Abstract
Cyclin-dependent kinase 5 (Cdk5) is known as a unique member of the cyclin-dependent family of serine/threonine kinases. Previously, we demonstrated Cdk5 to be an important regulator of T cell function and that disruption of Cdk5 expression ameliorates T cell mediated neuroinflammation. Here, we show a novel role of Cdk5 in the regulation of Foxp3 expression in murine CD4+ T cells. Our data indicate that disruption of Cdk5 activity in T cells abrogates the IL-6 suppression of Foxp3 expression. This effect is achieved through Cdk5 phosphorylation of the signal transducer and activator of transcription 3 (Stat3) specifically at Serine 727 in T cells, and we show this post-translational modification is required for proper Stat3 DNA binding to the Foxp3 gene on the enhancer II region. Taken together, our data point to an essential role for Cdk5 in the differentiation of T cells as it regulates Foxp3 gene expression through phosphorylation of Stat3.
Keywords: Cyclin dependent kinase, regulatory T cell, phosphorylation, post-translational, signaling
Introduction
Cyclin-dependent kinase 5 (Cdk5) is a unique member of the cyclin-dependent kinase (CDK) family of proline-directed serine/threonine kinases. In contrast to other CDKs, Cdk5 has no significant role in regulating cell cycle progression and does not require binding to cyclins to achieve kinase activity(Dhavan and Tsai 2001). Functionally, Cdk5 associates with its binding partners, either p35 or p39, whose constitutive expression is largely restricted to cells of neural crest origin, leading to the presumption that Cdk5 is principally a neuronal-specific regulator of processes including neuronal migration, synapse formation and neuronal survival. For example, the germ-line deletion of Cdk5 in mice results in altered cortical neuronal layering during development, clearly indicating an essential role for Cdk5 in neuronal development and function.
However, aberrant Cdk5 activity is also associated with several neurodegenerative and neuroinflammatory disorders(Lopes and Agostinho 2011). Furthermore, there are now several reports demonstrating the presence of Cdk5 activity in a number of non-neuronal tissues(Arif 2012), which suggest the importance of Cdk5 activity in the normal function of pancreatic beta-cells, monocytes, neutrophils, leukocytes, myocytes, epithelial cells, endothelial cells, and adipocytes(Rosales and Lee 2006, Feldmann, Mishra et al. 2010). Consequently, Cdk5 has been implicated in the pathogenesis of a number of diseases including cancer, diabetes and inflammation-mediated disorders(Arif 2012).
We recently reported our observations that the expression of both Cdk5 and p35 increases following activation of normal, non-transformed T cells, and that this increase associates with the induction of Cdk5 kinase activity. More importantly, disruption of Cdk5 expression in T cells was protective in the murine model of experimental autoimmune encephalomyelitis (EAE), a T cell-mediated autoimmune pre-clinical model of multiple sclerosis (MS)(Pareek, Lam et al. 2010). Thus, we hypothesized that Cdk5 may act through a number of different mechanisms to influence both T cell function and differentiation into specific T cell subsets. For example, the reduced susceptibility of mice harboring a deletion of the Cdk5 gene in T cells may result from an imbalance between T effector and T regulatory (Treg) cells. In the EAE model of MS, Foxp3+ Treg cells play a central role in controlling inflammation in the central nervous system (CNS) and ultimately their function may be important to improving clinical outcome(O’Connor, Malpass et al. 2007, O’Connor and Anderton 2008, Anderton 2010) as even a transient impairment in Tregs can alter the severity of EAE pathology.
A number of factors influence the differentiation of regulatory T cells and the induction of Foxp3, the principal transcriptional regulator of Treg differentiation. For example, the induction of Foxp3 expression that accompanies T cell receptor (TCR) stimulation in the presence of transforming growth factor-beta (TGF-β) is inhibited by a number of pro-inflammatory cytokines including IL-6, IL-27 and IL-21(Huber, Steinwald et al. 2008). These cytokines all share the ability to activate the signal transducer and activator of transcription 3 (Stat3) pathway and Stat3 is known to inhibit Foxp3 gene expression through binding to a conserved enhancer (enhancer II) region on the Foxp3 gene promoter, effectively preventing proper binding of the activated TGF-β intermediate pSmad3 to the enhancer I region(Xu, Kitani et al. 2010).
Stat3 is a DNA-binding transcription factor that is associated with a wide range of physiological processes. As with other members of the Stat family of proteins, Stat3 is activated by phosphorylation at a tyrosine residue close to the carboxy-terminus (Y705). Additionally, Stat3 activity is also regulated by the phosphorylation of serine 727 located in the transactivation domain of the protein(Levy and Lee 2002). Interestingly, this specific Stat3 Ser727 phosphorylation in T cells has been highlighted as having a distinct role in dictating CD4+ T cell differentiation into Th17 and Treg cells(Maitra, Davis et al. 2009). Previous reports have shown Stat3 as a Cdk5 substrate in medullary thyroid cancer cells(Lin, Chen et al. 2007), prostate cancer cells(Hsu, Chen et al. 2013), myotubes(Fu, Fu et al. 2004) and neuronal cells(Wen, Yu et al. 2008). However, to our knowledge, there is no evidence suggesting post-translational modification of Stat3 by Cdk5 is a critical step in regulating Stat3 function in T cells. Thus, we hypothesized that Cdk5 may regulate the expression of the Foxp3 transcription factor and impair Treg differentiation through post-translational modification of Stat3.
In this study, we examine how disruption or Cdk5 expression and activity influences the expression of Foxp3 in primary murine CD4+ T cells. Our data indicate that the suppression of Cdk5 signaling abrogates IL-6 inhibition of Foxp3 expression. Furthermore, we mechanistically link Cdk5 activity to the regulation of Stat3 signaling and its influence on Foxp3 expression in T cells. Here we show the inhibition of Cdk5 activity (either using a Cdk5-specific inhibitory peptide or through the disruption of Cdk5 gene expression in T cells) leads to decreased Stat3 phosphorylation at Ser727, and this in turn disrupts Stat3 binding to the Foxp3 gene on the enhancer region II. Taken together, our data reveal a novel role for Cdk5 in the differentiation of T cells, as it effectively regulates Foxp3 gene expression and Treg development through specific phosphorylation of Stat3 at Serine 727.
