivirus MAs. This similarity is reflected in the fact that EIAV MA has five long helices and one 310 helical loop, each of which has an equivalent in SIV and HIV-1 (13). However, the HIV-1 MA seems to bind to PI(4,5)P2 via helix 2 (32). K29 and K31, which are determinants for PI(4,5)P2 binding, are shown in Fig. 6C. The structural alignment of the HIV-1 MA and EIAV MA revealed that helix 1 of the EIAV MA seemed to be closer to loop 2 (Fig. 6D). Overall, despite the high structural similarity between EIAV MA and HIV-1 MA, sequence differences and several subtle distinctions may influence their binding affinity for phosphoinositides and impact targeting to the viral assembly site.
FIG 6.
3D structure analysis of EIAV and HIV-1 MA. (A) The overall structure of the EIAV MA is shown in schematic representation. Some of the secondary structural elements of the α/β domain are labeled. The K9, W7, and K49 residues that are essential for Gag assembly are highlighted. The primary region that binds to phosphoinositide is labeled in loop 2. K9 and K49 localized on the MA surface are shown in red. (B) A closeup view of the detailed interaction between EIAV MA helix 1 and loop 2 is highlighted. The residues involved in the interaction are labeled. Red dashed lines represent hydrogen bonds. (C) HIV-1 MA structure. K29 and K31 that could induce PI(4,5)P2 binding with MA are shown in a stick representation. (D) The structures of EIAV MA and HIV-1 MA were aligned by Pymol. The conformations are similar, but helix 2 of EIAV MA seems to be closer to the other structural elements.
MVB and microtubules are dispensable for EIAV particle egress.
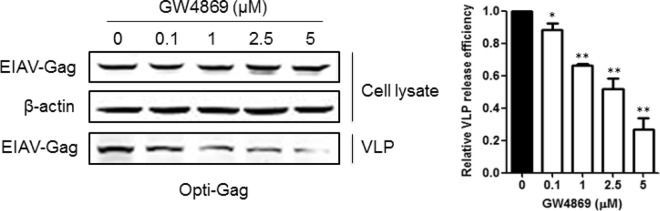
We have demonstrated that EIAV Gag proteins mainly accumulate at the internal cellular membrane. To determine how EIAV particles are exported to the extracellular environment, ED cells were infected with EIAVFDDV3-8, an ED-adapted EIAV strain (34, 35). Five days later, the cells were harvested for transmission electron microscopy analysis. We observed a large number of mature viral particles in intracellular vesicles-like component (data not show), suggesting that EIAV particles are transported by intracellular vesicles. To validate this finding, 293T cells were treated with GW4869 at 0.1 to 5 μM (which reduces vesicular-derived exosome release) after transfection with EIAV Gag (36). Gag expression in the cell lysate was not reduced, but Gag release was severely blocked under the treatment of GW4869 with a dose-dependent manner (Fig. 7).
FIG 7.
The export of EIAV particles is suppressed by treatment of GW4869. 293T cells were transfected with EIAV Gag. After 8 to 10 h, the medium was replaced with culture medium containing different concentrations (0, 0.1, 1, 2.5, and 5 μM) of GW4869 (Sigma). At 48 h posttransfection, the cell lysate and Gag VLPs in the supernatant were prepared as described above. Gag in the cell lysate and supernatant was detected with an anti-p26 antibody. The cell lysate was also used to detect β-actin expression to ensure that the same amount of protein was loaded. The relative virus release efficiency was calculated as the amount of VLP-associated Gag as a fraction of total Gag present in cell and VLP lysates and normalized to the virus release efficiency in a non-drug-treatment group (0 μM). **, P < 0.01; *, P < 0.05.
