Abstract
BACKGROUND
The prevalence and spectrum of predisposing mutations among children and adolescents with cancer are largely unknown. Knowledge of such mutations may improve the understanding of tumorigenesis, direct patient care, and enable genetic counseling of patients and families.
METHODS
In 1120 patients younger than 20 years of age, we sequenced the whole genomes (in 595 patients), whole exomes (in 456), or both (in 69). We analyzed the DNA sequences of 565 genes, including 60 that have been associated with autosomal dominant cancer-predisposition syndromes, for the presence of germline mutations. The pathogenicity of the mutations was determined by a panel of medical experts with the use of cancer-specific and locus-specific genetic databases, the medical literature, computational predictions, and second hits identified in the tumor genome. The same approach was used to analyze data from 966 persons who did not have known cancer in the 1000 Genomes Project, and a similar approach was used to analyze data from an autism study (from 515 persons with autism and 208 persons without autism).
RESULTS
Mutations that were deemed to be pathogenic or probably pathogenic were identified in 95 patients with cancer (8.5%), as compared with 1.1% of the persons in the 1000 Genomes Project and 0.6% of the participants in the autism study. The most commonly mutated genes in the affected patients were TP53 (in 50 patients), APC (in 6), BRCA2 (in 6), NF1 (in 4), PMS2 (in 4), RB1 (in 3), and RUNX1 (in 3). A total of 18 additional patients had protein-truncating mutations in tumor-suppressor genes. Of the 58 patients with a predisposing mutation and available information on family history, 23 (40%) had a family history of cancer.
CONCLUSIONS
Germline mutations in cancer-predisposing genes were identified in 8.5% of the children and adolescents with cancer. Family history did not predict the presence of an underlying predisposition syndrome in most patients. (Funded by the American Lebanese Syrian Associated Charities and the National Cancer Institute.)
The frequency of germline mutations in cancer-predisposition genes in children and adolescents with cancer and the implications of such mutations are largely unknown. Previous studies have relied mainly on candidate-gene approaches, which are, by design, limited. To better determine the contribution of germline predisposition mutations to childhood cancer, we used next-generation sequencing, including whole-genome and whole-exome sequencing, to analyze the genomes of 1120 children and adolescents with cancer. We describe the prevalence and spectrum of germline variants among 565 cancer-associated genes, with an emphasis on the analysis of 60 genes that have been associated with autosomal dominant cancer-predisposition syndromes. We also reviewed records of patients with mutations and those without mutations in these 60 genes for information on family history of cancer.
METHODS
ENROLLMENT OF THE PATIENTS
The 1120 patients included in this study represented the major subtypes of pediatric cancer (Fig. 1; and Table S1 in Supplementary Appendix 1, available with the full text of this article at NEJM.org). Whole-genome, whole-exome, or both types of sequencing data were generated with the use of germline DNA for 595, 456, and 69 patients, respectively, as part of the St. Jude–Washington University Pediatric Cancer Genome Project (PCGP; www.ebi.ac.uk/ega/search/site/PCGP). To verify predictions of aberrant splicing caused by variants affecting splice junctions, we sequenced the RNA transcripts extracted from 522 samples of tumor tissue obtained from 522 patients. The study was approved by the institutional review board at St. Jude Children’s Research Hospital. Written informed consent was provided by a parent or guardian of each child or by a patient who was 18 years of age or older.
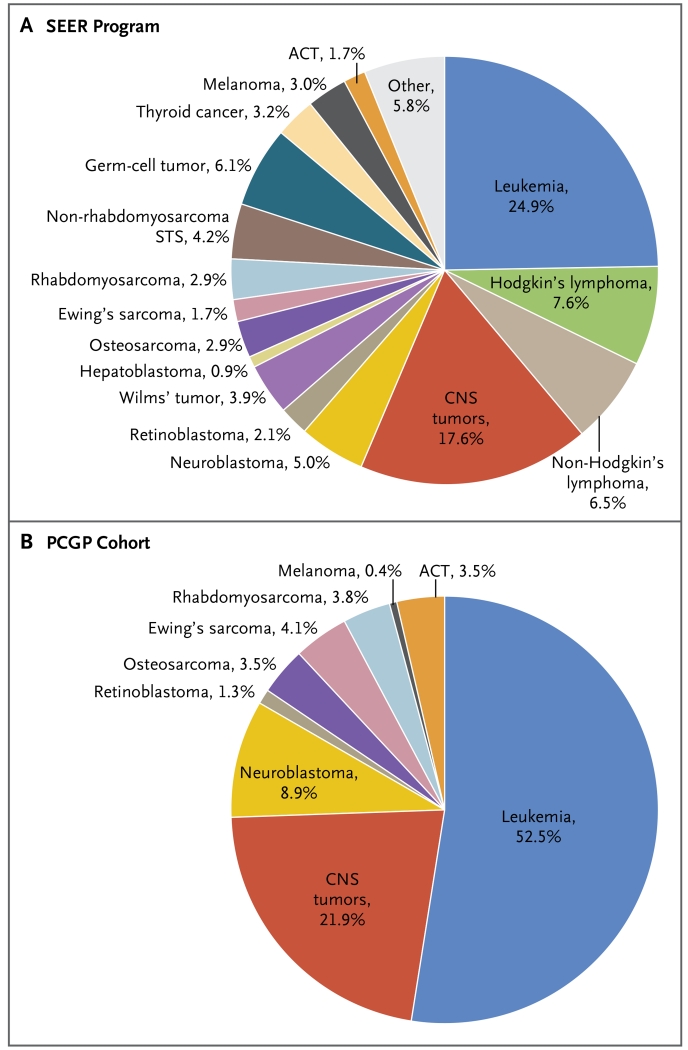
Figure 1. Frequency of Pediatric Cancer Types among Patients Younger than 20 Years of Age.
