Abstract
An increasing number of studies have indicated that the deregulation of microRNAs (miRNAs) contributes to tumorigenesis and metastasis. In the present study, significant upregulation of miR-362-5p was identified in the breast cancer MDA-MB-231 and MCF7 cell lines compared with the control CCD-1095Sk cell line. The inhibition of miR-362-5p was demonstrated to significantly inhibit the cell proliferation, migration and invasion of human breast cancer MCF7 cells. In addition, the knockdown of miR-362-5p induced G1 arrest and promoted apoptosis in the breast cancer cells. Mechanistic investigations confirmed that the tumor suppressor gene CYLD is a direct target of miR-362-5p. The ectopic expression of miR-362-5p represses CYLD expression, whereas miR-362-5p inhibitor treatment induces CYLD protein expression and decreases NF-κB expression in the downstream signaling pathway. Thus, these findings may provide novel insights into the molecular mechanisms through which miR-362-5p regulates breast cancer cell proliferation, migration and invasion. This study also suggests that miR-362-5p may act as a novel potential therapeutic target for the treatment of breast cancer.
Keywords: miR-362-5p, breast cancer, proliferation, invasion, migration
Introduction
Breast cancer is the leading cause of cancer-associated mortality in women worldwide (1). Breast cancer is a heterogeneous disease that is characterized by various molecular subtypes, which exhibit distinct molecular characteristics and clinical behaviors (2,3). Although improvements in the early detection and treatment of breast cancer have decreased the associated mortality rates in previous years, clarification of the molecular and cellular mechanisms underlying the development and progression of breast cancer continues to be required (2).
An increasing number of studies have revealed that malignant tumor progression involves multiple genetic and epigenetic changes, each of which may result in the deregulation of important etiology-specific pathways involved in complex cellular processes, such as proliferation, migration, invasion and apoptosis (4–6). The accumulation of these genetic and epigenetic alterations confers a malignant phenotype (7).
microRNAs (miRNA) are a class of small, endogenously expressed non-coding RNAs comprised of 18–25 nucleotides. miRNAs induce the silencing of the cognate target genes by either degrading the target messenger RNA (mRNA) or repressing translation (8). Previous studies have revealed that miRNA plays a critical role in the regulation of various cellular processes, including cell growth and metastasis, which indicated that miRNA may function either as an oncogene or as a tumor suppressor (9,10).
The cylindromatosis gene CYLD was initially identified as a mutated gene in familial cylindromatosis (11). CYLD contains an ubiquitin C-terminal hydrolase domain, which allows the protein to function as a deubiquitinating enzyme (12). CYLD has been revealed to play a central role in regulating various signaling pathways, including transforming growth factor-β, Wnt/β-catenin, c-Jun N-terminal kinase and nuclear factor-κB (NF-κB) signaling, and thus regulates the promotion of cancer development and progression (13–16). Furthermore, the downregulation of CYLD has been reported in several malignancies (17–20). Previous studies have revealed that CYLD expression is downregulated in breast cancer tissues compared with normal breast tissues (2,17). In addition, the downregulation of CYLD promotes cell survival and migratory activities through NF-κB activation (21). However, the precise molecular mechanisms underlying the deregulation of CYLD expression in breast cancer are not fully understood.
In the present study, miR-362-5p was investigated as it is known to be overexpressed in breast cancer cells, and CYLD was revealed to be a target of miRNA-362-5p (miR-362-5p) by TargetScan. To the best of our knowledge, the roles of miR-362-5p and the targets of miR-362-5p in breast cancer have not yet been reported. In this study, the inhibition of miR-362-5p was revealed to reduce the proliferation, migration and invasion of breast cancer cells. Therefore, the present results suggest a potential medical value of the miR-362-5p/CYLD axis in effective breast cancer therapy.
Materials and methods
Cell lines and cell culture
The human breast cancer MDA-MB-231 and MCF7, normal breast fibroblast CCD-1095Sk and human embryonic kidney HEK293 cell line were purchased from the American Type Culture Collection (Manassas, VA, USA). The MCF-7 cells were cultured in RPMI-1640 medium, and the MDA-MB-231 and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Little Chalfont, UK).
RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR) analysis
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. For miRNA analysis, equal amounts of RNA were reverse transcribed using a miRNA-specific primer (Hsa-miR-362-5p; Qiagen, Venlo, Limburg, Netherlands) and then subjected to RT-PCR, according to the manufacturer's protocol for the miScript Reverse Transcription and miScript SYBR Green PCR kits (Qiagen). RNU6B small nuclear RNA was used as an internal control. For CYLD mRNA analysis, RNA was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and random primers (Roche, Basel, Switzerland). RT-PCR was performed using SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd., Dalian, Liaoning, China). The PCR primers for CYLD were as follows: Forward, 5′-TCAGGCTTATGGAGCCAAGAA-3′; and reverse, 5′-ACTTCCCTTCGGTACTTTAAGGA-3′. GAPDH mRNA levels were used as an internal normalization control. The cycle threshold method was used to analyze the expression of miR-362-5p and CYLD relative to GAPDH expression.
Cell proliferation, cell cycle, apoptosis analysis and colony formation assays
The cell proliferation was measured using an MTT assay. The cells were seeded in 96-well plates at a density of 4,000 cells per well and maintained in a culture medium containing 5% fetal bovine serum for 48 h. The absorbance was then measured at 490 nm. The cell cycle analysis was conducted with a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) using a propidium iodide cell cycle detection kit (Beyotime Institute of Biotechnology, Beijing, China). The apoptosis assay was performed using the Annexin V-phycoerythrin (PE) Apoptosis Detection kit I (BD Biosciences), according to the manufacturer's instructions, and apoptosis was analyzed using a FACSCalibur flow cytometer. The apoptotic cells were indicated by high Annexin V-PE fluorescence signals. For the colony formation assays, 100 cells were placed into each well of a 6-well plate and cultured for two weeks at 37°C. The numbers of colonies per dish were counted subsequent staining with 0.1% crystal violet. All experiments were conducted with three replicates.
Wound-healing migration and invasion assays
For the wound-healing migration assay, the cells were seeded onto 35-mm dishes coated with fibronectin. Once the cells reached 100% confluency, a sterile p200 pipette tip was used to create a scratch (~500 µm) through the confluent monolayer. The medium was changed to fresh serum-free medium to remove the cellular debris, and the cells were cultured for another 48 h. Serial images were obtained at 0, 24 and 48 h. For the invasion assay, 1×104 cells were placed into the upper chamber of the Matrigel Transwell chamber (BD Biosciences) in serum-free medium. Medium containing 10% FBS in the lower chamber acted as the chemoattractant. Subsequent to 64 h of incubation at 37°C, the invasive cells attached to the lower membrane of the inserts were fixed, stained and then counted using a counting chamber and microscope (CX31; Olympus, Tokyo, Japan).
Western blotting
The cell cytoplasm or nucleus lysates subsequent to transfection with the miR-362-5p mimic, inhibitor or negative control miRNA (Shanghai GenePharma Co., Ltd., Shanghai, China) were separated by SDS-PAGE and then transferred onto a polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA). The blotted membranes were incubated with rabbit anti-human polyclonal CYLD (cat. no. 11110-1-AP; dilution, 1:1,000; ProteinTech Group, Inc., Chicago, IL, USA), rabbit anti-human polyclonal NF-κB P65 (cat. no. sc-372; dilution, 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), rabbit anti-human monoclonal anti-Lamin B1 (cat. no. 12586; dilution, 1:1,500; Cell Signaling Technology, Inc., Danvers, MA, USA) and monoclonal mouse anti-human β-actin (cat. no. A5316; dilution, 1:5,000; Sigma-Aldrich, St. Louis, MO, USA) antibodies. The membrane was then incubated with the secondary goat anti-rabbit horseradish peroxidase-conjugated antibody (cat. no. sc-2004; dilution, 1:5,000; Santa Cruz Biotechnology, Inc.). The immunoreactive protein bands were developed using the Enhanced Chemiluminescence System (Pierce Biotechnology, Inc., Rockford, IL, USA).
Dual-luciferase reporter assay
A fragment of the 3′-UTR of the CYLD gene that contained the miR-362-5p target sequence (CAAGGAT) was inserted into XhoI/NotI-digested psiCHECK-2 vectors (Promega, Madison, WI, USA). The insertion of the sequences was confirmed by sequencing. The psiCHECK-2 constructs and miR-362-5p mimic or negative control miRNA were cotransfected into HEK293 cells using the Lipofectamine 2000 transfection reagent (Invitrogen), according to the manufacturer's instructions. Subsequent to 48 h, the cells were lysed, and the luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay kit (Promega), according to the manufacturer's instructions. The results were expressed as the ratio of Renilla luciferase activity to firefly luciferase activity.