Results
Disruption of Cdk5 activity prevents suppression of Foxp3 in T cells following stimulation with TGF-β and IL-6
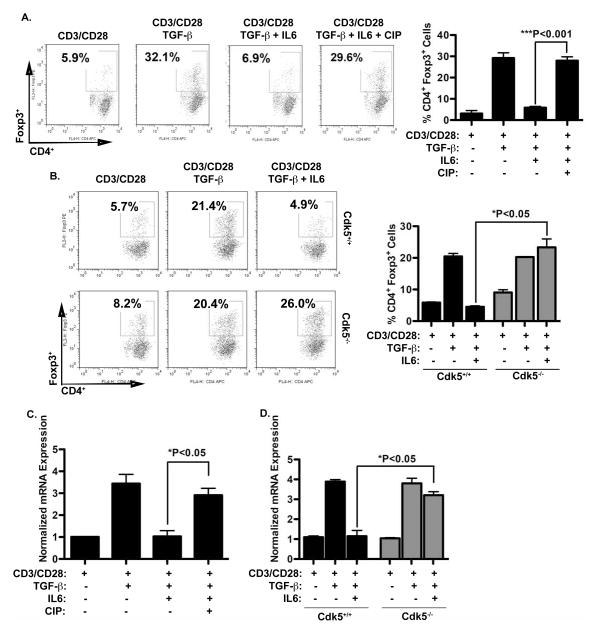
To determine whether Cdk5 activity contributes to the suppression of Foxp3 expression in T cells, we utilized primary murine T cells isolated from either wild type mice (Cdk5+/+) or mice with a T cell lineage-restricted Cdk5 gene deletion (Cdk5−/−) and culture conditions specific for Treg induction. Specifically, CD4+ T cells harvested from splenocytes of either Cdk5+/+ mice or Cdk5−/− mice were plated with anti-CD3/CD28 antibody in the presence or absence of TGF-β and IL-6. In cultures of Cdk5+/+ T cells, we also used the CIP peptide inhibitor to achieve specific inhibition of Cdk5 activity and to corroborate data from cultures of T cells from Cdk5−/− mice. As shown in flow cytometry analyses in Figure 1A, TGF-β expectedly increased the percentage of CD4+CD25+Foxp3+ cells, whereas the addition of IL-6 effectively suppressed this induction of Foxp3+ cells by TGF-β. Consistent with our hypothesis, suppression of Cdk5 activity by CIP prevented the suppression in Foxp3 expression observed when CD4+ cells were treated with both TGF-β and IL-6. These data are supported by analyses in Cdk5−/− CD4+ T cells (Fig 1B), as IL-6 also failed to suppress the induction of CD4+CD25+Foxp3+ T cells when Cdk5−/− CD4+ T cells were activated with anti-CD3/CD28 in the presence or absence of TGF-β and IL-6. The changes in Foxp3 protein expression correlated with changes in Foxp3 mRNA expression (Figure 1C and 1D). When either Cdk5+/+ T cells or Cdk5−/− T cells were activated in the presence of TGF-β, each demonstrated a significant increase in the expression of Foxp3 mRNA as measured by real-time RT-PCR. Correlating with data obtained by flow cytometry, the presence of IL-6 in cultures results in a significant suppression of Foxp3 message only in Cdk5+/+ T cells (Figure 1C), but not in Cdk5−/− T cells (Figure 1D).
Figure 1. Cdk5 is essential for IL-6 suppression of TGF-β induced Foxp3 expression in CD4+ T cells.
A) Naïve CD4+ T cells were isolated from wild-type mice and stimulated with anti-CD3 and anti-CD28, with or without TGF-β, in the presence or absence of IL-6 and the Cdk5 specific inhibitor CIP as indicated for 72 hours. Foxp3 protein expression and numbers of Foxp3+ cells were quantified by flow cytometry. B) Wild-type and Cdk5-deficient T cells were stimulated with anti-CD3 and anti-CD28, with or without TGF-β and IL-6 as indicated for 72hrs. Foxp3 protein expression and numbers of Foxp3+ cells were quantified by flow cytometry. C) Foxp3 mRNA expression levels in T cells activated in the presence or absence of TGF-β, IL-6 and CIP for 16hrs were analyzed with real time RT-PCR. D) Both Cdk5+/+ T cells and Cdk5−/− T cells were treated with or without TGF-β and IL-6 for 16hrs, and subsequently Foxp3 mRNA expression levels were examined using real time RT-PCR analysis.
Cdk5 activity does not inhibit TGF-β dependent activation of Smad proteins in T cells
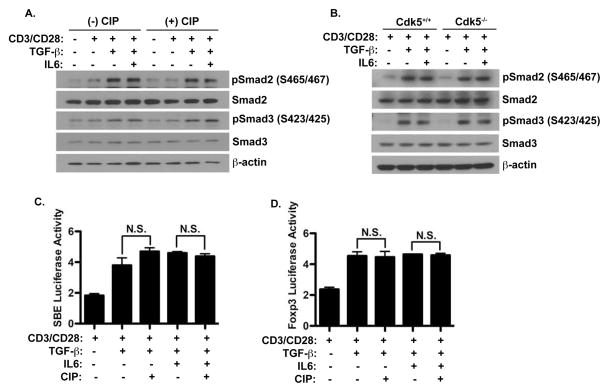
The role of Smad-dependent TGF-β signaling in the induction of Foxp3 has been estabished(Xu, Kitani et al. 2010). Thus we chose to examine whether Cdk5 influenced Smad2/3 signaling through direct phosphorylation Smad proteins. Cdk5+/+ T cells were activated with anti-CD3/CD28 in the presence of TGF-β and IL-6, either with or without the CIP peptide inhibitor of Cdk5. We observed a significant increase in phospho-Smad proteins (Figure 2A) following TGF-β treatment, whether added alone or in combination with IL-6. T cells treated under the same conditions but in the presence of CIP showed similar phosphorylation of the Smad proteins. Moreover, both Cdk5+/+ T cells and Cdk5−/− T cells (Figure 2B) activated in the presence of both TGF-β, either alone or in combination with IL-6 showed a similar induction in the phosphorylation status of Smad2 and Smad3 proteins, again indicating that Cdk5 does not interfere with Smad2/3 phosphorylation in response to TGF-β in T cells.
