Abstract
Doxorubicin has been widely used in the treatment of cancer. However, acquired doxorubicin resistance severely hinders the application of the drug. In the present study, doxorubicin resistance was investigated in lung carcinoma. microRNA-155 (miR-155) was found to be upregulated in the doxorubicin-resistant A549/dox cell line. Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-π, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AKT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-κB and activator protein-1 was also downregulated. In summary, the present results indicate that miR-155 may participate in doxorubicin resistance in lung carcinoma. The current study provides a novel target for lung carcinoma treatment.
Keywords: microRNA-155, resistance reversal, human lung cancer
Introduction
Lung cancer has become the cancer with the highest associated mortality rate worldwide (1). The two main types of lung cancer are small cell lung cancer (SCLC) and non-SCLC (NSCLC) (2). In total, >80% of lung cancer patients are diagnosed with NSCLC, such as squamous cell carcinoma, adenocarcinoma, adenosquamous cell carcinoma and large cell carcinoma (3). Although advanced molecular biology techniques have considerably increased the understanding of the biological mechanisms that underlie lung cancer development, the 5-year survival rate of lung cancer is 15%, which has not markedly improved from the survival rate of 13% (4,5). A major challenge in treating lung cancer is to identify novel therapeutic targets that may complement current chemotherapy regimens (6).
Doxorubicin targets DNA and topoisomerase II (Topo II) to inhibit DNA synthesis and transcription, arrest tumor cell growth and induce apoptosis (7,8). Clinical studies have found that doxorubicin in combination with other chemotherapeutic drugs acts as a treatment for lung carcinoma (9). In general, the efficacy of chemotherapeutic drugs is limited in the treatment of lung cancer and the carcinoma cells may readily develop resistance to the drugs in clinical practice, significantly reducing the therapeutic efficacy of chemotherapy. Therefore, the identification and development of novel methods for reversing drug resistance demonstrate an extremely important significance in improving the clinical benefit of treatment (10).
microRNAs (miRNAs) are 18–24 bases in length and are a class of endogenously expressed small non-coding ribonucleic acids that are able to regulate gene expression at the post-transcriptional level through binding with the complementary sequences of the target mRNA, resulting in the degradation of the target mRNA (11–13). Numerous studies have provided certain evidence on the close correlation between miRNAs and tumors (14). These studies revealed that particular miRNAs were able to regulate the uncontrolled growth of tumor cells, apoptosis, migration and invasion, and even the response of tumor cells to drug treatment, through the regulation of target genes (15,16). miRNA-155 (miR-155) is one of the miRNAs that are overexpressed in multiple cancers, as demonstrated in numerous studies (17). In gastric cancer cells, the level of miR-155 has been reported as significantly increased, and the overexpression of miR-155 promoted cell proliferation and invasion, while silencing miR-155 inhibited cell proliferation and enhanced apoptosis (18). A similar phenomenon was observed in lung cancer cells. Yan et al compared the expression profiles of miRNA in malignant and non-malignant bile duct epithelial cells (19). It was found that miR-155, miR-141 and miR-200b were overexpressed in malignant biliary epithelial cells, while the inhibition of miR-155 and miR-200b increased the sensitivity of cancer cells to gemcitabine (20). The study by Li et al also confirmed the cancer-promoting role of miR-155 in cholangiocarcinoma (21). However, studies investigating the role of miR-155 in the drug resistance of lung carcinoma cells are limited.
Materials and methods
Cell culture and transfection
The NSCLC A549 cell line was obtained from American Type Culture Collection (Manassas, VA, USA) and was grown in RPMI-1640 medium supplemented with 10% fetal bovine serum (Corning, Manassas, VA, USA). The doxorubicin-resistant A549/dox cell line was established from the A549 cells over 6 months by cultivation in medium with doxorubicin (Sigma-Aldrich, St. Louis, MO, USA); the medium was refreshed every 3 days. Transient transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
MTT assay
The half-maximal inhibitory concentration (IC50) values were calculated by colorimetric measurements of mitochondrial metabolic activity with the CellTiter MTS/PES assay following the manufacturer's instructions (Promega, Madison, WI, USA).