Statistical analysis
The data are expressed as the mean ± standard error of the mean from at least three independent experiments. Unless otherwise noted, the differences between groups were analyzed using two-sided Student's t-tests for two groups or by one-way analysis of variance (ANOVA) when more than two groups were compared. P<0.05 was considered to indicate a statistically significant difference.
Results
Inhibitory effects of anti-miR-362-5p on the proliferation of MCF7 cells
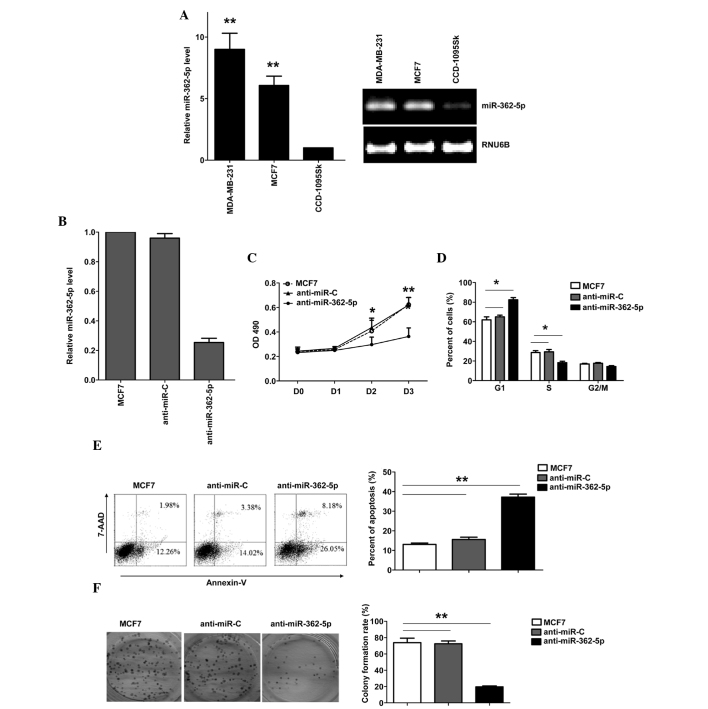
To evaluate the expression and importance of miR-362-5p in breast cancer, the expression of miR-362-5p was identified in human breast cancer MDA-MB-231 and MCF7 cell lines, and normal breast fibroblast CCD-1095Sk cell line. miR-362-5p was upregulated in breast cancer cell lines compared with CCD-1095Sk cells (Fig. 1A). To explore the biological significance of miR-26a in breast cancer cells, a miR-362-5p inhibitor (anti-miR-362-5p) was transfected into the human breast cancer MCF7 cell line. The expression of miR-362-5p was verified by RT-qPCR (Fig. 1B). Downregulation of miR-362-5p in the MCF7 cells resulted in a significant suppression of cell proliferation, G1 arrest and apoptosis induction. As shown in Fig. 1C, the results of the MTT assay revealed that downregulation of miR-362-5p significantly suppressed MCF7 cell proliferation (Fig. 1C). The cell cycle was arrested in the G1 phase, with 82.3% of the anti-miR-362-5p-transfected cells in the G0/G1 phase, compared with 65% of the control cells (Fig. 1D). In addition, the transfection of anti-miR-362-5p induced apoptosis, with 37.2% of anti-miR-362-5p-transfected cells demonstrating apoptosis, compared with 15.5% of the cells in the control group (Fig. 1E). Additional analysis of the effects of anti-miR-362-5p on the clonogenicity of breast cancer cells was performed, and it was found that the inhibition of miR-362-5p significantly decreased the colony formation of MCF7 cells (Fig. 1F). These results demonstrate that miR-362-5p regulates the proliferation of MCF7 cells.
Figure 1.
Inhibitory effect of anti-miR-362-5p on the proliferation, apoptosis and colony formation of MCF7 cells. (A) The expression level of miR-362-5p in normal breast fibroblast cells and breast cancer cell lines was examined by RT-qPCR. (B) RT-qPCR analysis of miR-362-5p expression in MCF7 cells transfected with inhibitory anti-miR-362-5p or control anti-miR-C miRNA. (C) The effect of the anti-miR-362-5p inhibitor miRNA on cell proliferation was measured using an MTT assay. (D) Analysis of the cell cycle distribution in untransfected MCF7 cells and cells transfected with inhibitory anti-miR-362-5p or control anti-miR-C miRNA. (E) Analysis of apoptosis in untransfected MCF7 cells and cells transfected with ihibitory anti-miR-362-5p or control anti-miR-C miRNA. (F) Colony formation assay of untransfected MCF7 cells and cells transfected with inhibitory anti-miR-362-5p or control anti-miR-C miRNA. The number of colonies was counted and compared. Data are expressed as the mean ± standard error of the mean. *P<0.05; **P<0.01. anti-miR-362-5p, anti-microRNA-362-5p; anti-miR-C, control microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; OD, optical density; 7-AAD, 7-aminoactinomycin D.