Figure 2. Cdk5 activity neither alters Smad activation nor binding to the Foxp3 gene.
A) Naïve primary T cells were stimulated with anti-CD3/CD28, with or without TGF-β, IL-6 and CIP for 72hrs. Whole cell protein lysates were prepared from these samples and western blot analyses were performed to probe for p-Smad2 (Ser465/467), p-Smad3 (Ser423/425) and total Smad2 and total Smad3. B) Similarly, both Cdk5+/+ T cells and Cdk5−/− T cells were stimulated and treated under conditions noted in (A) and probed for the presence of p-Smad2/3 and total Smad2/3. C) Luciferase activity was examined in EL4 T cells transfected with the Smad-binding element (SBE) reporter construct and E) the Foxp3 promoter and enhancer I reporter construct. Cells were stimulated with anti-CD3/CD28 with or without TGF-β, IL-6 and CIP as indicated.
Although we observed no detectable change in Smad2/3 activation with disruption of Cdk5 activity, this did not rule out the possibility that Cdk5 activity could influence Smad protein binding to DNA through modification of other co-activator or co-repressor proteins. To address this question, we first utilized a luciferase reporter controlled by a general Smad binding element, which we expressed in the EL4 T cell line (Figure 2C). Following TCR stimulation in the presence of TGF-β, either alone or with IL-6, we observed a significantly higher level of luciferase activity. Inhibition of Cdk5 activity in these cultures with the CIP inhibitory peptide did not alter luciferase activity. To take this analysis one step further, we analyzed the specific effect Cdk5 activity might have on Smad binding on the Foxp3 gene promoter by utilizing a luciferase reporter driven by the Foxp3 promoter linked in tandem to the enhancer I region (which is known to contain the Smad binding site). As seen in Figure 2D, in EL4 cells transfected with this reporter, the basal reporter activity increased significantly in conditions containing TGF-β treatment when compared to either unstimulated EL4 cells or to EL4 cells stimulated only with CD3/CD28. This induction was neither affected by the addition of IL-6, nor by exposure of cells to the CIP inhibitor of Cdk5. Taken together, these data clearly indicate that Cdk5 activity does not directly influence the activation of Smad proteins nor their ability to bind to DNA. Thus, the effect of Cdk5 on Foxp3 expression must be mediated through modulation of a non-Smad pathway that indirectly controls TGF-β induced Foxp3 expression.
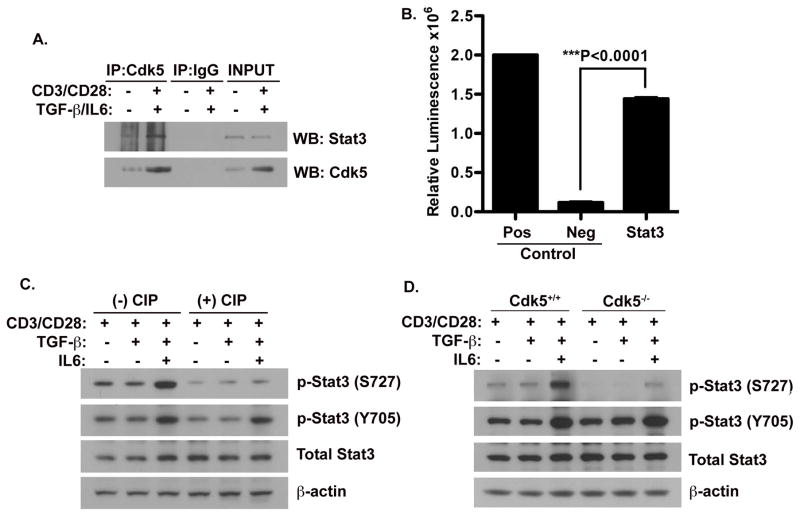
Cdk5 induces Stat3 function in T cells by physical interaction and Stat3 phosphorylation at Serine 727
Stat3 has been widely accepted as an essential signaling molecule, selectively interacting with enhancer elements in the promoter region of different genes, thereby affecting a wide variety of physiological processes and responses. The Stat3 molecule is normally a latent cytoplasmic transcription factor that will become activated following cellular stimulation by various extracellular signaling molecules and subsequently translocates into the nucleus ultimately altering transcription of select genes(Levy and Lee 2002). Stat3 is activated by tyrosine phosphorylation (Y705) at a residue close to the carboxy-terminus as well as by a serine phosphorylation (S727) located within the transactivation domain. Several groups have shown the ability for Cdk5 to interact with Stat3 and furthermore, the ability of Cdk5 to directly phosphorylate Stat3 at the Serine 727 site. However, these previous reports have not assessed the relevance of Stat3 modulation by Cdk5 in the context of T cell activation. Therefore, to determine whether Cdk5 modulates Stat3 function in T cells, we first immunoprecipitated Cdk5 from protein lysates prepared from T cells under various stimulation conditions, including either with or without anti-CD3/anti-CD28, in the presence or absence of TGF-β and IL-6, either alone or in combination. Probing Stat3 in immunoprecipitates of Cdk5 (Figure 3A), we confirmed the presence of a physical interaction between Stat3 and Cdk5. Moreover, the greater abundance of Stat3 observed in immunoprecipitates of TCR stimulated T cells (CD3/CD28) is consistent with the known increase in Cdk5 expression that occurs following TCR stimulation. More importantly, this interaction of Cdk5 with the Stat3 protein leads directly to Stat3 phosphorylation in T cells, as shown by an in vitro Cdk5 kinase assay, in which we combined Cdk5 and Stat3 immunoprecipitates obtained from T cells (Figure 3B). Since Stat3 phosphorylation mainly occurs on either the tyrosine 705 or serine 727 residues, we examined how Cdk5 activity influences phosphorylation of both residues. As shown in Figure 3C, Stat3 Ser727 phosphorylation induced by T cell activation is significantly attenuated by treatment with the CIP peptide inhibitor of Cdk5. However, Cdk5 inhibition had no effect on the phosphorylation of Stat3 on the Tyr705 site and did not alter the total expression of Stat3 (Figure 3C). These findings were corroborated in analyses of Cdk5−/− T cells activated under similar conditions (Figure 3D). There was a significant decrease in Stat3 serine727 phosphorylation in lysates of Cdk5−/− T cells, when compared to Cdk5+/+ T cells and once again, Y705 phosphorylation and total Stat3 levels were not affected. In total, these data implicate Stat3 as an important substrate of Cdk5 in T cells, with Cdk5 selectively phosphorylating Stat3 at the Ser727 residue.