Apoptosis
The apoptosis rate was examined using the Annexin V/propidium iodide double staining method (BD Biosciences, Franklin Lakes, NJ, USA). The cells were treated for 24 h with 5 µm doxorubicin, 48 h subsequent to transfection with antisense oligonucleotides against miR-155 (ASO-miR) or negative control antisense oligonucleotides (ASO-NC). The cells were then trypsinized and harvested for the assay, according to the manufacturer's instructions.
Cell cycle analysis
The A549 and A549/dox cells were washed 3 times with phosphate-buffered saline (PBS), trypsinized with 0.255 Trypsin-EDTA solution and pelleted by centrifugation at 300 × g for 5 min. Subsequent to 2 washes with PBS, the cells were fixed in 70% ethanol overnight at 4°C. The cells were washed 3 times with PBS prior to incubation for 1 h with 50 mg/ml propidium iodide and 50 mg/ml RNAse A in PBS. Fluorescence-activated cell sorting analysis was performed using FACSCalibur (BD Biosciences), using linear scale representation of forward and side scatter during flow analysis. A total of 10,000 events were measured per sample.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RT-qPCR was performed in triplicate in ABI StepOne Plus Real-Time PCR System (Applied Biosystems Life Technologies, Foster City, CA, USA) and normalized with U6 and β-actin endogenous control. miR-155 levels were measured using the TaqMan microRNA assay kit, and the endogenous mRNA levels of other genes were detected using the SYBR Green PCR Master Mix kit, according to the manufacturer's instructions (Applied Biosystems Life Technologies). The PCR primers used were as follows: multidrug resistance protein 1 (MDR1) forward, 5′-TTGCTGCTTACATTCAGGTTTCA-3′ and reverse, 5′-AGCCTATCTCCTGTCGCATTA-3′; multidrug resistance-associated protein 1 (MRP1) forward, 5′-CTCTATCTCTCCCGACATGACC-3′ and reverse, 5′-AGCAGACGATCCACAGCAAAA-3′; breast cancer resistance protein (BCRP) forward, 5′-CAGGTGGAGGCAAATCTTCGT-3′ and reverse, 5′-ACCCTGTTAATCCGTTCGTTTT-3′; glutathione S-transferase-π (GST-π) forward, 5′-CCCTACACCGTGGTCTATTTCC-3′ and reverse, 5′-CAGGAGGCTTTGAGTGAGC-3′; Survivin forward, 5′-AGGACCACCGCATCTCTACAT-3′ and reverse, 5′-AAGTCTGGCTCGTTCTCAGTG; B-cell lymphoma-2 (Bcl-2) forward, 5′-GGTGGGGTCATGTGTGTGG-3′ and reverse, 5′-CGGTTCAGGTACTCAGTCATCC-3′; caspase 8 forward, 5′-TTTCTGCCTACAGGGTCATGC-3′ and reverse, 5′-GCTGCTTCTCTCTTTGCTGAA-3′; caspase 3 forward, 5′-CATGGAAGCGAATCAATGGACT-3′ and reverse, 5′-CTGTACCAGACCGAGATGTCA-3′; and β-actin forward, 5′-GTCTGCCTTGGTAGTGGATAATG-3′ and reverse, 5′-TCGAGGACGCCCTATCATGG-3′.
Western blotting
Total lysate sample (50 mg per lane) was resolved by SDS-PAGE and immunoblotted with primary antibodies. The antibodies used against various proteins were as follows: Rabbit monoclonal anti-MDR1 (1:2,000; ab170904, Abcam, Cambridge, UK); mouse monoclonal anti-MPR1 (1:2,000; ab24102, Abcam); rat monoclonal anti-BCRP (1:2,000; ab24114, Abcam); mouse monoclonal anti-GST-π (1:2,000; ab47709, Abcam); goat monoclonal anti-Survivin (1:2,000; sc-8807, Santa Cruz Biotechnology, Inc., Dallas, TX, USA); rabbit monoclonal anti-Bcl-2 (1:2,000; sc-492, Santa Cruz Biotechnology, Inc.); mouse monoclonal anti-caspase 3 (1:2,000; sc-65496, Santa Cruz Biotechnology, Inc.); mouse monoclonal anti-caspase 8 (1:2,000; sc-81656, Santa Cruz Biotechnology, Inc.); rabbit monoclonal anti-AKT, anti-phosphorylated AKT (p-AKT; 1:2,000; 13038, Cell Signaling Technology, Inc., Danvers, MA, USA); rabbit monoclonal anti-extracellular signal-regulated kinase (ERK; 1:2,000; 4695, Cell Signaling Technology, Inc.); rabbit monoclonal anti-phosphorylated ERK (p-ERK; 1:2,000; 4370, Cell Signaling Technology, Inc.); and mouse monoclonal anti-Tubulin (1:3,000; 10198, Sigma-Aldrich, St. Louis, MO, USA).