Inhibition of miR-362-5p reduces the migration and invasion of MCF7 cells
To further determine the biological significance of miR-362-5p in breast cancer cell metastasis, wound-healing migration and Transwell assays were performed using the MCF7 cells. The mobility of the MCF7 cells in the wound-healing assay was significantly decreased subsequent to transfection with anti-miR-362-6p miRNA (Fig. 2A). The Transwell assay with Matrigel demonstrated that the inhibition of miR-362-5p markedly inhibited the invasive capacity of MCF7 cells compared with the control cells (Fig. 2B). These results suggest that anti-miR-362-5p significantly inhibits the migration and invasion of MCF7 cells.
Figure 2.
Inhibitory effect of the anti-miR-miR-362-5p inhibitor miRNA on the invasion and migration of MCF7 cells. (A) Wound-healing assay of MCF7 cells transfected with inhibitory anti-miR-362-5p or control anti-miR-C miRNA. (B) Invasion assay of MCF7 cells transfected with inhibitory anti-miR-362-5p or control anti-miR-C miRNA. Representative images of MCF7 cells that invaded through the Matrigel. The number of invasive MCF7 cells was quantified. *P<0.05; **P<0.01. anti-miR-362-5p, anti-microRNA-362-5p; anti-miR-C, control microRNA.
CYLD is the direct downstream target of miR-362-5p
The aforementioned phenotypic data indicate that the inhibition of miR-362-5p reduces the growth and migration of breast cancer cells. TargetScan was used to predict the mRNA targets of miR-362-5p, to identify potential mRNA targets that may contribute to its tumor-associated function. The present analysis revealed that CYLD was a potential target of miR-362-5p. The 3′-UTR of CYLD mRNA was found to contain a complementary site for the seed region of miR-362-5p (Fig. 3A). To determine whether CYLD is a direct target of miR-362-5p, a human CYLD 3′-UTR fragment containing the wild-type (WT 3′-UTR) or mutant (mutated 3′-UTR) miR-362-5p-binding sequences was cloned into the psiCHEK-2 vector (Fig. 3A). As shown in Fig. 3B, the relative luciferase activity of the reporter containing WT 3′-UTR was significantly suppressed following miR-362-5p transfection. However, site-directed deletion of the miR-362-5p-binding site within the 3′-UTR of CYLD completely abolished this suppression (Fig. 3B), suggesting that miR-362-5p directly binds to this site.
Figure 3.
Direct targeting of CYLD by miR-362-5p in breast cancer cells. (A) Sequence alignment between miR-362-5p and the 3′-UTR of CYLD. (B) The histogram bars represent the relative dual luciferase activity subsequent to the transfection of HEK293 cells with miR-362-5p mimics or control miRNAs (miR-C). **P<0.01 relative to the miR-C control. (C and D) The mRNA and protein expression levels of CYLD were examined by RT-qPCR and western blotting in CCD-1095Sk cells transfected with miR-362-5p mimics or control miRNAs (miR-C). (E and F) mRNA and protein expression levels of CYLD were examined by RT-qPCR and western blotting in MCF7 cells transfected with miR-362-5p inhibitor or control anti-miR-C. anti-miR-362-5p, anti-microRNA-362-5p; anti-miR-C, control microRNA; miR-NC, negative control microRNA; 3′-UTR, 3′-untranslated region; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Furthermore, the ability of miR-362-5p to regulate CYLD expression in human breast cells was tested. Normal breast fibroblast CCD-1095Sk cells, which demonstrate a low miR-362-5p expression level, were transfected with the miR-362-5p mimic or control miRNA and were cultured for 48 h. The results demonstrated that the CYLD mRNA and protein levels were decreased in CCD-1095Sk cells transfected with the miR-362-5p mimic (Fig. 3C and D). The ability of endogenous miR-362-5p to regulate CYLD expression was then assessed in breast cancer cells. Compared with the miRNA control, the CYLD mRNA and protein levels were upregulated when miR-362-5p was knocked down using anti-miR-362-5p in MCF7 cells (Fig. 3E and F). Overall, these results strongly indicate that CYLD is a direct target of miR-362-5p in breast cancer cells.