Figure 3. Cdk5 physically interacts with and phosphorylates Stat3 at Serine 727.
A) Protein lysates were isolated from primary T cells before and after TCR activation in the presence of TGF-β, IL-6 and CIP for 72hrs. Samples were then immuno-precipitated with anti-Cdk5 antibody and immunoprecipitates were then probed for the presence of Stat3 and Cdk5. B) A Cdk5-specific in vitro kinase assay was performed to determine the capacity of Cdk5 to phosphorylate Stat3 (the substrate) immunoprecipitated from T cells. Neurofilament-H (NFH) as a substrate, was combined with Cdk5 immunoprecipitates isolated from wild type T cells as a positive control (Pos), whereas Cdk5 immuprecipitates isolated from Cdk5−/− T cells served as the negative control (Neg). C) Western blot analysis of protein lysates isolated from T cells with or without anti-CD3/CD28 activation and treatment with or without TGF-β, IL-6 and CIP for 72hrs. Expression of total Stat3, p-Stat3 (Y705) and p-Stat3 (S727) were determined by Western blot, and β-actin expression was probed as a control. D) Lysates from both Cdk5+/+ T cells and Cdk5−/− T cells stimulated under similar treatment conditions were analyzed by Western blot to determine protein expression of total Stat3, p-Stat3 (Y705), p-Stat3 (S727) and β-actin.
Inhibition of Cdk5 activity in T cells decreases Stat3 binding to the enhancer region of the Foxp3 gene
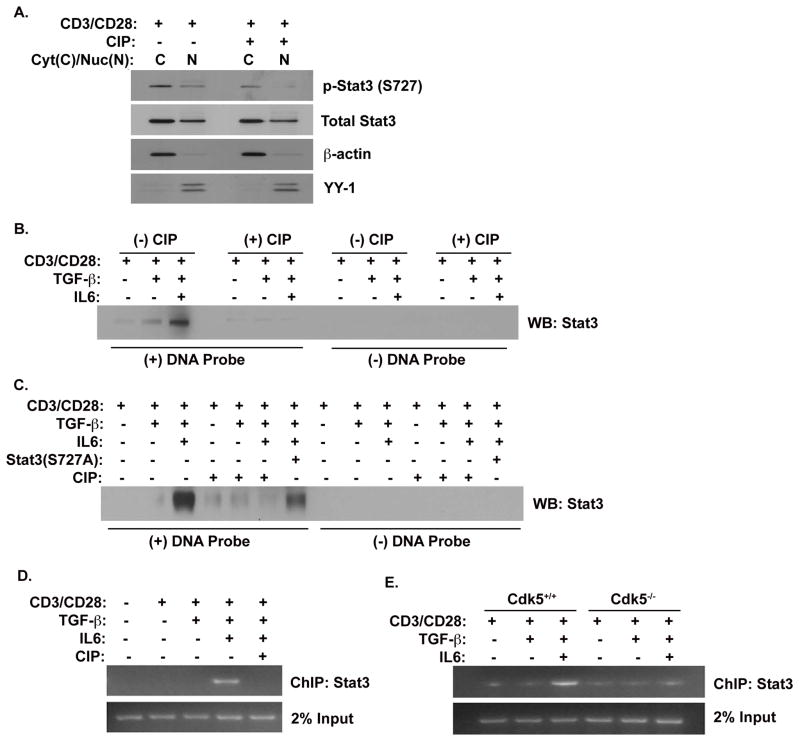
Having established the potential for Stat3 to serve as a substrate of Cdk5 in T cells, we next examined whether Cdk5 might affect Stat3 entry into the nucleus. Western blot analyses of nuclear and cytoplasmic fractions of T cells for the presence of p-Stat3 (S727) revealed a marked decrease in the abundance of nuclear Stat3 when Cdk5 activity was inhibited (Figure 4A). However, inhibition of Cdk5 had no discernable effect on either phospho-Stat3(Y705) or total Stat3 expression. Thus, while treatment with CIP suppresses Serine 727 phosphorylation in Stat3, this had no affect on the abundance of Stat3 or on Stat3 entry into the nuclear compartment.
Figure 4. Cdk5 is required to induce Stat3 binding to the Foxp3 gene.
A) Protein lysates were isolated from the cytoplasmic and nuclear compartments of activated T cells with or without TGF-β and CIP treatment for 48hrs. Western blot analyses were performed probing for p-Stat3 (S727), p-Stat3 (Y705), total Stat3 and Cdk5. Both β-actin and YY-1 were probed as cytoplasmic and nuclear loading controls, respectively. B) Nuclear protein lysates were isolated from primary T cells activated by anti-CD3/CD28, with or without TGF-β, IL-6 and CIP treatment for 72hrs. Lysates were incubated with DNA probes that corresponded to the specific Stat3 binding region on the enhancer region II of the Foxp3 gene and allowed to complex in a DNA pull down assay. Western blot analyses were then performed for Stat3 bound to the DNA probes. C) Nuclear protein lysates were prepared from EL4 T cells stimulated under similar conditions, and from EL4 T cells expressing the Stat3 (S727A) mutant protein. Lysates were subjected to a DNA pull-down using DNA probes corresponding to the Stat3 binding enhancer II region of the Foxp3 gene. D) ChIP analysis was performed with lysates from primary T cells that were similarly stimulated with or without anti-CD3/CD28, with or without TGF-β, in the presence or absence of IL-6 and CIP. PCR was performed to amplify the specific crosslinked DNA corresponding to the Stat3 region on the Foxp3 enhancer II region. E) Similarly, ChIP analyses were performed with lysates from both Cdk5+/+ T cells and Cdk5−/− T cells activated in the presence or absence of either TGF-β and/or IL-6.