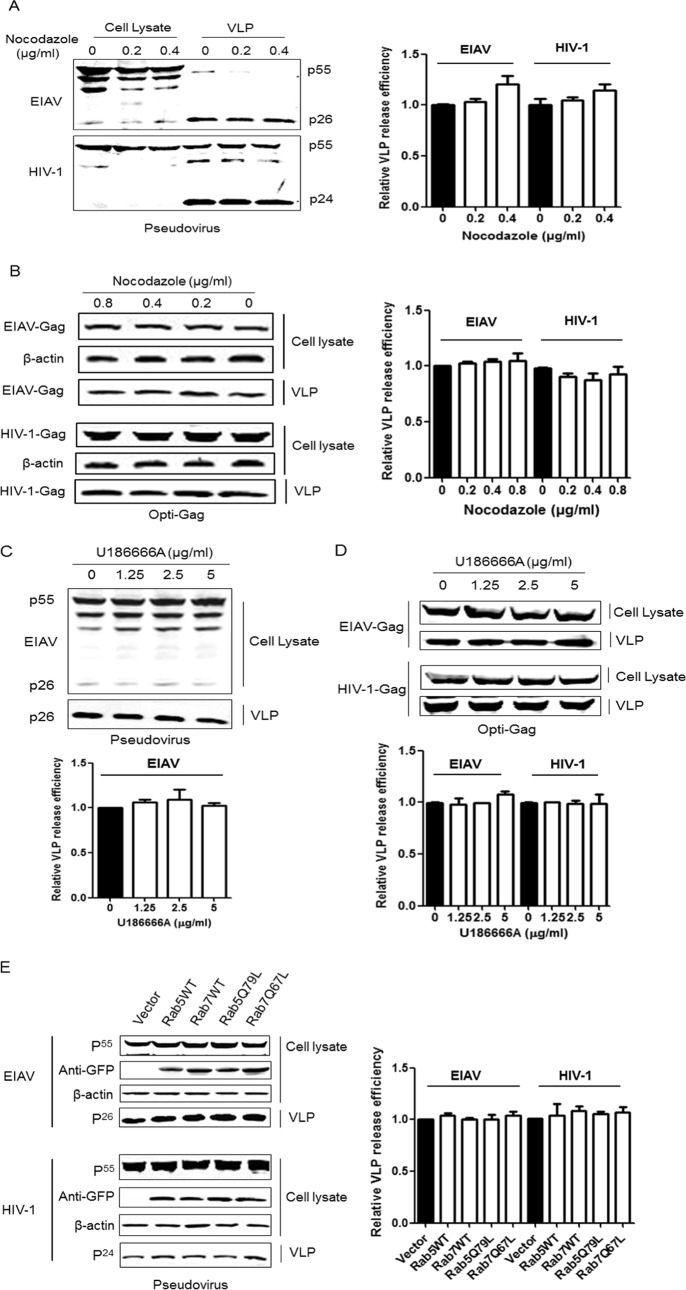
Cells are known to form large amounts of variant vesicles. Some types of vesicles will bud into endosomes to form MVBs and are released as exosomes into extracellular fluids by fusion of MVBs with the cell surface (37). To determine whether EIAV transportation in the cell interior is associated with the formation and motility of endosomes, 293T cells were transfected with EIAV Gag/Pol or the optimized Gag plasmid and then treated with different concentrations of nocodazole, which can block endosome movement by inducing microtubule depolymerization (23, 38, 39). The results revealed that EIAV Gag processing was slightly affected by nocodazole, but the drug did not inhibit the egress of EIAV particles (Fig. 8A). The expression and release of single EIAV Gag proteins were unaffected by nocodazole (Fig. 8B). Additionally, HIV-1 particles were not blocked by nocodazole, which was consistent with a previous report (Fig. 8A and B). As a complementary experiment, a different drug (U18666A) was used to arrest late endosome movement. U18666A can cause cholesterol accumulation in late endosomal membranes and leads to defects in kinesin recruitment (23, 39). The results show that neither EIAV nor HIV-1 particles were inhibited by U18666A treatment (Fig. 8C and D). Thus, these results suggest that the motility of endosomes in 293T cells is not required for EIAV and HIV-1 Gag assembly and release.
FIG 8.