Luciferase reporter assay
The cells were seeded into 24-well plates overnight prior to transfection, and then co-transfected with the control ASO-NC or ASO-miR (Sigma-Aldrich), the nuclear factor-κB (NF-κB) luciferase reporter or activator protein-1 (AP-1) and the pTK-Renilla luciferase reporter (Promega Corporation, Madison, WI, USA), which was used as an internal control, using Lipofectamine 2000. Luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega) 24 h subsequent to transfection.
Statistical analysis
Quantitative data are expressed as the mean ± standard deviation, unless otherwise indicated. Analysis of variance with Student's t-test was used to identify significant differences in multiple comparisons. For all statistical analyses, P<0.05 was considered to indicate a statistically significant difference.
Results
miR-155 is upregulated in doxorubicin-resistant tumor cells
In order to investigate the role of miR-155 in the doxorubicin resistance of lung cancer, the doxorubicin-resistant A549/dox cell line was established by feeding A549 cells with increasing concentrations of doxorubicin for 6 months. The IC50 of doxorubicin was determined in the novel A549/dox cell line. The IC50 for doxorubicin in A549/dox cells increased between 1.4 and 16.4 µm (Fig. 1A). The expression of miR-155 was then examined in the A549 and A549/dox cell lines. Notably, miR-155 expression was found to be significantly upregulated in A549/dox cells compared with the A549 cells (Fig. 1B). These results suggest that miR-155 may participate in doxorubicin resistance in lung cancer.
Figure 1.
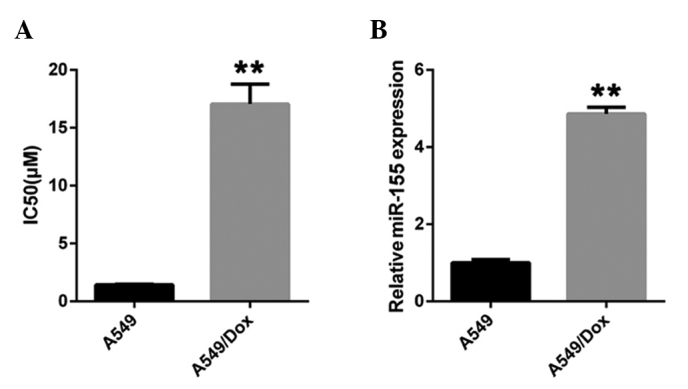
miR-155 is upregulated in doxorubicin-resistant lung cancer A549/dox cells. (A) The IC50 values of A549 and A549/dox cells were calculated using an MTS/PES assay. (B) miR-155 expression in A549 and A549/dox cells was measured by reverse transcription-quantitative polymerase chain reaction. The error bars represent the standard deviation of three independent measurements. **P<0.01. miR-155, microRNA-155; IC50, half-maximal inhibitory concentration.