The negative regulation of Foxp3 expression by Stat3 has been attributed to transcriptional repression exerted via binding to the enhancer II region of the Foxp3 gene. Therefore, we next examined whether Cdk5-dependent Stat3 phosphorylation might affect Stat3 DNA binding at the enhancer II region. Lysates prepared from T cells stimulated under various conditions (CD3/CD28, +/− IL-6, +/− TGF-β) were subjected to in vitro DNA pull-down assays (Figure 4B). Western blot analysis shows Stat3 protein bound to the enhancer II DNA probe in lysates from T cells treated with a combination of CD3/CD28, TGF-β and IL-6 together. Consistent with our hypothesis, the binding of Stat3 protein to the enhancer II DNA probe was suppressed in samples from T cells stimulated with CD3/CD28, TGF-β and IL-6 together in the presence of the CIP inhibitor of Cdk5 activity. These data suggest Cdk5 activity is necessary for Stat3 to properly bind to the enhancer II region of the Foxp3 gene. Next, we performed a DNA pull-down analysis on samples isolated from the EL4 T cell line (Figure 4C). We observed the same DNA-protein interaction as previously mentioned with Stat3 binding to the DNA probe under conditions of CD3/CD28, TGF-β and IL-6 stimulation. Once again, this binding decreased when Cdk5 activity was inhibited by the addition of CIP peptide. To further confirm whether this effect of Cdk5 on Stat3 is dependent on specific Serine 727 phosphorylation, we transfected the EL4 T cell line with a mutant form of Stat3 in which the serine 727 residue is substituted with an alanine residue. This substitution effectively suppressed Stat3 protein-DNA interaction, even under stimulation conditions including CD3/CD28, TGF-β and IL-6. The low level of Stat3 protein-DNA interaction seen in cells expressing Stat3S727A (lane 7, Figure 4C) is in fact the endogenous, normal Stat3, whose binding has been effectively competed by the Stat3S727A mutant (i.e. relative to lane 3). In total, these data clearly implicate a requirement for the phosphorylation of Stat3 at the Serine 727 residue by Cdk5 for proper DNA-Stat3 protein interaction.
Finally, to examine the effect of Cdk5 activity on Stat3-DNA binding in the in-vivo setting, we performed a ChIP assay designed to measure the ability of Stat3 to occupy the specific enhancer region of the Foxp3 gene under different conditions. As seen in Figure 4E, binding of Stat3 to the enhancer II region only occurred in conditions in which CD3/CD28 and TGF-β were combined with IL-6. Most importantly, the addition of the specific Cdk5 inhibitor CIP resulted in a loss of Stat3 binding. Similarly, in ChIP assays performed with Cdk5−/− T cells, we observed a lack of Stat3 protein occupying the enhancer II region when compared to ChIP assays with wild type T cells. Together, these results reinforce the concept that Cdk5 plays a central role in regulating the IL-6 suppression of Foxp3 expression in CD4+ cells through a direct regulation of Stat3 phosphorylation and consequent DNA binding.
Material and Methods
Animals
C57BL/6 mice, ages 6–8 weeks, were purchased from The Jackson Laboratory and used for T cell isolations. T cell specific Cdk5-deficient mice used in these studies were generated in our research facility. All animals were housed in micro-isolator cages and maintained in climate/light controlled rooms with free access to food and water. Studies performed were in compliance with the procedures approved by the Case Western Reserve University School of Medicine’s Institutional Animal Care and Use Committee.
Antibodies
The Cdk5 antibody (C8), p-Stat3(Ser727) antibody and rabbit IgG antibody were purchased from Santa Cruz Biotechnology and used for both immunoprecipitation as well as western blotting. The antibody used for total Stat3 was purchased from BD Biosciences.
Cdk5-Inhibitory Peptide (CIP)
CIP was generously provided by the laboratory of Dr. Harish Pant. This Cdk5-specific inhibitor is derived from p35 (amino acid residues 154–279) and has been shown to specifically inhibit Cdk5 activity in vitro(Zheng, Li et al. 2002, Kesavapany, Zheng et al. 2007).
Cdk5-conditional (T-cell specific) gene knock-out mice
The Letterio laboratory developed and characterized a new mouse model harboring a conditional lineage restricted deletion of the Cdk5 gene in T cells by crossing Cdk5 flox/flox mice with CD4-Cre mice(where Cre expression is initiated under control of the CD4 promoter at the double positive stage of T cell development). For this model we used a Cdk5 gene construct (provided by P. Greengard) that leaves neighboring alleles completely untouched(Hawasli, Benavides et al. 2007). T-cell specific conditional Cdk5 gene knock out mice show a complete loss of the Cdk5 gene in T cells as analyzed by Cdk5 mRNA expression, protein expression and Cdk5 specific kinase activity assay (data not shown, manuscript in preparation). Cre-negative littermate mice were utilized as Cdk5+/+ controls.
Cdk5 Kinase Assay
Cdk5 kinase activity assays were performed as previously described(Pareek, Lam et al. 2010), and the ADP-Glo kinase assay detection kit used was purchased from Promega to detect the occurrences of phosphorylation events. In brief, total protein cell lysates were prepared and then incubated overnight at 4°C with Cdk5 antibody. Afterwards, protein A-Agarose beads were added to samples for 3–5 hours at 4°C. Following immuno-precipitation, samples were combined with desired substrates and incubated at 30°C after the addition of ATP.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation (ChIP) analysis was performed according to the manufacturer’s instructions (Upstate Biotechnology) with T cells stimulated under the indicated treatment conditions. Immunoprecipitated samples (DNA) were subjected to semi-quantitative RT-PCR analyses of Foxp3 enhancer region II using primers 5′-agaacttgggttttgcatgg-3′ and 5′-gccagatttttctgccattg-3′.