MVBs and microtubules are dispensable for EIAV particle egress. (A and B) EIAV and HIV-1 Gag-Pol or codon-optimized Gag were transfected into 293T cells. After 8 to 10 h, the medium was replaced with fresh cell culture medium containing different concentrations (0, 0.2, 0.4, and 0.8 μg/liter) of nocodazole. EIAV and HIV-1 viral proteins in the cell lysate and supernatant were prepared as described in Fig. 2 and detected using the anti-p26 antibody or anti-p24 antibody. (C and D) The same experiments were performed except that U18666A was used in place of nocodazole. (E) 293T cells were cotransfected with EIAV Gag-Pol and Rab5, Rab7, or their defective mutants. After 48 h, viral proteins in the cell lysate and supernatant were detected using an anti-p26 antibody. The Rabs and their mutants were detected with anti-GFP antibodies. The same experiments were performed for HIV-1 Gag-Pol, except that the HIV-1 viral proteins were detected with an anti-p24 antibody. The relative virus release efficiency was calculated as described in Fig. 7. **, P < 0.01; *, P < 0.05.
Rab5 and Rab7 regulate individual steps in the transport of newly endocytosed vesicles from the plasma membrane to early endosomes (Rab5) and from late endosomes to lysosomes (Rab7) (40–42). To determine whether the assembly of EIAV and HIV-1 particles is associated with endosomes, 293T cells were transfected with the EIAV and HIV-1 Gag/Pol plasmids, together with Rab5, Rab7, and their mutants. The detection of Gag production by Western blotting assays showed that the assembly and release of EIAV and HIV-1 were unaffected by overexpression of the wild type or dominant-negative versions of Rab5 and Rab7 (Fig. 8E). Thus, we concluded that endosomes are not required for EIAV and HIV-1 assembly and release.
Chen et al. reported that F-actin was associated with different stages of EIAV production (43, 44); therefore, we investigated the influence of the actin-depolymerizing drug cytochalasin D on EIAV production. 293T cells were transfected with EIAV Gag-Pol or optimized Gag; then, 6 h later, the cells were treated with different concentrations of cytochalasin D. The results showed that the expression of the EIAV Gag-Pol protein in the cell lysate was not reduced after treatment with cytochalasin D, but the quantities of released viral particles were reduced under 10 μM or higher cytochalasin D treatment (Fig. 9A). However, the released optimized EIAV Gag VLPs were only reduced by 125 μM cytochalasin D treatment (Fig. 9B). This discrepancy may be due to the distinct expression level of the EIAV Gag-Pol and optimized Gag plasmids. These data suggest that dynamic actin depolymerization may be associated with EIAV production.
FIG 9.
EIAV export is inhibited by cytochalasin D. 293T cells were transfected with EIAV Gag-Pol (A) or optimized Gag (B) plasmids. After 8 to 10 h, the medium was replaced with fresh cell culture media containing different concentrations of cytochalasin D. After 48 h, the EIAV proteins in the cell lysate and supernatant were prepared as described in Fig. 2 and detected with an anti-p26 antibody. The relative virus release efficiency was calculated. **, P < 0.01; *, P < 0.05.
DISCUSSION
Intracellular EIAV replication is a complicated process that has not been clearly defined to date. Previous studies have demonstrated that EIAV binds to PI(3,5)P2, which is enriched in endocytic compartments, and utilizes the cellular ESCRT pathway to complete assembly and release (45). However, the detailed cellular trafficking pathways that facilitate EIAV release remain unknown. In the present study, we compared differences in the EIAV and HIV-1 Gag protein cellular assembly sites (Fig. 1A and B). We found that W7A and K9A severely impaired EIAV Gag release but did not alter the distribution of Gag at the cellular membrane (Fig. 2D). Then, we confirmed that the MA global head of EIAV and HIV-1 Gag was essential for their assembly and determined their membrane targeting (Fig. 3). Furthermore, we demonstrated that EIAV did not utilize MVBs and microtubules to complete its particle egress (Fig. 4 and 8). Interestingly, we observed that EIAV Gag accumulated in the intracellular trans-Golgi network rather than in endosomes (Fig. 4A and B). Finally, we demonstrated that EIAV hijacked the cellular vesicle pathway to export viral particles (Fig. 7).