Panel A shows the distribution of pediatric cancer types on the basis of data from the Surveillance, Epidemiology, and End Results (SEER) program. Panel B shows the distribution of cancer types analyzed by the Pediatric Cancer Genome Project (PCGP). ACT denotes adrenocortical tumor, CNS central nervous system, and STS soft-tissue sarcoma.
Whole-exome sequencing data from two control cohorts of persons without known cancer were analyzed. The first data set, a raw whole-exome sequencing data set from 966 unrelated adults who were part of the 1000 Genomes Project (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3), was analyzed by the same approach that was used in our pediatric cancer cohort. The second data set consisted of genotype files of 515 persons with autism and 208 persons without autism (median age, 6 years; range, 1 to 37) from the National Database for Autism Research (https://ndar.nih.gov/study.html?id=307). Analyses of this second data set did not involve variant detection owing to a lack of access to raw sequence data, and we excluded two cancer-predisposition genes, NF1 and PTEN, which are known to be associated with an autism spectrum phenotype (Supplementary Appendix 1).
CANCER-PREDISPOSITION GENES SELECTED FOR ANALYSIS
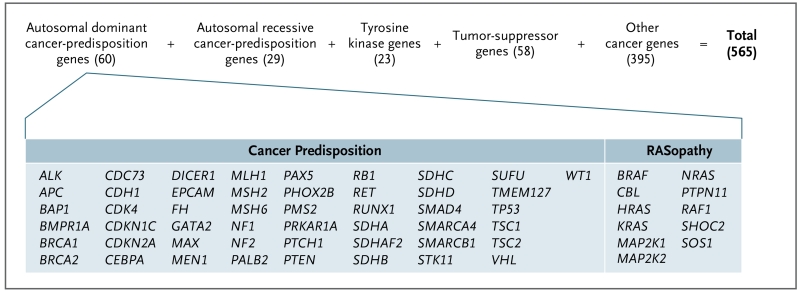
A total of 565 genes were chosen for analysis on the basis of review of the American College of Medical Genetics and Genomics (ACMG) gene list and the medical literature1-4 and were divided into five nonoverlapping categories (Fig. 2; and Table S3 in Supplementary Appendix 1 and Table S2 in Supplementary Appendix 2, available at NEJM.org). The first category included genes that have been associated with autosomal dominant cancer-predisposition syndromes, and it consisted of 49 classical genes (including 23 genes from the ACMG gene list5) and 11 genes that have been implicated in genetic syndromes associated with RAS mutations (sometimes called RASopathies; these include the cardiofaciocutaneous syndrome, Costello’s syndrome [also called the faciocutaneoskeletal syndrome], Noonan’s syndrome, and the multiple lentigines syndrome). These 60 genes were selected because of the potential effect of germline mutations on clinical practice, including avoidance of radiation therapy, choice of surgical approach for tumor resection, donor testing and selection for hematopoietic stem-cell transplantation, possible or proven benefits of tumor surveillance and early cancer detection, and risk-reductive surgery.3 Variants detected in these 60 genes were confirmed experimentally by an independent sequencing assay (Supplementary Appendix 1).
Figure 2. Categories of the 565 Cancer Genes Analyzed for Germline Mutations.
The number of genes in each category is shown in parentheses. Genes that have overlapping categories are listed only once. Gene names in the other categories are shown in Figure S9 in Supplementary Appendix 1. RASopathies are genetic syndromes that include the cardiofaciocutaneous syndrome, Costello’s syndrome (also called the faciocutaneoskeletal syndrome), Noonan’s syndrome, and the multiple lentigines syndrome.