miR-362-5p promotes cell proliferation, migration and invasion through the NF-κB signaling pathway
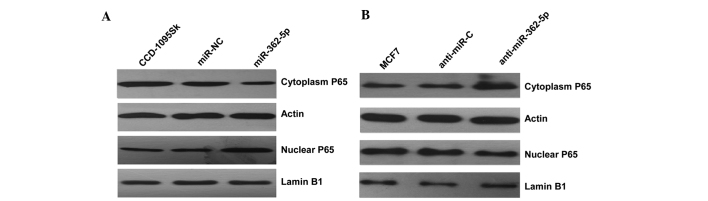
Since CYLD plays a predominant role in the negative regulation of NF-κB (21), the mechanism underlying the observed cellular phenotypic changes was explored by evaluating the impact of CYLD on NF-κB activation by detecting the level of nucleus NF-κB P65 protein in the two cell lines transfected with miR-362-5p mimics/inhibitors. Subsequent to transfection with miR-362-5p mimics, the results indicated that the level of nucleus NF-κB P65 protein was clearly increased compared with the negative control (Fig. 4A). By contrast, the expression level of nuclear P65 was clearly decreased in miR-362-5p-suppressed MCF7 cells (Fig. 4B). Overall, the present findings indicate that miR-362-5p likely promotes proliferation, migration and invasion through the NF-κB signaling pathway.
Figure 4.
Effect of miR-362-5p on NF-κB activation. (A) Analysis of the expression of the NF-kB P65 protein in CCD-1095Sk cells transfected with miR-362-5p mimic or anti-miR-C miRNA. (B) Analysis of the expression of the NF-kB P65 protein in MCF7 cells transfected with the miR-362-5p inhibitor or anti-miR-C miRNA. anti-miR-362-5p, anti-microRNA-362-5p; anti-miR-C, control microRNA; miR-NC, negative control microRNA; NF-κB, nuclear factor-κB.
Discussion
An increasing number of studies have demonstrated that the dysregulation of miRNA contributes to tumorigenesis (22). Numerous studies have confirmed that miRNA plays a critical role in tumor cell survival, invasion and metastasis. Xia et al identified that miR-362 induces cell proliferation and apoptosis resistance in gastric cancer (23), but the impact on breast cancer cell growth and metastasis remains unclear. In the present study, miR-362-5p was focused on, and it was found that miR-362-5p was clearly upregulated in human breast cancer cell lines. To the best of our knowledge, the present study is the first to report the upregulation of miR-362-5p in breast cancer cells. Since the upregulation of miR-362-5p in breast cancer has not been previously described, the role of miR-362-5p in breast cancer cells was analyzed. The present study provides the first evidence that the downregulation of miR-362-5p significantly inhibits cell proliferation, migration and invasion, induces cell cycle arrest, and promotes apoptosis in MCF7 cells. These results indicate that miR-362-5p may be a novel oncogene that plays an important role in the regulation of breast cancer cell growth and metastasis.
The function of miRNA is to regulate the target genes by direct cleavage of the mRNA or the inhibition of protein synthesis (24). To further explore the molecular mechanism underlying miR-362-5p function, direct target genes of miR-362-5p were identified through bioinformatics analysis. It was found that CYLD possesses a putative miR-362-5p-binding site within its 3′-UTR. CYLD was identified as a direct target of miR-362-5p in breast cancer cells, and this conclusion was supported by the following findings. Firstly, the complementary sequence of miR-362-5p was identified in the 3′-UTR of CYLD mRNA. In addition, miR-362-5p overexpression was found to suppress CYLD 3′-UTR luciferase reporter activity, and this effect was abolished by the deletion of the miR-362-5p seed binding site. The overexpression of miR-362-5p also led to a significant reduction in CYLD at the mRNA and protein levels. Finally, the inhibition of miR-362-5p increased the CYLD expression level in breast cancer cells.
CYLD is known to be a tumor suppressor gene in various types of cancer (25,26). Previous studies have revealed that CYLD expression is downregulated in breast cancer tissues. In addition, the downregulation of CYLD promoted cell survival and migration through the NF-κB signaling pathway (21). However, the exact regulatory mechanism responsible for the decrease of CYLD expression in breast cancer cells remains unclear. Notably, in the present study, miR-362-5p was revealed to be associated with CYLD and the downstream NF-κB signaling pathway. The current results revealed that miR-362-5p was likely to repress the expression of CYLD, which, in turn, promoted the proliferation, migration and invasion of breast cancer cells by regulating the NF-κB pathway.