Cytokines
TGF-β1 used was purchased from PeproTech. Working concentrations of 5ng/mL were used to stimulate T cells in all experiments. IL-6 cytokines were purchased from PeproTech and were used to stimulate T cells at a working concentration of 100ng/mL.
DNA Pull-down Assays
Biotin labeled oligonucleotides (5′-gccacttctcggaacgaaa-3′ and 5′tttcgttccgagaagtggc-3′) were dimerized and used to perform DNA pull-down assays. For each reaction, 1.5 μg of dimer was incubated for 15 min at room temperature with 50 μl of Dynabeads® M-280 streptavidin (Invitrogen) washed twice with 2 X B&W buffer (10 mM Tris-Cl (pH 7.5), 1 mM EDTA, 2 M NaCl). After conjugation in 1 X B&W buffer, oligo-conjugated beads were washed three times with 1 X B&W buffer to remove unconjugated oligonucleotides, and resuspended with ice-cold DNAP buffer (10 mM HEPES, pH 7.9, 100 mM KCl, 5 mM MgCl2, 10 % glycerol, 0.5 % NP-40, 1 mM EDTA) containing 1 mM DTT added freshly. 100 μg nuclear protein was incubated with oligo-conjugated beads and reaction volume was adjusted up to 500 μl with 1 X DNAP containing Complete EDTA-free Protease inhibitor Mixture (Roche), 1mM sodium orthovanadate, 1 mM phenymethylsulfonyl floride, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, and 1 mM DTT. Polydeoxyinosinic-deoxycytidylic acid (5 μg) was added to the reaction tube, which was then incubated for 4 h at 4 °C with gentle mixing on a rotator. Beads were washed three times on ice with DNAP containing 1 mM DTT, eluted with 45 μl of 1 X SDS buffer by treating for 5 min at 85 °C. Elutes were subjected to Western blot analysis.
FACS analysis
Antibodies (CD4, Foxp3) used in FACS analyses were purchased from BD Pharmingen and labeling was performed according to manufacturer’s protocol. After stimulating cells as described, cells were then collected and analyzed using the BD FacsCalibur cytometer.
Immunoprecipitation assays
Protein lysates were first diluted to a concentration of 1μg/μL and subsequently pre-cleared with protein A-Agarose beads (Santa Cruz) and then incubated overnight with rotation at 4°C with immunoprecipiating antibody. Lysates were then incubated with protein A-Agarose beads for 3–5 hours with rotation at 4°C. After incubation, protein A-Agarose beads were collected and washed with lysis buffer. Samples were then subjected to western blot analysis.
Quantitative Real-time RT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen). For RT-PCR, a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used according to the manufacturer’s protocol. Samples were then amplified using TaqMan Fast Universal PCR Master Mix (Applied Biosystems) and samples were normalized with GAPDH mRNA levels.
Semi-quantitative RT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen) and complementary DNA was then prepared using a cDNA first-strand synthesis kit (Invitrogen). PCR was performed with a Px2 Thermal Cycler (Thermo).
Statistical Analyses
Statistical evaluations were performed using the Prism computer program (GraphPad Software). Significant differences between experiments were assessed by comparing the means of data sets using the Student t test, with a p value of <0.05 considered significant.
T cell receptor stimulation
Anti-CD3 (3μg/mL) and anti-CD28 (1μg/ML) antibodies (BD Biosciences) diluted in PBS were allowed to incubate to become plate bound overnight in 4°C. Prior to use, plates were washed with fresh PBS twice.
T lymphocyte isolation
A mixed population of cells was collected by first isolating cells from both spleen and regional lymph nodes of mice and creating single cell suspensions by passing these tissues through a 40μm cell strainer (BD Biosciences). Cells were then incubated with ACK lysis buffer (Lonza) on ice for 5 minutes to deplete the mixed cell population of erythrocytes. T lymphocytes were purified through a process of negative isolation by passing mixed cells through MACS separation columns using a pan T isolation kit (MiltenyiBiotec) in accordance to the manufacturer’s protocol.
Western blot analysis
Protein samples were prepared from cellular lysates made in RIPA buffer (Thermo Fisher Scientific) containing a protease inhibitor cocktail tablet (Roche) in addition to a phosphatase inhibitor phosSTOP (Roche). Proteins were denatured by heating for 10 min at 94°C in sample loading buffer (2% SDS, 10% glycerol 80mM Tris, pH 6.8, and 1mM DTT). 50μg of total protein was separated by electrophoresis in 4–20% Tris-Glycine gels (Invitrogen). Proteins were transferred to a 0.2μm nitrocellulose membrane (Invitrogen) and subsequently blocked for 1 hour in blocking buffer (TBS containing 5% non-fat dry milk and 0.05% Tween 20). Membranes were incubated overnight at 4°C with primary antibody and probed with horse radish peroxidase-conjugated secondary antibody for 1 hour at room temperature.
Discussion
Here we provide the first demonstration that the activity of Cdk5 modulates the expression of the Foxp3 transcription factor in CD4+ T cells, through post-translational modification of Stat3. Either the pharmacologic inhibition of Cdk5 activity or the targeted deletion of the Cdk5 gene in T cells is sufficient to abrogate the suppression of Foxp3 gene expression by IL-6, which requires activation of Stat3 at the Serine 727 site. Our data not only demonstrate that this phosphorylation of Stat3 at Serine 727 requires Cdk5, but also indicate that this phosphorylation of Stat3 at Serine 727 is not necessary for Stat3 entry into the nucleus in T cells. However, nuclear Stat3 lacking this modification at Serine 727 does fail to bind to the enhancer region of the Foxp3 gene, and thus cannot effectively mediate suppression of Foxp3 in response to IL-6 signaling. Our data therefore provide evidence regarding the relevance of Cdk5 as a molecular mediator of regulatory T cell expansion in the context of chronic inflammation.