Gag assembly is an essential step during most retroviral life cycles. It is widely accepted that PM is the site of HIV-1 assembly and budding, but HIV-1 and MLV particles can also be observed in the late endosome or MVBs in some cell types (25, 26). Several groups have reported that EIAV Gag proteins localize to both the cell interior and the plasma membrane (6, 15). EIAV MA binds PI(3)P with higher affinity than PI(4,5)P2 (15). In the present study, we observed the primary accumulation of EIAV particles at an intracellular membrane adjacent to the RER, and we demonstrated that this intracellular membrane was related to the TGN (Fig. 1A and Fig. 4A and B). Whether EIAV Gag targets the TGN to facilitate its assembly and is dependent on binding to a specific phosphoinositide in the TGN membrane remains to be determined. Previously, our lab reported that the overexpression of equine viperin inhibited EIAV Gag release (46). Viperin, which is an evolutionarily conserved interferon-inducible protein that localizes to the ER, can reduce the secretion of some soluble proteins and reduces the rate of ER-to-Golgi traffic (47). The cellular distribution of viperin and its colocalization with EIAV Gag would positively support our finding that the TGN is involved in EIAV Gag assembly. Previous studies have shown that the HIV-1 Vpu protein is predominantly transported to RER/Golgi complex compartments (33). In our study, EIAV particles localized adjacent to the RER, and EIAV Gag multimers were surrounded by Vpu proteins (Fig. 4C), suggesting that that the ER/Golgi downstream membrane system (TGN) may be involved in EIAV Gag assembly. The formation of infectious EIAV particles requires the recruitment of the EIAV envelope protein (Env) into the viral membrane during budding; however, whether or how the EIAV Env and Gag proteins are corecruited to the TGN to form infectious particles requires further investigation.
HIV-1 Gag myristoylation is essential for PM binding. Mutations of the N-terminal Gly completely block HIV-1 Gag budding, and Gly is known to be required for the N-myristoyl transferase that attaches this fatty acid to the nascent protein (30). However, we observed that G2A of EIAV Gag only slightly blocked Gag release (Fig. 2D), supporting the hypothesis that EIAV Gag was not myristoylated, and myristoylation was not involved in EIAV Gag assembly. Moreover, we found that W7A and K9A localized to helix 1 of EIAV MA severely impaired Gag release (Fig. 2D and 6A). A previous study reported that K49 was located in the middle of the major PIP2 interaction region; indeed, the K49A mutation influenced MA PIP2 binding and severely inhibited VLP release (14, 15). K7 and K49 both localize to the MA surface; however, whether the inhibition of Gag release due to the K7A mutation resulted from the impairment of MA binding to PIP2 remains to be determined. W7 was hidden inside the molecule, but the W7A mutant completely blocked Gag release without changing the distribution of Gag at the cellular membrane (Fig. 2D and 6A). In the structure, we found that W7 and W51 formed hydrogen bonds via D50 (Fig. 6B). D50 has been reported as one of the residues involved in the PIP2-induced chemical shift (14). Thus, we hypothesize that the W7A mutant influences the interaction between W7, D50, and W51 and thereby affects loop 2 binding to phosphoinositide, ultimately blocking Gag release. Additionally, we speculate that the inhibition of Gag release mediated by T6A and S8A may result from affecting the function of W7 or K9.