The second category included 29 genes that have been associated with autosomal recessive cancer-predisposition syndromes, with a focus on identifying biallelic pathogenic mutations. Variants detected in the 89 genes that have been associated with autosomal dominant or autosomal recessive cancer-predisposition syndromes were reviewed by a multidisciplinary panel for classification and reporting.5-8
An additional 476 genes were chosen for evaluation on the basis of their recurrent somatic mutation in cancer (http://cancer.sanger.ac.uk/cancergenome/projects/census and www.pediatriccancergenomeproject.org/site). These genes were classified into three categories: tumor-suppressor genes (58 genes),9 tyrosine kinase genes (23), and other cancer genes (395). Our analyses of the genes in these three categories focused on known hotspot-activating mutations in genes encoding kinases and on truncation mutations in genes encoding tumor-suppressor proteins and in other cancer genes.
DATA ANALYSIS
The sequencing data were analyzed for the presence of single-nucleotide variants and small insertions and deletions10 and for evidence of germline mosaicism (Supplementary Appendix 1). Germline copy-number variations and structural variations were identified with the use of the Copy Number Segmentation by Regression Tree in Next Generation Sequencing (CONSERTING)11 and Clipping Reveals Structure (CREST)12 algo-rithms. Common germline structural variations and structural variations that did not affect coding exons were excluded from the analysis.
Nonsilent coding variants that passed quality-control and minor-allele population frequency checks were classified as pathogenic, probably pathogenic, of uncertain significance, probably benign, or benign. Classification criteria included information from curated databases, computational predictions of mutational effect on protein function, and recent ACMG guidelines for interpretation.13 Full details of the data analysis and interpretation are provided in Figure S1 in Supplementary Appendix 1.
RESULTS
CHARACTERISTICS OF THE PATIENTS
The PCGP cohort included 588 children and adolescents with leukemia (52.5%), 245 with central nervous system (CNS) tumors (21.9%), and 287 with non-CNS solid tumors (25.6%) (Fig. 1, and Table S1 in Supplementary Appendix 1). The median age of the patients was 6.9 years (range, 8 days to 19.7 years). The cancers that were selected for sequencing included those that have been associated with a poor clinical outcome (e.g., hypodiploid leukemia)14 and those without a clearly defined oncogenic cause (e.g., diffuse intrinsic pontine glioma).15 Our cohort included more patients with leukemia and adrenocortical tumors than was expected on the basis of the population in the Surveillance, Epidemiology, and End Results (SEER) program (http://seer.cancer.gov/iccc) (Fig. 1). Lymphoma, Wilms’ tumor, germ-cell tumors, non-rhabdomyosarcoma soft-tissue sarcoma, and hepatoblastoma were not included because of an inadequate number of samples for high-risk subtypes.
GERMLINE MUTATIONS IN CANCER-PREDISPOSITION GENES
In the 60 genes that have been associated with autosomal dominant cancer-predisposition syndromes, we identified 633 nonsilent germline variants, of which 78 (12%) were deemed to be pathogenic, 17 (3%) probably pathogenic, 226 (36%) of uncertain significance, 273 (43%) probably benign, and 39 (6%) benign (Table S4A in Supplementary Appendix 2). The 95 variants that were deemed to be pathogenic or probably pathogenic included 54 missense mutations, 14 nonsense mutations, 12 frameshift mutations, 9 splice-site mutations, and 1 in-frame deletion, as well as 5 copy-number alterations (Fig. S2 in Supplementary Appendix 1).
The 95 variants that were deemed to be pathogenic or probably pathogenic were detected in 21 of the 60 genes in 94 patients (Fig. 3A, and Fig. S3 in Supplementary Appendix 1). TP53 was most commonly involved (in 50 patients), followed by APC (in 6), BRCA2 (in 6), NF1 (in 4), PMS2 (in 4), RB1 (in 3), and RUNX1 (in 3). One patient (Patient HGG111) with café au lait spots and a high-grade glioma had 2 distinct PMS2 truncation mutations, which indicated a diagnosis of biallelic mismatch-repair deficiency that was corroborated by the somatic hypermutation observed in the genome of the high-grade glioma.15 The most common cancer types that were associated with germline TP53 mutations included adrenocortical tumors (in 27 of 39 patients [69%]), hypodiploid acute lymphoblastic leukemia (in 9 of 47 [19%]), and choroid plexus carcinoma (in 1 of 4 [25%]) — findings that were consistent with those in previous reports.14 As anticipated, the tumors from all 37 of these patients had a loss of heterozygosity at the TP53 locus (Table S4 in Supplementary Appendix 2), including 1 patient who had a germline deletion of 8.7 kb that removed TP53 exons 2 through 5 (Fig. S2 in Supplementary Appendix 1).