miR-155 suppression reverses doxorubicin resistance in A549/dox cells
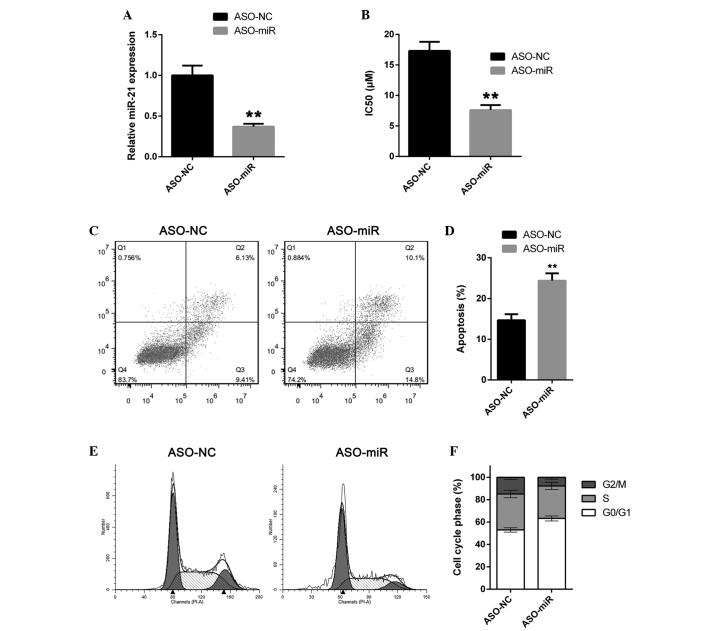
To explore the role of miR-155 in A549/dox cells, the expression of miR-155 was knocked down in A549/dox cells using the antisense oligonucleotide ASO-miR. The A549/dox cells were transfected with either ASO-miR or the negative control antisense oligonucleotide ASO-NC. The cells were then subjected to RT-qPCR. ASO-miR was found to efficiently suppress miR-155 expression (Fig. 2A). The IC50 of doxorubicin in A549/dox cells was then determined. As expected, miR-155 depletion significantly increased the sensitivity of A549/dox cells to doxorubicin. The IC50 of doxorubicin in A549/dox cells was decreased to 7.61 µm (Fig. 2B). The effect of miR-155 on doxorubicin-induced apoptosis in A549/dox cells was then examined. In agreement with previous results, miR-155 downregulation significantly increased doxorubicin-induced apoptosis in the A549/dox cells (Fig. 2C and D). The effect of miR-155 on the cell cycle transition in A549/dox cells was then considered, and a cell cycle distribution assay was performed. As expected, miR-155 depletion induced cell cycle arrest in A549/dox cells (Fig. 2E and F). Overall, these results indicate that miR-155 is an inducer of doxorubicin resistance.
Figure 2.
miR-155 suppression reverses doxorubicin resistance in A549/dox cells. (A) The A549/dox cells were transfected with ASO-miR or ASO-NC and miR-155 expression was then measured by reverse transcription-quantitative polymerase chain reaction. (B) A549/dox cells were transfected with ASO-miR or ASO-NC. The IC50 of doxorubicin in the cells was then measured by MTS assay. (C and D) The A549/dox cells were transfected with ASO-miR or ASO-NC and then treated with 5 µm doxorubicin for 24 h. The apoptosis rate was determined. (E and F) The cell cycle phase distribution of A549/dox cells transfected with ASO-miR or ASO-NC was determined. The error bars represent the standard deviation from three independent measurements. **P<0.01. miR-155, microRNA-155; ASO-miR, antisense oligonucleotides against miR-155; ASO-NC, negative control antisense oligonucleotides; IC50, half-maximal inhibitory concentration.
miR-155 depletion affects the expression of drug resistance-associated genes in A549/dox cells
It has previously been indicated that miR-155 is an inducer of doxorubicin resistance (22). However, the molecular mechanism underlying this effect remains unclear. Adenosine triphosphate (ATP)-binding cassette (ABC) transporters, also termed ATP-dependent transporters, have been implicated in drug resistance (23). To investigate whether the role of miR-155 in drug resistance is mediated by ABC transporters, the expression of MDR1, MRP1 and BCRP was examined in A549/dox cells transfected with ASO-miR or ASO-NC. Western blot analysis revealed that miR-155 depletion decreased all three ABC transporters in A549/dox cells (Fig. 3A). RT-qPCR confirmed these results (Fig. 3B). GST-π is a drug metabolism protein that is associated with cancer drug resistance. Western blot analysis and RT-qPCR revealed that miR-155 knockdown suppressed GST-π expression in A549/dox cells. These results suggest that miR-155 induces doxorubicin resistance through drug transportation and drug metabolism.