In the present study, we demonstrated that swapping EIAV MA with HIV-1 MA did not reduce cellular p55 expression and facilitated recombination of the Gag protein accumulating on the PM domain (Fig. 3A and B). However, a strong release inhibition of the recombinant Gag was observed. Thus, we speculate that the PM is not the suitable sites for EIAV Gag assembly and release. Similar results were observed by substituting HIV-1 MA with EIAV MA, suggesting that the interior cellular membranes are not the suitable sites for HIV-1 Gag assembly and release. Jouvenet et al. reported that replacing HIV-1 MA with the FYVE domain that specifically recognizes PI(3)P resulted in its targeting to endosomes and blocked HIV-1 particle release (23). These results indicate that EIAV and HIV-1 might use different cellular membrane sites that are appropriate for each virus to promote viral particle assembly. Our results also demonstrated that MA was the determinant that guided Gag to the assembly site, and the difference between the EIAV and HIV-1 MAs determined their distinct viral assembly features. human T-cell lymphotropic virus type 1 was reported to possess assembly characteristics that were distinct from HIV-1 (48). However, whether these different assembly features are driven by viral evolution is unknown. Recently, it was reported that HIV-1 MA binding to particular tRNAs regulated Gag targeting to the PM (16). Six lysines in the N-terminal MA were the tRNA binding determinants; the MAK4T and MAK6T mutations caused decreases in the level of tRNA binding and particle release (16). There are four lysines in the N terminus of the EIAV MA; however, whether EIAV MA binds to tRNAs remains to be determined. If EIAV MA also possesses the ability to bind tRNAs, questions remain concerning whether EIAV MA will bind to a distinct variety tRNAs compared to the HIV-1 MA and whether the different tRNA binding affinity will influence their distinct assembly features. These questions require intensive investigation.
A previous study observed the colocalization of HIV-1 particles with MVBs and suggested that HIV-1 could also use MVBs as its assembly site. In the present study, HIV-1 pseudovirus and Gag assembly were not blocked by treatment with nocodazole and U18666A, which inhibit the function of MVBs by separate mechanisms (Fig. 8A, B, and D). This result is identical to the results of the study of Jouvenet et al (23). Jouvenet et al. also suggested that endocytosis or phagocytosis may account for the observation of the endosomal localization of HIV-1 particles. Here, we observed that EIAV Gag assembly occurred in the TGN. MVBs are a type of cellular vesicle known to be responsible for exosome export (49). However, no colocalization of Gag and MVB was observed, and EIAV release was not blocked by MVB inhibitors (Fig. 4A and B and Fig. 8), suggesting that EIAV utilizes other cellular vesicles (i.e., apoptotic blebs, shedding vesicles, or microparticles) to export viral particles and does not directly bud into the extracellular environment. This result was supported by the GW4869 treatment experiment because the released EIAV Gag VLPs were severely decreased under 5 μM GW4869 treatment (Fig. 7). Furthermore, the partial localization of EIAV Gag with CD63, which is primarily associated with the membranes of intracellular vesicles, may also support the transportation of some EIAV Gag VLPs by vesicles (Fig. 4A and B). Therefore, we conclude that EIAV assembly occurs in the TGN, and viral particles are then exported through intracellular vesicles. Additionally, different expression characteristics of Gag (as demonstrated by the three bands detected in the cell lysate and the detection of only the P26 [CA] band in the cell culture supernatant) were detected by the anti-p26 MAb (Fig. 8A and E), suggesting that virus particle maturation might occur during vesicle transportation. In animals, there are three principal types of coated vesicles: clathrin-coated vesicles, coat protein I (COPI)-coated vesicles, and COPII-coated vesicles (49). The EIAV L domain has been reported to interact with AP-2, which is one of the adaptor proteins of the vesicle complex (6). However, the detailed export mechanism underlying the transport of EIAV particles through vesicles requires investigation.
In summary, we demonstrate that EIAV MA is the determinant that directs Gag to the TGN to facilitate viral assembly, while HIV-1 MA guides Gag to the PM to complete viral assembly. MVBs are not involved in EIAV and HIV-1 assembly and release. EIAV hijacks the cellular vesicle pathway for viral particle maturation and egress. These findings provide new understanding of the EIAV intracellular life cycle and will be useful for future studies with EIAV and other lentiviruses.
ACKNOWLEDGMENTS
We thank Walther Mothes from Yale University for providing the plasmids TGN38-YFP, Lamp1-YFP, and CD63-YFP.
This study was supported by grant 31372451 from the National Natural Science Foundation of China to J.M. and grant 31222054 to X.W.