Figure 3. Distribution of Germline Mutations in Different Gene Categories and Cancer Subtypes.
Panels A and B include only mutations that were deemed to be pathogenic or probably pathogenic and that affect genes that have been associated with autosomal dominant cancer-predisposition syndromes, according to tumor subtype. Panel A shows the distribution of mutations in each gene among patients with various cancers included in the PCGP cohort. Panel B shows the prevalence of the mutations in each cancer subtype. Five patients with melanoma without mutations are not shown, and one patient (HGG027) who had a CNS tumor with biallelic mutation in an autosomal recessive gene (ATM) is not included in the summary. Panel C shows the number of patients who had germline mutations considered to be pathogenic or probably pathogenic in genes that have been associated with autosomal dominant (60 genes) and autosomal recessive (29) cancer susceptibility, according to cancer subtype. Panel D shows the total number of patients who had truncation mutations in tumor-suppressor genes, tyrosine kinase genes, and other cancer genes, according to cancer subtype.
Four germline mutations were mosaic, with the detected level of the mutant allele less than a single copy (mutant allele fraction, 0.08 to 0.30). One patient with retinoblastoma had a mosaic RB1 mutation, and three patients with hypodiploid acute lymphoblastic leukemia had a mosaic TP53 mutation (Table S4A in Supplementary Appendix 2). The mutant allele fraction in matching tumor specimens ranged from 0.76 to 0.90, a finding that is consistent with the presence of a second hit within the tumors. Validation by means of deep sequencing at more than 2000× coverage verified the mutant allele fraction within the germline and tumor samples in each patient (Fig. 4).
Figure 4. Distinguishing Mosaicism from Tumor Contamination.
Panel A shows that in the tumor-contaminated germline sample of Patient 1 (E2A019), most somatic mutations were observed at a lower frequency in the germline than in the tumor. Nine genes were selected to show this point. Panel B shows that in the case of mosaicism in Patient 2 (HYPO055), only one TP53 mutation was observed at a lower frequency in the germline than in the tumor. Other somatic mutations in the tumor were absent in the matched germline sample. Panel C shows that MiSeq sequencing confirmed that the mutant allele fraction (MAF) of TP53 c.C374G (coding for p.T125R) in the germline sample of Patient 2 was still low (0.20; only 487 reads of 2383 reads had the mutation), a finding that is consistent with germline mosaicism. Two minor peaks supporting C and G alleles (arrows) were seen in the Sanger-sequencing chromatograph.
In the first control data set, from the 1000 Genomes Project, we identified 11 pathogenic or probably pathogenic mutations in the 60 genes that have been associated with autosomal dominant cancer-predisposition syndromes; mutations were found in APC (in one person), BRCA1 (in one), BRCA2 (in four), MSH6 (in one), SDHA (in one), SDHB (in one), and TP53 (in two) (Table S6 in Supplementary Appendix 1). The prevalence of mutations was 1.1%, which was significantly lower than the 8.4% prevalence observed in the PCGP cohort (P = 5.9 × 10−16 by Fisher’s exact test). A similar trend was observed in the second control set, which involved participants from the autism study (frequency, 0.6%; P = 7.4 × 10−16 for the comparison with the PCGP cohort) (Table S7 in Supplementary Appendix 1).
The PCGP cohort included a greater-than-expected proportion of patients with hypodiploid acute lymphoblastic leukemia and those with adrenocortical tumors (Fig. 1). However, after these two subtypes were excluded, the prevalence of germline mutations of 5.6% was still significantly higher than the prevalence in the two control cohorts (P<10−7 by Fisher’s exact test for both comparisons).