Figure 3.
miR-155 depletion affects drug resistance-associated genes expression in doxorubicin-resistant lung cancer A549/dox cells. (A and B) mRNA and protein expression of drug resistance-associated genes in A549/dox cells transfected with ASO-miR or ASO-NC. (C and D) mRNA and protein expression of apoptosis-associated genes in A549/dox cells transfected with ASO-miR or ASO-NC. The error bars represent the standard deviation from three independent measurements. *P<0.05 and **P<0.01.miR-155, microRNA-155; ASO-miR, antisense oligonucleotides against miR-155; ASO-NC, negative control antisense oligonucleotides; IC50, half-maximal inhibitory concentration; Bcl-2, B-cell lymphoma 2; MDR1, multi-drug resistance protein 1; MRP1, multidrug resistance-associated protein 1; BCRP, breast cancer resistance protein; GST-π, glutathione S-transferase-π.
Since miR-155 regulates doxorubicin-induced apoptosis, the regulation of apoptosis-associated genes by miR-155 was investigated. Bcl-2 and Survivin belong to the Bcl-2 family, which promotes cell survival. A549/dox cells transfected with ASO-miR demonstrated increased Bcl-2 and Survivin expression compared with cells transfected with ASO-NC (Fig. 3C). The expression of caspases 3 and 8 was then examined. It was found that miR-155 depletion upregulates the expression of caspases 3 and 8 (Fig. 3C). RT-qPCR analysis confirmed the aforementioned results (Fig. 3D). These findings indicate that miR-155 participates in the regulation of apoptosis.
miR-155 depletion inhibits the AKT and ERK pathway in A549/dox cells
The promotion of doxorubicin resistance by miR-155 was investigated by activating signal transduction pathways in A549/dox cells. The AKT and ERK pathways each demonstrate aberrant activation in lung cancer (24). The present study found that, subsequent to miR-155 inhibition, p-AKT and p-ERK were downregulated in A549/dox cells (Fig. 4A). Furthermore, the transcription activities of NF-κB and AP-1 were also determined. As shown in Fig. 4B, miR-155 depletion suppressed the transcription activities of NF-κB and AP-1 in A549/dox cells.
Figure 4.
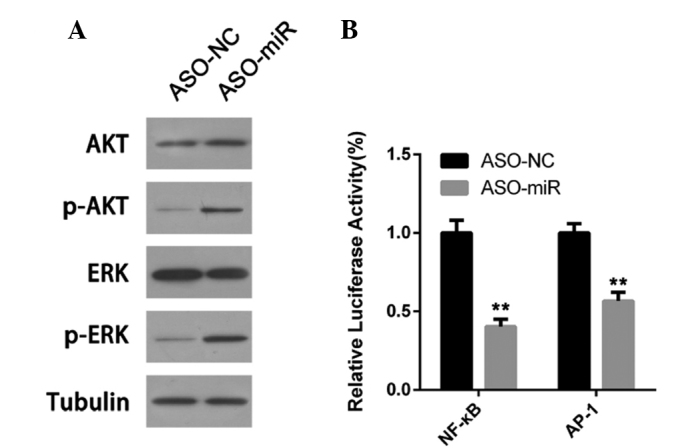
miR-155 depletion activates the AKT and ERK pathway in doxorubicin-resistant lung cancer A549/dox cells. (A) AKT and ERK activation was determined by western blot analysis. (B) The transcription activity of NF-κB and AP-1 was measured by a luciferase assay. Error bars represent the standard deviation of three independent measurements. **P<0.01. miR-155, microRNA-155; ASO-miR, antisense oligonucleotides against miR-155; ASO-NC, negative control antisense oligonucleotides; NF-κB, nuclear factor-κB; AP-1, activator protein-1; p-AKT, phosphorylated AKT; ERK, extracellular signal-regulated kinase; p-ERK, phosphorylated ERK.