REFERENCES
- 1.Leroux C, Cadore JL, Montelaro RC. 2004. Equine infectious anemia virus (EIAV): what has HIV's country cousin got to tell us? Vet Res 35:485–512. doi: 10.1051/vetres:2004020. [DOI] [PubMed] [Google Scholar]
- 2.Nagarajan MM, Simard C. 2007. Gag genetic heterogeneity of equine infectious anemia virus (EIAV) in naturally infected horses in Canada. Virus Res 129:228–235. doi: 10.1016/j.virusres.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 3.Stephens RM, Casey JW, Rice NR. 1986. Equine infectious anemia virus gag and pol genes: relatedness to visna and AIDS virus. Science 231:589–594. doi: 10.1126/science.3003905. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz PD. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Vincent O, Jin J, Weisz OA, Montelaro RC. 2005. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J Biol Chem 280:40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- 6.Puffer BA, Watkins SC, Montelaro RC. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J Virol 72:10218–10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanzi GO, Piefer AJ, Bates P. 2003. Equine infectious anemia virus utilizes host vesicular protein sorting machinery during particle release. J Virol 77:8440–8447. doi: 10.1128/JVI.77.15.8440-8447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieniasz PD. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 9.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol 13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saad JS, Loeliger E, Luncsford P, Liriano M, Tai J, Kim A, Miller J, Joshi A, Freed EO, Summers MF. 2007. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J Mol Biol 366:574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, Parent LJ, Wills JW, Resh MD. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol 68:2556–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J Virol 82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatanaka H, Iourin O, Rao Z, Fry E, Kingsman A, Stuart DI. 2002. Structure of equine infectious anemia virus matrix protein. J Virol 76:1876–1883. doi: 10.1128/JVI.76.4.1876-1883.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K, Bachtiar I, Piszczek G, Bouamr F, Carter C, Tjandra N. 2008. Solution NMR characterizations of oligomerization and dynamics of equine infectious anemia virus matrix protein and its interaction with PIP2. Biochemistry 47:1928–1937. doi: 10.1021/bi701984h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes F, Chen K, Ehrlich LS, Jin J, Chen MH, Medina GN, Symons M, Montelaro R, Donaldson J, Tjandra N, Carter CA. 2011. Phosphoinositides direct equine infectious anemia virus gag trafficking and release. Traffic 12:438–451. doi: 10.1111/j.1600-0854.2010.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. 2014. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65. doi: 10.1016/S0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Serrano J, Zang T, Bieniasz PD. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med 7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 19.Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci U S A 98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, Chen C, Puffer BA, Montelaro RC. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J Virol 76:1569–1577. doi: 10.1128/JVI.76.4.1569-1577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott DE, Coren LV, Gagliardi TD, Nagashima K. 2005. Heterologous late-domain sequences have various abilities to promote budding of human immunodeficiency virus type 1. J Virol 79:9038–9045. doi: 10.1128/JVI.79.14.9038-9045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jouvenet N, Bieniasz PD, Simon SM. 2008. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol 4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houzet L, Gay B, Morichaud Z, Briant L, Mougel M. 2006. Intracellular assembly and budding of the murine leukemia virus in infected cells. Retrovirology 3:12. doi: 10.1186/1742-4690-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelchen-Matthews A, Kramer B, Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol 162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Allen PG, Pypaert M, Cunningham JM, Mothes W. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 27.Ott DE, Coren LV, Sowder RC II, Adams J, Nagashima K, Schubert U. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J Virol 76:3038–3044. doi: 10.1128/JVI.76.6.3038-3044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voeltz GK, Rolls MM, Rapoport TA. 2002. Structural organization of the endoplasmic reticulum. EMBO Rep 3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berka U, Hamann MV, Lindemann D. 2013. Early events in foamy virus-host interaction and intracellular trafficking. Viruses 5:1055–1074. doi: 10.3390/v5041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryant M, Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A 87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chukkapalli V, Ono A. 2011. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J Mol Biol 410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tedbury PR, Mercredi PY, Gaines CR, Summers MF, Freed EO. 2015. Elucidating the mechanism by which compensatory mutations rescue an HIV-1 matrix mutant defective for gag membrane targeting and envelope glycoprotein incorporation. J Mol Biol 427:1413–1427. doi: 10.1016/j.jmb.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pacyniak E, Gomez ML, Gomez LM, Mulcahy ER, Jackson M, Hout DR, Wisdom BJ, Stephens EB. 2005. Identification of a region within the cytoplasmic domain of the subtype B Vpu protein of human immunodeficiency virus type 1 (HIV-1) that is responsible for retention in the Golgi complex and its absence in the Vpu protein from a subtype C HIV-1. AIDS Res Hum Retrovir 21:379–394. doi: 10.1089/aid.2005.21.379. [DOI] [PubMed] [Google Scholar]
- 34.Ma J, Wang SS, Lin YZ, Liu HF, Wei HM, Du C, Wang XF, Zhou JH. 2013. An attenuated EIAV strain and its molecular clone strain differentially induce the expression of Toll-like receptors and type-I interferons in equine monocyte-derived macrophages. Vet Microbiol 166:263–269. doi: 10.1016/j.vetmic.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Ma J, Shi N, Jiang CG, Lin YZ, Wang XF, Wang S, Lv XL, Zhao LP, Shao YM, Kong XG, Zhou JH, Shen RX. 2011. A proviral derivative from a reference attenuated EIAV vaccine strain failed to elicit protective immunity. Virology 410:96–106. doi: 10.1016/j.virol.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 36.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 37.Cocucci E, Racchetti G, Meldolesi J. 2009. Shedding microvesicles: artifacts no more. Trends Cell Biol 19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Aniento F, Emans N, Griffiths G, Gruenberg J. 1993. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol 123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebrand C, Corti M, Goodson H, Cosson P, Cavalli V, Mayran N, Faure J, Gruenberg J. 2002. Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J 21:1289–1300. doi: 10.1093/emboj/21.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Numrich J, Ungermann C. 2014. Endocytic Rabs in membrane trafficking and signaling. Biol Chem 395:327–333. [DOI] [PubMed] [Google Scholar]
- 41.Vanlandingham PA, Ceresa BP. 2009. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem 284:12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, Ming Z, Xiaochun W, Hong W. 2011. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal 23:516–521. doi: 10.1016/j.cellsig.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, Weisz OA, Stolz DB, Watkins SC, Montelaro RC. 2004. Differential effects of actin cytoskeleton dynamics on equine infectious anemia virus particle production. J Virol 78:882–891. doi: 10.1128/JVI.78.2.882-891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, Jin J, Rubin M, Huang L, Sturgeon T, Weixel KM, Stolz DB, Watkins SC, Bamburg JR, Weisz OA, Montelaro RC. 2007. Association of Gag multimers with filamentous actin during equine infectious anemia virus assembly. Curr HIV Res 5:315–323. doi: 10.2174/157016207780636542. [DOI] [PubMed] [Google Scholar]
- 45.Sandrin V, Sundquist WI. 2013. ESCRT requirements for EIAV budding. Retrovirology 10:104. doi: 10.1186/1742-4690-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang YD, Na L, Zhu CH, Shen N, Yang F, Fu XQ, Wang YH, Fu LH, Wang JY, Lin YZ, Wang XF, Wang X, Zhou JH, Li CY. 2014. Equine viperin restricts equine infectious anemia virus replication by inhibiting the production and/or release of viral Gag, Env, and receptor via distortion of the endoplasmic reticulum. J Virol 88:12296–12310. doi: 10.1128/JVI.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinson ER, Cresswell P. 2009. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem 284:4705–4712. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inlora J, Chukkapalli V, Derse D, Ono A. 2011. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J Virol 85:3802–3810. doi: 10.1128/JVI.02383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson DG, Pimpl P. 2014. Clathrin and post-Golgi trafficking: a very complicated issue. Trends Plant Sci 19:134–139. doi: 10.1016/j.tplants.2013.10.008. [DOI] [PubMed] [Google Scholar]