In our analysis of 29 autosomal recessive cancer-predisposition genes, we observed only one instance of biallelic pathogenic mutations in 1 patient (Table S8 in Supplementary Appendix 1). Combining data from this single patient, who had ataxia telangiectasia caused by biallelic mutations in ATM (Fig. S4 in Supplementary Appendix 1), with data from the 94 patients who had pathogenic mutations in the 60 autosomal dominant cancer-predisposition genes, we observed an 8.5% prevalence (95 of 1120 patients) of germline mutations that were pathogenic or probably pathogenic in the sample we analyzed. A total of 61 of the 93 patients (66%) with monoallelic germline mutations had a second hit within the tumor genome (Table S4 in Supplementary Appendix 2), as shown by loss of heterozygosity (in 57 patients) or mutational inactivation of the second allele (in 4). These data are available on our pediatric cancer data portal (http://pecan.stjude.org) (Figs. S5 and S6 in Supplementary Appendix 1)
PREVALENCE OF GERMLINE MUTATIONS ACROSS TUMOR TYPES
The prevalence of germline mutations that were pathogenic or probably pathogenic was greatest among patients with non-CNS solid tumors (48 of 287 patients [16.7%]), followed by those with CNS tumors (21 of 245 [8.6%], including the patient with biallelic loss of ATM) or leukemia (26 of 588 [4.4%]) (Fig. 3B). The prevalence of germline mutations varied among patients with different subtypes of non-CNS solid tumors, such as adrenocortical tumor (69.2%), osteosarcoma (17.9%), retinoblastoma (13.3%), Ewing’s sarcoma (10.9%), rhabdomyosarcoma (7.0%), and neuroblastoma (4.0%) (Fig. 3B). The histologic subtypes of CNS tumor that were most often associated with germline mutations included choroid plexus carcinoma (in 1 of 4 patients [25%]), medulloblastoma (in 5 of 37 [13.5%]), high-grade glioma (in 9 of 99 [9.1%]), low-grade glioma (in 3 of 38 [7.9%]), and ependymoma (in 4 of 67 [6.0%]). Overall, patients with leukemia had the lowest prevalence of germline mutations (4.4%), despite the inclusion of patients with hypodiploid acute lymphoblastic leukemia, a subtype with a high frequency of germline mutation.14
CORRELATION BETWEEN GERMLINE GENOTYPE AND TUMOR PHENOTYPE
The correlation of patient genotype with tumor phenotype revealed several known associations as well as some new ones. The known associations included the association of TP53 mutations with classic Li–Fraumeni syndrome–associated component cancers such as rhabdomyosarcomas, osteosarcomas, adrenocortical tumors, CNS tumors, and leukemia; NF1 mutations with CNS tumors; RB1 mutations with retinoblastoma and osteosarcoma; and ALK mutations with neuroblastoma (Fig. 3A). New associations included the association of germline TP53, PMS2, and RET mutations with Ewing’s sarcoma; APC and SDHB mutations with neuroblastoma; and a variety of mutations (APC, VHL, CDH1, PTCH1, or SDHA) with leukemia.
A total of eight children had germline mutations in the adult-onset cancer–predisposition genes BRCA1, BRCA2, and PALB2. The spectrum of cancers observed in these children included leukemia, CNS tumors, neuroblastoma, osteosarcoma, and rhabdomyosarcoma. Although biallelic mutations of BRCA1/2 and PALB2 are known to cause Fanconi’s anemia,16-19 there were no germline mutations or deletions involving the second alleles of these genes in any of the affected patients.
MEDICAL AND FAMILY HISTORY
Medical records were available for review for 75 of the 95 patients with mutations that were deemed to be pathogenic or probably pathogenic. The records showed that only 12 patients had undergone clinical genetic testing previously. Clinical testing did not identify the predisposing genetic lesions in 2 patients. Of these 2 patients, 1 had an adrenocortical tumor tested for TP53 (TP53 p.I332F in Patient ACT001) and 1 had retinoblastoma that was tested for RB1 (mosaic RB1 p.R445* in Patient RB002); both lesions were identified by means of the next-generation sequencing approaches used in this study.
A total of 58 of the 75 records (77%) contained information regarding family history, and only 23 of 58 records (40%) indicated a family history of cancer (defined here as one or more first- or second-degree relatives with cancer) (Fig. S7 in Supplementary Appendix 1). Furthermore, among these 23 patients, only 13 (57%) had a history that was consistent with the underlying genetic syndrome, including 8 patients with TP53 mutations (and thus the Li–Fraumeni syndrome), 2 with APC mutations (familial adenomatous polyposis), 2 with BRCA2 mutations (hereditary breast and ovarian cancer; the pedigrees are shown in Fig. S8 in Supplementary Appendix 1), and 1 with PMS2 mutations (hereditary nonpolyposis colorectal cancer, also known as the Lynch syndrome). The 8 patients with the Li–Fraumeni syndrome all met the revised Chompret criteria regarding family history.20
We completed a similar analysis of a comparison cohort of 100 randomly selected patients who did not have germline mutations in the 60 autosomal dominant cancer-predisposition genes. We observed that the percentage of patients with a family history of cancer (42%; 18 of 43 records with family-history information) was similar to that observed among persons with germline mutations (40%; 23 of 58 records).