The Foxp3 transcription factor is essential for control over the differentiation, function and survival of Treg cells. Although a number of factors orchestrate the expression of Foxp3, transforming growth factor-β (TGF-β) is an essential regulator of the inducible Treg population(Zheng, Gray et al. 2002). Induction of Foxp3 by TGF-β requires the action of the Smad3 transcription factor(Tone, Furuuchi et al. 2008). Activated by the TGF-β receptor, Smad3 binds to a specific enhancer site of the Foxp3 gene and becomes an integral part of the enhanceosome complex required for Treg development(Ruan, Kameswaran et al. 2009). Our data reveal that suppression of Foxp3 by Cdk5 is not mediated through a direct effect on the Smad2/3 pathway. Indeed, Smad2/3 activation by TGF-β is not altered in Cdk5-deficient T cells nor in wild type T cells activated in the presence of a Cdk5 specific inhibitor (CIP peptide). Moreover, utilizing a luciferase reporter assay linked to a Smad-binding element, we confirmed that treatment of T cells with the Cdk5 inhibitory peptide CIP did not alter the ability for Smad proteins to properly bind to DNA. Finally, our data from assays with a luciferase reporter driven by the Foxp3 enhancer I region show that Cdk5 inhibition does not alter the ability for the Smad proteins to specifically interact with the proper DNA region of the Foxp3 promoter.
In addition to TGF-β, several other factors are known to modulate Foxp3 expression. In particular, IL-6 and IL-27 have been found to be strong inhibitors of Foxp3 expression in T cells activated through the TCR in the presence of TGF-β, and do so by activating Stat3. Xu et al, recently demonstrated a novel enhancer region (enhancer II) of the Foxp3 gene to which activated Stat3 was found to bind(Xu, Kitani et al. 2010). These data support a model in which Stat3 acts as an inhibitor of Foxp3 expression by preventing the binding of pSmad3 to enhancer I.
Cdk5-mediated Stat3 phosphorylation has been demonstrated in previous reports(Fu, Fu et al. 2004, Courapied, Sellier et al. 2010). By performing a Cdk5 kinase assay using Cdk5 and Stat3 immunoprecipitated from primary T cell lysates, we clearly confirm Stat3 as a Cdk5 substrate in T cells. Consistent with the demonstrated preference towards phosphorylating proline-directed serine/threonine residues, Cdk5 has been shown to phosphorylate Stat3 at the Serine727 site in specific tissues including myotubes, neurons, colorectal cancer cells and prostate cancer cells. Our data suggest that this modification of Stat3 by Cdk5 plays a very important role in controlling T cell fate by modulating the response to extracellular signaling molecules that control T cell differentiation. Moreover, our data show that either inhibition of Cdk5 activity in wild-type T cells or the disruption of Cdk5 gene expression in T cells leads to a significant decrease in pStat3(S727), but fails to influence either pStat3(Y705) or total Stat3 levels. The data implicate Cdk5 as a required mediator of the Foxp3 repression downstream of IL-6 receptor signaling, and in a manner that cannot be compensated by other signaling intermediates.
Although phosphorylation of Stat3 on Tyrosine Y705 is clearly known to be important for Stat3 dimerization, nuclear translocation and DNA binding, the role of serine phosphorylation of Stat3 is somewhat controversial. There is evidence supporting both a positive and negative role for serine phosphorylation, thus indicating the potential for this modification of Stat3 to define context-dependent function of this transcription factor. In support of this possibility, disruption of Cdk5 activity has no discernible effect on total Stat3 and the decrease in pStat3(S727) caused by disruption of Cdk5 activity does not alter the ability of Stat3 to translocate into the nucleus.
Prior reports have suggested that Cdk5-dependent Stat3 serine phosphorylation alters the ability of this transcription factor to properly bind to DNA(Fu, Fu et al. 2004). Thus, although Stat3 nuclear translocation was not affected by Cdk5 inhibition in T cells, we explored the possibility that Cdk5 post-translational modification of Stat3 might influence Stat3 DNA binding. In T cells, DNA binding assays revealed phosphorylation of Stat3 at S727 is essential for Stat3 binding to the enhancer region II of the Foxp3 gene, and is thereby critical to the repression of Foxp3 expression in T cells exposed to cytokines like IL-6 and IL-27. Thus, our data not only reinforce the role of Cdk5 as an important post-translational modulator of transcription factor activity (Dhavan and Tsai 2001, Lin, Chen et al. 2007, Courapied, Sellier et al. 2010, Kazmierczak, Czapski et al. 2011, Lam, Pareek et al. 2015), but also demonstrate the importance of this function in controlling T cell fate. In summary, the observations presented here reveal a novel role of Cdk5 in controlling Foxp3 gene expression and suggest that targeting Cdk5 activity may overcome the ability of inflammatory cytokines to repress TGF-β-induced Foxp3 expression. In the future, approaches that target this function of Cdk5 may be exploited in the therapy of autoimmune and chronic inflammatory diseases.
Acknowledgments
We would like to acknowledge support from the Jane and Lee Seidman Chair in Pediatric Cancer Innovation (J.J.L.). Work in this report has been funded by grants from the NIH (R01HL111682-02 and R01 EY022937-01), The Hyundai Hope on Wheels Foundation and the St. Baldrick’s Foundation for Pediatric Cancer Research.