Discussion
NSCLC is the leading cause of cancer-associated mortality in the USA. In 2014, the estimated numbers of novel cancer cases and estimated mortalities due to NSCLC in the USA were 224,210 and 159,260, respectively (5). Doxorubicin has previously been used in NSCLC treatment (25). However, acquired drug resistance has been widely observed. A primary method through which tumor cells develop drug resistance to chemotherapeutic drugs is by increasing drug efflux and reducing drug absorption (26). In the ABC family, MDR1, MRP1 and BCRP are able to pump drugs out of cells and are often overexpressed (27). However, the regulation of this family remains unclear. In the present study, silencing miR-155 downregulated the levels of the MDR1, MRP1 and BCRP proteins and the level of mRNA expression. These results indicate that miR-155 is a novel ABC family regulator and provide a novel therapeutic target for the ABC family. GST-π is a member of the GST family that is able to catalyze the binding of hydrophobic and electrophilic compounds, including cisplatin, with reduced glutathione, causing the former to lose toxicity (28). This represents an additional mechanism of tumor cell resistance to chemotherapeutic drugs (29,30). The current study revealed that the GST-π expression level decreased significantly in A549/dox cells following miR-155 silencing, indicating the involvement of GST-π in miR-155-mediated drug resistance.
Survivin is an inhibitive gene of apoptosis that is only expressed in tumor and embryonic tissues (31). Survivin inhibits the apoptosis of tumor cells and enhances proliferation and angiogenesis, presenting a potential target for cancer therapy (32,33). Bcl-2 is another protein that exerts key actions in apoptosis and is able to inhibit tumor cell apoptosis. The overexpression of Bcl-2 in numerous tumors is associated with tumor drug resistance (34). In the present study, the silencing of miR-155 downregulated the expression of Survivin and Bcl-2 in A549/dox cells, which was consistent with the increase in the apoptosis rate of tumor cells due to miR-155 silencing. Caspases 3 and 8 are proteins that promote tumor apoptosis (35). In contrast to Survivin and Bcl-2, the expression of caspases 3 and 8 in the present study increased accordingly subsequent to the silencing of miR-155. Overall, these results suggest that tumor cell apoptosis is partly mediated by miR-155.
The majority of studies have reported that miR-155 promotes the proliferation of tumor cells, but silencing miR-155 arrests the tumor cell cycle (36). For example, the pancreatic cancer cell cycle was arrested in the G0/G1 phase (37). In the present study, silencing miR-155 resulted in a significantly increased G0/G1 phase ratio in A549/dox cells, which prevented cell proliferation to a certain extent. However, the effect of miR-155 silencing on the reversal of drug resistance in these cells was less evident, and the effect was associated with the mechanism of the effect of doxorubicin. Since Topo II is the main target of doxorubicin and also plays an important role in the S and G2/M phases (38), the cells in these phases may be more sensitive to doxorubicin.
The regulation of the tumor cell cycle and apoptosis mediated by miR-155 may be accomplished by regulating the two key signal transduction pathways, phosphoinositide 3-kinase (PI3K)/AKT and mitogen-activated protein kinase (MAPK)/ERK (39). Certain studies have already revealed that miR-155 enhances the activation of the PI3K/AKT and MAPK/ERK signaling pathways in prostate cancer DU145 cells, and also stimulated tumor cells to secrete angiogenic factors (40). This effect may be associated with the actions of miR-155, which targets phosphatase and tensin homolog (PTEN) to remove the inhibition of these two signaling pathways (41). The present study observed that the phosphorylation levels of AKT and ERK representative of signaling pathway activation decreased subsequent to miR-155 silencing, but it has yet to be determined whether the mechanism involves the regulation of PTEN expression. However, it has been revealed that the role of miR-155 in tumors involves multiple aspects and miR-155 interacts with the intracellular transcription factors regulating gene transcription (42,43). The present results demonstrated the ability of miR-155 to positively regulate the activity of NF-κB and AP-1. Although the specific mechanisms have yet to be elucidated in additional studies, this type of regulation may be an important pathway through which silencing miR-155 reverses drug resistance in A549/dox cells.