GERMLINE MUTATIONS IN OTHER CANCER-ASSOCIATED GENES
We identified 4348 nonsilent coding mutations in the remaining 476 genes that were analyzed. These included 114 heterozygous truncation mutations, in 109 patients, that involved tumor-suppressor genes, tyrosine kinase genes, or other cancer genes (Fig. 3D, and Table S5 in Supplementary Appendix 1 and Table S4 in Supplementary Appendix 2). The most commonly affected tumor-suppressor genes included CHEK2 (in 4 patients), PML (in 4), and BUB1B (in 3). A total of 18 patients who did not have pathogenic mutations in genes that have been associated with cancer-predisposition syndromes had protein-truncating mutations in tumor-suppressor genes. Two known hotspots of somatic activating mutations in EGFR, T790 and H773, were identified once each in the germline of 2 patients with leukemia (Fig. S6 in Supplementary Appendix 1).
DISCUSSION
In this study involving 1120 children and adolescents with cancer, we found that 8.5% of the patients had predisposing gene mutations. Sequence coverage exceeded 10× for more than 95% of the coding exons and 20× for more than 85% of the coding exons in the genes of interest (Table S3 in Supplementary Appendix 1), which was sufficient for genotype accuracy.21 However, the prevalence may be underestimated. First, we included mutations that were pathogenic or probably pathogenic in 60 genes that have been associated with clinically relevant autosomal dominant cancer-predisposition syndromes, and we did not include other genes that, when mutated, may contribute toward a patient’s susceptibility to cancer. In this regard, we observed that an additional 38 patients (3.4%) had heterozygous mutations that were pathogenic or probably pathogenic in 29 genes that are known to be associated with autosomal recessive cancer-predisposition syndromes (Table S4 in Supplementary Appendix 2). Moreover, 109 children (9.7%) had germline-truncating mutations in other cancer-associated genes, although non-hotspot missense mutations in these genes were not fully characterized, some of which may eventually be considered to be cancer-susceptibility genes.
Second, among the 226 variants of uncertain significance that were identified in the 60 genes that have been associated with autosomal dominant cancer-predisposition syndromes, 119 (52.7%) were predicted to be deleterious by at least two computational methods, and some of these could, in fact, confer susceptibility to cancer. Third, as sequencing depth increases, additional mosaic germline mutations will be discovered. Finally, as we learn more about how certain genetic alterations (e.g., structural variations, changes in noncoding regions, and epigenetic modifications) influence cellular function, new cancer-predisposing lesions in these and other newly discovered cancer-associated genes will be identified.
We found several unexpected germline mutations in patients with Ewing’s sarcoma, neuroblastoma, osteosarcoma, or leukemia. Although Ewing’s sarcoma has been recognized as a second cancer in children who have been treated for retinoblastoma,22-25 it has not, to our knowledge, been associated with other cancer-predisposition syndromes.26 We found that six patients with Ewing’s sarcoma had pathogenic germline mutations in TP53 (in four patients), PMS2 (in one), or RET (in one), although we cannot state with certainty that each mutation had a bearing on the patient’s cancer. Additional studies are needed to determine the role, if any, that these germline mutations played in the development of Ewing’s sarcoma or these other tumors.
Eight patients had heterozygous mutations in BRCA1, BRCA2, or PALB2. These genes are not normally examined in children because they are thought to be predisposition genes for adult cancer. Magnusson et al. described a high prevalence of childhood cancer in families with germline BRCA2 mutations,27 and Brooks et al. reported 20 cases of pediatric cancer among 379 families, members of which had a mutation in either BRCA1 or BRCA2.28 These reports suggest that pathogenic mutations in BRCA1 and BRCA2 are more common in pediatric cancer than has been recognized previously and that they potentially underpin a broader spectrum of cancer phenotypes.28-38
Family history is commonly used to identify persons with a possible heritable predisposition, especially within the pediatric cancer population.39 However, only 40% of our patients with germline mutations that were pathogenic or probably pathogenic and that could be evaluated had a family history of cancer. In addition, only half of those had a family history that was consistent with a known cancer-predisposition syndrome. This low frequency probably resulted from multiple factors, including incomplete information on family history, de novo mutations, and incomplete penetrance. Furthermore, parents and other first- or second-degree relatives of our pediatric patients are often young, and cancer may not have developed yet. A review of 100 randomly selected patients who did not have germline cancer-predisposition gene mutations revealed that 42% had a family history of cancer. Conceivably, some of these patients have mutations in genes that were not analyzed in the current study. Nonetheless, on the basis of these observations, family history cannot be the sole indication used to guide the provision of genetic testing.
This study has several limitations. First, several subtypes of pediatric cancers were not examined. In addition, our cohort included greater-than-expected proportions of patients with hypodiploid acute lymphoblastic leukemia and those with adrenocortical tumors. However, after excluding these two subtypes, the prevalence of germline mutations of 5.6% was still significantly higher than the prevalence in two control cohorts (P<10−7 by Fisher’s exact test for both comparisons). Observation of pathogenic mutations in the controls may indicate uncharacterized cancer phenotypes in the people enrolled, rather than refuting the pathogenicity of these mutations in cancer predisposition (Tables S6 and S7 in Supplementary Appendix 1). Moreover, by adjusting for the distribution of cancer subtypes observed in the SEER program and by applying previously reported mutation frequencies for those not included in this study, we predicted an overall mutation prevalence of 7.3 to 9.8% in the SEER pediatric cancer population (Table S10 in Supplementary Appendix 2).