In conclusion, the present study provides novel evidence that downregulation of miR-362-5p expression suppresses the proliferation, migration and invasion of human breast cancer cells. These data suggest that miR-362-5p and its downstream targets may be potential therapeutic targets in human breast cancer.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant nos. 81272258 and 31300715), Anhui Province Natural Science Foundation (grant no. 1308085QH136) and Anhui Medical University Training Program of National Outstanding Youth Foundation (grant no. GJYQ-1401). The study was also supported by a grant from the Grants for Scientific Research of bo shi ke yan of Anhui Medical University (grant no. XJ201316) awarded to Dr Fang Ni.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ. Panel members: Strategies for subtypes - dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giordano S, Columbano A. MicroRNAs: New tools for diagnosis prognosis, and therapy in HCC? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 5.Sanyal AJ, Yoon SK, Lencioni R. The etiology of hepatocellular carcinoma and consequences for treatment. Oncologist. 2010;15(Suppl 4):14–22. doi: 10.1634/theoncologist.2010-S4-14. [DOI] [PubMed] [Google Scholar]
- 6.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273.e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasinski AL, Slack FJ. Epigenetics and genetics. Histopathology. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Dvinge H, Git A, Gräf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 10.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 12.Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 13.Lim JH, Jono H, Komatsu K, Woo CH, Lee J, Miyata M, Matsuno T, Xu X, Huang Y, Zhang W, et al. CYLD negatively regulates transforming growth factor-beta-signalling via deubiquitinating Akt. Nat Commun. 2012;3:771. doi: 10.1038/ncomms1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tauriello DV, Haegebarth A, Kuper I, Edelmann MJ, Henraat M, Canninga-van Dijk MR, Kessler BM, Clevers H, Maurice MM. Loss of the tumor suppressor CYLD enhances Wnt/betacatenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37:607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 15.Pannem RR, Dorn C, Ahlqvist K, Bosserhoff AK, Hellerbrand C, Massoumi R. CYLD controls c-MYC expression through the JNK-dependent signaling pathway in hepatocellular carcinoma. Carcinogenesis. 2014;35:461–468. doi: 10.1093/carcin/bgt335. [DOI] [PubMed] [Google Scholar]
- 16.Wang WY, Lim JH, Li JD. Synergistic and feedback signaling mechanisms in the regulation of inflammation in respiratory infections. Cell Mol Immunol. 2012;9:131–135. doi: 10.1038/cmi.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautheron J, Luedde T. A novel player in inflammation and cancer: The deubiquitinase CYLD controls HCC development. J Hepatol. 2012;57:937–939. doi: 10.1016/j.jhep.2012.07.039. [DOI] [PubMed] [Google Scholar]
- 18.Font-Burgada J, Seki E, Karin M. CYLD and HCC, When being too sensitive to your dirty neighbors results in self-destruction. Cancer Cell. 2012;21:711–712. doi: 10.1016/j.ccr.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urbanik T, Köhler BC, Boger RJ, et al. Down-regulation of CYLD as a trigger for NF-κB activation and a mechanism of apoptotic resistance in hepatocellular carcinoma cells. Int J Oncol. 2011;38:121–131. [PubMed] [Google Scholar]
- 20.Hayashi M, Jono H, Shinriki S, et al. Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res Treat. 2014;143:447–457. doi: 10.1007/s10549-013-2824-3. [DOI] [PubMed] [Google Scholar]
- 21.Sun SC. CYLD: A tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Liu J, Wang G. The role of microRNAs in human breast cancer progression. Tumor Biol. 2014;35:6235–6244. doi: 10.1007/s13277-014-2202-8. [DOI] [PubMed] [Google Scholar]
- 23.Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL, He YL, Chen D, Li W. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-κB signaling. J Transl Med. 2014;12:33. doi: 10.1186/1479-5876-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated posttranscriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Deng LL, Shao YX, Lv HF, Deng HB, Lv FZ. Over-expressing CYLD augments antitumor activity of TRAIL by inhibiting the NF-κB survival signaling in lung cancer cells. Neoplasma. 2012;59:18–29. doi: 10.4149/neo_2012_003. [DOI] [PubMed] [Google Scholar]
- 26.Massoumi R. CYLD: A deubiquitination enzyme with multiple roles in cancer. Future Oncol. 2011;7:285–297. doi: 10.2217/fon.10.187. [DOI] [PubMed] [Google Scholar]