References
- 1.Niu X, Liu S, Jia L, Chen J. Role of MiR-3619-5p in β-catenin-mediated non-small cell lung cancer growth and invasion. Cell Physiol Biochem. 2015;4:1527–1536. doi: 10.1159/000438520. [DOI] [PubMed] [Google Scholar]
- 2.Won YK, Lee JY, Kang YN, Jang JS, Kang JH, Jung SL, Sung SY, Jo IY, Park HH, Lee DS, et al. Stereotactic radiosurgery for brain metastasis in non-small cell lung cancer. Radiat Oncol J. 2015;3:207–216. doi: 10.3857/roj.2015.33.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 4.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer, Epidemiology, risk factors, treatment and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 6.Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer-is it becoming a reality? Nat Rev Clin Oncol. 2010;7:401–414. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Müller I, Niethammer D, Bruchelt G. Anthracycline-derived chemotherapeutics in apoptosis and free radical cytotoxicity (Review) Int J Mol Med. 1998;1:491–494. doi: 10.3892/ijmm.1.2.491. [DOI] [PubMed] [Google Scholar]
- 9.Zhu T, Li LL, Xiao GF, Luo QZ, Liu QZ, Yao KT, Xiao GH. Berberine increases doxorubicin sensitivity by suppressing STAT3 in lung cancer. Am J Chin Med. 2015;27:1–16. doi: 10.1142/S0192415X15500846. [DOI] [PubMed] [Google Scholar]
- 10.Li ZL, Chen C, Yang Y, Wang C, Yang T, Yang X, Liu SC. Gamma secretase inhibitor enhances sensitivity to doxorubicin in MDA-MB-231 cells. Int J Clin Exp Pathol. 2015;5:4378–4387. [PMC free article] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Y, Yao YF, Hu SN, Gao J, Zhang LL. MiR-133a is functionally involved in doxorubicin-resistance in breast cancer cells MCF-7 via its regulation of the expression of uncoupling protein 2. PLoS One. 2015;6:e0129843. doi: 10.1371/journal.pone.0129843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandellini P, Profumo V, Folini M, Zaffaroni N. MicroRNAs as new therapeutic targets and tools in cancer. Expert Opin Ther Targets. 2011;15:265–279. doi: 10.1517/14728222.2011.550878. [DOI] [PubMed] [Google Scholar]
- 16.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27((Suppl 2)):S52–S57. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Zhou J, Zhang Y, Wang Y, Cheng L, Bai Y, Ma H. The value of microRNA-155 as a prognostic factor for survival in non-small cell lung cancer, A meta-analysis. PLoS One. 2015;8:e0136889. doi: 10.1371/journal.pone.0136889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, Pospisil V, Stopka T. Oncogenic microRNAs, MiR-155, miR-19a, miR-181b and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer. 2014;14:448. doi: 10.1186/1471-2407-14-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Z, Che S, Wang J, Jiao Y, Wang C, Meng Q. miR-155 contributes to the progression of glioma by enhancing Wnt/β-catenin pathway. Tumour Biol. 2015;7:5323–5331. doi: 10.1007/s13277-015-3193-9. [DOI] [PubMed] [Google Scholar]
- 20.Hongliang C, Shaojun H, Aihua L, Hua J. Correlation between expression of miR-155 in colon cancer and serum carcinoembryonic antigen level and its contribution to recurrence and metastasis forecast. Saudi Med J. 2014;35:547–553. [PubMed] [Google Scholar]
- 21.Li T, Yang J, Lv X, Liu K, Gao C, Xing Y, Xi T. MiR-155 regulates the proliferation and cell cycle of colorectal carcinoma cells by targeting E2F2. Biotechnol Lett. 2014;36:1743–1752. doi: 10.1007/s10529-014-1540-3. [DOI] [PubMed] [Google Scholar]
- 22.Merhautova J, Hezova R, Poprach A, Kovarikova A, Radova L, Svoboda M, Vyzula R, Demlova R, Slaby O. miR-155 and miR-484 are associated with time to progression in metastatic renal cell carcinoma treated with sunitinib. Biomed Res Int. 2015;2015:941980. doi: 10.1155/2015/941980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Liu XJ, Yang P, Zhao M, Lv LX, Zhang GD, Wang Q, Zhang L. Alkylglyceronephosphate synthase (AGPS) alters lipid signaling pathways and supports chemotherapy resistance of glioma and hepatic carcinoma cell lines. Asian Pac J Cancer Prev. 2014;15:3219–3226. doi: 10.7314/APJCP.2014.15.7.3219. [DOI] [PubMed] [Google Scholar]