Second, we did not study the parents or relatives of the patients in our cohort and hence could not assess whether variants were new or segregated with a cancer phenotype among family members. This information could have augmented the evidence of pathogenicity in some cases. Nonetheless, the discovery of four mosaic germline mutations in TP53 and RB1 indicates that a fraction of the mutations that were identified in this study were de novo. Third, although the 60 autosomal dominant cancer-predisposition genes that were the focus of this report have been well characterized, information regarding the penetrance of many mutations that were identified within these genes is lacking. Additional family, epidemiologic, and functional studies are warranted to better understand the cancer risks associated with each of these variants.
In conclusion, germline mutations in cancer-predisposing genes were identified in 8.5% of the children and adolescents with cancer who participated in this study. Family history did not predict the presence of an underlying predisposition syndrome in most patients. The germline mutations identified in this study may provide insights into the causes of cancer. Knowledge of their presence may influence clinical management by directing cancer care, enabling presymptomatic genetic testing of relatives, guiding family-planning measures, and facilitating the institution of potentially lifesaving measures for cancer prevention and surveillance.
Supplementary Material
Acknowledgments
Supported by funding from the American Lebanese Syrian Associated Charities to St. Jude Children’s Research Hospital and by a Cancer Center Support (Core) grant (CA21765) from the National Cancer Institute to St. Jude Children’s Research Hospital.
We thank the staff of the Pediatric Cancer Genome Project Laboratory and the Molecular Pathology Laboratory at St. Jude’s Children’s Research Hospital for next-generation sequencing and subsequent validation of identified pathogenic variants.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindor NM, McMaster ML, Lindor CJ, Greene MH. Concise handbook of familial cancer susceptibility syndromes — second edition. J Natl Cancer Inst Monogr. 2008;(38):1–93. doi: 10.1093/jncimonographs/lgn001. [DOI] [PubMed] [Google Scholar]
- 3.Pizzo PA, Poplack DG, editors. Principles and practice of pediatric oncology. 6th Wolters Kluwer-Lippincott Williams & Wilkins Health; Philadelphia: 2011. [Google Scholar]
- 4.Pui C-H. Childhood leukemias. 3rd Cambridge University Press; Cambridge, United Kingdom: 2012. [Google Scholar]
- 5.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bombard Y, Offit K, Robson ME. Risks to relatives in genomic research: a duty to warn? Am J Bioeth. 2012;12:12–4. doi: 10.1080/15265161.2012.699157. [DOI] [PubMed] [Google Scholar]
- 7.Offit K, Groeger E, Turner S, Wadsworth EA, Weiser MA. The “duty to warn” a patient’s family members about hereditary disease risks. JAMA. 2004;292:1469–73. doi: 10.1001/jama.292.12.1469. [DOI] [PubMed] [Google Scholar]
- 8.Storm C, Agarwal R, Offit K. Ethical and legal implications of cancer genetic testing: do physicians have a duty to warn patients’ relatives about possible genetic risks? J Oncol Pract. 2008;4:229–30. doi: 10.1200/JOP.0858504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao M, Sun J, Zhao Z. TSGene: a Web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edmonson MN, Zhang JH, Yan CH, Finney RP, Meerzaman DM, Buetow KH. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics. 2011;27:865–6. doi: 10.1093/bioinformatics/btr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Gupta P, Wang J, et al. CONSERTING: integrating copy-number analysis with structural-variation detection. Nat Methods. 2015;12:527–30. doi: 10.1038/nmeth.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Mullighan CG, Easton J, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods. 2011;8:652–4. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–52. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brain-stem high-grade glioma. Nat Genet. 2014;46:444–50. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alter BP, Kupfer G. Fanconi anemia. University of Washington; Seattle: 2002. [PubMed] [Google Scholar]
- 17.Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 18.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–4. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 19.Sawyer SL, Tian L, Kahkonen M, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–42. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bougeard G, Sesboüé R, Baert-Desurmont S, et al. Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J Med Genet. 2008;45:535–8. doi: 10.1136/jmg.2008.057570. [DOI] [PubMed] [Google Scholar]