Abbreviations
- Treg
regulatory T cell
- STAT3
signal transducer and activator of transcription
- Cdk5
Cyclin dependent kinase 5
- ChIP
chromatin immunoprecipitation
- IL-6
Interleukin 6
Footnotes
Disclosures
Authors declare no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderton SM. Treg and T-effector cells in autoimmune CNS inflammation: a delicate balance, easily disturbed. Eur J Immunol. 2010;40(12):3321–3324. doi: 10.1002/eji.201041100. [DOI] [PubMed] [Google Scholar]
- Arif A. Extraneuronal activities and regulatory mechanisms of the atypical cyclin-dependent kinase Cdk5. Biochem Pharmacol. 2012;84(8):985–993. doi: 10.1016/j.bcp.2012.06.027. [DOI] [PubMed] [Google Scholar]
- Courapied S, Sellier H, de Carne Trecesson S, Vigneron A, Bernard AC, Gamelin E, Barre B, Coqueret O. The cdk5 kinase regulates the STAT3 transcription factor to prevent DNA damage upon topoisomerase I inhibition. J Biol Chem. 2010;285(35):26765–26778. doi: 10.1074/jbc.M109.092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhavan R, Tsai LH. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2(10):749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- Feldmann G, Mishra A, Hong SM, Bisht S, Strock CJ, Ball DW, Goggins M, Maitra A, Nelkin BD. Inhibiting the cyclin-dependent kinase CDK5 blocks pancreatic cancer formation and progression through the suppression of Ras-Ral signaling. Cancer Res. 2010;70(11):4460–4469. doi: 10.1158/0008-5472.CAN-09-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu AK, Fu WY, Ng AK, Chien WW, Ng YP, Wang JH, Ip NY. Cyclin-dependent kinase 5 phosphorylates signal transducer and activator of transcription 3 and regulates its transcriptional activity. Proc Natl Acad Sci U S A. 2004;101(17):6728–6733. doi: 10.1073/pnas.0307606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawasli AH, Benavides DR, Nguyen C, Kansy JW, Hayashi K, Chambon P, Greengard P, Powell CM, Cooper DC, Bibb JA. Cyclin-dependent kinase 5 governs learning and synaptic plasticity via control of NMDAR degradation. Nat Neurosci. 2007;10(7):880–886. doi: 10.1038/nn1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FN, Chen MC, Lin KC, Peng YT, Li PC, Lin E, Chiang MC, Hsieh JT, Lin H. Cyclin-dependent kinase 5 modulates STAT3 and androgen receptor activation through phosphorylation of Ser(7)(2)(7) on STAT3 in prostate cancer cells. Am J Physiol Endocrinol Metab. 2013;305(8):E975–986. doi: 10.1152/ajpendo.00615.2012. [DOI] [PubMed] [Google Scholar]
- Huber M, Steinwald V, Guralnik A, Brustle A, Kleemann P, Rosenplanter C, Decker T, Lohoff M. IL-27 inhibits the development of regulatory T cells via STAT3. Int Immunol. 2008;20(2):223–234. doi: 10.1093/intimm/dxm139. [DOI] [PubMed] [Google Scholar]
- Kazmierczak A, Czapski GA, Adamczyk A, Gajkowska B, Strosznajder JB. A novel mechanism of non-Abeta component of Alzheimer’s disease amyloid (NAC) neurotoxicity. Interplay between p53 protein and cyclin-dependent kinase 5 (Cdk5) Neurochem Int. 2011;58(2):206–214. doi: 10.1016/j.neuint.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Kesavapany S, Zheng YL, Amin N, Pant HC. Peptides derived from Cdk5 activator p35, specifically inhibit deregulated activity of Cdk5. Biotechnol J. 2007;2(8):978–987. doi: 10.1002/biot.200700057. [DOI] [PubMed] [Google Scholar]
- Lam E, Pareek TK, Letterio JJ. Cdk5 controls IL-2 gene expression via repression of the mSin3a-HDAC complex. Cell Cycle. 2015;14(8):1327–1336. doi: 10.4161/15384101.2014.987621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109(9):1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Chen MC, Chiu CY, Song YM, Lin SY. Cdk5 regulates STAT3 activation and cell proliferation in medullary thyroid carcinoma cells. J Biol Chem. 2007;282(5):2776–2784. doi: 10.1074/jbc.M607234200. [DOI] [PubMed] [Google Scholar]
- Lopes JP, Agostinho P. Cdk5: multitasking between physiological and pathological conditions. Prog Neurobiol. 2011;94(1):49–63. doi: 10.1016/j.pneurobio.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Maitra U, Davis S, Reilly CM, Li L. Differential regulation of Foxp3 and IL-17 expression in CD4 T helper cells by IRAK-1. J Immunol. 2009;182(9):5763–5769. doi: 10.4049/jimmunol.0900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RA, Anderton SM. Foxp3+ regulatory T cells in the control of experimental CNS autoimmune disease. J Neuroimmunol. 2008;193(1–2):1–11. doi: 10.1016/j.jneuroim.2007.11.016. [DOI] [PubMed] [Google Scholar]
- O’Connor RA, Malpass KH, Anderton SM. The inflamed central nervous system drives the activation and rapid proliferation of Foxp3+ regulatory T cells. J Immunol. 2007;179(2):958–966. doi: 10.4049/jimmunol.179.2.958. [DOI] [PubMed] [Google Scholar]
- Pareek TK, Lam E, Zheng X, Askew D, Kulkarni AB, Chance MR, Huang AY, Cooke KR, Letterio JJ. Cyclin-dependent kinase 5 activity is required for T cell activation and induction of experimental autoimmune encephalomyelitis. J Exp Med. 2010;207(11):2507–2519. doi: 10.1084/jem.20100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales JL, Lee KY. Extraneuronal roles of cyclin-dependent kinase 5. Bioessays. 2006;28(10):1023–1034. doi: 10.1002/bies.20473. [DOI] [PubMed] [Google Scholar]
- Ruan Q, Kameswaran V, Tone Y, Li L, Liou HC, Greene MI, Tone M, Chen YH. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31(6):932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9(2):194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- Wen Y, Yu WH, Maloney B, Bailey J, Ma J, Marie I, Maurin T, Wang L, Figueroa H, Herman M, Krishnamurthy P, Liu L, Planel E, Lau LF, Lahiri DK, Duff K. Transcriptional regulation of beta-secretase by p25/cdk5 leads to enhanced amyloidogenic processing. Neuron. 2008;57(5):680–690. doi: 10.1016/j.neuron.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 2010;33(3):313–325. doi: 10.1016/j.immuni.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SG, Gray JD, Ohtsuka K, Yamagiwa S, Horwitz DA. Generation ex vivo of TGF-beta-producing regulatory T cells from CD4+CD25- precursors. J Immunol. 2002;169(8):4183–4189. doi: 10.4049/jimmunol.169.8.4183. [DOI] [PubMed] [Google Scholar]
- Zheng YL, Li BS, Amin ND, Albers W, Pant HC. A peptide derived from cyclin-dependent kinase activator (p35) specifically inhibits Cdk5 activity and phosphorylation of tau protein in transfected cells. Eur J Biochem. 2002;269(18):4427–4434. doi: 10.1046/j.1432-1033.2002.03133.x. [DOI] [PubMed] [Google Scholar]