- 24.Fruman DA, Rommel C. PI3K and cancer: Lessons challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendijani F. JavanmardS hH, Rafiee L and Sadeghi-Aliabadi H: Effect of human Wharton's jelly mesenchymal stem cell secretome on proliferation, apoptosis and drug resistance of lung cancer cells. Res Pharm Sci. 2015;2:134–142. [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao YY, Yu L, Liu BL, He XJ, Zhang BY. Downregulation of P-gp, Ras and p-ERK1/2 contributes to the arsenic trioxide-induced reduction in drug resistance towards doxorubicin in gastric cancer cell lines. Mol Med Rep. 2015;5:7335–7343. doi: 10.3892/mmr.2015.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keppler D. Multidrug resistance proteins (MRPs, ABCC s): Importance for pathophysiology and drug therapy. Handb Exp Pharmacol. 2011:299–323. doi: 10.1007/978-3-642-14541-4_8. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Jiao JW, Sun KX, Zong ZH, Zhao Y. MicroRNA-133b targets glutathione S-transferase π expression to increase ovarian cancer cell sensitivity to chemotherapy drugs. Drug Des Devel Ther. 2015;9:5225–5535. doi: 10.2147/DDDT.S87526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surowiak P, Materna V, Kaplenko I, Spaczyński M, Dietel M, Lage H, Zabel M. Augmented expression of metallothionein and glutathione S-transferase pi as unfavourable prognostic factors in cisplatin-treated ovarian cancer patients. Virchows Arch. 2005;447:626–633. doi: 10.1007/s00428-005-1228-0. [DOI] [PubMed] [Google Scholar]
- 30.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 31.de Moraes Nestal G, Delbue D, Silva KL, Robaina MC, Khongkow P, Gomes AR, Zona S, Crocamo S, Mencalha AL, Magalhães LM, Lam EW, Maia RC. FOXM1 targets XIAP and Survivin to modulate breast cancer survival and chemoresistance. Cell Signal. 2015;12:2496–2505. doi: 10.1016/j.cellsig.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Ryan BM, O'Donovan N, Duffy MJ. Survivin A new target for anti-cancer therapy. Cancer Treat Rev. 2009;35:553–562. doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Altieri DC. Survivin and IAP proteins in cell-death mechanisms. Biochem J. 2010;430:199–205. doi: 10.1042/BJ20100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yip KW, Reed JC. Bcl-2 family proteins and cancer. Oncogene. 2008;27:6398–6406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 35.Ghavami S, Hashemi M, Ande SR, Yeganeh B, Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ, Los M. Apoptosis and cancer, Mutations within caspase genes. J Med Genet. 2009;46:497–510. doi: 10.1136/jmg.2009.066944. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty C. GeorgeP riya Doss C and Bandyopadhyay S: miRNAs in insulin resistance and diabetes-associated pancreatic cancer: The 'minute and miracle' molecule moving as a monitor in the 'genomic galaxy'. Curr Drug Targets. 2013;10:1110–1117. doi: 10.2174/13894501113149990182. [DOI] [PubMed] [Google Scholar]
- 37.Zhang E, Wu Y. Dual effects of miR-155 on macrophages at different stages of atherosclerosis: LDL is the key? Med Hypotheses. 2014;83:74–78. doi: 10.1016/j.mehy.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;23:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Cai ZK, Chen Q, Chen YB, Gu M, Zheng DC, Zhou J, Wang Z. microRNA-155 promotes the proliferation of prostate cancer cells by targeting annexin 7. Mol Med Rep. 2015;1:533–538. doi: 10.3892/mmr.2014.2744. [DOI] [PubMed] [Google Scholar]
- 41.Xue P, Zheng M, Diao Z, Shen L, Liu M, Gong P, Sun H, Hu Y. miR-155* mediates suppressive effect of PTEN 3′-untranslated region on AP-1/NF-κB pathway in HTR-8/SVneo cells. Placenta. 2013;8:650–656. doi: 10.1016/j.placenta.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Farooqi AA, Qureshi MZ, Coskunpinar E, Naqvi SK, Yaylim I, Ismail M. MiR-421, miR-155 and miR-650: Emerging trends of regulation of cancer and apoptosis. Asian Pac J Cancer Prev. 2014;15:1909–1912. doi: 10.7314/APJCP.2014.15.5.1909. [DOI] [PubMed] [Google Scholar]
- 43.Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, Carasi S, Palmieri D, Costinean S, Shapiro CL, et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci USA. 2014;111:4536–4541. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]