- 21.Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev. 2011;32:177–95. [PMC free article] [PubMed] [Google Scholar]
- 22.Cope JU, Tsokos M, Miller RW. Ewing sarcoma and sinonasal neuroectodermal tumors as second malignant tumors after retinoblastoma and other neoplasms. Med Pediatr Oncol. 2001;36:290–4. doi: 10.1002/1096-911X(20010201)36:2<290::AID-MPO1067>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Mittal R, Al Awadi S, Sahar O, Behbehani AM. Ewing’s sarcoma as second malignant neoplasm after retinoblastoma: a case report. Med Princ Pract. 2008;17:84–5. doi: 10.1159/000109597. [DOI] [PubMed] [Google Scholar]
- 24.Spunt SL, Rodriguez-Galindo C, Fuller CE, et al. Ewing sarcoma-family tumors that arise after treatment of primary childhood cancer. Cancer. 2006;107:201–6. doi: 10.1002/cncr.21962. [DOI] [PubMed] [Google Scholar]
- 25.Tahasildar N, Goni V, Bhagwat K, Tripathy SK, Panda BB. Ewing’s sarcoma as second malignancy following a short latency in unilateral retinoblastoma. J Orthop Traumatol. 2011;12:167–71. doi: 10.1007/s10195-011-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randall RL, Lessnick SL, Jones KB, et al. Is there a predisposition gene for Ewing’s sarcoma? J Oncol. 2010;2010:397632. doi: 10.1155/2010/397632. 10.1155/2010/397632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magnusson S, Borg A, Kristoffersson U, Nilbert M, Wiebe T, Olsson H. Higher occurrence of childhood cancer in families with germline mutations in BRCA2, MMR and CDKN2A genes. Fam Cancer. 2008;7:331–7. doi: 10.1007/s10689-008-9195-7. [DOI] [PubMed] [Google Scholar]
- 28.Brooks GA, Stopfer JE, Erlichman J, Davidson R, Nathanson KL, Domchek SM. Childhood cancer in families with and without BRCA1 or BRCA2 mutations ascertained at a high-risk breast cancer clinic. Cancer Biol Ther. 2006;5:1098–102. doi: 10.4161/cbt.5.9.3167. [DOI] [PubMed] [Google Scholar]
- 29.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 30.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 31.Armes JE, Trute L, White D, et al. Distinct molecular pathogeneses of early-onset breast cancers in BRCA1 and BRCA2 mutation carriers: a population-based study. Cancer Res. 1999;59:2011–7. [PubMed] [Google Scholar]
- 32.Cavalli LR, Singh B, Isaacs C, Dickson RB, Haddad BR. Loss of heterozygosity in normal breast epithelial tissue and benign breast lesions in BRCA1/2 carriers with breast cancer. Cancer Genet Cytogenet. 2004;149:38–43. doi: 10.1016/S0165-4608(03)00282-6. [DOI] [PubMed] [Google Scholar]
- 33.Dworkin AM, Spearman AD, Tseng SY, Sweet K, Toland AE. Methylation not a frequent “second hit” in tumors with germline BRCA mutations. Fam Cancer. 2009;8:339–46. doi: 10.1007/s10689-009-9240-1. [DOI] [PubMed] [Google Scholar]
- 34.Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet. 2012;91:97–108. doi: 10.1016/j.ajhg.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucas AL, Shakya R, Lipsyc MD, et al. High prevalence of BRCA1 and BRCA2 germline mutations with loss of heterozygosity in a series of resected pancreatic adenocarcinoma and other neoplastic lesions. Clin Cancer Res. 2013;19:3396–403. doi: 10.1158/1078-0432.CCR-12-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osorio A, de la Hoya M, Rodriguez-Lopez R, et al. Loss of heterozygosity analysis at the BRCA loci in tumor samples from patients with familial breast cancer. Int J Cancer. 2002;99:305–9. doi: 10.1002/ijc.10337. [DOI] [PubMed] [Google Scholar]
- 37.Staff S, Isola JJ, Johannsson O, Borg A, Tanner MM. Frequent somatic loss of BRCA1 in breast tumours from BRCA2 germ-line mutation carriers and vice versa. Br J Cancer. 2001;85:1201–5. doi: 10.1054/bjoc.2001.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willems AJ, Dawson SJ, Samaratunga H, et al. Loss of heterozygosity at the BRCA2 locus detected by multiplex ligation-dependent probe amplification is common in prostate cancers from men with a germline BRCA2 mutation. Clin Cancer Res. 2008;14:2953–61. doi: 10.1158/1078-0432.CCR-07-5237. [DOI] [PubMed] [Google Scholar]
- 39.Knapke S, Nagarajan R, Correll J, Kent D, Burns K. Hereditary cancer risk assessment in a pediatric oncology follow-up clinic. Pediatr Blood Cancer. 2012;58:85–9. doi: 10.1002/pbc.23283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.